Abstract
Neurokinin-1 receptor (NK1R) signaling can be immunomodulatory and it can lead to preferential transmigration of CD14+CD16+ monocytes across the blood brain barrier, potentially promoting the development of inflammatory neurological diseases, such as neuroHIV. To evaluate how NK1R signaling alters monocyte biology, RNA sequencing was used to define NK1R-mediated transcriptional changes in different monocyte subsets. The data show that NK1R activation induces a greater number of changes in CD14+CD16+ monocytes (152 differentially expressed genes), than in CD14+CD16− monocytes (36 genes), including increases in the expression of NF-κB and components of the NLRP3 inflammasome pathway. These results suggest that NK1R may alter the inflammatory state of CD14+CD16+ monocytes, influencing the development of neuroinflammation.
Keywords: neurokinin-1 receptor, substance P, monocyte subsets, CD16, RNA sequencing
1. Introduction
Activation of the neurokinin-1 receptor (NK1R) by its primary ligand, the neuropeptide substance P (SP), has a broad spectrum of biologic effects including immunomodulation. In the CNS, SP promotes neurogenic inflammation and increased blood brain barrier (BBB) permeability (Martinez and Philipp, 2016). In the periphery, increased concentrations of plasma SP are observed in viral infections, such as HIV, and are associated with increased levels of inflammatory mediators (Spitsin et al., 2018).
In monocytes, NK1R activation can induce the production of pro-inflammatory cytokines, such as IL-1, IL-6 and TNF-α. SP also enhances transmigration of CD14+CD16+ monocytes in an in vitro model of the human BBB (Spitsin, Pappa, 2018, Spitsin et al., 2017), highlighting the potential impact of this neuropeptide on CNS inflammation. Although CD14+CD16+ monocytes comprise only 5–15% of the total circulating monocyte population (Patel et al., 2017, Williams et al., 2014), they are significantly expanded in HIV and they preferentially transmigrate across the BBB, thus becoming central to the establishment of CNS viral reservoirs and the development of HIV-associated neurocognitive disorders (HAND) (Williams et al., 2015, Williams, Veenstra, 2014).
Activation of NK1R in monocytes is poorly understood. It is thought to be connected to inflammatory disease progression (Ho et al., 1996) (Lai et al., 2001) through NK1R-mediated calcium mobilization or PKCδ and ERK phosphorylation, which increase NF-κB activity (Lai et al., 2008, Spitsin, Pappa, 2018). To better define the inflammatory impact of NK1R activation in monocytes, we examined the quantitative and qualitative differences in inflammatory gene expression mediated by NK1R activation in both CD14+CD16− (CD16−) and CD14+CD16+ (CD16+) monocytes using RNA sequencing.
2. Materials and Methods
2.1. Monocytes
Fresh, de-identified monocytes (purity: 92.3 – 96.4 %, mean 95.0 % as confirmed by flow cytometry, see representative cell subsets on Fig. 1), isolated with negative selection (RosetteSep™ Human Monocyte Enrichment Cocktail, StemCell Technologies) from apheresis products from 8 healthy, HIV-negative donors (age 25 – 52, 3 male / 5 female) were purchased from the Human Immunology Core Facility of the University Of Pennsylvania School Of Medicine. Monocytes were maintained in RPMI 1640, 10% FBS, 1% Pen/Strep and 1% MEM Non-Essential Amino Acids solution. For SP treatment, monocytes were incubated in polypropylene tubes for 3 hours at 37°C with 10 µM SP (Sigma-Aldrich) or 50 µM acetic acid (vehicle, Sigma-Aldrich).
Figure 1. Cell sorting of SP-treated and vehicle-treated control monocytes.
