Abstract
Human immunodeficiency virus type 1 (HIV-1) produces two polyproteins, Pr55Gag and Pr160Gag-Pol, that are cleaved into mature functional subunits by the virally encoded protease. Drugs that inhibit this protease are an important part of anti-HIV therapy. We studied the ordered accumulation of Gag and Gag-Pol processing intermediates by variably blocking the protease with HIV-1 protease inhibitors (PIs). Variable protease inhibition caused accumulation of a complex pattern of processing intermediates, which was the same after incubating HIV-1-infected cells with increasing concentrations of either one of the peptidomimetic inhibitors indinavir, saquinavir (SQV), ritonavir (RTV), nelfinavir, and SC-52151 or one of the nonpeptidomimetic inhibitors DMP450, DMP323, PNU-140135, and PNU-109112 for 3 days. The patterns of Gag and Gag-Pol processing intermediate accumulation were nearly identical when the following were compared: cell- versus virion-associated proteins, HIV-1-infected transformed cell lines versus primary human peripheral blood mononuclear cells (PBMCs) and HIV-1MN versus HIV-1IIIB virus strains. RTV was a more potent inhibitor of p24 production in PBMCs than SQV by approximately 7-fold, whereas SQV was a more potent inhibitor in transformed cells than RTV by approximately 30-fold. Although the antiretroviral potency of HIV-1 PIs may change as a function of cell type, the polyprotein intermediates that accumulate with increasing drug concentrations are the same. These results support sequential processing of Gag and Gag-Pol polyproteins by the HIV-1 protease and may have important implications for understanding common cross-resistance pathways.
HIV-1 encodes two polyproteins, Pr55Gag and Pr160Gag-Pol, that are cleaved by the virally encoded aspartyl protease. The active protease is capable of recognizing and cleaving a diverse array of amino acid sequences; at least 11 cleavage sites within the Gag and Gag-Pol polyproteins have been reported (28). The processing of Pr55Gag and Pr160Gag-Pol into functional, mature subunits is a complex and essential step in human immunodeficiency virus type 1 (HIV-1) maturation (21). Gag is cleaved to yield six proteins (p17 [matrix], p24 [capsid], p2 [Sp1], p7 [nucleocapsid], p1 [Sp2], and p6), and Pol is cleaved to yield three enzymatic proteins (p10 [protease], p66/51 [reverse transcriptase {RT}], and p32 [integrase]).
The HIV-1 protease is an important target for antiretroviral drug therapy. Peptidomimetic and nonpeptidomimetic competitive inhibitors with selective toxicity for the HIV-1 aspartyl protease have been synthesized by mimicking a unique cleavage site that human proteases do not cleave efficiently (6). Of these, only peptidomimetic inhibitors are approved for current clinical use. Previous reports indicated that high concentrations of protease inhibitors (PIs) failed to completely block the processing of Gag and Gag-Pol polyproteins (27, 31). In the presence of PIs, the concentration of mature, fully processed viral proteins decreases and the concentration of proteins that are not fully processed increases. We refer to the latter proteins as processing intermediates. These processing intermediates could be the result of either failure of the protease to cleave an established site or aberrant cleavage events.
Processing intermediates that accumulate in the presence of PIs are not the result of cleavage by cellular proteases. We show herein that cells harboring HIV-1 constructs that lack a functional protease do not produce the same intermediates; in addition, others have shown that no mammalian aspartyl proteases can efficiently cleave the Gag polyprotein (10). In our study, we systematically compared accumulation of processing intermediates after inhibiting the viral protease with increasing concentrations of numerous PIs.
Four clinically available PIs were used: indinavir (IDV), nelfinavir (NFV), ritonavir (RTV), and saquinavir (SQV). IDV powder was a gift from Merck Research Laboratories (West Point, Pa.); NFV powder was a gift from Agouron Laboratories (Torrey Pines, Calif.); RTV powder was a gift from Abbott Laboratories (Abbott Park, Ill.); SQV powder was a gift from Roche Discovery (Welwyn Garden City, United Kingdom). The concentrations of drugs used in our inhibition experiments were based on reported 50% inhibitory concentrations (IC50s), i.e., concentrations reported to inhibit HIV-1 replication by 50 percent. The specific IC50s used to design experiments were based on data from Lazdins et al. (23) and were 10 nM for SQV, 20 nM for IDV, and 20 nM for RTV. The IC50 used in NFV experiments (40 nM) was based on data from Patick et al. (32). The IC50 for PNU-140135 experiments was 100 nM (B. J. Bruce [Pharmacia and Upjohn, Inc., Kalamazoo, Mich.], personal communication). The IC50s that we calculated from experimental data were the concentrations of drugs required to inhibit the p24/p55 ratio of protein production by 50% calculated by using NIH Image software (National Institutes of Health, Bethesda, Md.). In addition, we obtained the investigational PI SC-52151 from Searle Laboratories (Skokie, Ill.) (2). DMP450 and DMP323 were kindly provided by Susan Erickson-Viitanen, Dupont Pharmaceuticals, Wilmington, Del. PNU-140135 and PNU-109112 were kindly provided by Barbara J. Bruce, Pharmacia and Upjohn, Inc.
