Abstract
Patients with hereditary haemorrhagic telangiectasia (HHT), also known as Rendu–Osler–Weber syndrome, suffer from the consequences of abnormal vessel structures. These structures can lead to haemorrhages or shunt effects in liver, lungs and brain. This inherited and rare disease is characterized by mutations affecting the transforming growth factor-β (TGF-β)/Bone Morphogenetic Protein (BMP) pathway that results in arteriovenous malformations and studies indicate an impaired immune response. The mechanism underlying this altered immune response in HHT patients is still unknown. TGF-β interacts with hypoxia inducible factors (HIF), which both orchestrate inflammatory and angiogenic processes. Therefore, we analysed the expression of HIF and related genes in whole blood samples from HHT patients. We could show significantly decreased expression of HIF-1α on the mRNA and protein level. However, commonly known upstream regulators of HIF-1α in inflammatory responses were not affected, whereas HIF-1α target genes were significantly downregulated. There was no correlation between HIF1A or HIF2A gene expression and the severity of HHT detected. Our results represent a rare case of HIF-1α downregulation in a human disease, which underlines the relevance of HIFs in HHT. The study indicates an interaction of the known mutation in HHT and the dysregulation of HIF-1α in HHT patients, which might contribute to the clinical phenotype.
Subject terms: Diseases, Medical research, Molecular medicine
Introduction
Patients with hereditary haemorrhagic telangiectasia (HHT), also known as Rendu–Osler–Weber syndrome, suffer from recurrent hemorrhages as the consequences of abnormal vessel structures. This inherited, rare disease is characterized by multisystemic arteriovenous malformations, particularly in liver, lung, intestine and brain1. The underlying mutations affect genes from the transforming growth factor-β (TGF-β)/Bone Morphogenetic Protein (BMP) pathway, thus impairing its signalling2. TGF-β is a regulatory cytokine involved in cell growth, apoptosis, smooth muscle cell differentiation, vascular remodeling and the immune response3. In addition to vascular malformations, few studies indicate impaired immune response in HHT patients, the precise mechanism of the immune dysregulation is still unknown4,5. In this context, several publications indicate an interaction of TGF-β with hypoxia inducible factors (HIFs)6–9. Further, Endoglin (the gene responsible for HHT type 1) expression is regulated by transcriptional cooperation between the HIF and TGF-β pathways10. In general, HIFs are responsible for cellular oxygen sensing and the further adaption to hypoxic conditions via the induction of target genes involved in vascularization, angiogenesis, cell metabolism, cell survival and tumourgenesis11. HIFs are heterodimeric transcription factors, which consist of an oxygen sensitive HIF-α subunit, with three identified isoforms [HIF-1α, HIF-2α and HIF-3α (with a minor role)], and a HIF-β subunit12. Both subunits are expressed constitutively but the expression pattern of HIF-1α and HIF-2α differ: HIF-1α is ubiquitously expressed among all tissues, while HIF-2α expression is limited for example to endothelium, kidney, pancreas, liver, heart, lungs, intestine, brain and immune cells13–15. Regulation of HIF-α expression by cellular oxygen sensing occurs mainly by a post-translational mechanism. Under normoxic conditions, HIF-α subunits are hydroxylated by oxygen-sensitive hydroxylase enzymes [prolyl hydroxylase domain-containing enzymes (PHDs)], afterwards ubiquitinylated through the von Hippel–Lindau protein ubiquitin ligase complex, and degraded in the proteasome16. Since PHDs require oxygen for their enzymatic activity, hypoxic conditions result in reduced hydroxylase activity of their substrates, thus leading to HIF-α subunits accumulation, increased translocation to the nucleus, and augmented formation of a heterodimeric complex with the HIF-β subunit.These dimers bind to genes with hypoxia response elements and thus promote cell responsiveness to diminished oxygen availability17. As a type of self-regulatory system, PHDs are HIF target genes and therefore upregulated following HIF activation. HIFs accumulate not only in hypoxia but also in inflammation and perform key functions in the immune response18,19. Due to the paramount role played by HIFs in modulation of both angiogenesis and the immune response, as well as in the transcriptional regulation of Endoglin, VEGF and Erythropoietin, gaining insight into HIFs expression profile in HHT may help characterization of the pathogenic mechanisms in this rare disease20. For the first time, this study has analysed HIFs and their corresponding target genes in whole blood samples from HHT patients.
Results
Characteristics of patients with HHT and non-HHT control individuals
The study included 66 patients with 29 (44%) males and 37 (56%) females. The age range was 38–66 years (m ± SD: 52 ± 14 years). Genetic testing was performed in 23 (35%) patients with most patients suffering from HHT type 1 (n = 7.11%). More than 90% of patients suffered from epistaxis and multiple typical telangiectasia, and reported a positive family history for HHT (Table 1). Patients with HHT and non-HHT donors were of comparable age and sex.
Table 1.