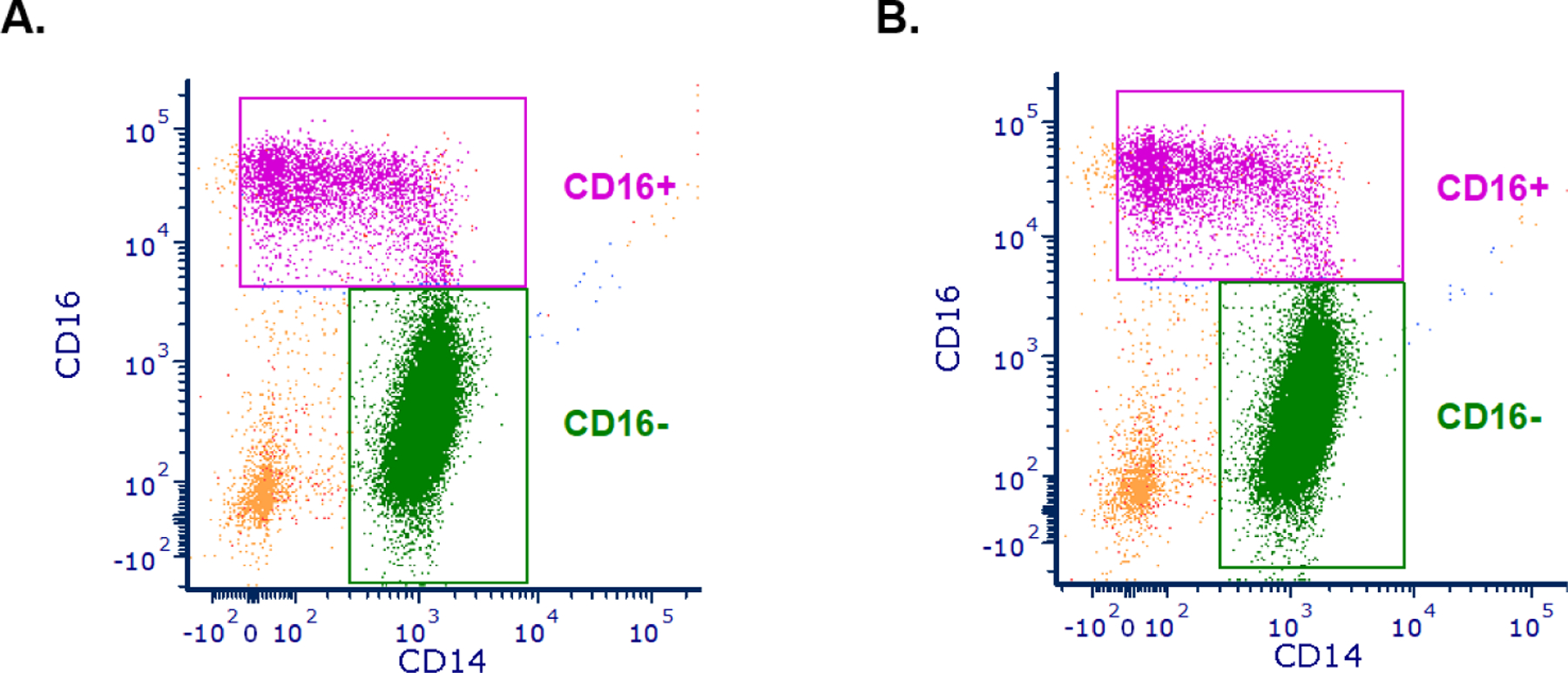
Monocytes were sorted based on the CD14 and CD16 surface markers into CD14+CD16− (CD16−) and CD14+CD16+ (CD16+) monocytes. Representative images for A. SP-treated and B. vehicle-treated control samples are shown. We observed no significant differences in the abundance of CD16+ monocytes between SP-treated and vehicle-treated control samples (Mann Whitney test, p > 0.99).
2.2. Cell sorting
SP-treated and vehicle-treated control monocytes were washed with RPMI 1640 without Phenol Red, containing 50% Accumax (Millipore), 1% FBS, 1% Pen/Strep, 10 mM HEPES and 0.5 mM EDTA. Sorted cells were stained with the antibodies in Table 1, with the FITC channel used for negative selection of contaminating cells. Monocytes were sorted into CD16− and CD16+ populations using the BD FACSJazz™ cell sorter in the Flow Cytometry Core at the Children’s Hospital of Philadelphia Research Institute (CHPRI) (Fig. 1) and collected in RPMI 1640, 20% FBS, 1% Pen/Strep, 10 mM HEPES, 1 U/μl Anti-RNase (Thermo Fisher Scientific).
Table 1.