To examine the effect of PIs on HIV-1 polyprotein processing, cell lines were each washed twice with medium and then plated at a concentration of 105 cells per ml in medium containing the appropriate concentration of PI. The medium was replaced with fresh medium containing the same concentration of PI after 24 h and thereafter every 48 h. Cells were cultured for 3 or 7 days before cell-associated and viral-associated proteins were isolated. For all experiments, we used PI concentrations of 500 to 0.03 times the published IC50s of the drugs. The highest concentration of NFV was 250 times the published IC50 because only limited quantities of pure drug were available. PNU-140135 and PNU-109112 were used in the range of 300 to 0.03 times the published IC50 because of limited solubility. Virion-associated proteins were isolated and examined by immunoblotting (25). For a positive control, we used an H9 cell line which contained a mutated HIV-1 with a nonfunctional protease to show complete inhibition of the protease (26). We also used chronically infected cell lines with no inhibition of the protease as a positive control. For a negative control, we used mock-infected cell lines.
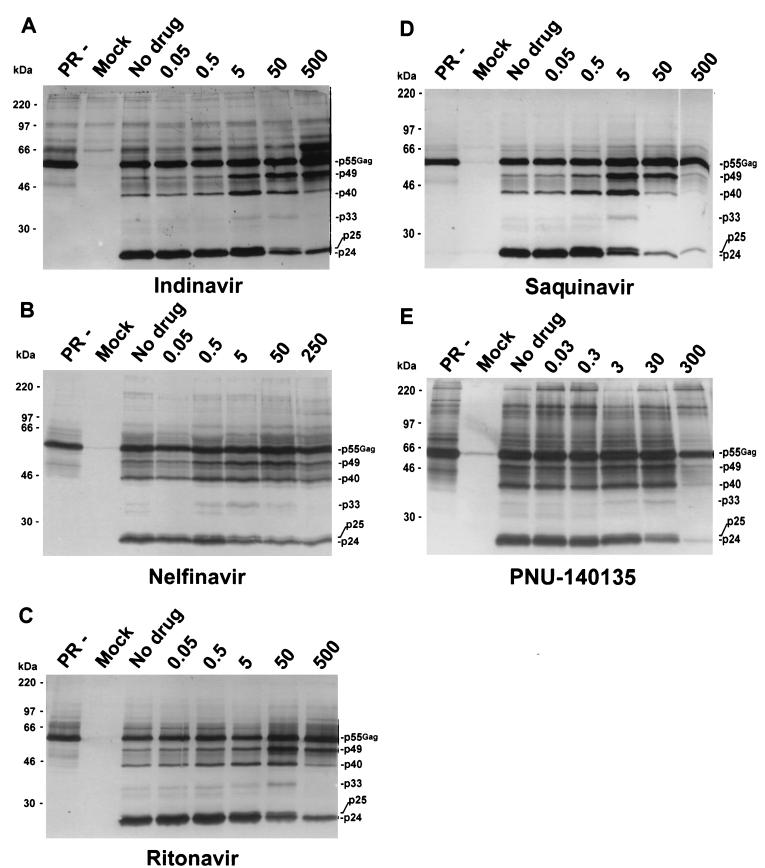
With increasing inhibition of the HIV-1 protease, HIV-1-specific Gag (Fig. 1) and Gag-Pol (data not shown) processing intermediates appeared in released virions and fully processed protein concentrations decreased. These observations were consistent with previous reports (5, 34, 39). The assigned nomenclature for the processing intermediates was based on their estimated molecular weights and on the previous nomenclatures developed by Pettit et al. (35) and by Lindhofer et al. (27).
FIG. 1.
Western blot analysis of Pr55Gag virion-associated proteins. H9 cells chronically infected with HIV-1IIIB were incubated with IDV (A), NFV (B), RTV (C), SQV (D), or PNU-140135 (E) for 3 days. Concentrations were normalized to the respective drug's reported IC50, and the values shown above the five rightmost lanes of each panel indicate concentrations relative to the IC50 of each drug. Proteins were processed and detected with anti-HIV-1 human serum as described in the text. PR−, H9 cells infected with an HIV-1 provirus construct that has an inactive protease (see text); Mock, mock-infected cells; No drug, untreated cells. Molecular weight markers are indicated on the left. Gag-specific proteins are indicated on the right of each blot.
Using purified virions from a chronically infected cell line, we found that different processing intermediates accumulated as a function of the magnitude of protease inhibition. The pattern of accumulated intermediates was similar for all drugs tested. Figure 1 shows the patterns of accumulation of proteins in released virions after inhibition of the HIV-1 protease with IDV, NFV, RTV, SQV, and PNU-140135. The immunoblots were probed with HIV-1-positive human serum. Numerous proteins demonstrate the similarity of processing intermediate accumulation with different drugs. Protein intermediates that are highlighted include 49-kDa (p49), 40-kDa (p40), 33-kDa (p33), and 25-kDa (p25) proteins. As an example of similar intermediate accumulation, p33 is not detectable at 500× indinavir, 250× nelfinavir, 500× ritonavir, 50× saquinavir, or 300× PNU-140135 (for each drug, the 1× concentration is the published IC50); however, it is present at a 10-fold-lower concentration of each drug (Fig. 1). The pattern of accumulation of processing intermediates was not entirely identical for all drugs tested. These slight differences could be the result of either the drug concentrations tested or small differences in protease inhibition.
