Abstract
The effects of the everninomicin antibiotic evernimicin (SCH27899) on growing Staphylococcus aureus cells were investigated. Cellular growth rates and viable cell numbers decreased with increasing antibiotic concentrations. The rate of protein synthesis, measured as 35S-amino acid incorporation, declined in parallel with the growth rate. Significantly, the formation of the 50S ribosomal subunit was inhibited in a dose-dependent fashion as well. 30S ribosomal subunit synthesis was not affected over the same concentration range. Evernimicin did not stimulate the breakdown of mature ribosomal subunits. Pulse-chase labeling experiments revealed a reduced rate of 50S subunit formation in drug-treated cells. Two erythromycin-resistant strains of S. aureus that carried the ermC gene were as sensitive as wild-type cells to antibiotic inhibition. In addition, two methicillin-resistant S. aureus organisms, one sensitive to erythromycin and one resistant to the macrolide, showed similar sensitivities to evernimicin. These results suggest a use for this novel antimicrobial agent against antibiotic-resistant bacterial infections.
In the struggle to keep up with the current increase in the number of antibiotic-resistant infectious organisms, both new antimicrobial agents and new cellular targets must be found (10). A compound identified 35 years ago has recently been reinvestigated as a new and potentially effective antibiotic. The everninomicins are a group of complex, sugar-derived antibiotics isolated from Micromonospora carbonacea (25, 26). They were described and characterized many years ago, but very few studies have been conducted to examine their modes of action (12, 21; A. K. Ganguly and A. K. Saksena, Communications to the editor, J. Antibiot. (Tokyo) 28:707–709, 1975). Avilamycin, a polysaccharide antibiotic with similarities to the everninomicins, was shown to affect protein synthesis by interacting with the 30S ribosomal subunit (27). This compound has been used as an antimicrobial agent in animal feed (1).
Recently, another everninomicin, evernimicin (SCH 27899), has been examined in more detail (13, 14, 24). Everninomicin B (evernimicin) has been shown to be very effective against a number of different gram-positive organisms (16, 23; R. S. Hare and F. J. Sabatelli, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-119, p. 204, 1998; Y. Sato, A. Kuga, R. Okamoto, and M. Inoue, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother. abstr. E-112, p. 202, 1998), including methicillin-resistant Staphylococcus aureus (MRSA) strains (19). It is also very effective against Legionella species (11). Recently, it was shown to specifically inhibit protein synthesis in growing cells (T. A. Black, W. Zhao, K. J. Shaw, and R. S. Hare, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-106, p. 99, 1998). In addition, resistant Streptococcus pneumoniae mutants which contained an altered 50S ribosomal subunit protein L16 (P. V. Adrian and K. P. Klugman, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-110, p. 99, 1998) or altered 23S rRNA nucleotides (P. V. Adrian, C. Mendrick, D. Loebenberg, K. J. Shaw, K. P. Klugman, R. S. Hare, and T. A. Black, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-845, p. 117, 1999) have been isolated.
We have identified a novel target for macrolide antibiotics in bacterial cells, the assembly of 50S ribosomal subunits (3–5). Macrolide and ketolide antibiotics have equivalent inhibitory effects on both translation and 50S subunit formation in S. aureus (7, 8). Since it has been suggested that evernimicin inhibits protein synthesis by interacting with the 50S subunit (Adrian and Klugman, 38th ICAAC), we decided to investigate its inhibitory effects on translation and subunit assembly in a systematic fashion. We found that both translation and 50S subunit formation were targets for inhibition in wild-type S. aureus cells and in both MRSA and erythromycin-resistant mutant strains. The significance of these findings are discussed in terms of the effects both on 50S subunit formation and on the potential clinical use of this antimicrobial agent.
MATERIALS AND METHODS
Measurements of cell growth, subunit assembly, and translation rates.
