Abstract
The movement of intercellular cargo, such as transcripts, proteins and organelles, is fundamental to cellular function. Neurons, due to their long axons and dendrites, are particularly dependent on proper intracellular trafficking and vulnerable to defects in the movement of intracellular cargo that are noted in neurodegenerative and neurodevelopmental disorders. Accurate quantification of intracellular transport is therefore needed for studying the mechanisms of cargo trafficking, the influence of mutations and the effects of potentially therapeutic pharmaceuticals. In this article we introduce an algorithm called “Kymolyzer”. The algorithm can quantify intracellular trafficking along a defined path, such as that formed by the aligned microtubules of axons and dendrites. Kymolyzer works as a semi-autonomous kymography software. It constructs and analyzes kymographs to measure the movement and distribution of fluorescently tagged objects along a user defined path. The algorithm can be used under a wide variety of experimental conditions and can extract a diverse array of motility parameters describing intracellular movement, including time spent in motion, percentage of objects in motion, percentage stationary, and velocities. This article serves as a user manual describing the design of Kymolyzer, providing a stepwise protocol for its use, and illustrating its functions with common examples.
Basic Protocol 1:
Kymolyzer, a semi-autonomous kymography tool to analyze intracellular motility.
Keywords: Intracellular trafficking, neuron, kymography, semi-autonomous software
INTRODUCTION
Intracellular movement defines fundamental aspects of cell biology. The advancement of imaging technologies, along with fluorescent tagging of proteins, has highlighted the intracellular milieu as a highly dynamic environment, crowded with macromolecular complexes, organelles and solutes (Leonard et al., 2015; Mourao, Hakim, & Schnell, 2014; Nixon-Abell et al., 2016; Shim et al., 2012). While Brownian motion can mediate short distance diffusion of solutes and macromolecules (Brangwynne, Koenderink, MacKintosh, & Weitz, 2009; Dlugosz & Trylska, 2011; Mourao et al., 2014; Mullineaux, 2017), the rapid long distance movement of organelles and large protein complexes is driven by active transport, mediated by molecular motors like Kinesin (Hirokawa, 1998; Miki, Okada, & Hirokawa, 2005; Verhey, Kaul, & Soppina, 2011), Dynein (Pilling, Horiuchi, Lively, & Saxton, 2006; Reck-Peterson, Redwine, Vale, & Carter, 2018; Roberts, Goodman, & Reck-Peterson, 2014; Schnapp & Reese, 1989), and Myosin (Hartman & Spudich, 2012; Kurth et al., 2017; Lalli, Gschmeissner, & Schiavo, 2003; Lombardo et al., 2019; Titus, 2018; Wu, Bowers, Rao, Wei, & Hammer, 1998).
The rates of both passive diffusion and active transport are tightly regulated and display a large degree of heterogeneity. A multitude of factors, such as the type of molecule or organelle under consideration, cell metabolism, signaling pathways, disease states, and intracellular localization, affect the rates of active and passive movements within a cell. Defects in intracellular movements, especially in the rates of active transport, are often correlated with pathologies. Neurons, owing to their extremely polarized shape, are highly dependent on active intracellular trafficking (Goldstein & Yang, 2000; Kapitein & Hoogenraad, 2011; Maday, Twelvetrees, Moughamian, & Holzbaur, 2014; Misgeld & Schwarz, 2017; Schwarz, 2013). Defective intracellular trafficking thus frequently culminates in neurodegenerative and neurodevelopmental diseases (Baloh, Schmidt, Pestronk, & Milbrandt, 2007; Collard, Cote, & Julien, 1995; Gunawardena & Goldstein, 2001; Hafezparast et al., 2003; Pigino et al., 2003; Szebenyi et al., 2003; Williamson & Cleveland, 1999). Studying intracellular transport is thus of vital importance to understanding fundamental and translational cell biology and to elucidating disease mechanisms and establishing disease biomarkers.
We introduce two new algorithms that serve as complementary approaches to quantitatively report organelle motility depending on the path and organization of intracellular cargo transport. For cells where transport occurs along linearly arranged microtubules or actin filaments, such as in neuronal axons and dendrites, we have developed the algorithm “Kymolyzer” that uses kymography to semi-autonomously measure movement and distribution of organelles along a defined path. For samples where microtubules are not parallel and cargoes may undergo shorter and less organized movement, such as in non-neuronal cell lines, we have developed “QuoVadoPro”, an algorithm that works autonomously to quantify organellar motility based on the temporal variance.