We also studied accumulation of Gag and Gag-Pol processing intermediates with SC-52151, an investigational peptidomimetic PI (data not shown), and with DMP323, DMP450, and PNU-109112, nonpeptidic PIs (data not shown). The results obtained with these drugs were identical to those found with the other PIs for Gag (Fig. 1) and Gag-Pol (data not shown) processing intermediates.
Although we normalized drug concentrations to their previously reported IC50s, there was an approximately 10-fold shift in apparent potency when comparing SQV with the other drugs. To determine if differences in drug potency were due to discrepancies between the reported IC50s and actual IC50s in our experimental system, we calculated IC50s by measuring PI effect on the p24/p55 ratio. The calculated IC50s were 20 nM, 500 nM, 500 nM, 900 nM and 1,500 nM for SQV, RTV, IDV, NFV, and PNU-140135 in the transformed cell line H9 infected with HIV-1IIIB. The calculated IC50s were 70 nM for SQV and 10 nM for RTV in peripheral blood mononuclear cells (PBMCs) infected with HIV-1IIIB. We therefore believe that apparent differences in potency were explained by differences between reported and calculated IC50s. Thus, inhibition of Gag and Gag-Pol processing in these experiments paralleled inhibition of p24 antigen production and, by inference, production of infectious virions. Previous reports demonstrate that IC50s for PIs can vary widely depending on the cell type, virus type, and protein concentrations used in experiments to obtain these values (19, 23, 32, 39).
Processing intermediates were not present in cell lines harboring a protease-negative provirus but were detectable even at the highest PI concentrations tested; 500× IDV, 250× NFV, 500× RTV, 500× SQV, and 300× PNU-140135 (Fig. 1). The most prominent intermediates were p40 and p49. The intermediates detected in the presence of the highest concentration of drug did not appear to represent residual proteins that had been present before drug was added, because these intermediates either were not present or were greatly diminished in untreated infected cultures (Fig. 1).
After 3 days of incubation with the highest drug concentrations, we could still detect both virion-associated p24 (Fig. 1) and cell-associated p24 (data not shown) in immunoblots of chronically infected T-cell lines and acutely infected PBMCs. Previous publications have reported varying levels of inhibition of p24 production with PIs in chronically infected cells (37, 39) and with different cell types (34, 36). To determine if this p24 was newly cleaved protein or residual protein that had been present prior to the addition of drug, we incubated cells with drug for an additional 4 days, while maintaining the same schedule for medium changes. The concentrations of virion-associated and cell-associated p24 after 7 days of incubation with 500× SQV or 50× RTV were greatly diminished compared to those observed after 3 days of incubation (data not shown). The p24 seen in virion-associated proteins after 3 or 7 days of drug exposure was unlikely to represent residual virions present in the supernatant before the addition of drug because cells were washed two times prior to the addition of drug and a complete medium change was performed at 24 h and thereafter every 48 h. The accumulation of Gag and Gag-Pol processing intermediates after 7 days of incubation with SQV and RTV was highly similar to that seen after 3 days of incubation with the same drugs (data not shown).
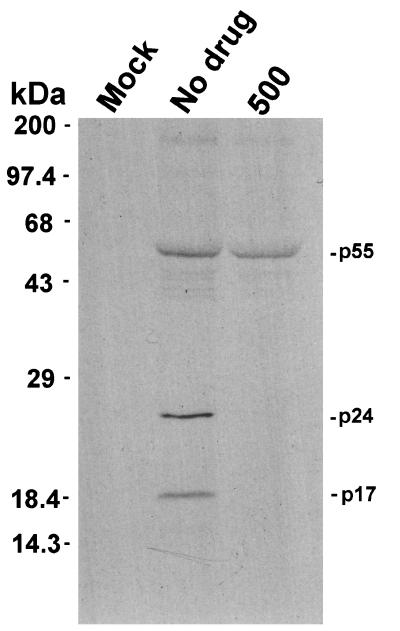
We also immunoprecipitated virion-associated proteins from H9 cells chronically infected with HIV-1IIIB that were incubated with 500× SQV for 30 min and then labeled with [35S]cysteine for 12 h. For radiolabeling and radioimmunoprecipitation (RIP) experiments, H9 and HIV-1IIIB-infected H9 cells (5 × 106) were washed with 1× phosphate-buffered saline (PBS) and resuspended at 106 cells per ml in cysteine-free RPMI 1640 media (Gibco, Gaithersburg, Md.). SQV was added at a final concentration of 5 μM (500× SQV). Replicate flasks contained no PI. Flasks were incubated at 37°C for 30 min and labeled with 100 μCi of [35S]cysteine (NEN, Boston, Mass.) per ml for 12 h. Virion-associated proteins were purified as previously described (25). Viral pellets were resuspended in 50 μl of RIPA buffer (0.15 M NaCl, 0.05 M Tris [pH 7.2], 1% Triton X-100, 1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). RIP was carried out overnight on a rotator shaker at 4°C. The RIP reaction mixture contained 10 μl of HIV-1-positive human serum, 15 μl of 10% staphylococcal protein A-Sepharose CL-4B (Sigma, St. Louis, Mo.) in 1× PBS, and 25 μl of the resuspended virion-associated proteins. Samples were washed five times with RIPA buffer without deoxycholate, resuspended in 30 μl of 1× running buffer, boiled for 3 min, and separated by SDS-polyacrylamide gel electrophoresis (PAGE). We could detect no newly created, radiolabeled p24 protein in released virions in the presence of 500× SQV (Fig. 2). These data suggest that the p24 observed with the highest concentrations of each inhibitor was residual protein that was present before addition of the drug. Residual, previously synthesized p24 remained in the cell and was released into the supernatant, associated with virions, for up to 7 days after addition of the PIs.