Clinical characteristics of 66 patients with hereditary haemorrhagic telangiectasia.
| All patients n = 66 (100) |
Men n = 29 (44) |
Women n = 37 (56) |
|||
|---|---|---|---|---|---|
| Age | Years | 52 ± 14 (min: 18, max: 82) | 56 ± 14 | 48 ± 14 | |
| Missing | 0 | 0 | 0 | ||
| Genetic testing | Yes | 23 (35) | 7 (24) | 16 (43) | |
| No | 43 (65) | 22 (76) | 21 (57) | ||
| Missing | 0 | 0 | 0 | ||
| Mutated gene | HHT 1 | 7 (11) | 0 | 7 (19) | |
| HHT 2 | 4 (6) | 2 (7) | 2 (5) | ||
| Others | 2 (3) | 1 (3) | 1 (3) | ||
| No mutation detected | 1 (2) | 0 | 1 (3) | ||
| Missing | 9 (39) | 4 (57) | 5 (31) | ||
| Positive FH | Yes | 60 (91) | 26 (90) | 34 (92) | |
| No | 5 (8) | 3 (10) | 2 (5) | ||
| Not known | 1 (2) | 0 | 1(3) | ||
| Yes | 64 (97) | 29 (100) | 35 (95) | ||
| TAE | No | 2 (3) | 0 | 2 (5) | |
| Missing | 0 | 0 | 0 | ||
| Visceral lesions | GI | Yes | 22 (33) | 12 (41) | 10 (27) |
| No | 27 (41) | 11 (38) | 16 (43) | ||
| Not known | 17 (26) | 6 (21) | 11 (30) | ||
| PAVM | Yes | 33 (55) | 12 (41) | 21 (57) | |
| No | 29 (44) | 15 (52) | 14 (38) | ||
| Not known | 4 (6) | 2 (7) | 2 (5) | ||
| HVM | Yes | 17 (26) | 6 (21) | 11 (30) | |
| No | 34 (52) | 16 (55) | 18 (49) | ||
| Not known | 15 (23) | 7 (24) | 8 (22) | ||
| CVM | Yes | 8 (12) | 5 (17) | 3 (8) | |
| No | 40 (61) | 16 (55) | 24 (65) | ||
| Not known | 18 (27) | 8 (28) | 10 (27) | ||
| Epistaxis | Yes | 64 (97) | 28 (97) | 36 (97) | |
| No | 2 (3) | 1 (3) | 1 (3) | ||
| Missing | 0 | 0 | 0 | ||
| ESS | Score | 5.5 ± 2.3 (min: 0, max: 9.26) | 5.4 ± 2.3 | 5.5 ± 2.4 | |
| Missing | 0 | 0 | 0 | ||
| Haemoglobin | g/dl | 12.5 ± 2.7 | 13.4 ± 2.7 | 11.8 ± 2.5 | |
| Missing | 0 | 0 | 0 | ||
TAE teleangiectasia, FH family history, GI gastrointestinal bleedings, PAVM pulmonary arteriovenous malformations, HVM hepatic vascular malformations, CVM cerebral vascular malformations, ESS Epistaxis Severity Score with a scale from 1 = mild epistaxis to 10 = severe epistaxis. Data is shown in number of patients [(n) and (percentage)] or mean ± SD.
Altered gene transcription of HIF and hypoxia related genes in whole blood of HHT patients
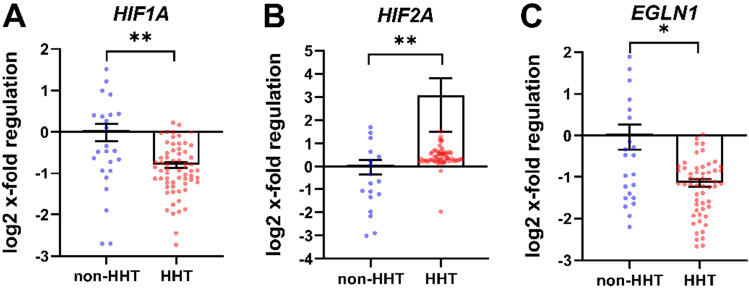
According to RT-PCR analysis, HIF1A gene expression was reduced by approximately 50% in blood cells of HHT patients compared to non-HHT controls (Fig. 1A). In contrast, HIF2A gene expression was upregulated in HHT patients compared to non-HHT controls (Fig. 1B). The HIF-1α target gene EGLN1 (encoding PHD2) (Fig. 1C), which is part of the hypoxic response, was also reduced by 50% in HHT patients compared to non-HHT controls.
Figure 1.
Hypoxia related gene transcription of whole blood in HHT compared to non-HHT controls. Patients with HHT showed decreased HIF1A (A) and increased HIF2A (B) gene expression. The HIF-1α target gene EGLN1 (C) is significantly decreased in HHT patients. Data are presented as log2 x-fold regulation in mean ± SEM. The Mann Whitney U test was used to compare the groups, *p < 0.05, **p < 0.01. n (non–HHT) = 26, n (HHT) = 66).
