Abstract
In December 2019, a new form of coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) started spreading in Wuhan, China. According to the situation report-95 published by the World Health Organization (WHO), the coronavirus disease spread rapidly to 213 countries and territories by April 24, 2020, with the number of confirmed cases and deaths of 26,26,321 and 1,81,938, respectively. The WHO declared coronavirus disease 2019 (COVID-19) as a pandemic on March 11, 2020. People living in many countries are in lockdown and staying at home because of this deadly virus. Patients of COVID-19 are reported to have single or multiple symptoms, while some patients do not have any remarkable symptom at all. Patients have reported symptoms of dry cough, sore throat, fever, fatigue, breathing problem, and gastrointestinal infection. COVID-19 may become very dangerous especially for aged people and people with any other disease such as diabetes, kidney problem, etc. In that case, the virus can cause acute respiratory distress syndrome and cytokine storm. The whole world is in lockdown because of this deadly virus. Currently, there is no particular cure for this disease; however, researchers are trying to find appropriate antiviral and repurposed drugs. This chapter provides a review on the different aspects of COVID-19 including the epidemiology, genomic sequence, and clinical characteristics; current medical treatment options; and development of vaccines and drugs.
Keywords: Clinical treatment, COVID-19, Data Analysis, Epidemiology, Genome, SARS-CoV-2, Transmission
1. Introduction
There are several types of coronaviruses pathogenic to humans. The most common types of pathogens or biological agents are bacteria, virus, fungi, and parasites. This pathogenic agent causes severe disease or illness to its hosts. There are numerous avian hosts such as bats, civets, camels, masked palm, mice, dogs, cats, etc. These hosts are responsible for bearing the coronavirus [1,2]. Many types of coronaviruses are related with mild clinical symptoms [3]. One is the severe acute respiratory syndrome coronavirus (SARS-CoV), a novel type of betacoronavirus. SARS-CoV appeared in Guangdong in November 2002 [4]. More than 8000 people were affected by SARS-CoV and 774 people died in 37 countries at that time [5]. Another type of coronavirus is the Middle East respiratory syndrome coronavirus (MERS-CoV) that was first detected in Saudi Arabia in 2012 [6]. Because of MERS-CoV, 858 people have died out of the 2494 laboratory detected and confirmed cases since the year 2012. In May 2015, about 186 confirmed MERS-CoV cases were found due to the virus transmission from a returned traveler from Saudi Arabia, and as a result, total 38 people were dead in South Korea where the fatality rate was 20.4% [7].
In late December 2019, many people around Huanan seafood market at Wuhan city of Hubei province of China became ill and their illness was similar to some sort of pneumonia. In Wuhan, the Huanan seafood market was selling live animals including various types of birds and rabbits. The pneumonialike disease was found to be caused by a novel, human-infecting coronavirus [8] provisionally named 2019-nCoV. This virus was identified by using next-generation sequencing. According to the report mentioned in the WHO website, there have been 83,597 confirmed cases and 3351 deaths in China from January 11, 2020, to April 13, 2020. There have been 0.058 cases per 1000 people. More than 180 countries [9], 200 territories, and 26 cruise ships [10] are affected by coronavirus disease 2019 (COVID-19). Most COVID-19-infected patients have high fever, dyspnea, pneumonia, and lung problems. Dyspnea means difficulty during breathing.
Currently there is no specific treatment options for COVID-19; doctors are treating patients based on their symptoms and clinical conditions. Unfortunately there is no preventive vaccine either. Scientists all over the world are trying to find appropriate vaccines and drugs to fight this disease. Hence many countries are adopting the lockdown approach and isolating patients to prevent the spread of the disease. Apart from the social distancing approach, everybody is asked to maintain personal hygiene. This includes washing hands frequently with water and soap for at least 20 s, not touching the face, covering coughs and sneezes with elbow, and cleaning surfaces regularly. Patients of this disease are being treated in hospitals while some others at home. In many cases, there has been huge pressure on the limited medical facilities. Currently there is shortage of appropriate masks and personal protective equipment (PPE) even for medical staff. The situation is worse in the developing countries. Besides, the lockdown approach is creating a huge impact on the global economy. Therefore COVID-19 is not only a health crisis but also has become an economic and a social crisis for humans. In order to overcome this global crisis, it is important to understand the characteristics of this virus and its impacts.
A number of research papers have reviewed COVID-19 in different perspectives [3,[11], [12], [13], [14], [15]]. These research results have advanced the understanding about the interaction of pathogens to human cells leading to the disease and other characteristics of COVID-19. One study [3] focuses on the causative agent, pathogenesis, diagnosis, treatment, prevention, and control of COVID-19. The influence of this disease on infants and children was also studied [11]. Understanding about how the novel coronavirus moves between species and how it infects hosts can be vital in order to predict when and where the virus can spread dangerously [12]. The transmission route of the virus via airborne droplets from human to human is depicted in a study [13]. Moreover, the starting dates of COVID-19 in several countries are depicted. Date-wise deaths are also shown graphically in the study [13]. A review of COVID-19 in the otorhinolaryngologic context is also reported [14]. It is an important matter of study why some patients develop ear, nose, and throat (ENT) and anosmia symptoms while some do not. COVID-19 causes otorhinolaryngologic manifestations, and so otolaryngologists and ENT surgeons are at a greater risk of getting SARS-CoV-2 infection [14]. Another study [15] discusses about COVID-19 focusing on pathogenesis and immunopathology.