FIG. 2.
RIP of virion-associated processing intermediates. H9 cells chronically infected with HIV-1IIIB were incubated with SQV for 30 min, after which the proteins were radiolabeled for 12 h with [35S]cysteine, isolated, immunoprecipitated with HIV-1 human antiserum, and separated by PAGE. Molecular weight markers are indicated on the left. Gag specific proteins are indicated on the right of the blot. Mock, mock-infected cells; No drug, untreated cells; 500, cells treated with 500× SQV.
These results indicate that HIV p24 is stable within the cell for at least 3 to 7 days after the addition of PIs and may be released into the supernatant for up to 7 days, probably in the form of immature virus particles. This result is in agreement with the findings of Lee and Yu. (26), who identified a stable virus assembly intermediate complex associated with the cell that contained cleaved viral proteins similar to those found in cell-free virions but distinct from released virions in terms of salt and protease K sensitivity. These subviral complexes could serve as a reservoir for rapid reemergence of infectious virions after PIs are removed from culture or after drug concentrations drop below some critical threshold in treated patients.
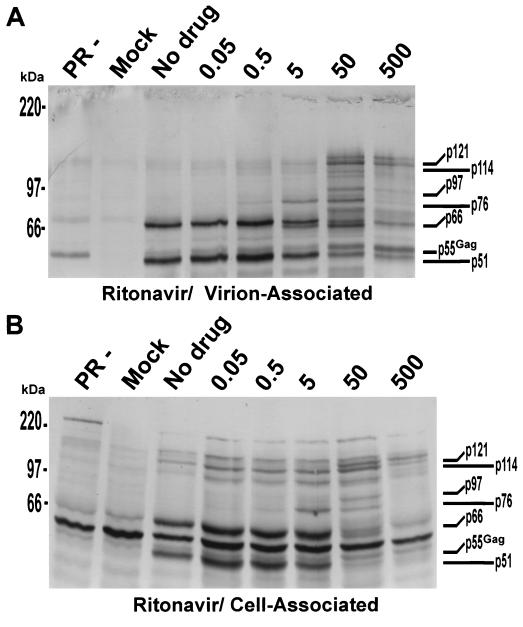
In conjunction with virion-associated proteins, we isolated and examined cell-associated proteins as previously described (25). Although the background level in the cell-associated protein immunoblots was higher, accumulation of HIV-1 Gag-Pol-specific processing intermediates was nearly identical to that observed with virions (Fig. 3). For example, the doublet p76-p97 seen in virion-associated proteins treated with 50× RTV (Fig. 3A) was conserved in cell-associated proteins treated with 50× RTV (Fig. 3B). The protein doublet p114-p121 was also conserved at 500× RTV (Fig. 3A and B). This indicates that the HIV-1 protease is active intracellularly and can cleave Pr55Gag and Pr160Gag-Pol to an extent similar to that observed when it is incorporated into the virus particle. This agrees with previous reports of Kaplan et al. (15, 16). Inhibition of processing in cell-associated proteins was similar to that in cell-free virions; this demonstrates that inhibition of processing by PIs may be initiated within the cell rather than the virion. Humphrey et al. (14) showed that removal of certain PIs from purified virions did not result in an increase in infectivity or the appearance of mature virion morphology, supporting this conclusion. The RT-specific monoclonal antibody used in our experiments had weak cross-reactivity with p24. This cross-reactivity explains the presence of Pr55Gag, as shown in Fig. 3.
FIG. 3.
Comparison of Pr160Gag-Pol cellular and virion-associated proteins by immunoblotting. H9 cells chronically infected with HIV-1IIIB were incubated with RTV for 3 days. Virion-associated (cell-free) (A) and cell-associated (B) proteins were separated, processed, and detected with anti-RT-specific monoclonal antibodies as described in the text. Molecular weight markers are indicated on the left. Gag-Pol specific proteins are indicated on the right of each blot. PR−, H9 cells infected with an HIV-1 provirus construct that has an inactive protease; Mock, mock-infected cells; No drug, untreated cells. The numbers above the five rightmost lanes of each panel indicate RTV concentrations.