Decreased gene transcription of inflammation related genes in whole blood of HHT patients
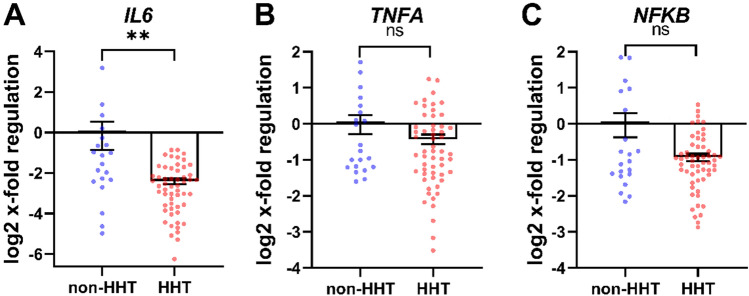
To confirm the biological relevance of altered HIF-regulation, the expression of different inflammation-related genes was analysed. The expression of HIF-1α target gene IL6 was significantly decreased in HHT patients compared to non-HHT controls (Fig. 2A). TNFA gene expression (Fig. 2B) and the gene expression of NFKB showed no significant difference between the analysed groups (Fig. 2C).
Figure 2.
Inflammation related gene transcription of whole blood in HHT compared to non-HHT controls. HIF-1α target gene IL6 (A) was significantly downregulated in HHT patients, whereas TNFA (B) and NFKB (C), the upstream regulator of HIF-1α, showed no difference in gene expression. Data are presented as log2 x-fold regulation in mean ± SEM. Mann Whitney test was used to compare the groups, ***p < 0.001, ns = not significant. n (non–HHT) = 26, n (HHT) = 66).
Gene transcription of angiogenesis related genes in whole blood of HHT patients is not affected
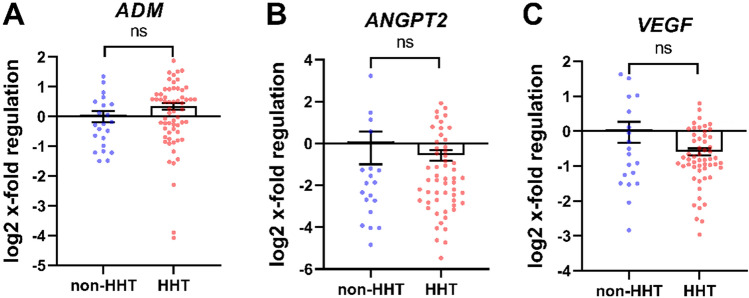
Further analysis was performed of HIF-regulated genes with biological relevance to angiogenesis. The expression of HIF-1 target gene ADM (Fig. 3A), ANGPT2 (Fig. 3B) and VEGF (Fig. 3C) was not affected, comparing HHT patients and non-HHT control individuals.
Figure 3.
Angiogenesis related gene transcription of whole blood in HHT compared to non-HHT controls. Patients with HHT showed no differences in the regulation of HIF target genes ADM (A). ANGPT2 (B) and VEGF (C). Data are presented as log2 x-fold regulation in mean ± SEM. The Mann–Whitney U test was used to compare the groups, ns = not significant. n (non-HHT) = 26, n (HHT) = 66.
Decreased HIF-1α protein in whole blood of HHT patients
To gain insight on the effects of downregulated HIF1A gene expression at the protein level, we assessed HIF-1α protein levels in blood cells of HHT patients and non-HHT control individuals by immunostaining. HIF-1α protein level was significantly reduced by about 50% in blood cells of HHT patients compared to non-HHT controls (Fig. 4).
Figure 4.
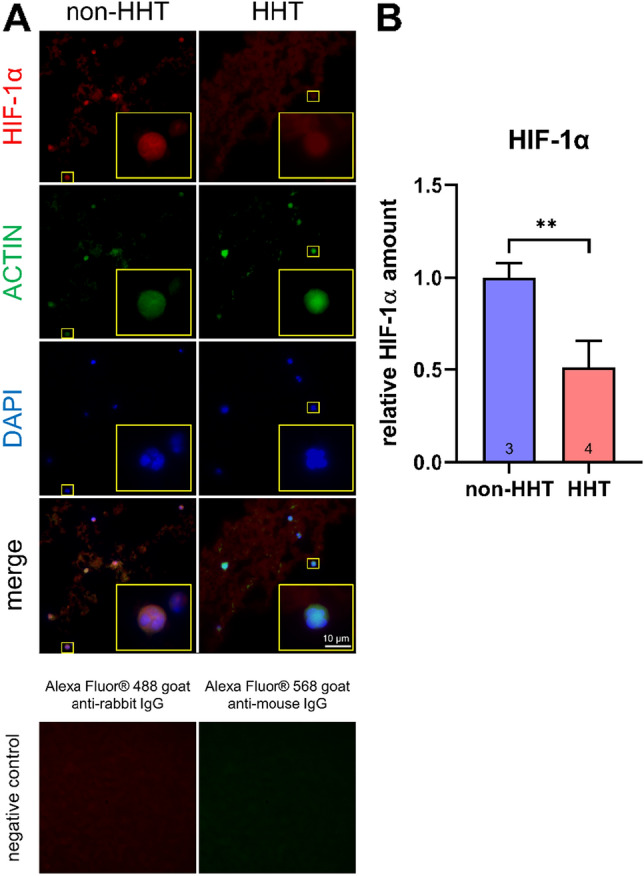
HIF-1α protein immunostaining in whole blood. Whole blood of HHT patients and non-HHT controls were immunostained. Examples of pictures from cells are shown in (A). HIF-1α is shown in red, ACTIN in green and the nuclei in blue. As negative control, staining with secondary antibody only is shown for non-HHT controls. Quantification of HIF-1α amount is shown in (B). HHT patients showed significantly less amounts of HIF-1α. Unpaired t-test was applied to determine significantly differences between the groups. Data are shown as mean ± SD, **p < 0.01.