Antibody panel for flow cytometry.
| Antigen | Fluorophore | Host species | reactivity | Clone | Company | Catalog number |
|---|---|---|---|---|---|---|
| CD14 | APC | Mouse | Human | M5E2 | BD Biosciences | 555399 |
| CD16 | PE/Cy7 | Mouse | Human | 3G8 | BioLegend | 302016 |
| CD235a | FITC | Mouse | Human | HIR2 | BD Biosciences | 559943 |
| CD15 | FITC | Mouse | Human | HI98 | BioLegend | 301904 |
| CD3 | FITC | Mouse | Human | UCHT1 | BioLegend | 300406 |
| CD56 | FITC | Mouse | Human | HCD56 | BioLegend | 318304 |
| CD19 | FITC | Mouse | Human | HIB19 | BD Biosciences | 555412 |
2.3. RNA sequencing
The RNeasy Mini Kit (Qiagen) was used to purify RNA from sorted monocytes. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer in the Nucleic Acid and PCR Core at the CHPRI. Approximately 200 ng of RNA was submitted to the High Throughput Sequencing Core/BGI at the CHPRI (2 samples) and the BGI Americas Corporation (6 samples). Libraries were constructed using the Illumina TruSeq stranded RNA sample preparation and Paired End 150 bp RNA sequencing was performed using the Illumina HiSeq 4000 Sequencing System (2 samples) and the NovaSeq 6000 (6 samples) with a coverage of 30 million reads. The Illumina HiSeq software was used to create RNA sequencing data files (GEO accession number: GSE109635, https://www.ncbi.nlm.nih.gov/geo/).
2.4. Statistical Analysis
For RNA sequencing analysis, STAR software (Dobin et al., 2013) was used to align RNA sequencing data to the Ensembl hg38 human reference genome. For differential gene expression, gene counts were estimated using the HTSeq Python package (Anders et al., 2015). The DESeq2 (Love et al., 2014) and DEXSeq (Anders et al., 2012) R packages were used to determine differentially expressed genes (DEGs) and for differential exon usage analysis. False discovery rate (FDR) adjusted p-values < 0.05 were considered statistically significant. The DAVID Functional Annotation tool (Huang da et al., 2009a, b) and the GOplot (Walter et al., 2015) R package were used for the Gene Ontology (GO) analysis.
3. Results
Human monocytes from 8 donors were treated with substance P (10 µM, SP) for 3 hours, then sorted into mature (CD16+) and immature (CD16−) subsets and processed for mRNA, which was analyzed by RNA sequencing. Differential gene expression analysis of these data shows that SP treatment led to transcriptional changes in both monocyte subsets. A greater number of changes were seen in CD16+ monocytes (152 differentially expressed genes (DEGs), supplementary Table 1) than in CD16− monocytes (36 DEGs, supplementary Table 2). DEGs were then used for a Gene Ontology (GO) analysis, which showed upregulation of annotations that were associated with the inflammation, glycolysis, cell adhesion and negative regulation of apoptosis in CD16+ monocytes (Fig. 2) (Fisher’s Exact test, p < 0.05). There were no significant GO annotations in the SP-treated CD16− subset (Fisher’s Exact test, p > 0.05). These results suggest that exposure to SP could mediate increases in inflammation in CD16+ monocytes through transcriptional changes.
Figure 2. Gene Ontology Biological Processes analysis in SP-treated CD16+ monocytes.