As there is no approved vaccines and drugs for COVID-19 at the moment, the understanding of the clinical signs, pathogenesis, and pathology [15] is important in order to save patients' lives. Moreover, the knowledge about this virus is frequently evolving and advancing; hence both original research and review reports on the latest condition of COVID-19 are important. This chapter provides a review on the origin, epidemiology, transmission, genomic sequence, phylogenetic analysis, clinical characteristics, and the development of vaccines and drugs. Besides, this chapter provides data visualization of the confirmed, death, and recovered cases around the world.
2. Origin of coronavirus
The recent novel coronavirus (SARS-CoV-2) is type of a β (beta) coronavirus. This β (beta) virus is an enveloped positive-sense RNA virus. This RNA virus is nonsegmented. The subgenus of this virus is Sarbecovirus and the subfamily is Orthocoronavirinae [16]. Coronaviruses can be categorized into four genera [16]:
-
(i)
α (alpha) CoV
-
(ii)
β (beta) CoV
-
(iii)
γ (gamma) CoV
-
(iv)
δ (delta) CoV
α (alpha) and β (beta) coronaviruses are capable of infecting mammals. On the contrary, γ- and δ- coronavirus tend to infect birds [17].
Alphacoronavirus genus has two types of human coronaviruses. These two types are
-
(i)
HCoV-NL63
-
(ii)
HCoV229E
Betacoronavirus genus has four types of human coronaviruses [17]. They are
-
(i)
HCoV-OC43,
-
(ii)
HCoVHKU1,
-
(iii)
SARS-CoV,
-
(iv)
MERS-CoV.
SARS became a pandemic in 2003. In 2020, the novel coronavirus infection has become a pandemic [19]. SARS-CoV and MERS-CoV are considered highly pathogenic [18]. According to Guan et al. [20], both SARS-CoV and MERS-CoV infected humans from bats using intermediate hosts of palm civets. According to Drosten [21], MERS-CoV was transmitted from dromedary camels. The genome sequence of SARS-CoV-2 is found to be 96.2% identical to a bat coronavirus RaTG13 type. It can be capable to share 79.5% identity to basic SARS coronavirus. According to the genome characteristics and the analysis of evolution, bats are thought to be the natural host of SARS-CoV-2, and this virus might be transmitted from bats to humans using intermediate hosts. From the research report by Zhou et al. [22], it can be said that ACE2 (angiotensin-converting enzyme 2), the same receptor as SARS-CoV is used by SARS-CoV-2 for infecting humans.
3. Epidemiology of coronavirus
Since December 12, 2019, coronavirus first broke out in Wuhan, China. It was an unknown acute respiratory tract infection. Some studies proposed that bats may be the probable reservoir of SARS-CoV-2 [23,24]. But, there is no confirmation as far as that the SARS-CoV-2 origin was from the seafood market. Relatively, bats are a wide variety natural reservoir of coronaviruses, as well as MERS-CoV-like and SARS-CoV-like viruses [[25], [26], [27]]. In the seafood market, bats were not available at that time for sale [28]. Moreover, phylogenetic analysis and protein sequence orientation [29,30] presented that similar residues were experimented in various species, which gave more probability for other transitional hosts, such as snakes, pangolin, and turtles. SARS-CoV-2 transmission occurs in humans to humans that is mainly among family members, i.e., friends and relatives who closely communicated with incubation carriers or patients. Zhu et al. [16] described that 72.3% of the patients communicated with some people from Wuhan between the nonresident patients of Wuhan and 31.3% of patients who currently moved to Wuhan. The National Health Commission of China reported that about 3.8% of COVID-19 infection is among healthcare workers. Healthcare workers' infections in 33%–42% of SARS cases as well as transmission among 62%–79% of patients was the very general route of infection in the case of MERS-CoV [31,32]. Direct connection with wild animals via consumption or with intermediate host animals was supposed to be the central route of transmission of SARS-CoV-2. On the other hand, transmission route(s) and the source(s) of SARS-CoV-2 are yet to be confirmed.
4. Transmission
The transmission mode for OC43, HCoV 229E, HKU1, and NL63 are not clearly identified, but as with the transmission of human coronaviruses, other respiratory virus transmission occurs via droplets, direct contact, or indirect contact. Aerosol transmission possibility is still under investigation. MERS-CoV spreads frequently through direct contact and droplets, but transmission by fecal-to-oral route, aerosols, or indirect contact is also feasible [35]. SARS-CoV mainly spreads via direct contact and droplets. Medical techniques that induce aerosol production, i.e., intubation or nebulizer treatment, are described to raise the transmission risk. Fecal-to-oral route may be potential, but only slight indication supports it. SARS-CoV-2 appears to share the transmission mode with MERS-CoV and SARS-CoV, as it spreads primarily via droplets or respiratory secretion. Current studies presented that SARS-CoV-2 can continue to be viable on different surfaces, i.e., cardboard, plastic, stainless steel, and glass, for at least some hours [36,37]. This specifies that SARS-CoV-2 transmission might be probable through polluted surfaces. Aerosol is presently not considered as the primary transmission mode, as no transmission of airborne particles has been described yet, although it looks possible in hospital locations where aerosol-producing processes occur. SARS-CoV-2 RNA has been identified in the stool, urine, and whole blood of COVID-19 patients, but whether transmission through such medium is probable is still unidentified [33,34].