After observing the same accumulation of processing intermediates with PIs in H9 cell lines chronically infected with HIV-1IIIB we wanted to confirm that this effect was neither cell line nor virus strain specific. We used SQV as a representative PI in these experiments. To study virus strain effects, we compared two H9 cell lines, one chronically infected with HIV-1IIIB and the other chronically infected with HIV-1MN. For cell line effects, we compared H9 cells chronically infected with HIV-1MN to CEM cells chronically infected with HIV-1MN. The accumulation of Pr55Gag and Pr160Gag-Pol processing intermediates was not dependent on the cell line or the laboratory virus strain used in chronically infected cells incubated with SQV (data not shown).
An apparent shift in PI potency was evident when patterns of accumulation of processing intermediates were compared for the same chronically infected cell line, H9, infected with different virus strains, HIV-1IIIB or HIV-1MN. The same protein intermediates accumulated, with a 10-fold-lower concentration of SQV, in H9 cells chronically infected with HIV-1IIIB as in cells infected with HIV-1MN. The pattern of processing intermediates that accumulated in the presence of 0.5×, 5×, and 50× SQV with H9 cells chronically infected with HIV-1IIIB was similar to that of those that accumulated in the presence of 5×, 50×, and 500× SQV, respectively, with HIV-1MN. One factor contributing to the shift may be that the enzyme cleavage sites in HIV-1MN could be more resistant to the effects of SQV than the enzyme cleavage sites in HIV-1IIIB. If this were true, more drug would be needed to inhibit processing of intermediates in HIV-1MN-infected cells than in HIV-1IIIB-infected cells. Another factor that could contribute to this shift in potency is differential accumulation of inhibitor in cells infected with the different virus strains due, perhaps, to shifts in the expression of drug transporters like P-glycoprotein.
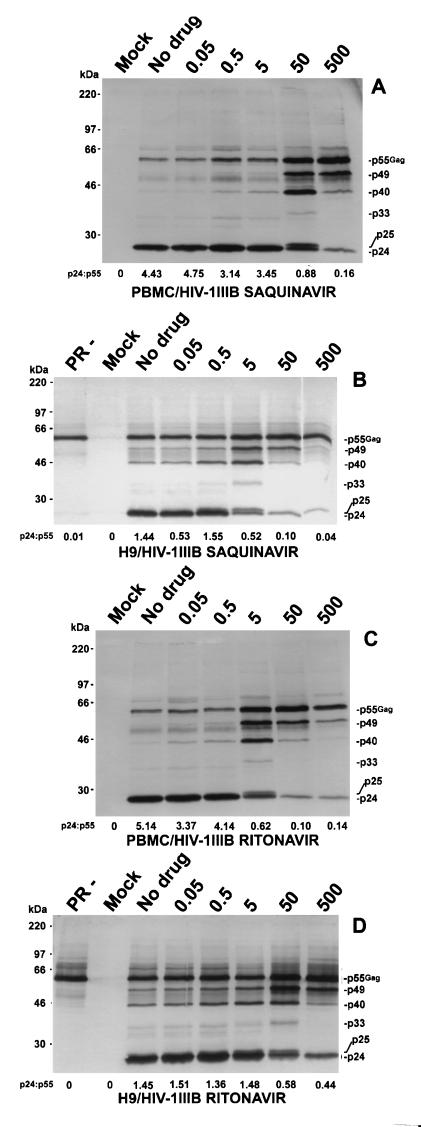
Antiretroviral drugs may behave differently in chronically infected cell lines and in acutely infected primary cells (34). We therefore studied the accumulation of processing intermediates in PBMCs that were acutely infected with HIV-1IIIB. We infected 2 × 105 PBMCs with 100,000 cpm of cell-free HIV-1IIIB as determined by RT assay; methods were as previously described (42). The relative accumulation of processing intermediates was nearly identical for acutely infected PBMCs with chronically infected H9 cells in the presence of SQV (Fig. 4A and B) or RTV (Fig. 4C and D). The protein intermediates p49, p40, p33, and p25, which were present in chronically infected H9 cells (Fig. 4B and D), were also present in acutely infected PBMCs (Fig. 4A and C).
FIG. 4.
Western blot analysis of virion-associated proteins by immunoblotting in acutely and chronically infected cells. Pr55Gag processing intermediates from PBMCs acutely infected for 10 days with HIV-1IIIB and from H9 cells chronically infected with HIV-1IIIB were incubated with five different concentrations each of SQV and RTV for 3 days (concentrations are indicated above the five rightmost lanes of each panel). Proteins were processed and detected with anti-HIV-1 human serum as described in the text. Molecular weight markers are indicated on the left. Gag specific proteins are indicated on the right of each blot. PR−, H9 cells infected with an HIV-1 provirus construct that has an inactive protease; Mock, mock-infected cells; No drug, untreated cells. (A) PBMCs incubated with SQV; (B) H9 cells incubated with SQV; (C) PBMCs incubated with RTV; (D) H9 cells incubated with RTV.