No correlation between HHT patients, HIF gene expression and the Epistaxis Severity Score
There was no significant correlation between HIF1A gene expression and the individual Epistaxis Severity Score of HHT patients (Fig. 5A). Furthermore, the correlation of HHT patients’ individual Epistaxis Severity Score and HIF2A gene expression showed no significant correlation (Fig. 5B).
Figure 5.
Correlation of HIF and Epistaxis Severity Score. Individual HHT patients’ Epistaxis Severity Score was correlated to individual relative HIF1A gene transcription (n = 62) (A) or individual relative HIF2A gene expression (n = 58) (B). Gene expression was calculated as 2-ΔCT expression with respect to ACTB. No significant correlation was observed. Data are presented as individual values; Spearman R test was used to assess the correlations. ns not significant.
Discussion
HHT patients suffer from vascular disorders, which lead to recurrent bleeding, as well as immunological abnormalities are indicated in few studies1,4,5,21. Angiogenesis and inflammation are closely related processes and HIF is a key player in both of them. HIF activation in both hypoxia and inflammation regulates the transcription of several genes related to angiogenesis, metabolism, cell proliferation and apoptosis17. For the first time, this study provides evidence that HHT might alter expression of HIF and, potentially, of some HIF-regulated downstream genes.
Recurrent bleeding cause lower haemoglobin levels (11.8 ± 2.5 g/dl, Table 1), which reduces the oxygen supply to the tissues resulting in hypoxia. Moreover, this hypoxia should be latent but global throughout the body. This phenomenon led us to assume that the amount of HIF-1α protein would have increased or—with respect to the mRNA level—at least not altered22. Unexpectedly, RNA expression analysis in whole blood revealed significantly decreased HIF-1α expression in HHT patients’ leukocytes (Fig. 1A). Even target genes related to cellular hypoxic response of HIF-1α showed decreased or unchanged expressions in HHT patients (Fig. 1C) which is consistent with a reduction of HIF-1α also at the protein level. We succeeded in performing HIF-1α staining in whole blood cryosections of human subjects and found reduced amounts of HIF-1α protein in HHT-patients’ leucocytes (Fig. 4).
Obviously, analysis of whole blood RNA and protein expression of HIF-α in particular will provide data for leucocyte expression because they are the only nucleated cells in the blood. Gene expression in leucocytes from HHT patients is also of interest due to their impaired immune response. Thus, it was of interest to see inflammation related HIF-1α target gene IL6 was significantly downregulated (Fig. 2A). On the other hand, there was no change in the expression of TNFα as a general marker of acute inflammation in HHT patients (Fig. 2B). This finding was corroborated by unaltered NFKB expression in HHT patients (Fig. 2C). This, again, is of interest with respect to HIF-1α expression because NFKB is believed to act upstream of HIF-1α gene expression; based on our result, no evidence exists for a role of NFKB to mediate the reduced expression of HIF-1α at the transcript level in HHT19,23.
As mentioned above, many studies describe abnormalities in the immune response of HHT patients, to the extent that patients had milder symptoms during COVID infections or are less prone to tumour development compared to the non-diseased population24,25. Both the development of symptoms in COVID-19 and the development of some tumours as an effect of chronic inflammation closely depend on the immune response, which in part is related to the increased expression of HIF target genes26,27. Of note, a very recent work by Taniguchi-Ponciano et al. showed that critically ill COVID-19 patients had significantly increased HIF-1α expression in leukocytes28. Thus, reduced HIF-1α expression in immune cells may protect HHT patients from the exacerbating activation of the immune system, as it occurs in COVID-19 infections.
How might reduced HIF-1α affect leukocyte function in HHT? Impaired migration of human HHT monocytes was found in a study by Han et al.29. In 2003, one of the first studies on the role of HIF-1α in immune cells by Cramer et al. already reported myeloid cell aggregation, motility and invasiveness to be impaired in mice with a conditional Hif1α knockout. Similarly, Kojima et al. suggested that HIF-1α deficiency depressed the function of cytotoxic T-lymphocytes and blocked B-cell development in bone marrow30,31. The observation by Han et al.29 may therefore now be understood as an effect of reduced HIF-1α. This may have contributed to an impaired motility. This is in agreement with our previous studies, where we could demonstrate impaired motility of mouse macrophages with a conditional Hif1α knock out32.