The first SARS case was reported on November 16, 2002, and was not confirmed a pandemic despite the 8096 confirmed cases prominent to 774 deaths during the 8-month period. Meanwhile, in April 2009, the H1N1pdm09 virus spread quickly all over the world generating more than 30,000 cases in 74 countries. The WHO declared the infection of H1N1pdm09 influenza virus as a pandemic [38]. The SARS-CoV-2 transmission is not exactly identified yet. A meta-analysis study between January 1, 2020, and February 7, 2020, proposes that the basic SARS-CoV-2 reproduction number (R0) ranges from 1.4 to 6.49, with a median value of 2.79 and a mean value of 3.23 [39]. Gathering of new epidemiologic data may adapt this value. However, R0 is estimated to be around between 2 and 3. The transmission of SARS-CoV occurs some days following the inception of symptoms [40]. SARS-CoV was efficiently controlled by isolating the patients presently after the inception of illness, as the rate of secondary attack was critically decreased by separating the index case within 5–6 days. Instead, the SARS-CoV-2 transmission point remains unidentified. Generally, when patients have some symptoms, respiratory viruses have the maximum transmissibility as may be the case for COVID-19. The respiratory viral load of 18 COVID-19 patients in China is described by Zou et al. [41]. Patients with coronavirus infection sometimes bear a resemblance to patients with influenza virus infection. The transmission of COVID-19 usually occurs during the first few days after the onset of symptoms.
5. Genome structure
COVID-19 virus is an enveloped particle of spherical size or pleomorphic. A positive-sense and single-stranded RNA is contained in it. The RNA is associated with a nucleoprotein (combination of nucleic acid and protein) within a capsid. The capsid consists of matrix protein. Matrix protein describes a protein forming layer on the inside of the viral envelope. The envelope bears glycoprotein projections. This projection is club shaped. Some coronaviruses also contain a hemagglutinin-esterase (HE) protein. Coronaviruses have the largest size of genomes among all known RNA viruses. Their genome size is 26.4–31.7 kb. SARS-CoV has a positive-sense RNA genome of size 27.9 kb, while MERS-CoV has a positive-sense RNA genome of 30.1 kb. The genome of coronaviruses contains a variable number of open-reading frames (ORFs) ranging from 6 to 11. Two-thirds of the viral RNA is mainly located in the first ORF (ORF1a/b). This major RNA portion translates two polyproteins. The polyproteins are pp1a and pp1ab. The viral RNA can also encode 16 nonstructural proteins. On the contrary, the remaining ORFs encode accessory and structural proteins [43,45]. When whole-genome sequence is taken into consideration, SARS-CoV-2 has some similarity with the SARS-like bat coronaviruses.
A number of proteins constitute this virus. These are membrane (M) protein, spike (S) glycoprotein, envelope (E) membrane protein, nucleoprotein (N), and HE protein [42,43]. The proteins S, M, E, and N are encoded by ORFs 10 and 11 [46,47]. Genome maintenance and virus replication are carried out by HE protein, 3a/b protein, and 4a/b protein [46].
Within the coronavirus particle, the RNA genome is wrapped by N for forming a coiled tubular structure. This helical nucleocapsid is surrounded by E protein. Two or three structural proteins are associated with E protein. The M protein is embedded in an envelope. The S protein attached with the envelope is the target of neutralizing antibodies. HE protein is found in several of the betacoronaviruses. There are five essential genes in coronaviruses. Among these, four are for structural proteins (N, E, M, S) and the rest is for viral replication or transcription (RNA-dependent RNA polymerase [RdRp]). The organization of the genome can be described in the following format: 5′-RdRp-S-E-M-N-3′ [44,45].
6. Phylogenetic analysis
Researchers have performed phylogenetic analysis to report that SARS-CoV-2 fell within the subgenus Sarbecovirus, with the genus Betacoronavirus. This SARS-CoV-2 has 88% similarity with two other coronaviruses, namely, bat-SL-CoVZC45 and bat-SL-CoVZXC21. It has 79% similarity with SARS-CoV and 50% similarity with MERS-CoV. This virus has amino acid variation at some key residues but still has a receptor-binding domain (RBD) structure similar to SARS-CoV [48,49]. This means the ACE2-binding mode of the novel coronavirus RBD is closely identical to that of the previously identified SARS-CoV RBD. ACE2 is also used as a cell surface receptor or membrane receptor in SARS-CoV.
7. Statistical analysis on COVID-19
In this section, data visualization [1] is provided on the confirmed, death, and recovered cases of COVID-19 in different countries using datasets available in Refs. [[76], [77], [78]]. Python programming language is used as the data visualization tool. Statistical analyses are illustrated in Table 25.1, Table 25.2, Table 25.3 . From Table 25.1, it can be seen that the United States has the highest number of confirmed cases followed by Spain and Italy. The United States also has the highest number of deaths, as shown in Table 25.2. Europe and North America have the most number of COVID-19 patients, as reported in Table 25.3. Country-wise correlation up to April 27, 2020, is illustrated in Fig. 25.1 . Fig. 25.2 shows the visualization of confirmed, active, death, and recovered cases of the top 10 countries.
Table 25.1.