In this article we report here the algorithm “Kymolyzer” (Pekkurnaz, Trinidad, Wang, Kong, & Schwarz, 2014). A common use of this algorithm is to quantify the movement of fluorescently tagged organelles or protein complexes along neuronal axons or other thin cellular processes. Kymography has been the traditional method of choice to depict and measure such intracellular movements that occur along a specific path (Guimaraes et al., 2015; Klinman & Holzbaur, 2016; Misgeld & Schwarz, 2017). A kymograph is a representation of motion along a path where time is displayed along the Y-axis and distance along the path on the X-axis. Kymograph mapping thus allows the visualization of objects moving along the defined path in a static image from which a variety of motility parameters can be extracted. Multiple tools to build and quantify kymographs in a manual or semi-autonomous manner already exist. However, most of them do not extract complex motility parameters like pause rates or reversal rates (Chaphalkar, Jain, Gangan, & Athale, 2016), require high signal to noise ratios (Chetta & Shah, 2011; Chiba, Shimada, Kinjo, Suzuki, & Uchida, 2014; Neumann, Chassefeyre, Campbell, & Encalada, 2017), have complex workflows, or are most suited to measure short unidirectional movements (Chaphalkar et al., 2016; Jakobs, Dimitracopoulos, & Franze, 2019).
Kymolyzer, the software described here, behaves as a semi-autonomous kymography tool that can extract multiple movement parameters from objects displaying long range bi-directional movements along linear tracks. The algorithm is highly versatile and can be easily adjusted to analyze intracellular trafficking in a variety of experimental paradigms, such as different experimental systems, organelles and microscopy setups (Gutnick, Banghart, West, & Schwarz, 2019; Pekkurnaz et al., 2014; Shlevkov et al., 2019). Kymolyzer works by means of a graphical interface that guides the user through a simple and intuitive workflow of building and analyzing kymographs. In the Kymolyzer workflow, the user is initially assisted to define the path of movement that is then converted to a kymograph depicting the movement of objects along the path. Kymolyzer then assists the user to indicate the movement tracks of individual objects on the kymograph and finally quantifies motility of the tracked objects based on user-defined thresholds. Kymolyzer is packaged as an ImageJ based macro-set that can read time-lapse movies acquired through most microscopy setups and output easily interpretable quantified motility data in form of CSV files.
BASIC PROTOCOL 1
Kymolyzer, a semi-autonomous kymography tool.
We developed Kymolyzer as a tool to semi-autonomously build and analyze kymographs from time-lapse images of fluorescently tagged proteins or organelles moving along a defined path, like neuronal axons. To initially build the kymograph, the user defines a central line along which object movement is monitored. Distance moved along the path by individual fluorescent objects is then plotted against time, thereby generating the kymograph. The user then indicates the tracks of individual objects on the kymograph. Each object track is then broken down along the time axis into individual segments and each segment is in turn analyzed for distance moved and direction. The final motility parameters for each object are then derived from the motility of individual segments. Kymolyzer then allows the user to group data from multiple kymographs either as individual objects or as averages of all objects in each kymograph.
Kymolyzer can be used to reliably track fluorescently tagged objects and provide a quantitative description of complex in vivo movement parameters including motility, pause frequency, reversal rates and speed. The thresholds that are used to define motility and direction in Kymolyzer are easily adjustable, making it applicable to tracking a wide variety of cellular cargoes at different temporal and spatial resolutions.
The Kymolyzer package contains a set of four macros that can be installed onto the open-source image analysis platform Fiji (a distribution of ImageJ with bundled plugins from the National Institutes of Health) and can be run sequentially through a user-friendly guided graphical interface.
Necessary Resources
Hardware:
Workstation with internet access
Software:
Please read Instructions_Readme file and download folder named as “Kymography_Macro_TLS_Lab_<date>” from https://github.com/ThomasSchwarzLab/KymolyzerCodes
Sample File:
https://drive.google.com/file/d/161uMXC3YP_pEzQjwhDzRhUE2ryFosHeY/view?usp=sharing
To use the sample file, please download the zipped file and extract all files within it to a new folder before use. The zipped file contains a time lapse image stack of a distal axon segment expressing a fluorescent tag that marks the mitochondria (mito-DsRed, addgene: 55838) and a cytosolic GFP (meGFP-N1, addgene: 54767) that helps visualize the axon. Both these channels are present as individual tif files that can be opened using ImageJ (or FIJI).