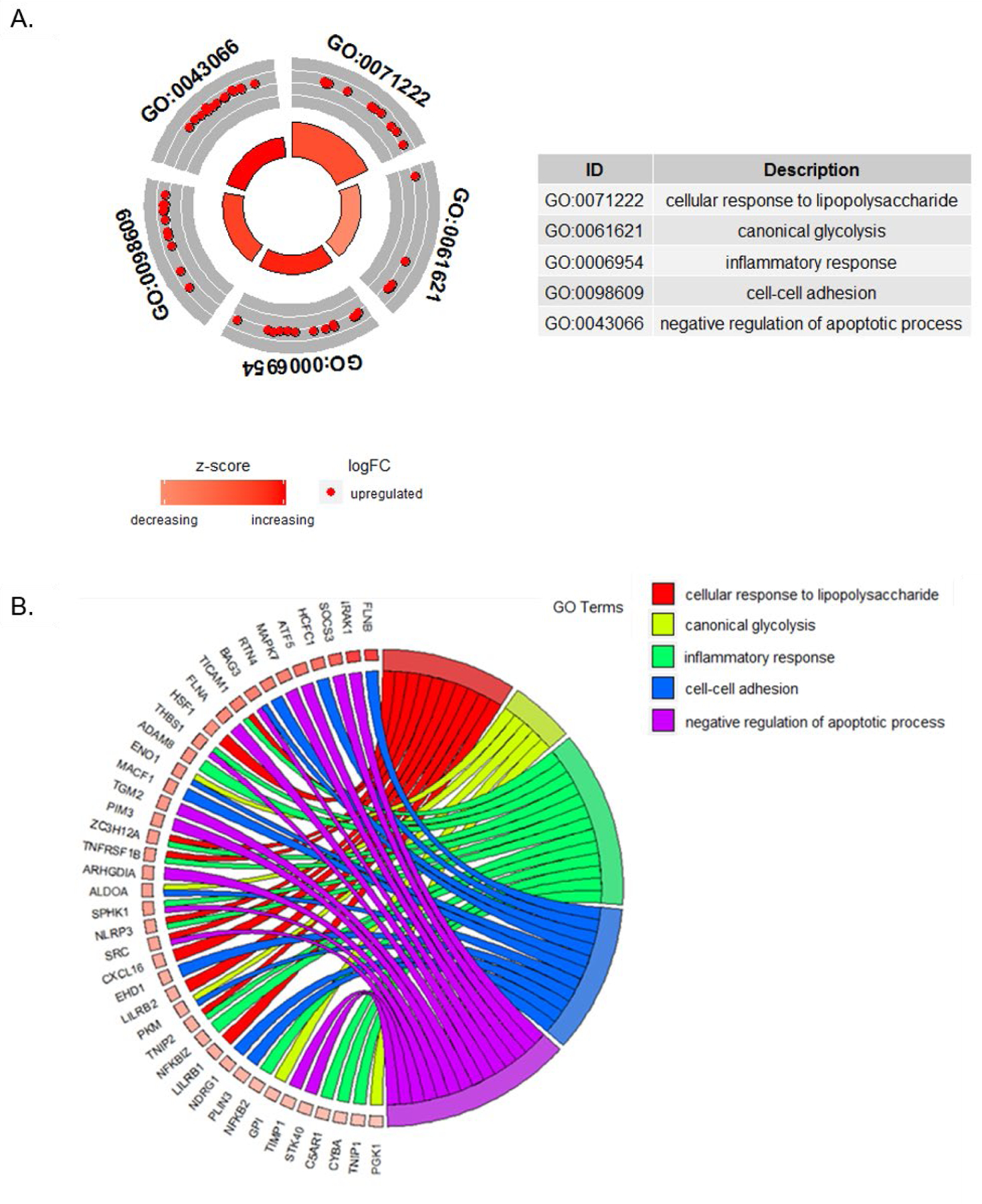
Gene ontology analysis based on 152 differentially expressed genes (DEGs) in SP-treated CD16+ monocytes shows 5 significantly upregulated Gene Ontology Biological Processes (GO BP). A. GOCircle showing the 5 significant GO BP annotations. Red circles show upregulated genes, whereas downregulated genes would have been depicted in blue. The size of the inner circle shows the statistical significance (−log10 adjusted P-value). The table shows the GO BP annotations and their descriptions. B. GOChord plot showing the relationship between DEGs and GO BP annotations. Genes are linked to their assigned terms.
The first GO term associated with SP-treated CD16+ monocytes was the “Cellular response to lipopolysaccharide (GO:0071222)” (Fig. 2). Our analysis showed that SP-treated CD16+ monocytes displayed significant upregulation of transcripts for NFKB2 (log2 Fold Change (log2 FC): 0.128, FDR-adjusted p = 0.049) and NLRP3 (log2 FC: 0.207, FDR-adjusted p = 0.017) (supplementary Table 1). In addition, analysis of differential exon usage was performed on the RNA sequencing data to assess the effect of SP on alternative splicing and it showed that in SP-treated CD16+ monocytes there was differential exon usage in caspase-1 (transcript: ENST00000527979, log2 FC: 0.16, FDRadjusted p = 0.004) (Fig. 3) (supplementary Table 3). These results suggest that SP treatment of CD16+ monocytes may increase the priming or activation of the NLRP3 inflammasome. Activation of inflammasome-mediated inflammation is further supported by GO data indicating that SP treatment of CD16+ monocytes has additional inflammatory effects, showing upregulated transcripts within the GO term “Inflammatory response (GO:0006954)” (Fig. 2). Notably, SP treatment of CD16+ monocytes increased transcription of IRAK1 (log2 FC: 0.3, FDR-adjusted p = 0.006), OSCAR (log2 FC: 0.189, FDR-adjusted p = 0.026) and CD300c (log2 FC: 0.153, FDR-adjusted p = 0.0491) (supplementary Table 1).