Of note, HIF2A expression showed the opposite effect from what we observed with HIF1A, namely an increased gene expression in HHT patients (Fig. 1B). In mice, the conditional knockout of Hif2a in cells of the myeloid lineage resulted in increased recruitment of neutrophils to deeper layers of the colon during colitis33. One could speculate that HHT immune cells with increased HIF-2α might react in the opposite way and not be able to migrate into tissues and activate the common immune response. Obviously, we have to take into account that HIF-1α is expressed in all immune cell types, while HIF-2α is more selectively found in neutrophils and NK cells under hypoxic conditions as reviewed in Palazon et al.19. Thus, differential composition of immune cells and in particular the higher numbers of neutrophils and NK-cells in HHT patients may explain the increase in the overall higher leukocyte HIF2A expression21.
Following several studies’ descriptions of the influence of immune cells on angiogenesis and the vascular abnormalities in HHT patients, we analysed HIF target genes influencing the angiogenesis34–37. Surprisingly, changes in angiogenesis associated genes were not detected in HHT patients’ leukocytes (Fig. 3). It appears that, at the very least, blood leucocytes do not contribute to the vascular phenotype in HHT by expressing and secreting significantly higher levels of angiogenic molecules. This stands in contrast to the report of increased plasma VEGF levels in HHT patients by Cirulli et al. and Sadick et al. but may simply indicate that tissues other than those that are immune secrete VEGF in this disease38,39.
As mentioned above, key molecules in HHT are components of the TGF-β/BMP-pathway2. Caused by the haploid deficiency of the TGF-β/BMP pathway, TGF-β levels are elevated in the plasma of HHT patients39. Following previous studies, this should lead to an increase in HIF-1α regulation6–8; thus TGF-β itself is unlikely to be the reason for the reduction of HIF-1α in HHT patients. However, when the impaired TGF-β/BMP pathway occurring in HHT was stimulated by suppressing TGF-β signalling with TGF-β antagonists, rat kidney cells showed decreased HIF-1α expression40. We therefore assume that the full functionality of the TGF-β/BMP pathway is required for both HIF-1α expression and protein accumulation. To prove this hypothesis, further studies should aim to reconstitute TGF-β signalling in HHT leucocytes in order to increase HIF1A expression to normal levels.
To evaluate the severity of the clinical presentation of the disease, clinicians are still searching for suitable biomarkers for HHT, which are missing in daily clinical use41–43. To that end, we correlated the results of HIF1A and HIF2A gene expression with the clinically determined Epistasis Severity Score of each individual HHT patient. Unfortunately, a significant correlation between disease severity and HIF gene expression was not detected (Fig. 5) which may indicate that the situation is more complex and cannot be fully explained by such a correlation. It was demonstrated that TGF-β/BMP pathway members and HIF-1α interact and form a multi-protein complex on the promoter of the TGF-β co-receptor (endoglin) and through that mechanism, influence vascular remodelling and angiogenesis9. This multi-protein complex might be disturbed in HHTs because of reduced HIF-1α. In addition to the known mutations in endoglin, HIF-1α could contribute and aggravate defects in TGF-β signalling leading to dysregulated angiogenesis in HHT patients.
Taken together, our findings show that the regulation of HIF-1α in HHT patients does not support the canonical regulation during hypoxia or inflammation. The results point towards a downregulation caused by the haploid impairment of the TGF-β/BMP pathway, but the role of different other factors remain to be elucidated While reduced expression of the HIF1A-gene and HIF-1α protein has, to our knowledge, never been described in any disease, future studies might reveal the underlying mechanism.
Materials and methods
Selection of participants and clinical parameters
The study was approved by the ethics committee of the University Duisburg-Essen (20-9162-BO). Participants were informed and gave written consent in accordance with the Declaration of Helsinki. The study is registered at Clinical trials.gov (ID NCT04469517).
Adult patients with confirmed HHT (fulfilled at least three out of four Curaçao Criteria and/or had a positive genetic testing) were included in this prospective study1,44. Alongside patients’ characteristics such as age and gender, factors that might also affect HIF production were documented. HIF affecting parameters include smoking habits, days spent at height (> 1000 m above sea level) and the median weekly time spent doing sports. The documented HHT related clinical parameters include frequency, duration and intensity of epistaxis, need for medical attention, transfusions and signs of anaemia (haemoglobin level), family history, mucocutaneous telangiectases and visceral arterio-venous malformations especially involving the liver, lungs, gastrointestinal tract and central nervous system. The recurrent nosebleeds were described using the Epistaxis Severity Score (ESS)45. In all cases, whole blood samples were taken from patients peripheral arm veins. All experiments were compared with blood from a non-HHT control group. The non-HHT control group exists of 26 volunteers (15 men, 11 women) recruited from university employees (mean age: 40 ± 13; min: 21, max: 63). The number of patients and non-HHT donors is listed in the figures for each experiment.