In addition, the zipped file also has a movie (in avi format) depicting both the channels, the axon segment and its orientation. The end of the axon segment closer to the cell body is marked as the negative (-ve) end while the end away from the cell body is marked as the positive (+ ve) end. This axon segment has been used as an example to illustrate the steps of analyzing movement by Kymolyzer (figures 1, 2 and 3) and to highlight the features and critical parameters (figure 5 and 6) of which the user should be aware while using Kymolyzer. For all the examples, the path of movement is traced from the -ve end to the +ve end. Any movement towards the +ve end is considered as anterograde movement and any movement towards the -ve end is considered as retrograde.
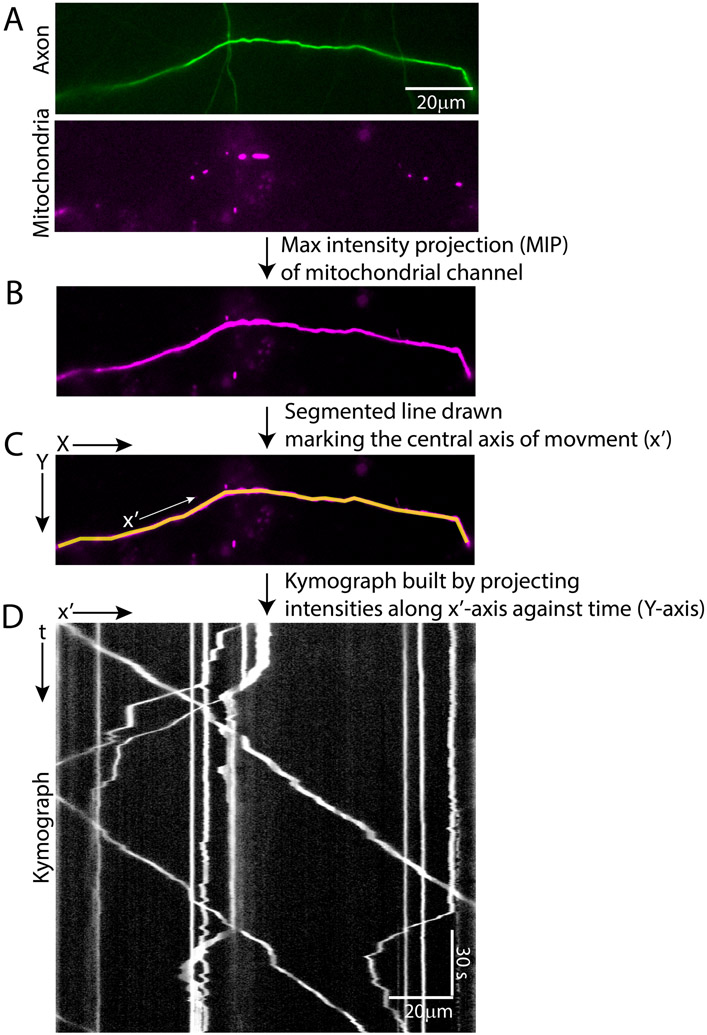
Figure 1. Example of fluorescently tagged mitochondrial movement in a distal axon segment, illustrating step 1 of Kymolyzer.
A distal axon segment expressing GFP (addgene: 54767) to mark the cytosol and having dsRED tagged mitochondria (addgene: 55838) is shown (A). A new image is made from the mitochondrial channel depicting maximum intensity projection (MIP) in time (B). As the density of mitochondria is relatively low in the distal axon, any single time frame of the mitochondrial channel cannot be used to visualize the entire axon segment. However, as the mitochondria are highly motile along the entire axon segment, the MIP of the mitochondrial channel allows the visualization of the whole segment. The MIP is then used to draw a segmented line along the axis of movement (x’) (C). Alternatively, the GFP channel can also be utilized to draw the axis of movement. The x’axis is drawn from left to right representing an orientation that starts nearer to the cell body and progresses away from it. The kymograph is built by plotting the intensities along the defined x’ axis against time (represented in the Y-axis) (D).
