Abstract
Biological treatment of waterborne viruses, specifically grazing of viruses by protists, can enhance microbial water quality while avoiding the production of toxic byproducts and high energy costs. However, tangible applications are limited by the lack of understanding of the underlying mechanisms. Here, we examined the feeding behavior of Tetrahymena pyriformis ciliates on 13 viruses, including bacteriophages, enteric viruses, and respiratory viruses. Significant differences in virus removal by T. pyriformis were observed, ranging from no removal (Qbeta, coxsackievirus B5) to ≥2.7 log10 (JC polyomavirus) after 48 h of co-incubation of the protist with the virus. Removal rates were conserved even when protists were co-incubated with multiple viruses simultaneously. Video analysis revealed that the extent of virus removal was correlated with an increase in the protists’ swimming speed, a behavioral trait consistent with the protists’ response to the availability of food. Protistan feeding may be driven by a virus’ hydrophobicity but was independent of virus size or the presence of a lipid envelope.
Keywords: waterborne virus, enveloped virus, protists, biological water treatment, swimming speed, grazing
Short abstract
Removal of waterborne viruses by Tetrahymena pyriformis is driven by neither virus size nor structure but is consistent with behavioral changes associated with the protists’ ability to recognize viruses as food.
Introduction
Among the many processes determining the fate of waterborne viruses in natural and engineered systems, biological virus control is the least understood.1 Previous studies have demonstrated that various microorganisms can contribute to virus removal,2−6 though mechanistic insights are scarce. Of particular interest are the interactions of viruses with protists, which are ubiquitous in our aquatic environment and can serve a dual role in virus fate. On the one hand, they can act as a virus reservoir7 and thereby shield viruses from inactivating stressors such as chemical disinfectants or UV light.8 On the other hand, they can serve as antiviral agents.9
Among protists, ciliates are an important group due to their diversity and abundance in both natural waters and wastewater.10 Ciliate concentrations of 103 cells mL–1 and 105 cells mL–1 have been reported in surface waters and in the mixed liquor of activated sludge, respectively.11,12 Ciliates have been found to graze on echoviruses, polioviruses, adenoviruses, and bacteriophages,9,13−15 though not all viruses are equally affected. Both virus and protist species determine whether or not a virus is susceptible to grazing,4,6,15,16 and a single protist species (Tetrahymena spp.) can exert varying effects on different viruses, including activation,17 protection,18 or inactivation.19
Virus susceptibility to grazing may be driven by a protist’s ability to sense and uptake their prey, or by its capacity to digest it. The ciliate Tetrahymena thermophila (T. thermophila) was found to more efficiently graze on bacteria exhibiting a low surface hydrophobicity.20 Alternatively, grazing rates may be driven by prey size. For ciliates, a linear relationship between the clearance rate and the prey size was found for prey ranging between 100 nm and 30 μm.21 Size and hydrophobicity thus stand out as prey properties of interest. Removal rates may furthermore be influenced by the presence of alternative prey. In a sample containing two viral strains, a preferential uptake of type A2 myxovirus influenza over coxsackievirus B5 by T. pyriformis was observed.15 Similarly, selective feeding on specific bacterial species by T. pyriformis was demonstrated.22 In contrast, T. pyriformis did not exhibit preferential uptake of bacteria over polystyrene latex particles23 or over clay.24
The objective of this study was to characterize the susceptibility of a suite of viruses to removal by the model ciliate T. pyriformis(25,26) and to investigate the determinants that drive this process. We hypothesized that virus susceptibility to removal increases with increasing virus size and decreasing virus hydrophobicity and that removal rates depend on the presence of alternative prey. We co-incubated 13 different viruses with T. pyriformis alone or in combination, and removal rates were monitored and evaluated with respect to virus size, presence of an envelope, and hydrophobicity. Finally, we assessed the role of virus sensing by protists in determining the extent of virus loss. Overall, this work provides novel insights into the efficacy and mechanism of virus removal by protists and advances our understanding of biological virus control.
Materials and Methods
Maintenance of Tetrahymena pyriformis Cultures
Cultures of Tetrahymena pyriformis (T. pyriformis) were obtained from the Culture Collection of Algae and Protozoa (CCAP no. 1630/1W) and axenically maintained in 75 cm2 cell culture flasks (TTP, Milian). The culture medium used was proteose peptone yeast extract (PPYE), which consisted of 20.0 g of proteose peptone (Bacto peptone, Difco) and 2.5 g of yeast extract (BioChemika) dissolved in 1 L of Milli-Q water, and autoclaved and stored at 4 °C. The cultures were maintained in a 28 °C incubator to promote ciliate growth.27 Subcultures were prepared every week by spiking 1 mL of the previous culture into 19 mL of fresh PPYE medium (total volume of 20 mL). General information on the use of Tetrahymena as a model organism are reported in Asai and Forney.26
Virus Propagation, Purification, and Enumeration
Ten mammalian viruses and three phages were included in this study. The viruses chosen cover a range of sizes and genome types and include both enveloped and non-enveloped species. An overview over the viruses used in this study and their properties is given in Table S1.
Mammalian viruses included echovirus 11 (E11), coxsackievirus A9 (CVA9), coxsackievirus B5 (CVB5), human mastadenovirus C type 2 (HAdV2), human rotavirus (RoV), JC polyomavirus (JCV), influenza A virus (IAV), murine norovirus (MNV), murine hepatitis virus (MHV), and murine sendai virus (SeV). Bacteriophages included T4, MS2, and Qbeta. The production and purification of viral stock solutions is described in the Supporting Information. Viral concentrations were quantified by (RT-)qPCR as described below and were expressed as genomic copies per milliliter (GC mL–1). In addition, infectious concentrations were determined for a subset of mammalian viruses (E11, CVA9, CVB5, HAdV2) by the most probable number method (MPN) as described previously,28 using five replicates and up to six dilutions per sample. Bacteriophages were enumerated by plaque assay as described elsewhere29 (T4, MS2, and Qbeta). The limits of quantification (LOQ) for each virus and phage by (RT-) qPCR, MPN, or plaque assay are reported in Tables S2 and S4.