RNA preparation and RT-PCR of whole blood
RNA from whole blood was isolated using the PAXgene® blood RNA collection system (Qiagen, Mississauga, Canada) according to the manufacturer’s protocol. 2.5 ml of whole blood was drawn from HHT patients or non-HHT controls into PAXgene® blood RNA tubes (Qiagen, Mississauga, Canada) and stored at – 20 °C.
Complementary DNA (cDNA) was synthesised using one µg of RNA and Moloney murine leukemia virus reverse transcriptase (Promega, Walldorf, Germany) according to the manufacturer’s instructions. Quantification of gene expression was performed by real-time PCR (RT-PCR) with SYBR green fluorescent dye (Eurogentec, Verviers, Belgium) and the CFX96™ Real Time System (Bio-Rad Laboratories GmbH, Munich, Germany). cDNA was amplified by 40 cycles of 95 °C for 15 s and 60 °C for 90 s with gene specific primers (Table 2) and normalized to ACTB (actin). Primer specificity was checked by Primer-BLAST and confirmed by size analysis of the PCR amplicons46. Expression was calculated with the 2-ΔCT method for statistical analysis and set as an induction relative to non-HHT controls in figures.
Table 2.
Human primer sequences.
| Gene | 5′Primer | 3′Primer |
|---|---|---|
| ACTB | TCACCCACACTGTGCCCATCTACGA | CAGCGGAACCGCTCATTGCCAATGG |
| ADM | AGTCGTGGGAAGAGGGAACT | ATCCGGACTGCTGTCTTCGG |
| ANPT2 | AACTTTCGGAAGAGCATGGAC | CGAGTCATCGTATTCGAGCGG |
| HIF-1α | TCACTGGGACTATTAGGCTCAGGT | CTCCATTACCCACCGCTGAA |
| HIF-2α | CGGAGGTGTTCTATGAGCTGG | ASCTTGTGTGTTCGCAGGAA |
| IL-6 | TGCATCTAGATTCTTTGCCTTTTT | CCACTCACCTCTTCAGAACGAA |
| NFκB | AACAGAGAGGATTTCGTTTCCG | TTTGACCTGAGGGTAAGACTTCT |
| EGLN1 | CCAGCTTCCCGTTACAGT | GCACGACACCGGGAAGTT |
| TNFα | GGCGTGGAGCTGAGAGATAAC | GGTGTGGGTGAGGAGCACAT |
| VEGF | GCAAGACAAGAAAATCCCTGTGGGCC | CCGCCTCGGCTTGTCACA |
Primer sequences of specific PCR products used for RNA quantifications of human whole blood via RT-PCR.
HIF-1α staining of whole blood
Following withdrawal, blood was immediately transferred in to PHEM buffer (3 mmol PIPES, 1 mmol/l HEPES, 0.5 mmol/l EGTA, 0.25 mmol/l MgCl2, 3.5 mmol/l KCl, pH = 7.2) containing 4% paraformaldehyde and 2.5% glutaraldehyde. After 1 h of fixation, cells were centrifuged (RT, 10 min, 1500 rcf) and the pellet was cryoconserved in Tissue-Tek® O.C.T.™ Compound (Sakura Finetek, Netherlands). 10 µm sections were stained with primary antibodies against HIF-1α (BD Transduction Laboratories™, Switzerland) and ACTIN (Sigma-Aldrich, Inc.). For secondary antibodies Alexa Fluor® 488 goat anti-rabbit IgG (Life Technologies) and Alexa Fluor® 568 goat anti-mouse IgG (Invitrogen AG) were used. To visualize nuclei, cells were covered with 2-[4-(Aminoiminomethyl)phenyl]-1H-Indole-6-carboximidamide hydrochloride (DAPI)-added Mowiol. At least 20 cells per subject were quantified.
Statistics
GraphPad Prism 8.4.3. (GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analysis. After elimination of outliers (ROUT-test), the groups were analysed for Gaussian distribution using the D’Agostino–Pearson test. An unpaired t-test was applied to Gaussian distributed data and Mann–Whitney U test to non-Gaussian distributed data. Statistical significance is displayed as *(p < 0.05), **(p < 0.01), ***(p < 0.001) or ****(p < 0.0001).
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Limitations of the study
As in many studies of rare diseases, we have to mention the limited size of the investigated patient cohort. Regarding gene expression only a small selection of HIF target genes was analysed, more genes could complement this.
Acknowledgements
We are very grateful to Michael Denniss, for proof reading as a native speaker. Further, we want to acknowledge all blood donors.