Processing intermediates accumulating in the cell line H9 chronically infected with HIV-1IIIB appeared at a 10-fold-lower concentration of SQV than they did in acutely infected PBMCs (Fig. 4). SQV was a more potent inhibitor of processing in H9 cells, whereas RTV was a more potent inhibitor of processing in PBMCs. For example, p33 was most prominent at 50× SQV in PBMCs (Fig. 4A) but at 5× SQV in H9 cells chronically infected with HIV-1IIIB (Fig. 4B). The reverse was true for RTV. Processing intermediates accumulating in H9 cells chronically infected with HIV-1IIIB required a 10-fold-higher RTV concentration than those accumulating in acutely infected PBMCs. For example, p33 was most prominent at 5× RTV in PBMCs (Fig. 4C) but at 50× RTV in H9 cells chronically infected with HIV-1IIIB (Fig. 4D). RTV was a more potent inhibitor of p24 production in PBMCs than SQV by approximately 7-fold, whereas SQV was a more potent inhibitor in transformed cells than RTV by approximately 30-fold. All experiments were repeated with similar results.
We do not know the mechanism underlying this differential potency of SQV and RTV in transformed cells and PBMCs. Our calculated IC50s for these drugs were different for the two types of cells and were consistent with processing inhibition seen on immunoblots. Intracellular drug concentrations might differ for these two drugs in different cell types. One possibility is differential expression and activity of the human multidrug resistance transporter, P-glycoprotein. Recent reports show that HIV-1 PIs are P-glycoprotein substrates; in addition, there is evidence that some of these drugs are inhibitors of P-glycoprotein (20, 24, 40). If, as suggested by Lee et al. (24) and Washington et al. (40), RTV inhibits P-glycoprotein, then the intracellular concentrations of RTV could be higher than concentrations of SQV in PBMCs. We are currently conducting experiments to investigate this possibility.
The ordered accumulation of processing intermediates as a function of drug concentration suggests that inhibition of processing events is not random. Random inhibition would have produced similar patterns of processing intermediates regardless of drug concentration and greater amounts of all intermediates with higher concentrations of drug. As shown in Fig. 1, patterns of accumulation of processing intermediates were similar and specific as a function of drug concentration. The accumulation of intermediates with peptidomimetic inhibitors was similar to that observed with four nonpeptidic, competitive inhibitors, DMP450, DMP323, PNU-109112, and PNU-140135 (data not shown). These results indicate that (i) the protease's ability to cleave target sites is similarly inhibited by all nine competitive inhibitors tested and (ii) cleavage is not random. The latter conclusion is consistent with previous reports on recombinant proteins that suggest the HIV-1 protease cleaves Gag and Gag-Pol sequentially (8, 35, 41). Although we did not identify by protein sequencing which Pr55Gag and Pr160Gag-Pol intermediates accumulated in our immunoblots, most of the Gag-specific proteins corresponded to intermediates identified by Pettit et al. (35) and most of the Gag-Pol specific proteins corresponded to intermediates identified by Lindhofer et al. (27). Our results support sequential cleavage of Gag and Gag-Pol; however, we cannot exclude the possibility that tested inhibitors had different Kis for the same cleavage sites and had identical access to all cleavage sites.
This research and previous studies (8, 35, 41) support sequential ordered substrate cleavage by the HIV-1 protease. If this is indeed true, there may exist a critical initial cleavage event that is required before all other cleavages can occur, and there may also exist a critical cleavage event or events that allow immature virions to become mature and infectious. The identification of such critical steps could allow for specific targeting of new inhibitors. Serio et al. (38) indirectly addressed this theory by developing anti-HIV agents that mimicked specific HIV-1 protease cleavage sites. The degree of inhibition of viral replication in these experiments depended on the cleavage site employed, with the cleavage site between p24 and p2 being the most efficient, capable of completely abolishing virus infectivity.
These findings may also be important in understanding the antiretroviral benefits of dual PI therapy in vivo. Combination therapy studies with RTV-SQV and other dual PI regimens show very promising clinical results (18, 30). The benefits of dual PI therapy may be the consequence of advantageous pharmacokinetic interactions, slower emergence of resistance, or differential inhibition of the protease by two different drugs. Our results suggest that the last explanation is least likely to be true. We have shown that the PIs tested do not have a differential effect on inhibition of the protease as a function of processing intermediate accumulation. Further, combining RTV and SQV in our experimental system did not alter the pattern of accumulation of processing intermediates observed with SQV or RTV alone (data not shown).
Our results may be important in understanding the emergence of cross-resistance to HIV-1 PIs. Previous reports showed that the majority of primary mutations leading to PI resistance were different for different drugs, whereas secondary mutations that confer cross-resistance had substantial overlap (1, 4). We have shown that, although these PIs are structurally diverse, all the tested drugs likely inhibit the protease's ability to cleave its target sites by an identical pathway. This may help explain why secondary mutations that confer a cross-resistant phenotype are so highly conserved among different PIs and why cross-resistance to these drugs is so broad. If all competitive inhibitors follow the same sequential pathway in blocking cleavage events, then the same compensatory changes in the protease enzyme could have an equivalent impact on these drugs and confer cross-resistance.