Figure 3. Transcript model of the differential exon usage within CASP1 in SP-treated CD16+ monocytes.
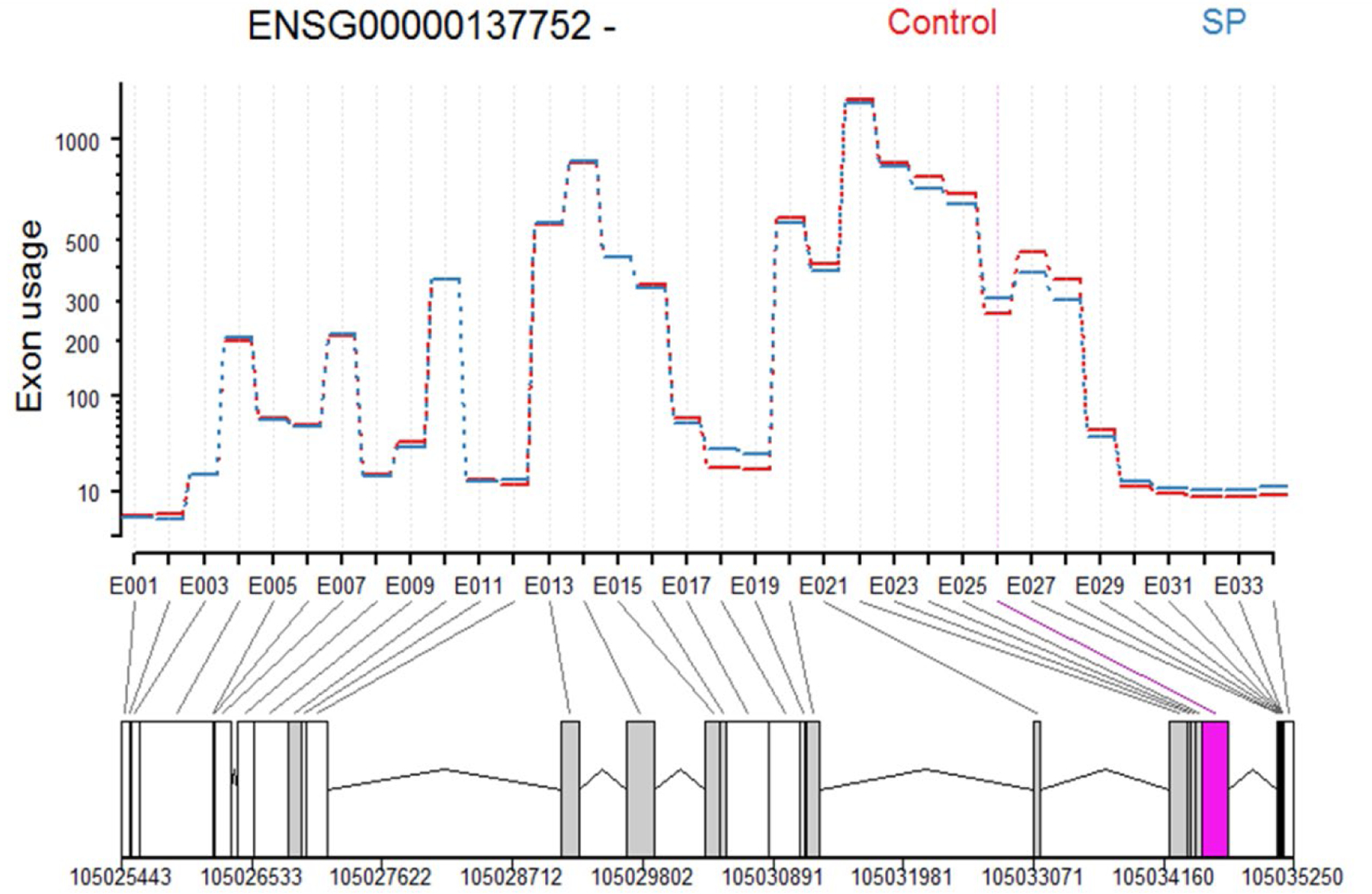
In CD16+ monocytes SP treatment results in alternative splicing of caspase-1 (CASP1), where the use of exon 026 shows an increase of 0.21 in log2 Fold Change. The upregulated isoform corresponds to the protein coding ENST00000527979.5 transcript.