Studies were conducted with S. aureus strains RN1786, RN4220 ermC (inducible macrolide-lincosamide-streptogramin B [MLSB] resistance), and SK983 ermC (constitutive MLSB resistance), MRSA A1018 ermA (constitutive MLSB resistance), and MRSA A1024, all of which were provided by J. Sutcliffe of Pfizer Central Research. Evernimicin (SCH 27899) and its placebo were generously provided by T. Black of Schering-Plough Corp. Evernimicin was made as a stock solution at 160 μg/ml in placebo and was diluted in placebo as needed. Cells were grown at 37°C in tryptic soy broth (TSB) in the presence and absence of evernimicin as described previously (7, 8). The erythromycin-resistant strains were grown with erythromycin at 50 μg/ml, and the MRSA organisms were grown with ampicillin at 50 μg/ml. Growth rates were measured by following the increase in cell density in a Klett-Summerson colorimeter. To examine ribosomal subunit assembly, the cells were labeled with [3H]uridine (1 μCi/ml; 2 μg/ml) and were allowed to grow for two doublings in the presence of the drug. Isotope incorporation was halted by adding uridine to a final concentration of 50 μg/ml, followed by a 30-min chase period. The rate of protein synthesis was determined in each culture during the chase period by adding [35S]methionine and cysteine (Tran 35S-label; ICN Pharmaceuticals) to 1 μCi/ml. Three samples of 0.2 ml were collected at 5-min intervals and were precipitated with 10% trichloroacetic acid, and the 35S-methionine and cysteine in the proteins were measured by liquid scintillation counting. At the end of the chase period, viable cell counts were measured by serial dilution of cells in A salts (18) followed by plating of 10 μl on square TSB agar plates by the method of Jett et al. (15). The colonies were counted after 48 h at 37°C.
Cell lysis and sucrose gradient sedimentation of ribosomal subunits were performed as described previously (7, 8). The absorbance at 254 nm for each gradient was detected with an ISCO model UA-5 absorbance monitor. Gradient fractions were mixed with 3 ml of ScintiSafe Gel, and the incorporation of [3H]uridine into RNA in ribosomal subunits was measured by liquid scintillation counting.
Uridine pulse and chase labeling.
Two 12-ml cultures of cells, one control and one with evernimicin at 0.32 μg/ml, were grown to a Klett reading of 40. The cells were pulse labeled with [3H]uridine (1 μCi/ml) for 90 s and were then chased with uridine at 25 μg/ml. At intervals, 2-ml samples were removed, collected by centrifugation, washed, and stored frozen until lysis for sucrose gradient centrifugation as described above.
Ribosomal subunit turnover measurement.
The effect of evernimicin on the degradation of mature ribosomal subunits was measured by growing duplicate 5-ml cultures of cells in [3H]uridine (1 μCi/ml; 2 μg/ml) for two generations. The cells were collected by centrifugation at room temperature, washed, and resuspended at a 10-fold dilution in fresh TSB. One culture received evernimicin at 0.32 μg/ml, and both were regrown for two doublings in the absence of [3H]uridine. The cells were collected, washed, and frozen, and the lysates were examined by sucrose gradient centrifugation as described above. The absorbance at 260 nm of each gradient fraction was determined to measure the relative amounts of both ribosomal subunits in each gradient. The fractions were then mixed with an equal volume of 20% trichloroacetic acid, and the precipitates were collected on glass fiber filters for liquid scintillation counting (3).
MIC determination.
The MIC of evernimicin for each strain used was determined by a dilution method as described elsewhere (7, 8). Tubes containing 1 ml of TSB received 50 μl of an overnight culture of S. aureus cells and the antibiotic over a concentration range of 0.06 to 16 μg/ml. The tubes were incubated at 37°C for 24 h, and the absorbance at 600 nm was recorded.
RESULTS
The effects of evernimicin on the growth of S. aureus cells were examined. An MIC of 0.8 μg/ml was found for the wild-type strain and for two erythromycin-resistant strains that carried the ermC gene. The two MRSA strains examined were slightly more sensitive to evernimicin (MIC, 0.4 μg/ml). The growth rates of the five strains examined were all reduced in liquid culture with evernimicin (Table 1). The decline in the cellular growth rate was paralleled by a decline in viable cell number for each organism. In addition, the growth rate of the wild-type cells in TSB was reduced with increasing concentrations of evernimicin. These results are presented in Table 1.
TABLE 1.