Top 10 countries with the most number of reported cases (up to April 27, 2020).
| Country/region | Confirmed cases |
|---|---|
| The United States | 988,469 |
| Spain | 232,148 |
| Italy | 199,414 |
| France | 165,977 |
| Germany | 158,758 |
| The United Kingdom | 158,348 |
| Turkey | 112,261 |
| Russia | 93,558 |
| Iran | 92,584 |
| China | 83,938 |
Table 25.2.
Top 10 countries with deaths reported (up to April 27, 2020).
| Country/region | Deaths |
|---|---|
| The United States | 56,253 |
| Italy | 26,977 |
| Spain | 23,822 |
| France | 23,327 |
| The United Kingdom | 21,157 |
| Belgium | 7331 |
| Germany | 6126 |
| Iran | 5877 |
| China | 4637 |
| Brazil | 4603 |
Table 25.3.
Continent-wise reported cases.
| Continent | Confirmed | Deaths | Recovered | Active | Incident rate | Mortality rate (per 100) |
|---|---|---|---|---|---|---|
| Africa | 33,372 | 1471 | 10,190 | 21,711 | 304.6353 | 4.41 |
| Asia | 486,909 | 17,708 | 239,068 | 230,133 | 1907.7668 | 3.64 |
| Australia | 8219 | 102 | 6814 | 1303 | 59.0212 | 1.24 |
| Europe | 131,283 | 124,615 | 452,015 | 735,553 | 9240.9101 | 9.50 |
| North America | 1,070,441 | 61,172 | 141,648 | 867,621 | 868.7687 | 5.71 |
| Others | 1758 | 34 | 850 | 874 | 1150.4528 | 1.93 |
| South America | 145,075 | 6792 | 51,544 | 86,739 | 384.6387 | 4.68 |
Figure 25.1.

Country-wise correlation up to April 27, 2020.
Figure 25.2.

Visualization of confirmed, active, death, and recovered cases of the top 10 countries.
Fig. 25.3 presents a visualization of the confirmed and death cases in the world for the period from January 22, 2020, to April 27, 2020. It can be seen that after around 55 days, that is, after mid-March, the number of confirmed cases has increased drastically. The increase in the confirmed cases has been exponential in nature. The number of deaths has also increased sharply after mid-March 2020. Fig. 25.4 shows the confirmed and death cases for China, the United States, and Spain during January 22, 2020, to April 27, 2020. It can be seen from Fig. 25.4A that after 40 days, that I,s after the beginning of March 2020, there has been little increase in the number of confirmed cases in China. This shows that China has managed the COVID-19 to a great extent since early March 2020. There has also been little increase in the number of deaths in China during this period. On the other hand, there has been significant increase in the number of confirmed and death cases in the United States and Spain after 60 days, that is, after March 22, 2020. Fig. 25.4B illustrates the increase in cases in the United States, while Fig. 25.4C demonstrates the same for Spain. However, as of 27 April, the number of confirmed cases in the United States is close to 0.988 million, which is more than four times that of Spain with 0.229 million cases. Fig. 25.5 presents the visualization of confirmed, death, recovered, and active cases in the Hubei state of China from January 22, 2020, to April 27, 2020. It can be seen that since early March 2020, there is little increase in the number of confirmed cases but significant decrease in the number of active cases. Moreover, Hubei state has experienced increase in the number of recovered patients since early March 2020. The number of deaths has also been low since March 2020 except a slight increase after mid-April 2020.
Figure 25.3.
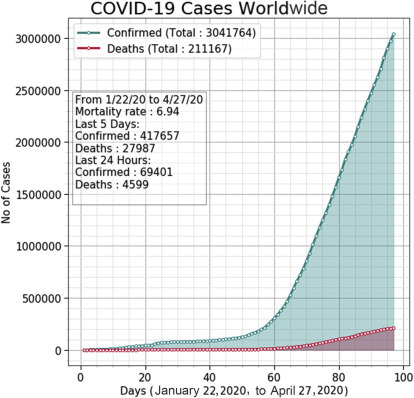
Visualization of confirmed cases and deaths all over the world from January 22, 2020, to April 27, 2020.
Figure 25.4.

Visualization of confirmed and death cases from January 22, 2020, to April 27, 2020, in (A) China, (B) the United States, and (C) Spain.
Figure 25.5.

Visualization of confirmed, death, recovered, and active cases in the Hubei state of China from January 22, 2020, to April 27, 2020.