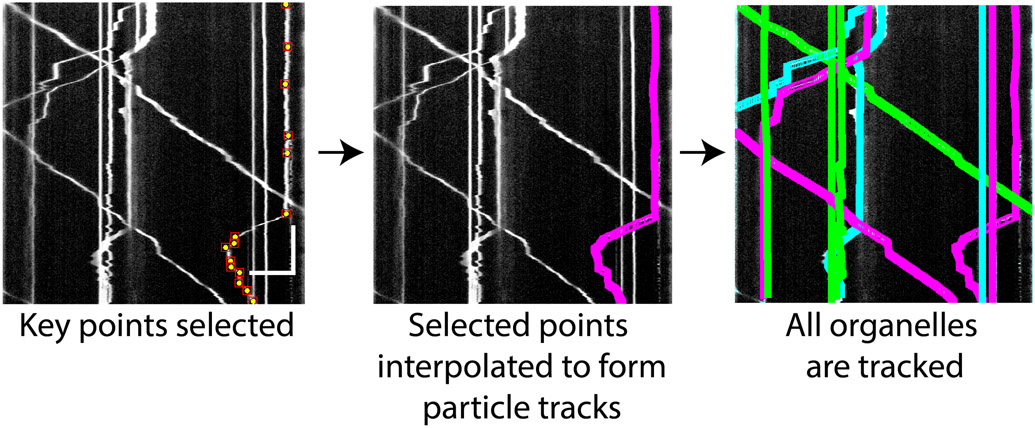
Figure 2. Example kymograph illustrating step 2 of Kymolyzer.
The user selects key points on the kymograph marking the movements of an object. These are shown as yellow dots on the kymograph (left panel). The key points indicate changes in the object’s speed and directionality. Kymolyzer then interpolates the selected points to effectively track the object in each frame (middle panel). After the user confirms the fidelity of tracking, the algorithm loops back to the previous step to mark another track. All marked tracks are displayed as colored paths on the kymograph. Through this re-iterative process, the user marks all the object tracks on the kymograph (right panel). Horizontal scale bar on kymograph depicts 10μm and vertical scale bar depicts 30s.
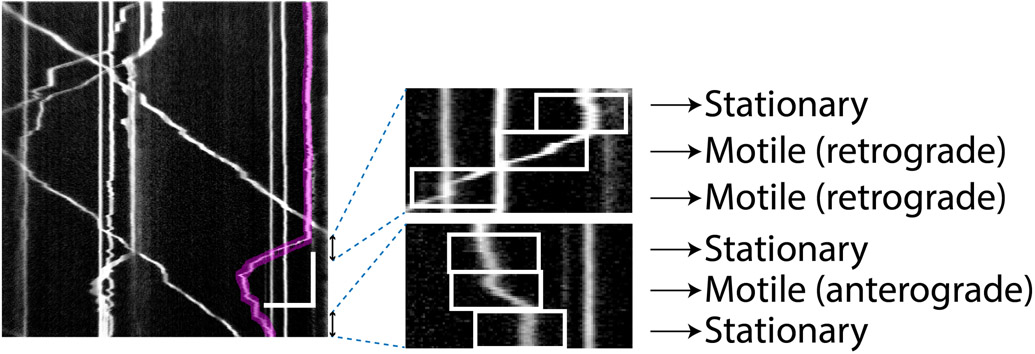
Figure 3. Example kymograph illustrating step 3 of Kymolyzer.
In step 3 of Kymolyzer, each object track is broken down into a defined number of temporal segments which are classified according to direction and speed. The motility parameters for each object are then derived from analyzing the velocities of the temporal segments that make up the object track. In this example, one such object track (marked in red) is shown on the kymograph (left panel). To quantify its motility, Kymolyzer breaks down the movement of the object into five-second segments, each of which is analyzed for speed and directionality and classified accordingly. Two sets of three consecutive segments of the track depicted in red are magnified and shown (right panels). The individual segments are boxed in white and their classifications are indicated. Horizontal scale bar on kymograph depicts 10μm and vertical scale bar depicts 30s.
Figure 5. Choosing optimal temporal resolution for reliable quantification by Kymolyzer.
Kymographs depicting movement of mitochondria in a neuronal axon (top panels, same example as that used in figure 1) with different temporal resolutions. When mitochondrial movement is imaged at a high temporal resolution (3.3Hz), the object tracks appear as un-broken lines in the kymograph and object tracks that cross can be easily differentiated (lower-left panel). To simulate the effects of a low temporal resolution, the same kymograph was plotted with only a subset of frames from the original movie to reduce the temporal resolution to 0.3Hz (lower-right panel). In this case, the motile objects form broken lines. At areas where multiple objects cross (arrow), the tracks cannot be assigned confidently to their proper objects. Thus, while a temporal resolution of 3.33 Hz is enough to track neuronal mitochondria reliably, a temporal resolution of 0.3 Hz is not optimal. Horizontal scale bars on both kymographs depict 10μm and vertical scale bars depict 30s.