(RT-)qPCR Analyses
Viral nucleic acids were extracted from 140 μL samples using the QIAamp Viral RNA Mini Kit (Qiagen) following the manufacturer’s instructions. Nucleic acids were eluted in 60 μL of AVE buffer and stored at −20 °C until analysis. Each batch of samples included a negative extraction control (NEC), which was always negative. (RT-)qPCR analyses were performed on a Mic Real-Time PCR system (Bio Molecular Systems). Primers and probes, reaction volumes, as well as the thermocycling protocols are detailed in Table S2. DNA was analyzed using the TaqMan Environmental PCR Master Mix (Applied Biosystems, cat. no. 4396838) or TB Green Advantage qPCR Premix (Takara, cat. no. 639676). For RNA, RNA Ultrasense One-Step Quantitative RT-PCR System (Invitrogen, cat. no.: 11732–927), or One Step SYBR PrimeScript RT-PCR Kit (Takara, cat. no. RR066A) were used. Standard curves for each viral target were generated by preparing 10-fold serial dilutions of gblock gene fragments over a range of 10 to 1 × 107 GC μL–1 (Integrated DNA Technologies) in TE buffer (Invitrogen). All (RT-)qPCR reactions were performed in duplicate, and each PCR run contained nontemplate controls (NTC) which were always negative. Previous work has shown the absence of PCR inhibition in our experimental protocol.30
For pooled virus experiments, (RT-)qPCR assays for each virus were checked against the other viruses included in a given experiment to ensure the specificity of the assay.
The micPCR software (version 2.10.0) was used to acquire Cq values and to quantify viral genomic copy concentrations. Pooled standard curves were analyzed in R using the Generic qPCR Limit of Detection (LOD)/Limit of Quantification (LOQ) calculator.31 PCR efficiency, LOQ, R2, slope, and intercept of the standard curves for each assay are reported in Table S2 along with a checklist of experimental details as requested by the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines32 (Table S3).
Co-incubation Experiments
Prior to experimental use, T. pyriformis was washed following the protocol of Pinheiro et al.,10 except that it was rinsed with sterile moderately hard synthetic freshwater (MHSFW). MHSFW was prepared following a previously published protocol33 and sterilized by autoclave or 0.22 μm filtration using a syringe filter (Millex-GV, 0.22 μm, PVDF, 33 mm, Gamma-Sterilized). After resuspension in sterile MHSFW, T. pyriformis were left starving at room temperature for 15 to 21 h before being used. Ciliate concentration was determined using a Neubauer chamber (Bright-line, Hausser Scientific) as the average of three distinct counts. For each biological replicate, 3 mL of T. pyriformis in sterile MHSFW solution at an average concentration of 4 × 104 cells mL–1 was transferred into a 50 mL plastic tube (Falcon, Greiner). A total of 100 μL of a given virus stock was then spiked into the solution to reach an initial virus titer ranging between 1 × 106 GC mL–1 and 3.2 × 107 GC mL–1. Given a clearance rate of T. pyriformis of 0.0001 mL h–1 ciliate–1,34 the entire solution passed through the ciliates approximately 60 times in this experimental setup. Controls consisted of 100 μL of the corresponding virus spiked into 3 mL of sterile MHSFW. From these reactors, samples were taken at 0 and 48 h, unless mentioned otherwise. Aliquots of 300 μL were filtered through a 0.22 μm syringe filter to remove T. pyriformis cells and were analyzed for viral infectivity and/or genomic copy content.
Net virus removal by T. pyriformis was quantified on a log10 scale, as either the loss in the infectious virus concentration (log10(C/C0)) or the loss in genomic copies (log10(N/N0)) over the duration of the experiment. The net removal was calculated as the removal in the experiment (log10(Cexp/Cexp,0) or log10(Nexp/Nexp,0)) corrected for the removal measured in the protist-free control (log10(Ccont/Ccont,0) or log10(Ncont/Ncont,0) (eq 1). The concentration of T. pyriformis in MHSFW was regularly checked over the course of the experiments and remained stable (variation <1 log10).
| 1 |
The net virus removal was compared among viruses by ordinary one-way ANOVA, under the assumptions of normal distribution, homogeneity in variances, and independent observations.
Co-incubation Experiments with Pooled Viruses
Two co-incubation experiments with multiple viruses were performed to assess the role of alternative viral prey on observed removal rates. Experiments were conducted as described above, except that either two (CVB5 and JCV) or three (Qbeta, MS2, and T4) viruses were pooled and added to reactors simultaneously. In parallel, co-incubations were carried out for each virus individually, using the same T. pyriformis batch and experimental solutions as for the pooled experiments. The concentration of each phage in the pooled experiment was lowered by a factor of 3 compared to individual experiments, such that the total phage concentration corresponded to that in individual phage experiments.
Stability Assessment of the Experimental System
Effect of Virus-to-Protist Ratio on Removal Efficiency
The effect of organism concentrations on virus removal was assessed in experiments with different virus-to-protist ratios. For these experiments, protists from a single batch were co-incubated with RoV or CVA9. For RoV, the initial virus titer was 4 × 107 or or 6 × 106 GC mL–1 , and the initial protist concentration was fixed to 6 × 104 cells mL–1 (ratios of ∼1000:1 and 100:1, respectively) ratio of 100:1). For CVA9, the initial virus titer was fixed to 1.8 × 106 GC mL–1, and the protist concentration was 4 × 104 cells mL–1 (ratio of ∼50:1) or 4 × 105 cells mL–1 (ratio of ∼5:1). Co-incubations were conducted over 48 h. Virus concentrations were measured before and after co-incubation, and virus removal was determined as described in eq 1.
Batch Variability
To determine the experimental variability arising from the use of different T. pyriformis batches, experiments with similar initial virus-to-protist ratios (100:1), but different protist batches, were performed. A batch consists of T. pyriformis originating from subcultures produced from a single culture flask on a given day, whereas different batches are produced on different days. Protist concentrations in the different batches ranged from 2 × 104 cells mL–1 to 8 × 104 cells mL–1, and virus concentrations were adjusted accordingly. These experiments were performed using four different viruses (E11, T4, JCV, and MHV) and 10 different protist batches, of which two or three were tested per virus. Co-incubations were conducted over 48 h (T4, JCV, and MHV) or 72 h (E11). Viruses were enumerated before and after co-incubation, and virus removal was determined as described in eq 1.
Experiments to Assess Virus Removal in the Absence of Live T. pyriformis
Contribution of Virus Sorption on T. pyriformis to Overall Removal
Experiments with inactivated ciliates were performed to control for viral sorption onto T. pyriformis. Two viruses (E11 and RoV) were individually co-incubated with killed T. pyriformis for 48 h (E11) or up to 120 h (RoV), and virus removal was quantified as described in eq 1. Two different approaches for T. pyriformis were used to ensure that results were not biased by changes in surface properties or protist integrity due to the killing mechanism. T. pyriformis were killed either by one cycle of −20 °C/22 °C freezing/thawing (E11) or by chemical inactivation with 10% neutral-buffered formalin (Sigma-Aldrich), at a dose of 1 mL formalin for 3 mL of T. pyriformis solution followed by 10 min incubation at 25 °C (RoV). No effect of formalin on virus quantification by qPCR was found.