8. Clinical features of COVID-19
The range of COVID-19 incubation period is 2–14 days, with a median period of 5.1 days [50,51]. The household transmission analysis has shown that respiratory symptoms and fever appeared 3–7 days since coverage to the virus. Fatigue, dry cough, and fever have been more regularly reported, whereas rhinorrhea, nasal sore throat, myalgia, and congestion are relatively rare [52]. Sometimes, nonrespiratory symptoms, i.e., palpitation, headache, or diarrhea, led the respiratory symptoms. Many patients are primarily afebrile. The COVID-19 clinical spectrum ranged from asymptomatic to fatal pneumonia. Asymptomatic infection rate is yet to be well-defined, as initially most of the asymptomatic infections finally turned symptomatic. According to the Chinese National Reporting System data, after February 20, 2020, the confirmed cases for median age is 51 years (2 days to 100 years), of which 77.8% are 30–69 years [53]. Totally 51.1% of men has been reported. According to that report, 80% of confirmed cases were without pneumonia or had slight to moderate pneumonia, around 15% had simple pneumonia, around 6% were under intensive care owing to multiple organ failure, shock, and respiratory failure. COVID-19 has a fatality rate of 3.8% in China, 5.8% in Wuhan city, and 0.7% in the rest of mainland China. Severe death or pneumonia with its risk factors include age 60 years or older, besides medical comorbidity, i.e., cardiovascular disease, diabetes mellitus, hypertension, malignancy, or chronic pulmonary disease. The confirmed COVID-19 cases indicated lymphopenia, leukopenia, and mild elevated C-reactive protein levels in laboratory tests. But the patients of severe pneumonia had increased creatinine kinase levels, neutrophil count, and leukocyte count. Computed tomography (CT) discovered interstitial infiltration or ground-glass appearance and multiple patchy consolidations in both lung fields [54].
A team of researchers analyzed 138 hospitalized patients based on the severity of pneumonia in Wuhan, China [55]. Among them, 36 patients required intensive-care unit (ICU) admission. According to the report [55], the ICU-admitted patients had a median age of 66 years, while others had an average age of 51 years. The patients of intensive-care group can bear some conditions such as hypertension and diabetes mellitus. The median time from onset of symptoms to dyspnea was 5 days and to acute respiratory syndrome was 8 days. After a few days, six patients had died, with an overall mortality of 4.3%. In fatal cases, elevations in total leukocyte count and neutrophil ratio and decrease in lymphocyte count were observed 7 days after the onset of symptoms. In laboratory test results, most of the patients had decreased or normal lymphocytopenia and white blood cell counts [30,56]. But in the simple patients, the D-dimer, creatinine levels, neutrophil count, and blood urea concentration were higher considerably, besides the lymphocyte counts were decreased.
9. Treatment with drugs
The current treatment of COVID-19 mainly focuses on respiratory support and symptomatic treatment according to the treatment and diagnosis of pneumonia affected by COVID-19 for the absence of actual antiviral therapy [57]. Approximately all patients recognized oxygen therapy, as well as WHO suggested extracorporeal membrane oxygenation (ECMO) to patients with refractory hypoxemia [58]. Antiviral systemic corticosteroid treatment and drugs normally used in clinical practice, containing neuraminidase inhibitors (zanamivir, peramivir, oseltamivir, etc.), acyclovir, ganciclovir, and methylprednisolone, as well as ribavirin [59], for influenza virus, are invalid for COVID-19 and not suggested. Hydroxychloroquine, antimalarial drug, is licensed for the treatment of malaria and chemoprophylaxis and as a disease-modifying antirheumatic drug [[60], [61], [62]]. It is reported that prophylaxis with hydroxychloroquine at permitted doses may prevent SARS-CoV-2 infection and ameliorate viral shedding [63]. Recently scientists in China have implemented clinical trials of hydroxychloroquine medicine for COVID-19. Chloroquine and hydroxychloroquine are relatively well tolerated as demonstrated by extensive experience in patients with systemic lupus erythematosus and malaria. However, both agents can cause rare and serious adverse effects [64,65]. There are also many other drugs in the trial stage, such as remdesivir [[66], [67], [68], [69], [70]], favipiravir [[71], [72], [73]], arbidol [74,75], etc. The trials of various drugs related to the fight against COVID 19 are summarized in Fig. 25.6 .
Figure 25.6.

Summary of some clinical outcomes.
There had been widespread hope that remdesivir could treat COVID-19. But it is declared that this drug has failed in its first randomized clinical trial according to draft reports accidentally published by the WHO. Based on the report, researchers studied 237 patients and administered the drug to 158 patients. Then their progress was compared with the remaining 79 who received a placebo. It is reported [83] that about 13.9% and 12.8% of patients died after 1 month of taking remdesivir and placebo, respectively. This means remdesivir has not shown significant benefits compared to placebo and hence requires further investigation to prove its effectiveness in treating COVID-19.
10. Progress on vaccines
As of January 2020, some vaccines are being introduced for preventing COVID-19. A number of vaccines including Moderna vaccine and Pfizer-BioNTech vaccine are approved by the national regulatory authorities for public use. The National Institute of Allergy and Infectious Diseases (NIAID) is leading the funding of federal research and response to COVID-19. The S protein is the major target for developing the COVID-19 vaccine [80]. Some of the potential drugs [79,81] are presented in Fig. 25.7 .
Figure 25.7.

Summary of vaccine progress.
It is also noted that researchers at the University of Oxford led by Professor Sarah Gilbert have begun testing a COVID-19 vaccine named ChAdOx1 nCoV-19 in human volunteers from the last week of April 2020 [82]. ChAdOx1 has been given to more than 320 people to date, and after taking the vaccine, it has been safe and was well tolerated. Although there are some temporary side effects, such as a temperature, headache, or sore arm.