Figure 6. Impact of temporal segment length on motility quantifications.
The track of a single mitochondrion (marked in red, A, left panel) from a kymograph of axonal mitochondria is analyzed with Kymolyzer using different temporal segmentations. A magnified portion of the object track is shown (A, right panel). An example of a processive movement is marked (arrowhead) along with a small non-processive movement, also referred to as a jitter or jiggle (arrow) (A). The movement is analyzed by breaking down the object track into temporal segments that are 5s, 15s, or 0.6s (B). Kymolyzer records the position of the object every 5s, 15s or 0.6s. With 5s temporal segmentation (B, left panel), movement due to small jitters is eliminated, while processive movement is retained and quantified. When the temporal segmentation is too large (B, middle panel), the net movement and speed are artificially reduced. Conversely, the 0.6s segmentation (B, right panel) results in a higher net movement and speed, as transient changes in behavior and small jitters in the object track are now included. Horizontal scale bar on kymograph depicts 10μm and vertical scale bar depicts 30s.
Movie S1 depicts the segment of the axon shown in the sample file along with a kymograph.
Protocol steps—Step annotations
1. Installation instructions:
Download and install FIJI from: https://imagej.net/Fiji/Downloads. After installing Fiji the user should update to the latest version by going to the following tab: help > update
Download the zipped folder named “Kymography_Macro_TLS_Lab_<date>” from https://github.com/ThomasSchwarzLab/KymolyzerCodes and extract it in a separate folder anywhere on the computer (preferably on the desktop, so that it can be accessed easily. This folder can be deleted after installation).
From FIJI, go to the following tabs: Plugins > Macros > run. In the file selection window that opens up, select the file named installer_Kymolyzer_HimanishBasu_SchwarzLab_ and run it.
Restart FIJI to complete macro installation. Following the restart, a new tab will appear as follows: Plugins > Macros > Kymolyzer.
2. Step 1: Generation of kymographs.
In step 1, Kymolyzer assists the user to trace the central axis of movement. To initiate step 1, Kymolyzer imports a time-lapse image sequence, supplied by the user. The user is then prompted to draw a segmented line over the path of movement. The segmented line is then used to build the kymograph by plotting the intensities along the selected line (x’-axis) against time (t, represented on the Y-axis) (figure 1).
As the central axis of movement may not be easily visualized in an individual time frame depicting the fluorescent objects (especially in cases where the object density is low, figure 1A, bottom panel), the user can choose to have Kymolyzer depict a maximum intensity projection (MIP) of each pixel over time, thereby highlighting the track of all fluorescent objects at all time points along the path of movement (Figure 1B). Alternatively, the user can also have Kymolyzer temporarily open another image (or channel) where the central axis of movement can be clearly visualized (for example, figure 1A, top panel). The user can then use the MIP or the separate image (or channel) to indicate the central axis of movement.
Instructions for running Step 1:
To run Step 1 of Kymolyzer go to the following tabs in FIJI: Plugins>Macros>Kymolyzer>Step1_MakeKymograph. A file selection window should appear.
-
In the file selection window, select the movie file to analyze. If it is a dual channel movie, the macro will ask the user to select any one channel. The user should select the channel which has the moving objects that are to be tracked.
Note: Kymolyzer can read most microscopy file formats. If the user encounters a file format that cannot be read by Kymolyzer, the user should convert or re-export the file as a multi-page tiff file beforehand.
Note: Instead of opening the image through the file selection window of step 1, the user can choose to open the time lapse through the native file reader inbuilt into FIJI (or imageJ) before running step 1. In this case when an image is already open before step 1 is run, no file selection window will appear, but rather Kymolyzer will process the already open image. This however is not a preferred method, as the native image reader on ImageJ or FIJI is often not successful in reading the metadata of the image (such as spatial and temporal resolutions) which are thus likely to be lost and would need to be re-entered in step 3.
-
The macro will then build the maximum intensity projection image (MIP) of the selected channel. Following this, the macro will ask the user to define the central axis along which the object movement will subsequently be analyzed, for example, to trace the neuronal axon along which object movement is to be recorded. To define the axis of movement, the user should trace the axis on the maximum intensity projection image.
Note: To trace the axis, the user can left click to add points on the maximum intensity projection image (MIP), that represent the axis of movement. The segmented line tool needed to do the selection will be automatically selected. The selected points are then interpolated to form a segmented line highlighting the axis of movement. To finish tracing the line, the user should double click on the last point to be added.