Virus Removal by Extracellular Protist Metabolites
To determine if metabolites produced by T. pyriformis over the course of an experiment had an effect on virus infectivity, organisms were washed and starved as described above, and the solution was filtered through a 0.22 μm syringe-filter to remove protists. Viruses (E11) were then exposed to the filtrate for 96 h, and virus removal was determined as described in eq 1.
Protist Movement
We used video analysis to investigate how protist swimming speed was affected by exposure to different viruses. Swimming speed was measured as the length of a trajectory traveled by a protist, divided by the duration of the observation. An increase in swimming speed has previously been associated with food availability in experiments using nonviral prey.35,36 Analogously, we here use swimming speed as an indicator that T. pyriformis senses a virus and recognizes it as food.37,38 We first incubated T. pyriformis populations overnight in sterile MHSFW to ensure cells were starved. We then co-incubated T. pyriformis with Qbeta, MS2, and T4, either individually or in combination. Because the video analysis had to be done in a biosafety level 1 lab, we restricted this analysis to bacteriophages only. Each condition was tested in triplicate, in 3 mL of starved T. pyriformis culture at an average concentration of 5.2 × 103 cells mL–1 (standard deviation (sd) = 1.9 × 103 cells mL–1). The total initial phage concentration in all experiments was fixed to an average value of 1.9 × 107 GC mL–1. The experimental solutions were sampled at eight different time points for the video analysis: prior to adding the phages (0 h), as well as 0.5, 1, 2, 3, 4, 5, and 6 h after adding the phage stocks to the protist populations.
We followed an existing video analysis protocol,25 using the same handling method as in previous experiments.25,40−42 Briefly, we sampled 200 μL from each experimental solution and transferred it to a microscope slide. The focal volume targeted had a fixed capacity of 34.4 μL, to ensure that the same volume was measured in every video. We then recorded a 20 s video (25 fps, for a total of 500 frames) using a Leica M165FC stereomicroscope with a top-mounted Hamamatsu Orca Flash 4.0 camera. Finally, we extracted from the video information on cell movement behavior.
Gross cell speed values (i.e., swimming speed) were standardized for each of the 15 populations by dividing the gross cell speed for each population by the corresponding speed prior to adding phages (value at 0 h), to obtain the change in movement speed relative to the pre-exposure value. We then fit a linear mixed model (nlme package,43 version 3.1–144) to determine how swimming speed changed over time for each of the phage treatments, using phage identity and time after phage exposure (factorial) as fixed effects. To account for multiple measurements of individual populations, we included population ID as a random effect in the model. To consider potential confounding effects due to differences in population density, protist density (numerical) was included as a covariate in the analysis. We report statistical output for the fixed effects of the model. Fixed effects were evaluated using a likelihood-ratio test,44 and we report model estimates, as well as χ2 values (along with the degrees of freedom associated with fitting the variable (dff) and the residual degrees of freedom (rdf), in the format χ2dff,rdf) and p values associated with each fixed effect.
Data Analysis
The Rstudio software45 (version 1.0.153) was used to compute the infectious virus titers and to analyze the videos using the BEMOVI R-package39 and nlme package,43 version 3.1–144. Statistics were performed in GraphPad Prism 9 (version 9.0.2).47 Statistical significance was defined at an α value of 0.05. Graphs were made using GraphPad Prism 9 software,47 version 9.0.2.
Results
Validation of the Experimental Setup
Virus removal may be influenced by the number of encounters between the virus and protist over the course of an experiment. Because the exact virus and protist concentrations used in co-incubation experiments can vary, we first evaluated the stability of our experimental system to fluctuations in the virus-to-protist ratio.
The observed virus loss was found to be independent of the virus-to-protist ratio under the experimental conditions studied. For a fixed initial virus titer, a 10-fold change in the T. pyriformis concentration did not significantly affect the observed loss of infectious CVA9 (unpaired t test, t(4) = 1.493, p value = 0.21; Figure S1). Similarly, for a fixed T. pyriformis concentration, a 10-fold change in the initial virus concentration caused only a minor difference (0.2 log10) in the loss of RoV genomic copies (Figure S1). While statistically significant (unpaired t test, t(4) = 4.031; p = 0.016), a 0.2 log10 difference is not biologically relevant and within the range of the RT-qPCR precision. Given the insensitivity of virus removal to the virus-to-protist ratio over the range considered, we used ratios between 100:1 and 1000:1 in subsequent experiments. We chose an initial T. pyriformis concentration ranging between 3 × 104 and 8 × 104 cells mL–1 and virus titers ranging from 1 × 106 GC mL–1 to 3.2 × 107 GC mL–1.
Virus removal may furthermore depend on the specific batch of ciliates used. Batch-to-batch variability of virus removal by T. pyriformis was assessed in co-incubation experiments with JCV, MHV, E11, or T4 at a constant virus-to-protist ratio. As shown in Figure S2, variability for JCV, MHV, and E11 was not statistically significant (Table S6) and did not exceed 0.5 log10. The sole exception was T4, which showed a significant batch-to-batch variability greater than 1 log10 (Table S6).
Virus Removal Requires the Presence of Biologically Active T. pyriformis
To assess if the presence of live T. pyriformis was required for virus removal, we tested if virus sorption to T. pyriformis or the presence of extracellular metabolites could contribute to the observed virus loss. Virus removal observed in the presence of T. pyriformis could not be attributed to sorption onto T. pyriformis. We observed no decrease in infectious E11 titers in samples containing mechanically killed T. pyriformis, while a ≥ 2 log10 loss of infectious E11 occurred over 48 h in samples containing live T. pyriformis (unpaired t test, two-tailed: t(4) = 5.059; p value = 0.0072; Figure S3A). Likewise, for RoV, the number of viral genomic copies in the samples containing the inert organisms remained constant, whereas they decreased over 120 h in the samples containing the live organisms (Figure S3B).
Similarly, we found that extracellular antiviral metabolites released by T. pyriformis alone could not explain the observed virus removal. The titers of E11 decreased by >1 log10 in solutions containing T. pyriformis, whereas the decrease was only minimal (mean = 0.2 log10) if solutions were filtered to remove the ciliates (Figure S2; at 48 h: unpaired t test, two-tailed: t(2) = 3.628; p value = 0.0683). While the absence of antiviral action of metabolites remains to be confirmed for the other viruses included in this study, our subsequent analysis is rooted in the assumption that metabolites do not significantly contribute to virus loss.
T. pyriformis Induced Removal Is Virus-Specific and Independent of Alternative Viral Prey
Virus removal by protists was determined by individually co-incubating each virus listed in Table S1 with T. pyriformis. The observed virus loss was independent of the measurement technique used. Both (RT-)qPCR and culturing methods yielded similar results if applied to the same virus (T4, unpaired t test, two-tailed t(10) = 0.2653, p value = 0.7961; E11, unpaired t test, two-tailed t(4) = 0.9439, p value = 0.3986; HAdV2, unpaired t test, two-tailed t(9) = 0.2016, p value = 0.8447), consistent with physical removal of the virions from solution.