REFERENCES
- 1.Boden D, Markowitz M. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 1998;42:2775–2783. doi: 10.1128/aac.42.11.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant M, Getman D, Smidt M, et al. SC-52151, a novel inhibitor of the human immunodeficiency virus protease. Antimicrob Agents Chemother. 1995;39:2229–2234. doi: 10.1128/aac.39.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong K T, Pagano P J. In vitro combination of PNU-140690, a human immunodeficiency virus type 1 protease inhibitor, with ritonavir against ritonavir-sensitive and -resistant clinical isolates. Antimicrob Agents Chemother. 1997;41:2367–2373. doi: 10.1128/aac.41.11.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 5.Craig J C, Duncan I B, Hockley D, Grief C, Roberts N A, Mills J S. Antiviral properties of Ro 31-8959, an inhibitor of human immunodeficiency virus (HIV) proteinase. Antivir Res. 1991;16:295–305. doi: 10.1016/0166-3542(91)90045-s. [DOI] [PubMed] [Google Scholar]
- 6.Debouck C. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res Hum Retrovir. 1992;8:153–164. doi: 10.1089/aid.1992.8.153. [DOI] [PubMed] [Google Scholar]
- 7.Deminie C A, Bechtold C M, Stock D, Alam M, Djang F, Balch A H, Chou T C, Prichard M, Colonno R J, Lin P F. Evaluation of reverse transcriptase and protease inhibitors in two-drug combinations against human immunodeficiency virus replication. Antimicrob Agents Chemother. 1996;40:1346–1351. doi: 10.1128/aac.40.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe D E, Tritch R, Hutchison III C A, Loeb D D, Swanstrom R. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retrovir. 1989;5:577–591. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 9.Flexner C. HIV protease inhibitors. N Engl J Med. 1998;338:1281–1292. doi: 10.1056/NEJM199804303381808. [DOI] [PubMed] [Google Scholar]
- 10.Flexner C, Broyles S S, Earl P, Chakrabarti S, Moss B. Characterization of human immunodeficiency virus gag/pol gene products expressed by recombinant vaccinia viruses. Virology. 1988;166:339–349. doi: 10.1016/0042-6822(88)90504-1. [DOI] [PubMed] [Google Scholar]
- 11.Graves M C, Lim J J, Heimer E P, Kramer R A. An 11-kDa form of human immunodeficiency virus protease expressed in Escherichia coli is sufficient for enzymatic activity. Proc Natl Acad Sci USA. 1988;85:2449–2453. doi: 10.1073/pnas.85.8.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene W C. The molecular biology of human immunodeficiency virus type 1 infection. N Engl J Med. 1991;324:308–317. doi: 10.1056/NEJM199101313240506. [DOI] [PubMed] [Google Scholar]
- 13.Hodge C N, Aldrich P E, Bacheler L T, Chang C H, Eyermann C J, Garber S, Grubb M, Jackson D A, Jadhav P K, Korant B, Lam P Y, Maurin M B, Meek J L, Otto M J, Rayner M M, Reid C, Sharpe T R, Shum L, Winslow D L, Erickson-Viitanen S. Improved cyclic urea inhibitors of the HIV-1 protease: synthesis, potency, resistance profile, human pharmacokinetics and X-ray crystal structure of DMP 450. Chem Biol. 1996;3:301–314. doi: 10.1016/s1074-5521(96)90110-6. [DOI] [PubMed] [Google Scholar]
- 14.Humphrey R W, Ohagen A, Davis D A, Fukazawa T, Hayashi H, Hoglund S, Mitsuya H, Yarchoan R. Removal of human immunodeficiency virus type 1 (HIV-1) protease inhibitors from preparations of immature HIV-1 virions does not result in an increase in infectivity or the appearance of mature morphology. Antimicrob Agents Chemother. 1997;41:1017–1023. doi: 10.1128/aac.41.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan A H, Manchester M, Swanstrom R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan A H, Swanstrom R. The HIV-1 gag precursor is processed via two pathways: implications for cytotoxicity. Biomed Biochim Acta. 1991;50:647–653. [PubMed] [Google Scholar]
- 17.Karacostas V, Nagashima K, Gonda M A, Moss B. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaufmann G R, Duncombe C, Cunningham P, Beveridge A, Carr A, Sayer D, French M, Cooper D A. Treatment response and durability of a double protease inhibitor therapy with saquinavir and ritonavir in an observational cohort of HIV-1-infected individuals. AIDS. 1998;12:1625–1630. doi: 10.1097/00002030-199813000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Kempf D J, Marsh K C, Denissen J F, McDonald E, Vasavanonda S, Flentge C A, Green B E, Fino L, Park C H, Kong X P, Wideburg N E, Saldivar A, Ruiz L, Kati W M, Sham H L, Robins T, Stewart K D, Hsu A, Plattner J J, Leonard J M, Norbeck D W. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA. 1995;92:2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim R B, Fromm M F, Wandel C, Leake B, Wood A J, Roden D M, Wilkinson G R. The drug transporter P-glycoprotein limits oral absorption and brain entry of HIV-1 protease inhibitors. J Clin Investig. 1998;101:289–294. doi: 10.1172/JCI1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohl N E, Emini E A, Schleif W A, Davis L J, Heimbach J C, Dixon R A, Scolnick E M, Sigal I S. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–4690. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer R A, Schaber M D, Skalka A M, Ganguly K, Wong-Staal F, Reddy E P. HTLV-III gag protein is processed in yeast cells by the virus pol-protease. Science. 1986;231:1580–1584. doi: 10.1126/science.2420008. [DOI] [PubMed] [Google Scholar]