Effect of evernimicin (SCH27899) on doubling time, total cell number, protein synthesis rate, and 30S and 50S subunit amounts in S. aureus cellsa
| Strain | Evernimicin concn (μg/ml) | Doubling time (h) | No. of cells (107)/ml | Protein synthesis (% of control) | % Total cpm
|
|
|---|---|---|---|---|---|---|
| 30S | 50S | |||||
| SK1786 | 0.0 | 0.8 | 22 | 100 | 20 | 38 (100)b |
| 0.02 | 1.3 | 21 | 59 | 21 | 34 (89) | |
| 0.04 | 2.5 | 18 | 35 | 23 | 33 (87) | |
| 0.08 | 2.9 | 18 | 24 | 22 | 31 (82) | |
| 0.16 | 3.5 | 13 | 19 | 22 | 30 (79) | |
| 0.32 | 4.7 | 6.3 | 17 | 22 | 24 (63) | |
| 0.64 | 7.8 | 3.9 | 14 | 18 | 16 (42) | |
| RN4220 (ermC) | 0.0 | 0.6 | 19 | 100 | 23 | 33 (100) |
| 0.32 | 3.3 | 6.3 | 17 | 22 | 25 (76) | |
| SK983 (ermC) | 0.0 | 0.6 | 10 | 100 | 23 | 35 (100) |
| 0.32 | 4.3 | 7.2 | 16 | 22 | 19 (54) | |
| A1018 (MRSA ermA) | 0.0 | 0.6 | 44 | 100 | 19 | 37 (100) |
| 0.04 | 1.4 | 20 | 33 | 23 | 33 (89) | |
| A1024 (MRSA) | 0.0 | 0.75 | 27 | 100 | 21 | 35 (100) |
| 0.04 | 1.9 | 15 | 23 | 22 | 26 (74) | |
The results for doubling times, cell numbers, and protein synthesis rates are the averages of three or more experiments, with the standard error of the mean being ±3.5% for control samples (n = 7). Protein synthesis rates for the different strains are based on the data in Fig. 1. The percentage of the total radioactivity in the 30S and 50S subunit regions of each sucrose gradient is indicated. The results for the ribosomal subunit synthesis experiments are the averages of at least three experiments, with the standard error of the mean being ±2.0% for controls (n = 6).
The percentage of the total gradient radioactivity in the 50S subunit fractions is given in parentheses.
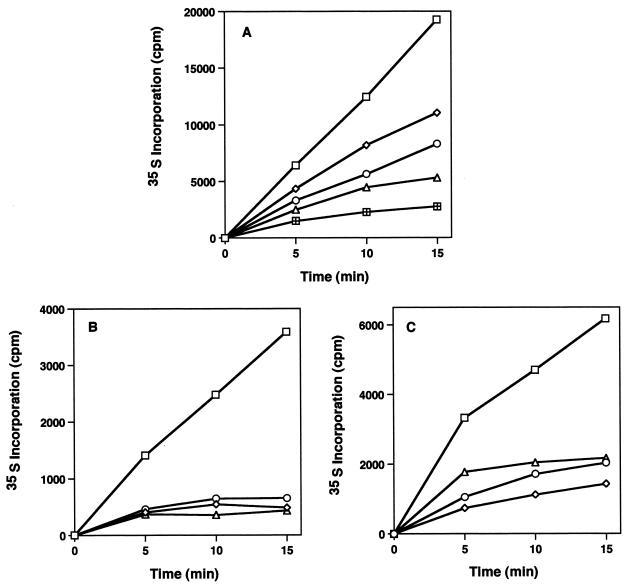
Evernimicin and avilamycin have both been shown to inhibit protein synthesis in cells (27; Black et al., 38th ICAAC). For each of the five strains studied, the rates of 35S-amino acid incorporation into cellular proteins were examined in the presence and the absence of the antibiotic. A dose-dependent decline in the rate of protein synthesis was observed for wild-type cells growing in the presence of evernimicin, as Fig. 1A shows. The growth rate and cell number declined in parallel with the reduction in protein synthesis rate (Fig. 2A), with a 50% inhibitory dose of 0.03 μg/ml. The two ermC strains showed similar sensitivities to the inhibitory effects of the drug (Fig. 1B). Protein synthesis in the two MRSA strains was also affected in a similar fashion, as Fig. 1C indicates. These effects of evernimicin on cellular protein synthesis are summarized in Table 1.
FIG. 1.