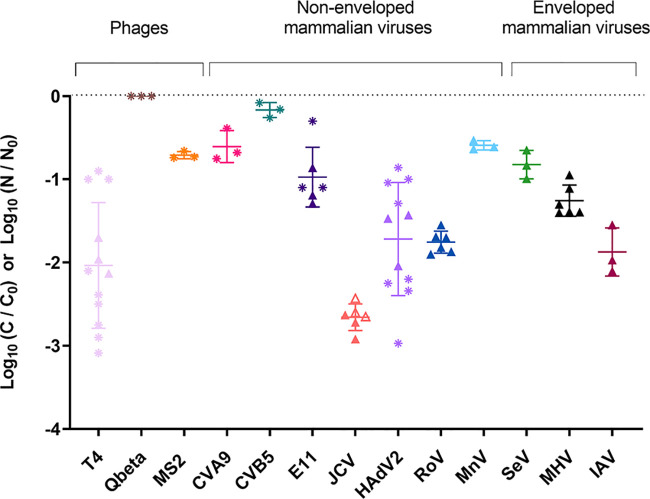
The different viruses exhibited significant differences in their susceptibility to removal by T. pyriformis (see Table S5 for full ANOVA results), with mean removal per virus ranging from no removal to 2.7 log10 over 48 h (Figure 1). Additional time points are shown in Figure S5 and Figure S6.
Figure 1.
Net removal (*, log10(C/C0); ▲, log10(N/N0)) of different viruses by T. pyriformis over 48 h. The virus-to-protist ratio ranged from 100:1 to 1000:1. The initial viral titer ranged between 1 × 106 GC mL–1 and 3.2 × 107 GC mL–1, except for T4 (1.6 × 108 GC mL–1). The initial T. pyriformis concentration ranged from 2 × 104 cells mL–1 to 8 × 104 cells mL–1. The horizontal bars represent the mean of multiple replicates, and the error bars represent the standard deviation. When the concentration at 48 h was below the LOQ, the concentration was set to the LOQ to determine the removal. The corresponding data points are presented as empty symbols.
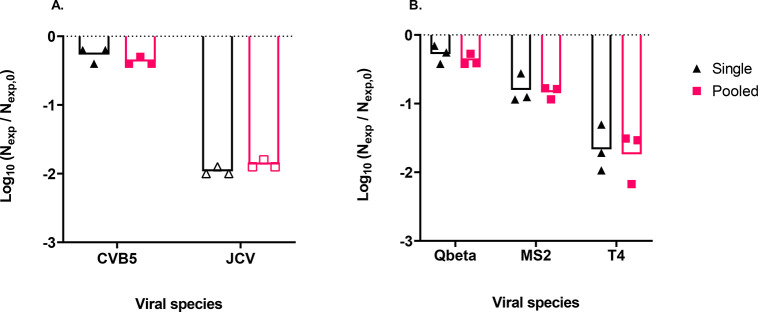
To investigate if the susceptibility of a given virus to removal by T. pyriformis is influenced by other viral prey, the removal of selected viruses was additionally measured in experiments containing multiple viruses. No statistically significant difference in removal was observed when CVB5 or JCV was incubated with T. pyriformis simultaneously or alone (Figure 2A). Similarly, the individual incubation of T4, Qbeta, and MS2 with T. pyriformis resulted in the same virus removal as observed in experiments where all three phages were pooled (Figure 2B). Full statistical results can be found in Table S7.
Figure 2.
Comparison of virus removal in co-incubation experiments with a single virus versus pooled viruses. (A) Log-reduction in CVB5 and JCV genomic copies after exposure to 9.7 × 104T. pyriformis cells mL–1. (B) Log-reduction in Qbeta, MS2, and T4 genomic copies after exposure to 6.8 × 104T. pyriformis cells mL–1. When the concentration at 48 h was below the LOQ, the concentration was set to the LOQ to determine the removal. The corresponding data points are presented as empty symbols. Vertical bars represent the mean removal value of three replicates.
Protists Move Faster in the Presence of MS2, T4, and Pooled Phages
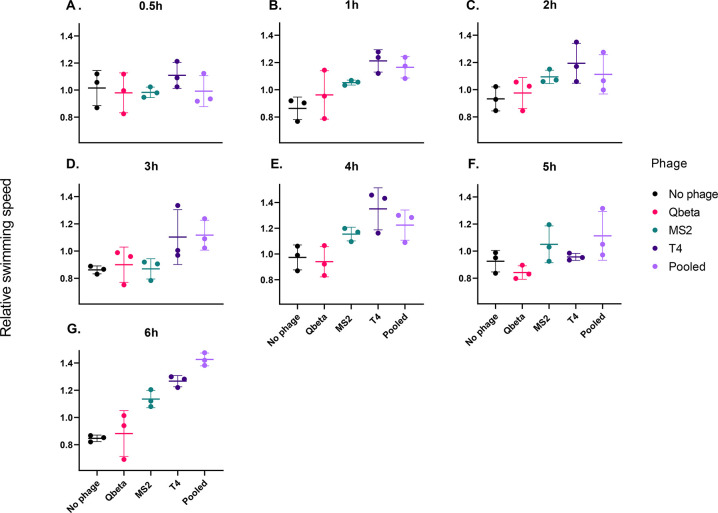
Movement analysis of T. pyriformis can inform on the ciliate’s recognition and ingestion of prey.48,49 Here, we used this tool to determine if the susceptibility of phages MS2, Qbeta, and T4 to T. pyriformis can be explained by the ciliate’s ability to recognize the different phages as food. To do so, we fit a linear mixed model to evaluate the change in ciliate movement over time and assessed how the presence of the different phages as well as exposure time to those phages affected the swimming speed of the ciliates. Swimming speed depended on time since phage addition to the T. pyriformis culture (p = 0.002, χ26,105 = 20.515), protist concentration (p = 0.004, χ21,105 = 8.196) and the interaction between phage exposure and time since phage addition (p < 0.001, χ224,105 = 153.789). Immediately after adding phages, swimming speed did not differ from the value prior to phage addition, for any of the phage treatments (Figure 3A). However, differences arose between the treatments over time. In the phage-free control, swimming speed was lower compared to the initial value, and significantly so at 1 h, 3 h, and 6 h (Figure 3B, D, and G, respectively) after we added the phages. Swimming speed remained unchanged over time for protists that were co-incubated with Qbeta individually. Exposure to MS2 caused a moderate increase in protist movement: swimming speed was significantly higher at 1, 2, 4, and 6 h (Figures 3B, C, E, and G, respectively) after MS2 addition. For co-incubation with T4, a stronger increase in swimming speed was observed, with significantly higher speeds at 1, 4, and 6 h (Figures 3B, E, and G, respectively), and marginally significantly at 2 h (Figures 3C). The most consistent change in movement was observed for the co-incubation with pooled phages, where a statistically significant increase in protist swimming speed was observed at 1, 2, 3, 4, 5, and 6 h of co-incubation (Figures 3A–G). The full statistical output can be found in Tables S8 and S9.