11. Conclusions
Currently, the entire world is fighting against the coronavirus disease. About 30,318,681 people are affected from COVID-19 already and 207,973 are dead according the situation report-100 of WHO as of April 29, 2020. The world has not experienced such a health crisis since the influenza outbreak in 1918. The mutation rate of the coronavirus is very fast. It becomes very dangerous for aged people and people suffering from any other diseases such as diabetes, high blood pressure, kidney problem, etc. SARS-CoV-2 is more severe than seasonal influenza in part due to having many more ways to stop cells from calling out to the immune system for help. At present, there is no prescribed drug or vaccine for this deadly COVID-19. Countries are applying social distancing measures to stop the spread of the disease, which in turn is hampering the global economic condition. Hence continuous research is required to understand the characteristics of SARS-CoV-2 and to overcome the COVID-19 pandemic soon.
Early detection is required for reducing the spread of COVID-19 [84]. Therefore some machine learning methods [[85], [86], [87], [88], [89], [90], [91]] can be helpful for predicting COVID-19. Moreover, deep learning methods [[92], [93], [94]] can be potentially used to detect COVID-19 at the initial stage by applying CT and X-ray images [[96], [97], [98], [99]].
References
- 1.Mondal M.R.H., Bharati S., Podder P., Podder P. Data analytics for novel coronavirus disease. Inform. Med. Unlocked. 2020;20:100374. doi: 10.1016/j.imu.2020.100374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su S., Wong G., Shi W., et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harapan H., Itoh N., Yufika A., Winardi W., Keam S., Te H., Megawati D., Hayati Z., Wagner A.L., Mudatsir M. Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Public Health. 2020;13(5):667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris J.S., Guan Y., Yuen K.Y. Severe acute respiratory syndrome. Nat. Med. 2004;10(Suppl. 12):S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan-Yeung M., Xu R.H. SARS: epidemiology. Respirology. 2003;8(Suppl. 1):S9–S14. doi: 10.1046/j.1440-1843.2003.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 7.Korea Centers for Disease Control and Prevention, Updates on COVID-19 in Republic of Korea (as of 26 March). https://is.cdc.go.kr/upload_comm/syview/doc.html?fn=158522019760700.pdf&rs=/upload_c omm/docu/0030/.
- 8.Bharati S., Podder P., Mondal M.R.H. Hybrid deep learning for detecting lung diseases from X-ray images. Inform. Med. Unlocked. 2020;20:100391. doi: 10.1016/j.imu.2020.100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU), ArcGIS, Johns Hopkins CSSE. (Retrieved 13 April 2020).
- 10.Mallapaty S. What the cruise-ship outbreaks reveal about COVID-19. Nature. 2020;580(7801):18. doi: 10.1038/d41586-020-00885-w. [DOI] [PubMed] [Google Scholar]
- 11.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unhale S.S., Ansar Q.B., Sanap S., Thakhre S., Wadatkar S., Bairagi R., et al. A review on corona virus COVID-19. World J. Pharm. Life Sci. 2020;6(4):109–115. [Google Scholar]
- 13.Kumar D., Malviya R., Sharma P.K. Corona virus: a review of COVID-19. Eurasian J. Med. Oncol. 2020;4(1):8–25. [Google Scholar]
- 14.Krajewska J., Krajewski W., Zub K., Zatoński T. COVID-19 in otolaryngologist practice: a review of current knowledge. Eur. Arch. Oto-Rhino-Laryngol. 2020 doi: 10.1007/s00405-020-05968-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhama K., Patel S.K., Pathak M., Yatoo M.I., Tiwari R., Malik Y.S., Singh R., Sah R., Rabaan A.A., Bonilla-Aldana D.K., Rodriguez-Morales A.J. An update on SARS-COV-2/COVID-19 with particular reference on its clinical pathology, pathogenesis, immunopathology and mitigation strategies – a review. Public Health. 2020 doi: 10.20944/preprints202003.0348.v1. Preprints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. Published online January 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. Published online January 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:E59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 21.Drosten C., Kellam P., Memish Z.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014;371:1359–1360. doi: 10.1056/NEJMc1409847. [DOI] [PubMed] [Google Scholar]
- 22.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giovanetti M., Benvenuto D., Angeletti S., Ciccozzi M. The first two cases of 2019-nCoV in Italy: where they come from? J. Med. Virol. 2020:1–4. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampton T. Bats may be SARS reservoir. J. Am. Med. Assoc. 2005;294(18):2291. doi: 10.1001/jama.294.18.2291. [DOI] [PubMed] [Google Scholar]
- 26.Banerjee A., Kulcsar K., Misra V., Frieman M., Mossman K. Bats and coronaviruses. Viruses. 2019;11(1):E41. doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 28.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., et al. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chowell G., Abdirizak F., Lee S., Lee J., Jung E., Nishiura H., et al. Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med. 2015;13:210. doi: 10.1186/s12916-015-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang C.K., Song K.H., Choe P.G., Park W.B., Bang J.H., Kim E.S., et al. Clinical and epidemiologic characteristics of spreaders of middle east respiratory syndrome coronavirus during the 2015 outbreak in Korea. J. Kor. Med. Sci. 2017;32(5):744–749. doi: 10.3346/jkms.2017.32.5.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J., et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. J. Am. Med. Assoc. 2020;323(15):1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.To K.K., Tsang O.T., Chik-Yan Yip C., Chan K.H., Wu T.C., Chan J.M.C., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020;71(15):841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ong S.W.X., Tan Y.K., Chia P.Y., Lee T.H., Ng O.T., Wong M.S.Y., et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. J. Am. Med. Assoc. 2020;323(16):1610–1612. doi: 10.1001/jama.2020.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2004973. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization, World Now at the Start of 2009 Influenza Pandemic. https://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/. (Accessed 12 March).