3. Discussion
These data indicate that in CD16+ monocytes, NK1R signaling has a pro-inflammatory transcriptional effect that may involve the NLRP3 inflammasome. The SP-treated CD16+ monocytes show upregulation of transcripts implicated in the cellular response to bacterial lipopolysaccharide (LPS) molecules, which activate toll-like receptor 4 (TLR4) on monocytes and macrophages. This further suggests that SP could predispose these cells to a more robust inflammatory response, as TLR4 activation provokes an immune response that includes activation of inflammasomes, which are multiprotein complexes that regulate the production of IL-1 cytokines (Swanson et al., 2019).
Activation of inflammasomes is usually a two-step process, involving an initial priming step followed by an activation step; one-step activation of NLRP3 inflammasomes can also occur. Priming involves NF-κB mediated transcriptional upregulation of inflammasome components, such as NLRP3, and proIL-1 cytokine transcripts. Activation is initiated by interaction with pathogen or damage-associated molecular patterns (PAMPs or DAMPs) and leads to the formation of the inflammasome complex, activation of caspase-1, and secretion of IL-1 cytokines, including IL-1β and IL-18 (Swanson, Deng, 2019). We observed that SP-treated CD16+ monocytes displayed upregulation of the NFKB2 and NLRP3 transcripts, as well as differential exon usage in caspase-1. This suggests that SP treatment of CD16+ monocytes could impact either the priming or activation of the NLRP3 inflammasome. This concept is supported by data showing that NK1R signaling activated NF-κB in HEK293 cells (Lai, Lai, 2008), and induced IL-1β secretion in conjunction with LPS in rat microglia (Martin et al., 1993).
The pro-inflammatory effect of NK1R signaling that we observed in CD16+ monocytes is also highlighted by the upregulation of genes involved in inflammatory responses. These include IRAK1, which is involved in IL-1 mediated activation of the transcription factor NF-κB (Liu et al., 2007), as well as OSCAR and CD300c, both of which interact with TLRs to induce the release of pro-inflammatory cytokines (Merck et al., 2006, Simhadri et al., 2013). In addition, we observed upregulation of transcripts involved in canonical glycolysis, cell adhesion and negative regulation of apoptosis (data not shown). Similar pathways (i.e. inflammation, lipid metabolism, cell adhesion and apoptosis) were impacted in a clinical trial that studied the effect of NK1R inhibition on plasma protein levels of HIV infected individuals (Spitsin, Tebas, 2017), which supports the importance of NK1R signaling in inflammation during HIV infection. Notably, glycolysis has also been implicated in the regulation of the NLRP3 inflammasome in human monocytes and macrophages (Lee et al., 2019) (Moon et al., 2015) (Finucane et al., 2019). Taken together, these data indicate that NK1R signaling may be involved in NLRP3 inflammasome activity and downstream inflammatory activity in mature monocytes but the precise nature of these effects remain undetermined.
4. Conclusion
We examined the effect of SP treatment on the transcriptional profile and alternative splicing of monocyte subsets. We found that NK1R signaling has a predominantly pro-inflammatory effect in CD16+ monocytes that may be mediated via activation of NF-κB and subsequent activation of the NLRP3 inflammasome pathway. These data support the hypothesis that activation of the NK1R pathway could play an important role in disease and/or conditions involving the expansion of CD16+ monocytes, such as HIV, especially in the context of neuroinflammation and HAND. This suggests that NK1R inhibition may have useful anti-inflammatory effects but additional studies are needed to evaluate the therapeutic potential of this pathway in CNS infection and neuroinflammation.
Supplementary Material
Acknowledgements:
We thank the members of the Douglas Laboratory, the staff of the Flow Cytometry Core, the High Throughput Sequencing Core and the Scientific Computing team of the Department of Biomedical and Health Informatics at the CHPRI for technical assistance.
Funding:
This work was supported by the National Institutes of Health [RO1 AI106509, UO1 MH090325, P30 MH097488, PO1 MH105303, T32-AI007632, R01 DA039005 and R21 DA049227].