Figure 3.
Phage treatment affects protist swimming speed. Subplots show the relative swimming speed of protists compared to the swimming speed prior to phage addition. A relative swimming speed value of 1 indicates no change in speed upon addition of phages. Each subplot shows data for a different time point (0.5, 1, 2, 3, 4, 5, and 6 h after phage addition). Horizontal bars represent the mean over three replicates, and the error bars represent the standard deviation.
Virus Susceptibility Is Not Correlated with Virus Size and Both Enveloped and Non-enveloped Viruses Can Be Susceptible
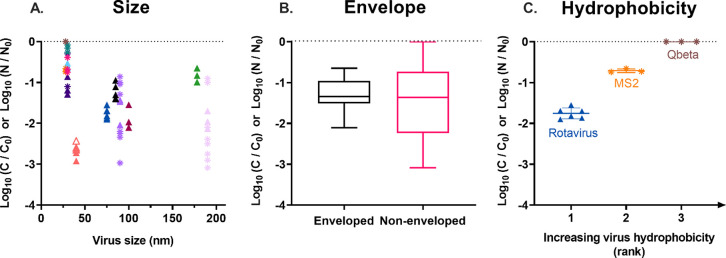
To determine which viral properties render a virus susceptible to removal by T. pyriformis, we evaluated the observed virus removal (Figure 1) with respect to different viral characteristics, specifically virus size, the presence of an envelope, and hydrophobicity. Only a very weak correlation was found between the extent of virus loss within 48 h as measured by either infectivity or genome copy loss and the virus size (Pearson r = 0.147; Figure 4A). Similarly, no difference in the median removal values was observed for enveloped and non-enveloped viruses (−1.3 log10; Figure 4B). In contrast, virus hydrophobicity appeared to be inversely correlated with virus removal (Figure 4C). Unfortunately, hydrophobicity measurements were only available for three of the viruses considered (Qbeta, MS2, and RoV).50,51 Of these, Qbeta is the most and RoV the least hydrophobic. Determining virus hydrophobicity indices for additional viruses was beyond the scope of this study.
Figure 4.
Net removal (*, log10(C/C0); ▲, log10(N/N0)) of viruses exposed to T. pyriformis for 48 h as a function of different virus properties. Removal data and symbol colors correspond to those in Figure 1. Data points for which the concentration at 48 h was below the LOQ are represented by empty symbols. (A) Net removal as a function of virus size. (B) Box plot of the net removal varlues of all enveloped and all non-enveloped viruses. Boxes indicate the 25th, 50th and 75th percentiles; error bars indicate the 5th and 95th percentiles. (C) Net removal as a function of virus hydrophobicity, ranked according to literature values.50,51 The bars indicate the mean and standard deviation of experimental replicates.
Discussion
Virus Removal Is Consistent with T. pyriformis’ Ability to Recognize a Virus As Food
Out of the 13 studied viruses, eight showed a susceptibility to T. pyriformis equal or greater to 1 log10 removal over 48 h (Figure 1). Virus removal could not be attributed to sorption onto the T. pyriformis surface nor to the production of extracellular metabolites. We therefore conclude that T. pyriformis’ virucidal action is rooted in grazing. Tetrahymena pyriformis has previously been found to graze on PhiX174,18 T4,19 adenovirus type 3,13 simian rotavirus SA11,52 vaccinia virus,53 influenza A and B,54 poliovirus type I and type 2,9,55 and echovirus type 30.55 We expand this list of susceptible viruses by JCV, SeV, and a coronavirus, MHV. Notably, susceptibility to a protist was not a virus family or genus trait. For example, among members of the Enterovirus genus, CVA9 and E11 were removed by protists, whereas CVB5 was resistant. Similarly, among the two members of the Leviviridae family investigated, only MS2 was susceptible to removal by protists, whereas Qbeta was not.
When simultaneously exposing T. pyriformis to multiple viral species, the individual susceptibility of each virus strain was conserved (Figure 2). Our findings on CVB5 and JCV showed that even when simultaneously fed to T. pyriformis, the removal rate of JCV remained high and that of CVB5 remained low. This is consistent with work by Teras15 who found type A2 myxovirus influenza, but not CVB5, inside T. pyriformis, confirming a limited removal of CVB5. Even in a more complex system with three phages, the presence of other viruses did not affect the individual virus removal rate. This suggests that virus removal by T. pyriformis is virus-specific and independent of the presence of alternative viral prey.
The observed differences in virus removal may stem from T. pyriformis’ ability to sense a given virus and recognize it as food. T. pyriformis possesses a feeding receptor in the cytosome, which may assist in differentiating between different virus species or strains.56 Alternatively, the different removal rates may result from the virus’ differing persistence toward digestion once taken up by T. pyriformis. These two possibilities are difficult to conclusively disentangle. However, the behavioral characteristics of T. pyriformis observed by video analysis were consistent with those previously associated with food availability,35,36 suggesting that the discrepancies in virus removal result from the ability of T. pyriformis to recognize viruses as food (Figure 3). Specifically, Qbeta, which was not removed by T. pyriformis, did not affect the swimming speed in individual co-incubation experiments. In contrast, T4, which was one of the most susceptible viruses studied herein, altered the swimming speed extensively. MS2 had both an intermediate removal and an intermediate effect on T. pyriformis movement. We emphasize that these observations are at a population level (effect of total viral particles removed) and thus do not reveal the underlying mechanisms by which the protists recognize and remove individual viral particles.
Viral Susceptibility to T. pyriformis May Be Determined by Surface Hydrophobicity but Does Not Depend on Virus Size or the Presence of an Envelope
Enveloped viruses are often hypothesized to be more susceptible to environmental stressors than non-enveloped viruses57 Here, we found no evidence that enveloped viruses are more readily removed by protists than non-enveloped viruses. The strain most susceptible to T. pyriformis was JCV, a non-enveloped icosahedral human virus that exhibited up to > 3 log10 removal over 48 h of co-incubation. In contrast, enveloped viruses such as SeV and MHV only showed an average removal of 1 log10, which is similar to E11, a non-enveloped virus. Among the enveloped viruses, influenza virus was the most susceptible with 2 log10 removal over 48 h, which is similar to the average value found for the non-enveloped virus HAdV2. Our findings are thus consistent with a recent review that demonstrates similar environmental persistence of enveloped and non-enveloped viruses.57
Fenchel proposed the existence of an optimal protist-to-prey size ratio for which the grazing rate is maximal.21 This suggests that prey size is an important factor in grazing efficiency. Here, however, no influence of size on susceptibility to T. pyriformis was observed (Figure 4A). The virus size range considered in the present study was 28 to 190 nm. The highest removals were obtained for JCV, which is 30 nm in diameter, and T4, which is 90 nm wide and 190 nm long. This virus size range may not be sufficiently large to trigger a size-specific response, especially as size may not be the only factor in prey preference. The latter is supported as even viruses belonging to the same species, such as E11, CVA9, and CVB5, which are highly similar in size and shape, exhibited vastly differing susceptibilities to protistan action.