If the analyzed structure has a known polarity, the user should be consistent in the direction in which the lines are drawn. For example, for a neuron, the user should consistently trace starting at the segment end closest to the cell body. As discussed later in step 3, any movement towards the end of the selection will be considered anterograde, while any movement towards the beginning of the selection, as retrograde.
-
To build the kymograph, the program plots intensities along the defined path on the X-axis of the kymograph (called the x’-axis), with each timepoint being stacked along the Y-axis (called the t-axis).
Note: In case of a multichannel image, at this stage, the user will have the option to temporarily open a different channel (or image) to define the central axis. The defined axis will then be automatically copied on to the channel representing the fluorescently tagged objects (selected in step1a), which will then be used to build the kymograph. This step is often useful when the central axis of movement is not apparent in the maximum projection image. For example, while processing a sample similar to that shown in figure 1, the user can choose to temporarily open the GFP channel (figure 1A, top panel) to trace the axon (as the axis of movement) in an unbiased manner. This trace will then be applied to the mitochondrial channel (dsRed channel) to generate the kymograph (figure 1D)
At the final stage of step 1, following the generation of the kymograph, the macro will prompt the user to adjust the brightness and contrast of the kymograph as necessary. The user can then choose to adjust the image parameters such that all the object tracks can be clearly visualized against the background. The resulting kymograph is then saved in a folder made in the same location as the image.
3. Step 2: Track assignment.
In step 2 of Kymolyzer, the algorithm assists the user in tracking the movement of objects using the kymograph generated in step 1. Step 2 runs in a semi-autonomous manner. For each object, the user selects key points along the movement track of the object (object track), following which Kymolyzer interpolates the points in between the selections (figure 2), thus building the complete tracks.
Note: Given the semi-autonomous nature of this step, the track assignment can be subjective, especially at points where two objects cross-over or undergo fission or fusion or in highly crowded areas of the kymograph. The variability resulting from the subjectivity is minimized when the imaging is done with high temporal resolution that allows the user to confidently assign the tracks.
Instructions for running Step 2:
To run Step 2 of Kymolyzer select the following tabs in FIJI: Plugins>Macros>Kymolyzer>Step2_Track. A folder selection window will open. In this window select the folder made in step 1 (This folder will have been made at the end of step 1 at the same location from where the original image was loaded. It will be named with the same name as the original image).
- The selected folder contains the kymograph made in step 1 along with other metadata from the original image. The macro will automatically find and open the kymograph file and ask the user the following in the form of a pop-up window:
- Puncta Start Count: All objects tracked by Kymolyzer are numbered as punctum 1, punctum 2, and so on. The input made by the user at this window indicates the punctum number from which the algorithm should start counting.
- At the start of each image this number should be 1, thus denoting it as the first object to be tracked.
- If, for some reason, step 2 on a given image has to be restarted when some objects have already been tracked in that image, then the user should change the punctum start count to the number of objects that have already been tracked +1 (so that it represents the present object number to be tracked).
-
Show box size: This is the size of the box used to mark the tracked object on the kymograph and in a pop-up movie of the time lapse, after the tracking is completed. These features allow the user to verify the correctness of the tracking. The show box size should ideally be just large enough to surround the object tracked. The “Show box size” denotes the size of the box created (in pixels) to mark the tracked object. The default value is set to 5 pixels, but the user should change it according to the maximum size of the objects to be tracked.Note: The show box size is exclusively for visualization purposes and has no bearing on the quantifications by itself.After the user inputs the two values mentioned above, the user should press the OK button on the pop-up window to start tracking the first object.
-
Object tracking is done on the kymograph window named “select”. After indicating the puncta start count and the show box size, this window (depicting the kymograph) will automatically be maximized and brought to the front of the screen. To track a given object on the kymograph window, the user should select the key points on the kymograph representing the movements of an object. The key points are points on the object track indicating a change in speed or direction (figure 2, left panel).
To indicate the key points on the movement track of an object, the user can perform one the following actions at any pixel on the kymograph window:- Shift+LeftClick to add points on the track of an object.
- Cntrl+leftClick to finish the track of an object.