Effect of evernimicin on cellular protein synthesis. (A) The rates of incorporation of 35S-amino acids into proteins were determined as described in Materials and Methods for wild-type S. aureus cells growing without evernimicin (□) and with evernimicin at 0.02 μg/ml (◊), 0.04 μg/ml (○), 0.08 μg/ml (▵), and 0.16 μg/ml (⊞). (B) Protein synthesis rates for wild-type cells growing without evernimicin (□) and with evernimicin at 0.32 μg/ml (◊) and for ermC strains RN4220 (○) and SK983 (▵) growing with evernimicin at 0.32 μg/ml. (C) Protein synthesis rates for wild-type cells growing without evernimicin (□) and with evernimicin at 0.04 μg/ml (▵) and for MRSA strains A1018 (○) and A1024 (◊) growing with evernimicin at 0.04 μg/ml.
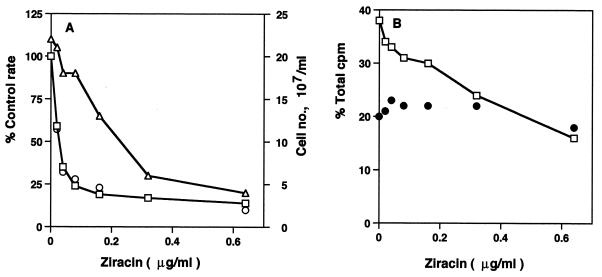
FIG. 2.
Effect of increasing evernimicin (Ziracin) concentrations on protein synthesis, growth rate, cell number, and ribosomal subunit synthesis in growing S. aureus cells. (A) Percentage of control protein synthesis rate (□) and control growth rate (○). ▵, viable cell number. (B) Percentage of total gradient radioactivity (in counts per minute) in 50S subunit (□) and 30S subunit (●) sucrose gradient fractions.
The assembly of the large 50S ribosomal subunit in S. aureus cells is a novel target for macrolide and ketolide antibiotics (2, 7). Evernimicin was tested to see if this drug would have a similar effect in these cells. 50S ribosomal subunit formation in each strain was susceptible to evernimicin inhibition, as Table 1 indicates. Compared with the untreated controls, evernimicin specifically reduced the percentage of the total [3H]uridine radioactivity found in the 50S subunit region of sucrose gradients, without affecting 30S subunit amounts. Table 1 also shows the effects of increasing antibiotic concentrations on 50S formation in wild-type cells. A dose-dependent inhibition in the formation of the larger subunit was found. This dose dependency for 50S assembly inhibition is displayed in Fig. 2B. A 50% inhibitory dose of 0.4 μg/ml was found, which is 13 times more than the value found for the effect on translation. 30S subunit formation was unaffected by evernimicin, except at the highest dose examined.
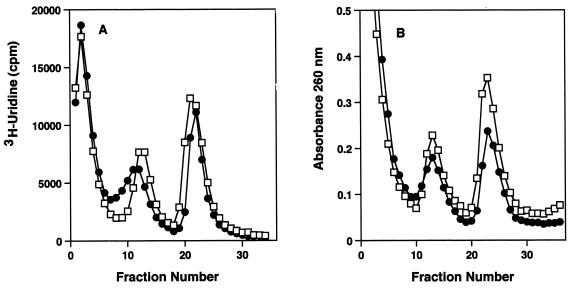
The macrolide antibiotics specifically prevent the formation of the 50S subunit and do not stimulate the breakdown of mature 50S particles (3, 9). Evernimicin was tested for a stimulatory effect on 50S subunit breakdown. Ribosomal subunits labeled with [3H]uridine in the absence of evernimicin were completely stable in the presence of the antibiotic, as Fig. 3A shows. There was no difference in the amount of [3H]uridine in 50S particles from cells grown with or without evernimicin. By contrast, the UV absorbance profile of the ribosomal subunits from evernimicin-treated cells showed a decline in the amount of 50S particles present (Fig. 3B).
FIG. 3.
Effect of evernimicin on ribosomal subunit breakdown. Cells were grown in the presence of [3H]uridine, washed, diluted, and regrown with and without evernimicin at 0.32 μg/ml. (A) [3H]uridine radioactivity profiles of sucrose gradients of cell lysates from cultures grown without (□) and with (●) evernimicin. (B) UV absorbance profiles of sucrose gradients of cell lysates from cultures grown without (□) and with (●) evernimicin.