The sole viral property for which we could identify a potential influence on susceptibility to protists is surface hydrophobicity. Qbeta, which is more hydrophobic than MS2,50 had a lower susceptibility to T. pyriformis. Similarly, MS2 is more hydrophobic51 and less susceptible to removal by protists compared to RoV. This trend is consistent with work by Gurijala and Alexander,20 who found that more hydrophobic bacteria were less susceptible to grazing by T. thermophila. This trend, however, was not confirmed for flagellates,58 which exhibited uptake rates of bacteria that were independent of bacterial cell hydrophobicity.
Our findings on differential removal of viruses by the model protist T. pyriformis in highly controlled settings is a first step toward a more general understanding of viral dynamics in natural freshwater systems as well as in reactors of activated sludge or similar settings. Importantly, both in natural systems as well as in activated sludge, we expect a much higher diversity of protists, and it is reasonable to assume that different protist species exhibit different feeding preferences. It has been shown that a higher diversity of protists has both positive and negative effects on ecosystem dynamics, depending on the variable of interest,59 and similar effects can be expected with respect to removal rates of viral particles in natural and engineered setting. As a next step, the interactions and possible additive effects of different protist species on viral removal should thus be studied, in order to transfer our findings to a more general and more realistic setting.
In summary, our findings demonstrate the capability of T. pyriformis to remove a range of mammalian viruses and bacteriophages, though the extent of removal varies between viruses. In addition, T. pyriformis movement analysis suggests that virus removal depends on the protists’ ability to recognize a given virus as food. In future work, these findings should be confirmed in environmentally relevant systems that include a diverse array of protists and bacteria, as well as nutrients, organic matter, and particles. Nevertheless, our results show that at environmentally relevant concentrations, protists can achieve a significant removal of viruses from the water column. Their contribution to the environmental fate of different viruses should therefore not be neglected.
Acknowledgments
This work was supported by the Swiss National Science Foundation (grants no. 31003A_182468 and PP00P3_179089). X.F.C. was a fellow of the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska–Curie Grant Agreement No. 754462. The authors thank Soile Blomqvist and Carita Savolainen-Kopra (Finnish National Institute for Health and Welfare) for providing environmental enterovirus isolates.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.1c05518.
Text describing virus origin, cell origin, propagation methods, and infectivity assays; tables of virus characteristics; detailed (RT-)qPCR procedures and LOQ values; MIQE table; infectivity LOQ values; results of statistical analyses; additional figures to validate the experimental setup; individual virus removal time courses; correlation between susceptibility to protists and virus properties (PDF)
The authors declare no competing financial interest.
Notes
All data and code used in this manuscript is available at https://doi.org/10.5281/zenodo.6225902.
Supplementary Material
References
- Feichtmayer J.; Deng L.; Griebler C. Antagonistic Microbial Interactions: Contributions and Potential Applications for Controlling Pathogens in the Aquatic Systems. Front. Microbiol. 2017, 8, 1–14. 10.3389/fmicb.2017.02192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliver D. O.; Herrmann J. E. Proteolytic and microbial inactivation of enteroviruses. Water Res. 1972, 6 (7), 797–805. 10.1016/0043-1354(72)90032-2. [DOI] [Google Scholar]
- Ward R. L.; Knowlton D. R.; Winston P. E. Mechanism of Inactivation of Enteric Viruses in Fresh-Water. Appl. Environ. Microbiol. 1986, 52 (3), 450–459. 10.1128/aem.52.3.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L.; Krauss S.; Feichtmayer J.; Hofmann R.; Arndt H.; Griebler C. Grazing of heterotrophic flagellates on viruses is driven by feeding behaviour. Environ. Microbiol Rep. 2014, 6 (4), 325–330. 10.1111/1758-2229.12119. [DOI] [PubMed] [Google Scholar]
- Chaudhry R. M.; Nelson K. L.; Drewes J. E. Mechanisms of pathogenic virus removal in a full-scale membrane bioreactor. Environ. Sci. Technol. 2015, 49 (5), 2815–2822. 10.1021/es505332n. [DOI] [PubMed] [Google Scholar]
- Olive M.; Gan C.; Carratalà A.; Kohn T.. Control of waterborne human viruses by indigenous bacteria and protists is influenced by temperature, virus type, and microbial species. Appl. Environ. Microbiol. 2020, 86 ( (3), ), 10.1128/AEM.01992-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh T. Y.; Gibson K. E. Interactions between human norovirus surrogates and Acanthamoeba spp. Appl. Environ. Microbiol. 2015, 81 (12), 4005–4013. 10.1128/AEM.00649-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkins M. A.; Dey R.; Ashbolt N. J. Interactions between Human Reovirus and Free-Living Amoebae: Implications for Enteric Virus Disinfection and Aquatic Persistence. Environ. Sci. Technol. 2020, 54 (16), 10201–10206. 10.1021/acs.est.0c02896. [DOI] [PubMed] [Google Scholar]
- Kim T. D.; Unno H. The roles of microbes in the removal and inactivation of viruses in a biological wastewater treatment system. Water Sci. Technol. 1996, 33 (10–11), 243–250. 10.2166/wst.1996.0681. [DOI] [Google Scholar]
- Pinheiro M. D. O.; Power M. E.; Butler B. J.; Dayeh V. R.; Slawson R.; Lee L. E. J.; Lynn D. H.; Bols N. C. Use of Tetrahymena thermophila to study the role of protozoa in inactivation of viruses in water. Appl. Environ. Microbiol. 2007, 73 (2), 643–649. 10.1128/AEM.02363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingel P.; NÕges T. Seasonal and annual population dynamics of ciliates in a shallow eutrophic lake. Fundam Appl. Limnol. 2010, 176 (2), 133–143. 10.1127/1863-9135/2010/0176-0133. [DOI] [Google Scholar]