Author contributions
Conceptualization, A.W. and F.D.; methodology, A.W., T.L. and F.D.; investigation, A.W. and T.L.; resources, A.W.; writing—original draft preparation, A.W.; writing—review and editing, A.W., F.D., T.L., U.G., J.J., S.L. and J.F.; visualization, A.W., T.L.; supervision, A.W.; project administration, A.W., F.D.; funding acquisition, S.L. and J.F. All authors have read and agreed to the published version of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Faughnan M, et al. International guidelines for the diagnosis and management of hereditary haemorrhagic telangiectasia. J. Med. Genet. 2011;48:73–87. doi: 10.1136/jmg.2009.069013. [DOI] [PubMed] [Google Scholar]
- 2.Bernabéu C, Blanco FJ, Langa C, Garrido-Martin EM, Botella LM. Involvement of the TGF-β superfamily signalling pathway in hereditary haemorrhagic telangiectasia. J. Appl. Biomed. 2010;8:169–177. doi: 10.2478/v10136-009-0020-x. [DOI] [Google Scholar]
- 3.Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu. Rev. Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirulli A, et al. Patients with hereditary hemorrhagic telangectasia (HHT) exhibit a deficit of polymorphonuclear cell and monocyte oxidative burst and phagocytosis: A possible correlation with altered adaptive immune responsiveness in HHT. Curr. Pharm. Des. 2006;12:1209–1215. doi: 10.2174/138161206776361336. [DOI] [PubMed] [Google Scholar]
- 5.Guilhem A, Malcus C, Clarivet B, Plauchu H, Dupuis-Girod S. Immunological abnormalities associated with hereditary haemorrhagic telangiectasia. J. Intern. Med. 2013;274:351–362. doi: 10.1111/joim.12098. [DOI] [PubMed] [Google Scholar]
- 6.Basu RK, et al. Interdependence of HIF-1α and TGF-β/Smad3 signaling in normoxic and hypoxic renal epithelial cell collagen expression. Am. J. Physiol. Ren. Physiol. 2011;300:F898–F905. doi: 10.1152/ajprenal.00335.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna C, et al. Hypoxia-inducible factor-2α and TGF-β signaling interact to promote normoxic glomerular fibrogenesis. Am. J. Physiol. Ren. Physiol. 2013;305:F1323–F1331. doi: 10.1152/ajprenal.00155.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozen-Zvi B, et al. TGF-β/Smad3 activates mammalian target of rapamycin complex-1 to promote collagen production by increasing HIF-1α expression. Am. J. Physiol. Ren. Physiol. 2013;305:F485–F494. doi: 10.1152/ajprenal.00215.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sánchez-Elsner T, et al. Synergistic cooperation between hypoxia and transforming growth factor-β pathways on human vascular endothelial growth factor gene expression. J. Biol. Chem. 2001;276:38527–38535. doi: 10.1074/jbc.M104536200. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez-Elsner T, Botella LM, Velasco B, Langa C, Bernabéu C. Endoglin expression is regulated by transcriptional cooperation between the hypoxia and transforming growth factor-β pathways. J. Biol. Chem. 2002;277:43799–43808. doi: 10.1074/jbc.M207160200. [DOI] [PubMed] [Google Scholar]
- 11.Fandrey J, Gorr TA, Gassmann M. Regulating cellular oxygen sensing by hydroxylation. Cardiovasc. Res. 2006;71:642–651. doi: 10.1016/j.cardiores.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Semenza, G. In Cold Spring Harbor Symposia on Quantitative Biology. 347–353 (Cold Spring Harbor Laboratory Press, 1979).
- 13.Ema M, et al. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 15.Wiesener MS, et al. Widespread, hypoxia-inducible expression of HIF-2α in distinct cell populations of different organs. FASEB J. 2003;17:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 16.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 17.Semenza GL. HIF-1, O2, and the 3 PHDs: How animal cells signal hypoxia to the nucleus. Cell. 2001;107:1–3. doi: 10.1016/s0092-8674(01)00518-9. [DOI] [PubMed] [Google Scholar]
- 18.Frede S, Berchner-Pfannschmidt U, Fandrey J. Regulation of hypoxia-inducible factors during inflammation. Methods Enzymol. 2007;435:403–419. doi: 10.1016/S0076-6879(07)35021-0. [DOI] [PubMed] [Google Scholar]
- 19.Palazon A, Goldrath AW, Nizet V, Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41:518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez-Elsner T, et al. A cross-talk between hypoxia and TGF-β orchestrates erythropoietin gene regulation through SP1 and Smads. J. Mol. Biol. 2004;336:9–24. doi: 10.1016/j.jmb.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 21.Droege F, et al. Impaired release of neutrophil extracellular traps and anemia-associated T cell deficiency in hereditary hemorrhagic telangiectasia. J. Clin. Med. 2020;9:767. doi: 10.3390/jcm9030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wiener CM, Booth G, Semenza GL. In vivo expression of mRNAs encoding hypoxia-inducible factor 1. Biochem. Biophys. Res. Commun. 1996;225:485–488. doi: 10.1006/bbrc.1996.1199. [DOI] [PubMed] [Google Scholar]
- 23.Albina JE, et al. HIF-1 expression in healing wounds: HIF-1α induction in primary inflammatory cells by TNF-α. Am. J. Physiol. Cell Physiol. 2001;281:C1971–C1977. doi: 10.1152/ajpcell.2001.281.6.C1971. [DOI] [PubMed] [Google Scholar]