- 23.Lazdins J K, Mestan J, Goutte G, Walker M R, Bold G, Capraro H G, Klimkait T. In vitro effect of alpha1-acid glycoprotein on the anti-human immunodeficiency virus (HIV) activity of the protease inhibitor CGP 61755: a comparative study with other relevant HIV protease inhibitors. J Infect Dis. 1997;175:1063–1070. doi: 10.1086/520352. [DOI] [PubMed] [Google Scholar]
- 24.Lee C G, Gottesman M M, Cardarelli C O, Ramachandra M, Jeang K T, Ambudkar S V, Pastan I, Dey S. HIV-1 protease inhibitors are substrates for the MDR1 multidrug transporter. Biochemistry. 1998;37:3594–3601. doi: 10.1021/bi972709x. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y-M, Tang X-B, Cimakasky L M, Hildreth J E, Yu X-F. Mutations in the matrix protein of human immunodeficiency virus type 1 inhibit surface expression and virion incorporation of viral envelope glycoproteins in CD4+ T lymphocytes. J Virol. 1997;71:1443–1452. doi: 10.1128/jvi.71.2.1443-1452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 27.Lindhofer H, von der Helm K, Nitschko H. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology. 1995;214:624–627. doi: 10.1006/viro.1995.0074. [DOI] [PubMed] [Google Scholar]
- 28.Mellors J W. HIV-1 protease inhibitors: historical perspective and basic science. Infect Med. 1996;13:9–15. [Google Scholar]
- 29.Merrill D P, Manion D J, Chou T C, Hirsch M S. Antagonism between human immunodeficiency virus type 1 protease inhibitors indinavir and saquinavir in vitro. J Infect Dis. 1997;176:265–268. doi: 10.1086/517263. [DOI] [PubMed] [Google Scholar]
- 30.Michelet C, Bellissant E, Ruffault A, Arvieux C, Delfraissy J F, Raffi F, Bazin C, Renard I, Sebille V, Chauvin J P, Dohin E, Cartier F. Safety and efficacy of ritonavir and saquinavir in combination with zidovudine and lamivudine. Clin Pharmacol Ther. 1999;65:661–671. doi: 10.1016/S0009-9236(99)90088-7. [DOI] [PubMed] [Google Scholar]
- 31.Overton H A, McMillan D J, Gridley S J, Brenner J, Redshaw S, Mills J S. Effect of two novel inhibitors of the human immunodeficiency virus protease on the maturation of the HIV gag and gag-pol polyproteins. Virology. 1990;179:508–511. doi: 10.1016/0042-6822(90)90326-m. [DOI] [PubMed] [Google Scholar]
- 32.Patick A K, Mo H, Markowitz M, Appelt K, Wu B, Musick L, Kalish V, Kaldor S, Reich S, Ho D, Webber S. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob Agents Chemother. 1996;40:292–297. doi: 10.1128/aac.40.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearl L H, Taylor W R. A structural model for the retroviral proteases. Nature. 1987;329:351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- 34.Perno C F, Newcomb F M, Davis D A, Aquaro S, Humphrey R W, Calio R, Yarchoan R. Relative potency of protease inhibitors in monocytes/macrophages acutely and chronically infected with human immunodeficiency virus. J Infect Dis. 1998;178:413–422. doi: 10.1086/515642. [DOI] [PubMed] [Google Scholar]
- 35.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pretzer E, Flasher D, Duzgunes N. Inhibition of human immunodeficiency virus type-1 replication in macrophages and H9 cells by free or liposome-encapsulated L-689,502, an inhibitor of the viral protease. Antivir Res. 1997;34:1–15. doi: 10.1016/s0166-3542(96)01017-0. [DOI] [PubMed] [Google Scholar]
- 37.Roberts N A, Martin J A, Kinchington D, et al. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 38.Serio D, Rizvi T A, Cartas M, Kalyanaraman V S, Weber I T, Koprowski H, Srinivasan A. Development of a novel anti-HIV-1 agent from within: effect of chimeric Vpr-containing protease cleavage site residues on virus replication. Proc Natl Acad Sci USA. 1997;94:3346–3351. doi: 10.1073/pnas.94.7.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vacca J P, et al. L-735,524: an orally bioavailable human immunodeficiency virus type 1 protease inhibitor. Proc Natl Acad Sci USA. 1994;91:4096–4100. doi: 10.1073/pnas.91.9.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washington C B, Duran G E, Man M C, Sikic B I, Blaschke T F. Interaction of anti-HIV protease inhibitors with the multidrug transporter P-glycoprotein (P-gp) in human cultured cells. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:203–209. doi: 10.1097/00042560-199811010-00001. [DOI] [PubMed] [Google Scholar]
- 41.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich H-G. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu X, Yuan X, McLane M F, Lee T H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]