- 39.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel. Med. 2020;27:taaa021. doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.American Academy of Pediatrics . In: Red Book®: 2018 Report of the Committee on Infectious Diseases. 31st ed. Kimberlin D.W., Brady M.T., Jackson M.A., Long S.S., editors. American Academy of Pediatrics; Itasca: 2018. Coronaviruses, including SARS and MERS; pp. 297–301. [Google Scholar]
- 41.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.http://ruleof6ix.fieldofscience.com/2012/09/a-new-coronavirus-should-youcare.html.
- 43.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Haan C.A.M., Kuo L., Masters P.S., Vennema H., Rottier P.J.M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72(8):6838e50. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woo P.C.Y., Huang Y., Lau S.K.P., Yuen K.-Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804e20. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3(6):e00473e12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Czub M., Weingartl H., Czub S., He R., Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23(17e18):2273e9. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome -coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19) Clin. Exp. Pediatr. 2020;63(4):119–124. doi: 10.3345/cep.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J. Med. Virol. 2020;92(6):568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.World Health Organization . World Health Organization; Geneva: 2020. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19)https://www.who.int/docs/default-source/coronaviruse/whochina-joint-mission-on-covid-19-final report.pdf (Internet) (Cited 28 February 2020). Available from: [Google Scholar]
- 54.Zhao W., Zhong Z., Xie X., Yu Q., Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. AJR Am. J. Roentgenol. 2020;214(5):1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 55.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323:1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kui L., Fang Y.Y., Deng Y., Liu W., Wang M.F., Ma J.P., et al. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin. Med. J. 2020 doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.National Health Commission of the People's Republic of China . 2020. Diagnosis and Treatment of Pneumonia Caused by 2019-nCoV (Version 6)http://www.gov.cn/zhengce/zhengceku/2020-02/19/content_5480948.htm [Google Scholar]
- 58.WHO, Clinical Management of Severe Acute Respiratory Infection When Novel Coronavirus (nCoV) Infection Is Suspected. https://www.who.int/publicationsdetail/clinical-management-of-severe-acute-respiratory-infection-when-novelcoronavirus-(ncov)-infection-is-suspected.
- 59.Li H., Wang Y.M., Xu J.Y., Cao B. Potential antiviral therapeutics for 2019 novel coronavirus. Chin. J. Tuberc. Respir. Dis. 2020;43(0):E002. doi: 10.3760/cma.j.issn.1001-0939.2020.0002. [DOI] [PubMed] [Google Scholar]
- 60.Bharati S., Podder P., Mondal M.R.H., Prasath V.B.S. Medical imaging with deep learning for COVID-19 diagnosis: A comprehensive review. Int. J. Comput. Inf. Syst. Ind. Manag. Appl. 2021;13:91–112. [Google Scholar]
- 61.Mitjà O., Cloteta B. Use of antiviral drugs to reduce COVID-19 transmission. Lancet Glob. Health. 2020;8(4):e488–e496. doi: 10.1016/S2214-109X(20)30114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao X., Ye F., Zhang M., et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa237. Published online March 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tett S.E., Cutler D.J., Day R.O., Brown K.F. Bioavailability of hydroxychloroquine tablets in healthy volunteers. Br. J. Clin. Pharmacol. 1989;27:771–779. doi: 10.1111/j.1365-2125.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalil A.C. Treating COVID-19—off-label drug use, compassionate use, and randomized clinical trials during pandemics. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.4742. [DOI] [PubMed] [Google Scholar]
- 65.Interview with David Juurlink Coronavirus (COVID-19) update: chloroquine/ hydroxychloroquine and azithromycin. J. Am. Med. Assoc. 2020 https://edhub.ama-assn.org/jn-learning/audio-player/18337225 (Accessed 3 April 2020) [Google Scholar]
- 66.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., et al. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sheahan T.P., Sims A.C., Leist S.R., et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11(1):222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang M., Cao R., Zhang L., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Tawfiq J.A., Al-Homoud A.H., Memish Z.A. Remdesivir as a possible therapeutic option for the COVID-19. Trav. Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101615. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.6019. Published online. [DOI] [PubMed] [Google Scholar]
- 71.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sissoko D., Laouenan C., Folkesson E., et al. JIKI Study Group Experimental treatment with favipiravir for Ebola virus disease (the JIKI Trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med. 2016;13(3):e1001967. doi: 10.1371/journal.pmed.1001967. (Published correction appears in PLoS Med. 2016;13(4): e1002009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020:107512. doi: 10.1016/j.pharmthera.2020.107512. Published online February 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. USA. 2017;114(2):206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khamitov R.A., SIa L., Shchukina V.N., Borisevich S.V., Maksimov V.A., Shuster A.M. Antiviral activity of arbidol and its derivatives against the pathogen of severe acute respiratory syndrome in the cell cultures [in Russian] Vopr. Virusol. 2008;53(4):9–13. [PubMed] [Google Scholar]
- 76.https://github.com/CSSEGISandData/COVID-19.
- 77.https://www.kaggle.com/sudalairajkumar/novel-corona-virus-2019-dataset.
- 78.https://www.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6.
- 79.https://www.clinicaltrials.gov/ct2/show/NCT04283461. (Accessed 24 April 2020).