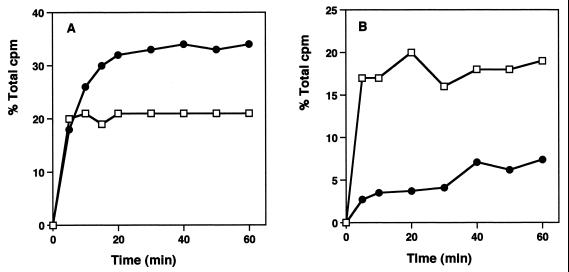
Pulse-and-chase labeling procedures have been used to examine the rates of ribosomal subunit formation in bacterial cells (22). The kinetics of ribosomal subunit formation were examined by measuring the level of incorporation of [3H]uridine into ribosomal particles after a 90-s pulse labeling. In the absence of antibiotic, synthesis of both subunits was complete in 20 min, with the characteristic 2:1 ratio of subunits found at this time (Fig. 4A). In evernimicin-treated cells, the kinetics of 30S formation were comparable to those for cells in the control culture. By contrast, 50S formation was substantially retarded, with a gradual, linear increase in subunits observed for up to 1 h following the uridine chase (Fig. 4B).
FIG. 4.
Pulse-chase labeling kinetics of ribosomal subunit formation in cells growing with and without evernimicin at 0.32 μg/ml. (A) Kinetics of 30S (□) and 50S (●) subunit formation in control cells. (B) Kinetics of 30S (□) and 50S (●) subunit formation in evernimicin-treated cells.
DISCUSSION
The results from this work indicate that evernimicin is a potent inhibitor of the growth and viability of S. aureus cells. It is effective at low concentrations and has an MIC for S. aureus which is equal to or lower than those of most macrolide antibiotics that we have examined (7, 8). The MICs of evernimicin for the strains that we have tested are very comparable to the MICs found by others for methicillin-susceptible S. aureus and MRSA organisms (19, 23). Viable cell numbers were reduced in parallel with the reduction in growth rate at increased evernimicin concentrations, indicating a bacteriocidal activity of the drug. Protein synthesis and specifically the 50S subunit have been indicated as the inhibitory targets for this compound by other work (Adrian and Klugman, 38th ICAAC; Black et al., 38th ICAAC). We have confirmed and extended these observations.
Inhibition of translation was found to be the preferential target for evernimicin in these cells, unlike our findings with macrolide antibiotics (4, 5). The inhibition of growth rate was directly related to the inhibition of cellular protein synthesis. The antibiotic's effect on 50S formation was less significant, with about 13 times as much drug needed to give an equivalent degree of inhibition. This suggests that the interaction of the antibiotic with the mature subunit is stronger than the interaction with the particle(s) in the assembly pathway. By contrast, three different macrolide antibiotics were shown to have equivalent inhibitory effects on translation and 50S subunit formation (4, 5). The difference in structure between the macrolide compounds and evernimicin suggests a different interaction with the macromolecules in the 50S particle. Evernimicin-resistant mutants of S. aureus with alterations in ribosomal protein L16 bind to the antibiotic with a six- to eightfold lower affinities (P. M. McNicholas, P. A. Mann, D. J. Najarian, L. Miesel, T. A. Black, R. S. Hare, and K. J. Shaw, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-846, p. 117, 1999). In S. aureus, evernimicin could interact weakly with 23S rRNA and proteins in the precursor particle to affect assembly and more strongly only when L16 is assembled into the mature 50S subunit. It is important in this regard that ribosomal protein L16 is not found in either 50S precursor particle in growing Escherichia coli cells (20).
It is significant that four antibiotic-resistant strains of S. aureus were as sensitive as wild-type cells to the inhibitory effects of this drug. The inducible and constitutive ermC strains (MLSB organisms) were as susceptible to evernimicin as an antibiotic-susceptible organism was. The MRSA strains also showed comparable sensitivities to evernimicin. The MRSA strains have been tested previously for their sensitivities to erythromycin and other macrolides (6). Their clear susceptibility to evernimicin suggests a potentially important clinical role for this everninomicin in the treatment of infectious diseases caused by antibiotic-resistant organisms.
Significantly, this work is the first to describe a nonmacrolide antibiotic which functions by inhibition of 50S ribosomal subunit formation in cells (3, 9). Evernimicin specifically prevents the complete formation of the large ribosomal subunit, apparently inhibiting both translation and assembly by 50S particle binding. Like the macrolides and ketolides, this antibiotic does not prevent 30S subunit formation except at very high concentrations and does not stimulate the breakdown of mature particles.