List of abbreviations:
- BBB
blood brain barrier
- CNS
central nervous system
- DAMP
damage-associated molecular pattern
- DEG
differentially expressed gene
- ERK
extracellular signal-regulated kinase
- FDR
false discovery rate GO: gene ontology
- GO BP
gene ontology biological process
- HAND
HIV-associated neurocognitive disorder
- HIV
human immunodeficiency virus
- IFN-γ
interferon gamma
- IL
interleukin
- IRAK1
interleukin 1 receptor associated kinase 1
- LPS
lipopolysaccharide
- NF-κB
nuclear factor-κB
- NK1R
neurokinin-1 receptor
- NLRP3
nucleotide-binding domain and leucine-rich repeat containing protein 3
- OSCAR
osteoclast-associated receptor
- PAMP
pathogen-associated molecular pattern
- PKC
protein kinase C
- SP
substance P
- TLR
toll-like receptor
- TNF-α
tumor necrosis factor alpha
Footnotes
Declarations of interest: none
References:
- Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res 2012;22:2008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane OM, Sugrue J, Rubio-Araiz A, Guillot-Sestier MV, Lynch MA. The NLRP3 inflammasome modulates glycolysis by increasing PFKFB3 in an IL-1beta-dependent manner in macrophages. Sci Rep 2019;9:4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WZ, Cnaan A, Li YH, Zhao H, Lee HR, Song L, et al. Substance P modulates human immunodeficiency virus replication in human peripheral blood monocyte-derived macrophages. AIDS Res Hum Retroviruses 1996;12:195–8. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009a;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009b;4:44–57. [DOI] [PubMed] [Google Scholar]
- Lai JP, Ho WZ, Zhan GX, Yi Y, Collman RG, Douglas SD. Substance P antagonist (CP-96,345) inhibits HIV-1 replication in human mononuclear phagocytes. Proc Natl Acad Sci U S A 2001;98:3970–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Lai S, Tuluc F, Tansky MF, Kilpatrick LE, Leeman SE, et al. Differences in the length of the carboxyl terminus mediate functional properties of neurokinin-1 receptor. Proc Natl Acad Sci U S A 2008;105:12605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MKS, Al-Sharea A, Shihata WA, Bertuzzo Veiga C, Cooney OD, Fleetwood AJ, et al. Glycolysis Is Required for LPS-Induced Activation and Adhesion of Human CD14(+)CD16(−) Monocytes. Front Immunol 2019;10:2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FC, Anton PA, Gornbein JA, Shanahan F, Merrill JE. Production of interleukin-1 by microglia in response to substance P: role for a non-classical NK-1 receptor. J Neuroimmunol 1993;42:53–60. [DOI] [PubMed] [Google Scholar]
- Martinez AN, Philipp MT. Substance P and Antagonists of the Neurokinin-1 Receptor in Neuroinflammation Associated with Infectious and Neurodegenerative Diseases of the Central Nervous System. J Neurol Neuromedicine 2016;1:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JS, Hisata S, Park MA, DeNicola GM, Ryter SW, Nakahira K, et al. mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell Rep 2015;12:102–15. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Patel AA, Zhang Y, Fullerton JN, Boelen L, Rongvaux A, Maini AA, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med 2017;214:1913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitsin S, Pappa V, Douglas SD. Truncation of neurokinin-1 receptor-Negative regulation of substance P signaling. J Leukoc Biol 2018. [DOI] [PubMed]
- Spitsin S, Tebas P, Barrett JS, Pappa V, Kim D, Taylor D, et al. Antiinflammatory effects of aprepitant coadministration with cART regimen containing ritonavir in HIV-infected adults. JCI Insight 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 2019;19:477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015;31:2912–4. [DOI] [PubMed] [Google Scholar]
- Williams DW, Anastos K, Morgello S, Berman JW. JAM-A and ALCAM are therapeutic targets to inhibit diapedesis across the BBB of CD14+CD16+ monocytes in HIV-infected individuals. J Leukoc Biol 2015;97:401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DW, Veenstra M, Gaskill PJ, Morgello S, Calderon TM, Berman JW. Monocytes mediate HIV neuropathogenesis: mechanisms that contribute to HIV associated neurocognitive disorders. Curr HIV Res 2014;12:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