- 80.Wu S. Progress and concept for COVID-19 vaccine development. Biotechnol. J. 2020;15:2000147. doi: 10.1002/biot.202000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.RAPS. Regulatory Focus, COVID-19 Tracker. (Accessed 23 April 2020). https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker.
- 82.COVID-19 Vaccine Begins Human Trial Stage. (Accessed 24 April 2020). https://www.cambridgenetwork.co.uk/news/covid-19-vaccine-begins-human-trial-stage.
- 83.Hopes Dashed as Coronavirus Drug Remdesivir ‘Fails First Trial’. (Accessed 24 April 2020). https://www.bbc.com/news/world-52406261.
- 84.Podder P., Khamparia A., Mondal M.R.H., Rahman M.A., Bharati S. Forecasting the spread of COVID-19 and ICU requirements. Int. J. Online Biomed. Eng. (iJOE) 2021 [Google Scholar]
- 85.Bharati S., Podder P., Mondal M.R.H. 2020 IEEE Region 10 Symposium (TENSYMP) IEEE; 2020. Diagnosis of polycystic ovary syndrome using machine learning algorithms; pp. 1486–1489. [Google Scholar]
- 86.Raihan-Al-Masud M., Mondal M.R.H. Data-driven diagnosis of spinal abnormalities using feature selection and machine learning algorithms. PLoS One. 2020;15(2):e0228422. doi: 10.1371/journal.pone.0228422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Madhavan M.V., Thanh D.N.H., Khamparia A., Pande S., Malik R., Gupta D. Recognition and classification of pomegranate leaves diseases by image processing and machine learning techniques. CMC Comput. Mater. Con. 2021;66(3):2939–2955. [Google Scholar]
- 88.Thanh D.N.H., Prasath V.B.S., Hien N.N. Melanoma skin cancer detection method based on adaptive principal curvature, colour normalisation and feature extraction with the ABCD rule. J. Digit. Imag. 2019:1–2. doi: 10.1007/s10278-019-00316-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Podder P., Bharati S., Mondal M.R.H. In: Artificial Intelligence for Data-Driven Medical Diagnosis. Gupta D., Kose U., Le Nguyen B., Bhattacharyya S., Nguyen B., editors. De Gruyter; Berlin, Boston: 2021. Automated gastric cancer detection and classification using machine learning; pp. 207–224. [DOI] [Google Scholar]
- 90.Bharati S., Robel M.R., Rahman M.A., Podder P., Gandhi N. International Conference on Innovations in Bio-Inspired Computing and Applications. Springer; Cham: 2019. Comparative performance exploration and prediction of fibrosis, malign lymph, metastases, normal lymphogram using machine learning method; pp. 66–77. [Google Scholar]
- 91.Bharati S., Rahman M.A., Podder P. 2018 4th International Conference on Electrical Engineering and Information & Communication Technology (iCEEiCT) IEEE; 2018. Breast cancer prediction applying different classification algorithm with comparative analysis using WEKA; pp. 581–584. [Google Scholar]
- 92.Khamparia A., Bharati S., Podder P., et al. Diagnosis of breast cancer based on modern mammography using hybrid transfer learning. Multidimens. Syst. Signal Process. 2021 doi: 10.1007/s11045-020-00756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bharati S., Podder P., Mondal M.R.H. Artificial neural network based breast cancer screening: a comprehensive review. Int. J.Comput. Inf. Syst. Ind. Manag. Appl. 2020;12:125–137. [Google Scholar]
- 94.Bharati S., Podder P. In: Artificial Intelligence for Data-Driven Medical Diagnosis. Gupta D., Kose U., Le Nguyen B., Bhattacharyya S., Nguyen B., editors. De Gruyter; Berlin, Boston: 2021. 1 Performance of CNN for predicting cancerous lung nodules using LightGBM; pp. 1–18. [Google Scholar]
- 95.Thanh D.N.H., Prasath V.B.S. A review on CT and X-ray images denoising methods. Informatica. 2019;43(2) [Google Scholar]
- 96.Mondal M.R.H., Bharati S., Podder P. Diagnosis of COVID-19 using machine learning and deep learning: A review. Curr. Med. Imag. 2021 doi: 10.2174/1573405617666210713113439. [DOI] [PubMed] [Google Scholar]
- 97.Bharati S., Podder P., Mondal M.R.H., Prasath V.B.S. CO-ResNet: Optimized ResNet model for COVID-19 diagnosis from X-ray images. Int. J. Hybrid Intell. Syst. 2021;17(1–2):71–85. doi: 10.3233/HIS-210008. [DOI] [Google Scholar]
- 98.Bharati S., Podder P., Mondal M.R.H., Gandhi N. In: Intell. Syst. Design Appl. Abraham A., Piuri V., Gandhi N., Siarry P., Kaklauskas A., Madureira A., editors. Vol. 1351. Springer, Cham; 2021. Optimized NASNet for diagnosis of COVID-19 from lung CT images; pp. 647–656. [Google Scholar]
- 99.Podder P., Bharati S., Mondal M.R.H., Kose U. In: Data Science for COVID-19. Kose U., Gupta D., de Albuquerque V.H.C., Khanna A., editors. Academic Press; 2021. Application of machine learning for the diagnosis of COVID-19; pp. 175–194. [Google Scholar]