The effect on 50S subunit formation is clearly seen in pulse-chase analysis of subunit formation. Control cells assembled subunits with kinetics very similar to those observed previously in E. coli cells, with 10 min required for 30S subunit formation and 20 min required for 50S subunit synthesis (2, 17, 22). Evernimicin had no effect on 30S subunit synthesis but substantially reduced the rate of 50S subunit formation in treated cells. About 16% of the expected amount of 50S particles were assembled after 1 h in the presence of the drug.
These observations help to substantiate the model that we have proposed that 50S subunit translational inhibitors can also interact with a precursor stage of the nascent 50S particle and prevent its further maturation (2). It will be interesting to compare the rRNA and protein compositions of the precursor targets for evernimicin and the macrolides in cells.
This work has identified two cellular targets for this novel antimicrobial agent. The inhibition of these two vital cellular activities is responsible for its killing effect on bacterial cells. Its effectiveness against two types of antibiotic-resistant S. aureus strains suggests that it will be a welcome addition to the diminishing list of compounds effective against antibiotic-resistant microorganisms.
ACKNOWLEDGMENTS
We are pleased to acknowledge Joyce Sutcliffe at Pfizer Central Research for strains and Todd Black at Schering-Plough Corp. for evernimicin.
This work was funded in part by a grant from Schering-Plough Corp.
REFERENCES
- 1.Aarestrup F M. Association between decreased susceptibility to a new antibiotic for treatment of human diseases, everninomycin (SCH 27899), and resistance to an antibiotic used for growth promotion in animals, avilamycin. Microb Drug Resist. 1998;4:137–141. doi: 10.1089/mdr.1998.4.137. [DOI] [PubMed] [Google Scholar]
- 2.Champney W S. Macrolide antibiotic inhibition of 50S ribosomal subunit formation in bacterial cells. Recent Res. Dev. Antimicrob Agents Chemother. 1999;3:39–58. [Google Scholar]
- 3.Champney W S, Burdine R. Macrolide antibiotics inhibit 50S ribosomal subunit assembly in Bacillus subtilis and Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:2141–2144. doi: 10.1128/aac.39.9.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champney W S, Burdine R. 50S ribosomal subunit synthesis and translation are equivalent targets for erythromycin inhibition in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1301–1303. doi: 10.1128/aac.40.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champney W S, Burdine R. Azithromycin and clarithromycin inhibition of 50S ribosomal subunit formation in Staphylococcus aureus cells. Curr Microbiol. 1998;36:119–123. doi: 10.1007/s002849900290. [DOI] [PubMed] [Google Scholar]
- 6.Champney W S, Burdine R. Macrolide antibiotic inhibition of translation and 50S ribosomal subunit assembly in methicillin-resistant Staphylococcus aureus cells. Microb Drug Resist. 1998;4:169–174. doi: 10.1089/mdr.1998.4.169. [DOI] [PubMed] [Google Scholar]
- 7.Champney W S, Tober C L. Inhibition of translation and 50S ribosomal subunit formation in Staphylococcus aureus cells by eleven different ketolide antibiotics. Curr Microbiol. 1998;37:418–425. doi: 10.1007/s002849900403. [DOI] [PubMed] [Google Scholar]
- 8.Champney W S, Tober C L, Burdine R. A comparison of the inhibition of translation and 50S ribosomal subunit formation in Staphylococcus aureus cells by nine different macrolide antibiotics. Curr Microbiol. 1998;37:412–417. doi: 10.1007/s002849900402. [DOI] [PubMed] [Google Scholar]
- 9.Chittum H S, Champney W S. Erythromycin inhibits the assembly of the large ribosomal subunit in growing Escherichia coli cells. Curr Microbiol. 1995;30:273–279. doi: 10.1007/BF00295501. [DOI] [PubMed] [Google Scholar]