- Warren A.; Salvado H.; Curds C. R.; Roberts D. M. L.. Protozoa in activated sludge. Microbial Ecology of Activated Sludge; Seviour R, Nielsen P. H., Eds.; IWA, 2010; Chapter 4. [Google Scholar]
- Sepp T.; Järvekülg L.; Saarma M. Investigations of virus-protozoa relationships in the model of the free-living ciliate Tetrahymena pyriformis and adenovirus type 3. Eur. J. Protistol. 1992, 28 (2), 170–174. 10.1016/S0932-4739(11)80046-5. [DOI] [PubMed] [Google Scholar]
- Pinheiro M. D. O.; Power M. E.; Butler B. J.; Dayeh V. R.; Slawson R.; Lee L. E. J.; Lynn D. H.; Bols N. C. Inactivation of the bacteriophage MS2 by the ciliated protozoan, Tetrahymena thermophila. Water Qual Res. J. Can. 2008, 43 (1), 69–76. 10.2166/wqrj.2008.009. [DOI] [Google Scholar]
- Teras J. H. Protozoal viruses and the interaction of protozoa with mammalian viruses. Int. J. Trop Insect Sci. 1986, 7 (03), 355–361. 10.1017/S1742758400009425. [DOI] [Google Scholar]
- Hsueh T. Y.; Gibson K. E. Interactions between human norovirus surrogates and Acanthamoeba spp. Appl. Environ. Microbiol. 2015, 81 (12), 4005–4013. 10.1128/AEM.00649-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro M. D. O.; Bols N. C. Activation of an Aquareovirus, Chum Salmon Reovirus (CSV), by the Ciliates Tetrahymena thermophila and T. canadensis. J. Eukaryot Microbiol. 2018, 65 (5), 694–704. 10.1111/jeu.12514. [DOI] [PubMed] [Google Scholar]
- Akunyili A. A.; Alfatlawi M.; Upadhyaya B.; Rhoads L. S.; Eichelberger H.; Van Bell C. T. Ingestion without inactivation of bacteriophages by Tetrahymena. J. Eukaryot Microbiol. 2008, 55 (3), 207–213. 10.1111/j.1550-7408.2008.00316.x. [DOI] [PubMed] [Google Scholar]
- Hennemuth W.; Rhoads L.; Eichelberger H.; Watanabe M.; Van Bell K. M.; Ke L.; Kim H.; Nguyen G.; Jonas J.; Veith D.; Van Bell C. T. Ingestion and Inactivation of Bacteriophages by. Tetrahymena. 2008, 55 (1), 44–50. 10.1111/j.1550-7408.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- Gurijala K. R.; Alexander M. Effect of growth rate and hydrophobicity on bacteria surviving protozoan grazing. Appl. Environ. Microbiol. 1990, 56 (6), 1631–1635. 10.1128/aem.56.6.1631-1635.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T. Suspension feeding in ciliated protozoa: Feeding rates and their ecological significance. Microb Ecol. 1980, 6 (1), 13–25. 10.1007/BF02020371. [DOI] [PubMed] [Google Scholar]
- Thurman J.; Parry J. D.; Hill P. J.; Laybourn-Parry J. The Filter-Feeding Ciliates Colpidium striatum and Tetrahymena pyriformis Display Selective Feeding Behaviours in the Presence of Mixed, Equally-Sized, Bacterial Prey. Protist 2010, 161, 577–588. 10.1016/j.protis.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Mueller M.; Röhlich P.; Törö I. Studies on Feeding and Digestion in Protozoa. VII. Ingestion of Polystyrene Latex Particles and its Early Effect on Acid Phosphatase in Paramecium multimicronucleatum and Tetrahymena pyriformis. Journal of Protozoology 1965, 12, 27–34. 10.1111/j.1550-7408.1965.tb01807.x. [DOI] [Google Scholar]
- Boenigk J.; Novarino G. Effect of suspended clay on the feeding and growth of bacterivorous flagellates and ciliates. Aquat Microb Ecol. 2004, 34 (2), 181–192. 10.3354/ame034181. [DOI] [Google Scholar]
- Altermatt F.; Fronhofer E. A.; Garnier A.; Giometto A.; Hammes F.; Klecka J.; Legrand D.; Mächler E.; Massie T. M.; Pennekamp F.; Plebani M.; Pontarp M.; Schtickzelle N.; Thuillier V.; Petchey O. L. Big answers from small worlds: A user’s guide for protist microcosms as a model system in ecology and evolution. Methods Ecol Evol. 2015, 6 (2), 218–231. 10.1111/2041-210X.12312. [DOI] [Google Scholar]
- Asai D.; Forney J.. Tetrahymena Thermophila. In Methods in Cell Biology, 1st ed.; Elsevier, 1999; Vol. 62. [Google Scholar]
- Thormar H. Effect of temperature on the reproduction rate of Tetrahymena pyriformis. Exp. Cell Res. 1962, 28, 269–279. 10.1016/0014-4827(62)90283-5. [DOI] [PubMed] [Google Scholar]
- Carratalà A.; Shim H.; Zhong Q.; Bachmann V.; Jensen J. D.; Kohn T. Experimental adaptation of human echovirus 11 to ultraviolet radiation leads to resistance to disinfection and ribavirin. Virus Evol. 2017, 3 (2), 1–11. 10.1093/ve/vex035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A. M.; Mazzocco A. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. Bacteriophages: Methods and Protocols, Vol. 1: Isolation, Characterization, and Interactions; Clokie M. R. J., Kropinski A. M., Eds.; Humana Press, 2009; pp 69–76, 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- Ismail N. S.; Olive M.; Fernandez-Cassi X.; Bachmann V.; Kohn T. Viral Transfer and Inactivation through Zooplankton Trophic Interactions. Environ. Sci. Technol. 2020, 54 (15), 9418–9426. 10.1021/acs.est.0c02545. [DOI] [PubMed] [Google Scholar]
- Merkes C. M.; Klymus K. E.; Allison M. J.; Goldberg C.; Helbing C. C.; Hunter M. E.; Jackson C. A.; Lance R. F.; Mangan A. M.; Monroe E. M.; Piaggio A. J.; Stokdyk J. P.; Wilson C. C.; Richter C. Generic qPCR Limit of Detection (LOD)/Limit of Quantification (LOQ) calculator. R Script. https://github.com/cmerkes/qPCR_LOD_Calc. Published online 2019. 10.5066/P9GT00GB. [DOI]
- Bustin S. A.; Benes V.; Garson J. A.; Hellemans J.; Huggett J.; Kubista M.; Mueller R.; Nolan T.; Pfaffl M. W.; Shipley G. L.; Vandesompele J.; Wittwer C. T. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009, 55 (4), 611–622. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Methods for Measuring the Acute Toxicity of Effluents and Receiving Waters to Freshwater and Marine Organisms, 5th ed.; United States Enviromental Protection Agency, 2002; p 266.