-
Alt+leftClick to drop a vertical line (depicting a stationary object) till the end of the movieAfter selecting the key points of an object’s track on the kymograph (i.e. when the last point is marked through the Cntrl+leftClick action or a vertical line is dropped by the Alt+leftClick action), Kymolyzer will automatically interpolate the points in between the selections, thereby effectively marking the whole track (figure 2, center panel). To confirm and display the fidelity of tracking, a movie will popup, where the tracked object in each frame is marked by a box. The user should use the slider on the movie window to go through the movie and ensure that the object is tracked (surrounded by a box) in each frame. Additionally, a dialog box will appear, asking the user to save the track or discard it. The user can click OK to save the track or choose to discard the track if it failed to follow the object faithfully and needs to be re-tracked.With either choice, a new dialog box will pop-up asking the user whether another object needs to be tracked. On this dialog box, named as “For another track”, the user can click “yes” to assign another track. If the user clicks “yes”, the algorithm will loop back to step 2c (mentioned above) and allow the user to mark the key points of another object track. If the user had chosen to save the previously marked track, it will now be indicated on the kymograph window as a colored track (figure 2, right panel).Note: The colors chosen to mark the tracks of previously tracked objects are randomly chosen as red, blue or green. Marking previously tracked objects prevents the user from tracking the same object twice.After tracking the final object, the user should click “No” on the “For another track” dialog box to end this step.
4. Step 3: Measurement.
To quantify the movement of objects whose tracks have been assigned in the previous step, in step 3 each track is broken down into user defined segments of time. The segments are then scored as motile or non-motile based on a defined speed threshold. The distance travelled in each motile segment is calculated and they are further sub-classified according to net anterograde or retrograde movement (figure 3). The directionality of movement is decided by the direction in which the central axis of movement (x’) is defined. Any movement towards the end of the x’ axis, i.e. in the positive direction is defined as anterograde movement, while any movement towards the beginning of the x’ axis, i.e. in the negative direction is defined as retrograde movement. The motility of each object is then quantified by analyzing the speed and directionality of movement in each temporal segment of that object’s track. The motility parameters for each object, for example speed, directionality, percent movement, pause frequency, reversal frequency etc. are expressed as a fraction of the temporal segments that span the object’s movement and show the relevant behavior.
Instructions for running Step 3:
To run Step 3 of Kymolyzer go to the following tabs in FIJI: Plugins>Macros>Kymolyzer>Step3_Measure. A folder selection window will open as in step 2. In this window select the folder made in step 1 (which has the same name as that of the original image).
- At this step, the selected folder should contain the kymograph, the saved object tracks and image metadata. After selecting the folder, Kymolyzer will automatically find and load the object tracks for further analysis. A dialog box will appear asking for the temporal and spatial resolution of the image. The macro tries to automatically fill these parameters from the meta-data of the image (already saved in the folder) if it is legible. These parameters should always be cross-checked and changed if necessary. The following image parameters are used for measurement:
- Pixel Scale (in μm/pixel): This is the length of each pixel in microns after magnification. The macro assumes a square pixel.
- Timescale (in seconds): This is the time interval between sequential frames of the video.
- Kymograph Width (in pixels): This is the length of the segmented line selection drawn by the user in step 1 to indicate the central axis of movement.
- Kymograph Height (in pixels): This represents the total number of time points in the video, as every time point is projected onto one pixel in the t axis of the kymograph.
- After setting the resolution parameters, the user will have to select the following calculation options in another pop-up dialog box:
- Number of frames per time step: The track of each object is divided into temporal segments. The “Number of frames per time step” value defines the length of each temporal segment in terms of frames (or pixels in the t-axis of the kymograph). For example, a value of 5 will divide the track of each object in a movie into segments consisting of 5 frames. If an object is tracked for 100 frames, its track will be divided into 20 segments. For velocity and direction quantification, the object position at the beginning and end of each temporal segment is considered (figure 3, right panel).