- 10.Chu D T, Plattner J J, Katz L. New directions in antibacterial research. J Med Chem. 1996;39:3853–3874. doi: 10.1021/jm960294s. [DOI] [PubMed] [Google Scholar]
- 11.Edelstein P H, Edelstein M A C. In vitro activity of SCH 27899 (Ziracin) against Legionella species. Diagn Microbiol Infect Dis. 1999;33:59–62. doi: 10.1016/s0732-8893(98)00106-0. [DOI] [PubMed] [Google Scholar]
- 12.Ganguly A K, Girijavallabhan V M, Miller G H, Sarre O Z. Chemical modification of everninomicins. J Antibiot (Tokyo) 1982;35:561–570. doi: 10.7164/antibiotics.35.561. [DOI] [PubMed] [Google Scholar]
- 13.Ganguly A K, McCormick J L, Chan T-M, Saksena A K, Das P R. Determination of the absolute stereochemistry at the C16 orthoester of everninomicin antibiotics: a novel acid-catalyzed isomerization of orthoesters. Tetrahedron Lett. 1997;38:7989–7992. [Google Scholar]
- 14.Ganguly A K, Pramanik B, Chan T M, Sarre O, Iiu T-T, Morton J, Girijavallabhan V. The structure of new oligosaccharide antibiotics, 13-384 components 1 and 5. Heterocycles. 1989;28:83–88. [Google Scholar]
- 15.Jett B D, Hatter K L, Huycke M, Gilmore M S. Simplified agar plate method for quantifying viable bacteria. BioTechniques. 1997;23:648–650. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 16.Jones R N, Barrett M S. Antimicrobial activity of SCH 27899, oligosaccharide member of the everninomycin class with a wide gram-positive spectrum. J Clin Microbiol Infect. 1996;1:35–43. doi: 10.1111/j.1469-0691.1995.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 17.Michaels G A. Ribosome maturation of Escherichia coli growing at different growth rates. J Bacteriol. 1972;107:385–387. doi: 10.1128/jb.110.3.889-894.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 19.Nakashio S, Iwasawa H, Dun F Y, Kanemitsu K, Shimada J. Everninimicin, a new oligosaccharide antibiotic: its antimicrobial activity, post-antibiotic effect and synergistic bactericidal activity. Drugs Exp Clin Res. 1995;21:7–16. [PubMed] [Google Scholar]
- 20.Nierhaus K H, Bordash K, Homan H E. Ribosomal proteins. XLIII. In vivo assembly of Escherichia coli ribosomal proteins. J Mol Biol. 1973;74:587–597. doi: 10.1016/0022-2836(73)90049-1. [DOI] [PubMed] [Google Scholar]
- 21.Reimann H, Jaret R S, Sarre O Z. The chemistry of the everninomicin antibiotics. II. The structure of everninocin and its identification with curacin. J Antibiot (Tokyo) 1969;22:131–132. doi: 10.7164/antibiotics.22.131. [DOI] [PubMed] [Google Scholar]
- 22.Turco E, Altruda F, Ponzetto A, Mangiarotti G. Ribosome biosynthesis in Escherichia coli. Concerning the limiting step. Biochemistry. 1974;13:4752–4757. doi: 10.1021/bi00720a011. [DOI] [PubMed] [Google Scholar]
- 23.Urban C, Mariano N, Mosinka-Snipas K, Wadee C, Chahrour T, Rahal J J. Comparative in-vitro activity of SCH 27899, a novel everninomicin, and vancomycin. J Antimicrob Chemother. 1996;37:361–364. doi: 10.1093/jac/37.2.361. [DOI] [PubMed] [Google Scholar]
- 24.Wagman G H, Weinstein M J. Antibiotics from Micromonospora. Annu Rev Microbiol. 1980;34:537–557. doi: 10.1146/annurev.mi.34.100180.002541. [DOI] [PubMed] [Google Scholar]
- 25.Weinstein M J, Luedemann G M, Oden E M, Wagman G H. Everninomycin, a new antibiotic complex from Micromonospora carbonacea. 1965. pp. 24–52. . Antimicrob. Agents Chemother. 1964. [PubMed] [Google Scholar]
- 26.Weinstein M J, Wagman G H, Oden E M, Luedemann G M, Sloane P, Murawski A, Marquez J. Purification and biological studies of everninomicin B. Antimicrob Agents Chemother. 1965;5:821–827. [PubMed] [Google Scholar]
- 27.Wolf H. Avilamycin, an inhibitor of the 30S ribosomal subunits function. FEBS Lett. 1973;36:181–186. doi: 10.1016/0014-5793(73)80364-3. [DOI] [PubMed] [Google Scholar]