- Lavin D. P.; Hatzis C.; Srienc F.; Fredrickson A. G. Size Effects on the Uptake of Particles by Populations of Tetrahymena pyriformis Cells. J. Protozool. 1990, 37 (3), 157–163. 10.1111/j.1550-7408.1990.tb01120.x. [DOI] [Google Scholar]
- Fronhofer E. A.; Kropf T.; Altermatt F. Density-dependent movement and the consequences of the Allee effect in the model organism Tetrahymena. J. Anim Ecol. 2015, 84 (3), 712–722. 10.1111/1365-2656.12315. [DOI] [PubMed] [Google Scholar]
- Deuer S. M.; Grünbaum D. Individual foraging behaviors and population distributions of a planktonic predator aggregating to phytoplankton thin layers. Limnol Oceanogr. 2006, 51 (1), 109–116. 10.4319/lo.2006.51.1.0109. [DOI] [Google Scholar]
- Harvey E. L.; Jeong H. J.; Menden-Deuer S. Avoidance and attraction: Chemical cues influence predator-prey interactions of planktonic protists. Limnol Oceanogr. 2013, 58 (4), 1176–1184. 10.4319/lo.2013.58.4.1176. [DOI] [Google Scholar]
- Cairns J.; Moerman F.; Fronhofer E. A.; Altermatt F.; Hiltunen T. Evolution in interacting species alters predator life-history traits, behaviour and morphology in experimental microbial communities. Proc. Royal Soc. B 2020, 287 (1928), 20200652. 10.1098/rspb.2020.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennekamp F.; Schtickzelle N.; Petchey O. L. BEMOVI, software for extracting behavior and morphology from videos, illustrated with analyses of microbes. Ecol Evol. 2015, 5 (13), 2584–2595. 10.1002/ece3.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronhofer E. A.; Altermatt F. Eco-evolutionary feedbacks during experimental range expansions. Nat. Commun. 2015, 6, 6. 10.1038/ncomms7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman F.; Fronhofer E. A.; Wagner A.; Altermatt F. Gene swamping alters evolution during range expansions in the protist Tetrahymena thermophila. Biol. Lett. 2020, 16 (6), 20200244. 10.1098/rsbl.2020.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman F.; Arquint A.; Merkli S.; Wagner A.; Altermatt F.; Fronhofer E. A. Evolution under pH stress and high population densities leads to increased density-dependent fitness in the protist Tetrahymena thermophila. Evolution. 2020, 74 (3), 573–586. 10.1111/evo.13921. [DOI] [PubMed] [Google Scholar]
- Pinheiro J.; Bates D.; DebRoy S.; Sarkar D.; R Core Team . Nlme: Linear and Nonlinear Mixed Effects Models. Published online 2020. [Google Scholar]
- Morrell C. H. Likelihood Ratio Testing of Variance Components in the Linear Mixed-Effects Model Using Restricted Maximum Likelihood. Biometrics. 1998, 54 (4), 1560. 10.2307/2533680. [DOI] [PubMed] [Google Scholar]
- Team. RC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- GraphPad Software. www.graphpad.com.
- Grønlien H. K.; Berg T.; Løvlie A. M. In the polymorphic ciliate Tetrahymena vorax the non-selective phagocytosis seen in microstomes changes to a highly selective process in macrostomes. J. Exp Biol. 2002, 205 (14), 2089–2097. 10.1242/jeb.205.14.2089. [DOI] [PubMed] [Google Scholar]
- Fronhofer E. A.; Kropf T.; Altermatt F. Density-dependent movement and the consequences of the Allee effect in the model organism Tetrahymena. J. Anim Ecol. 2015, 84 (3), 712–722. 10.1111/1365-2656.12315. [DOI] [PubMed] [Google Scholar]
- Armanious A.; Aeppli M.; Jacak R.; Refardt D.; Sigstam T.; Kohn T.; Sander M. Viruses at Solid-Water Interfaces: A Systematic Assessment of Interactions Driving Adsorption. Environ. Sci. Technol. 2016, 50 (2), 732–743. 10.1021/acs.est.5b04644. [DOI] [PubMed] [Google Scholar]
- Farkas K.; Varsani A.; Pang L. Adsorption of Rotavirus, MS2 Bacteriophage and Surface-Modified Silica Nanoparticles to Hydrophobic Matter. Food Environ. Virol. 2015, 7 (3), 261–268. 10.1007/s12560-014-9171-3. [DOI] [PubMed] [Google Scholar]
- Benyahya M.; Laveran H.; Bohatier J.; Senaud J.; Ettayebi M. Interactions between the Ciliated Protozoan Tetrahymena pyriformis and the Simian Rotavirus SA11. Eur. J. Protistol. 1997, 33, 211–213. 10.1016/S0932-4739(97)80038-7. [DOI] [Google Scholar]
- Pérez-Prieto S. G. G. A. Uptake of vaccinia virus by Tetrahymena pyriformis. Microbiol Esp.1 1981, 34, 29–43. [PubMed] [Google Scholar]
- Groupé V.; Pugh L. H. Inactivation of Influenza Virus and of Viral Hemagglutinin by the Ciliate Tetrahymena. Science. 1952, 115, 307–308. 10.1126/science.115.2986.307. [DOI] [PubMed] [Google Scholar]
- Danes L.; Cerva L. Poliovirus and echovirus survival in Tetrahymena pyriformis culture in vivo. J. Hyg Epidemiol. Microbiol. Immunol. 1984, 28 (2), 193–200. [PubMed] [Google Scholar]
- Dürichen H.; Siegmund L.; Burmester A.; Fischer M. S.; Wöstemeyer J. Ingestion and digestion studies in Tetrahymena pyriformis based on chemically modified microparticles. Eur. J. Protistol. 2016, 52, 45–57. 10.1016/j.ejop.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Silverman A. I.; Boehm A. B. Systematic Review and Meta-Analysis of the Persistence and Disinfection of Human Coronaviruses and Their Viral Surrogates in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7 (8), 544–553. 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- Matz C.; Jürgens K. Effects of Hydrophobic and Electrostatic Cell Surface Properties of Bacteria on Feeding Rates of Heterotrophic Nanoflagellates. Appl. Environ. Microbiol. 2001, 67 (2), 814–820. 10.1128/AEM.67.2.814-820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennekamp F.; Pontarp M.; Tabi A.; Altermatt F.; Alther R.; Choffat Y.; Fronhofer E. A.; Ganesanandamoorthy P.; Garnier A.; Griffiths J. I.; Greene S.; Horgan K.; Massie T. M.; Mächler E.; Palamara G. M.; Seymour M.; Petchey O. L. Biodiversity increases and decreases ecosystem stability. Nature. 2018, 563 (7729), 109–112. 10.1038/s41586-018-0627-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.