- 24.Marcos S, et al. SARS-CoV-2 infection in hereditary hemorrhagic telangiectasia patients suggests less clinical impact than in the general population. J. Clin. Med. 2021;10:1884. doi: 10.3390/jcm10091884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duarte CW, Black AW, Lucas FL, Vary CP. Cancer incidence in patients with hereditary hemorrhagic telangiectasia. J. Cancer Res. Clin. Oncol. 2017;143:209–214. doi: 10.1007/s00432-016-2272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaGory ELG, Amato J. The ever expanding role of hif in tumor and stromal biology. Nat. Cell Biol. 2016;18:356–365. doi: 10.1038/ncb3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jahani M, Dokaneheifard S, Mansouri K. Hypoxia: A key feature of COVID-19 launching activation of HIF-1 and cytokine storm. J. Inflamm. 2020;17:1–10. doi: 10.1186/s12950-020-00263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi-Ponciano K, et al. Increased expression of hypoxia-induced factor 1α mRNA and its related genes in myeloid blood cells from critically ill COVID-19 patients. Ann. Med. 2021;53:197–207. doi: 10.1080/07853890.2020.1858234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Z, et al. Reduction of endoglin receptor impairs mononuclear cell-migration. Explor. Med. 2020;1:136. doi: 10.37349/emed.2020.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kojima H, Sitkovsky MV, Cascalho M. HIF-1α deficiency perturbs T and B cell functions. Curr. Pharm. Des. 2003;9:1827–1832. doi: 10.2174/1381612033454388. [DOI] [PubMed] [Google Scholar]
- 31.Cramer T, et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber T, Quinting T, Dittmer U, Fandrey J, Sutter K. Hypoxia-inducible factor 1α is essential for macrophage-mediated erythroblast proliferation in acute friend retrovirus infection. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-17324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerber EL, et al. The importance of hypoxia-inducible factors (HIF-1 and HIF-2) for the pathophysiology of inflammatory bowel disease. Int. J. Mol. Sci. 2020;21:8551. doi: 10.3390/ijms21228551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salafutdinov II, et al. Influence of recombinant codon-optimized plasmid DNA encoding VEGF and FGF2 on co-induction of angiogenesis. Cells. 2021;10:432. doi: 10.3390/cells10020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikuchi-Taura A, et al. Bone marrow mononuclear cells activate angiogenesis via gap junction-mediated cell–cell interaction. Stroke. 2020;51:1279–1289. doi: 10.1161/STROKEAHA.119.028072. [DOI] [PubMed] [Google Scholar]
- 36.Ding LL, et al. Effect of interleukin-34 on secretion of angiogenesis cytokines by peripheral blood mononuclear cells of rheumatoid arthritis. Immunol. Investig. 2020;49:81–87. doi: 10.1080/08820139.2019.1649281. [DOI] [PubMed] [Google Scholar]
- 37.Cho KH, et al. Therapeutic mechanism of cord blood mononuclear cells via the IL-8-mediated angiogenic pathway in neonatal hypoxic-ischaemic brain injury. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-61441-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cirulli A, et al. Vascular endothelial growth factor serum levels are elevated in patients with hereditary hemorrhagic telangiectasia. Acta Haematol. 2003;110:29–32. doi: 10.1159/000072411. [DOI] [PubMed] [Google Scholar]
- 39.Sadick H, et al. Patients with hereditary hemorrhagic telangiectasia have increased plasma levels of vascular endothelial growth factor and transforming growth factor-beta1 as well as high ALK1 tissue expression. Haematologica. 2005;90:818–828. [PubMed] [Google Scholar]
- 40.Lan R, et al. Mitochondrial pathology and glycolytic shift during proximal tubule atrophy after ischemic AKI. J. Am. Soc. Nephrol. 2016;27:3356–3367. doi: 10.1681/ASN.2015020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadick H, et al. Does the genotype of HHT patients with mutations of the ENG and ACVRL1 gene correlate to different expression levels of the angiogenic factor VEGF? Int. J. Mol. Med. 2008;22:575–580. [PubMed] [Google Scholar]
- 42.Botella L-M, Albiñana V, Ojeda-Fernandez L, Recio-Poveda L, Bernabéu C. Research on potential biomarkers in hereditary hemorrhagic telangiectasia. Front. Genet. 2015;6:115. doi: 10.3389/fgene.2015.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steineger J, et al. Pentraxin 3 level is elevated in hereditary hemorrhagic telangiectasia and reflects the severity of disease-associated epistaxis. Laryngoscope. 2019;129:E44–E49. doi: 10.1002/lary.27548. [DOI] [PubMed] [Google Scholar]
- 44.Shovlin CL, et al. Diagnostic criteria for hereditary hemorrhagic telangiectasia (Rendu–Osler–Weber syndrome) Am. J. Med. Genet. 2000;91:66–67. doi: 10.1002/(sici)1096-8628(20000306)91:1<66::aid-ajmg12>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 45.Hoag JB, Terry P, Mitchell S, Reh D, Merlo CA. An epistaxis severity score for hereditary hemorrhagic telangiectasia. Laryngoscope. 2010;120:838–843. doi: 10.1002/lary.20818. [DOI] [PubMed] [Google Scholar]
- 46.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author.