-
Lower limit of speed: This value represents the lowest speed of an object within a particular temporal segment below which it is considered as stationary for that time segment.By default, the temporal segmentation parameter is set to 10 frames (i.e. the position of each object at every 10th time frame interval is used for quantification) and the velocity parameter is set to 0.5μm/s (i.e. the object is considered stationary during a temporal segment if its speed during that segment is less than 0.5μm/s). The user should empirically change these thresholds to accurately reflect the biology (see commentary). An efficient way to set the minimum velocities and time step thresholds is to track known moving and stationary objects, and set the length of the temporal segments and the lower limit of speed to the lowest possible values such that a known stationary object is indeed recorded to be stationary while movement is detected for the motile objects.Note: As discussed later, these parameters significantly affect the quantifications, and thus should be set to accurately reflect the biology. For example, setting the temporal segmentation feature too low, (i.e. if every frame is considered) would result in an over-estimation of motility as frame to frame jitters will come into effect. On the other hand, if the temporal segmentation parameter is set too high (i.e. too many frames are skipped in between two segments), it would result in an under-estimate of motility.The raw data for all objects in a kymograph can be found in the following excel file: “…pathTofolderWithImageName/RawPunctaFiles/Summary.xls”. To check the effect of different thresholds, the user can open this file and check the tab “Move %” to validate whether the stationary objects are indeed recorded to be stationary. If not, the user can repeat step 3 on the same folder with higher thresholds.Once the thresholds are decided upon by the user, these values should not be changed between different samples of the same experiment.
By analyzing the speed and directionality of each temporal segment of an object’s movement track, Kymolyzer extracts multiple motility parameters for each object. The motility parameters provide a thorough quantitative description of motility of the objects that have been tracked. They can be found in the excel file: “…pathTofolderWithImageName/RawPunctaFiles/Summary.xls”. The explanations and the guidelines to understanding and interpreting each motility parameter are listed later (see guidelines for understanding results).
5. Step 4: Data collation.
As a final step in Kymolyzer, the data from multiple kymographs are collated through an automated interface into one output file. The data from different kymographs can be collated as individual objects (object-wise collation) or as an average of all objects in each kymograph (kymograph-wise collation).
Instructions for running Step 4:
To collate the data as a kymograph-wise average, the user should go to the following tabs in FIJI: Plugins>Macros>Kymolyzer>Step4_ CollateDataMeansOnly. As described in the previous steps, a folder selection window will open. In this window the user should select the folder made in step 1 of Kymolyzer (i.e. the folder with the same name as that of the image). The macro will automatically find the quantification file in that folder and calculate the average of the motility parameters of all the tracked objects in that kymograph. A dialog box will then appear asking the user whether to add more image folders. If the user selects “yes”, the algorithm will automatically open a folder selection window where the user can choose to add another image folder. This process is repeated so that the user can re-iteratively collate all the individual quantification files from the desired image folders into one file. The macro will then ask the user for a name and location where the output file is to be saved. This creates a file in the desired location which has the average per kymograph of the various motility parameters.
If the single object measurements are needed instead of the mean of all the objects per kymograph, the object-wise raw data for each kymograph can also be collated into one excel document in a similar manner by running Plugins>Macros>Kymolyzer>Step4_ CollateData.
GUIDELINES FOR UNDERSTANDING RESULTS
Explanation of motility parameters quantified by Kymolyzer
To quantify different features of movement, Kymolyzer breaks down each object track into several segments in time. The position of the object at the beginning and end of each temporal segment is used to classify the object as motile or stationary during that temporal segment (figure 3). A motile temporal segment (referred to as a motile segment) is further sub classified as anterograde or retrograde depending on the direction of motility of the object (figure 3, right panel). The final motility parameters (got at the end of step 3) for each object are then calculated as the fraction of temporal segments that display a particular movement behavior. Kymolyzer calculates the following movement parameters for each object:
Total Segments: Total number of temporal segments that span a track
Total Move Length (μm): Total distance moved by the object. The “Total Move Length” is the summation of the absolute distances moved at every motile segment by an object regardless of direction. To calculate the “Total Move Length” both anterograde and retrograde movements are considered as positive movements and are summed.
Average Speed (μm/s): This value represents the average of the speeds of movement recorded at every temporal segment where an object was classified as motile. During the calculation of average speed, the direction of movement is not taken into consideration (i.e. it serves as an average of both anterograde and retrograde speeds).
Move (%): Percent of temporal segments of an object track that were classified as motile among all the segments spanning that object track.
Still (%): Percent of segments within an object track that were classified as stationary among all the segments spanning that object track.
Forward Length (μm): Summation of distances moved by an object in all its temporal segments that were classified as motile and anterograde (i.e. moving in the positive direction along the x’ axis).
Average Forward Speed (μm/s): Mean of the speeds at which an object moved in every temporal segment classified as motile and anterograde.
Forward (%): Percent of the temporal segments that were classified as motile and anterograde among all the segments spanning the track of an object.
Backward Length (μm): Summation of distances moved by an object in the segments classified as motile and retrograde (i.e. moving in the negative direction along the x’ axis).
Average Backward Speed (μm/s): Mean of the speeds at which an object moved in every motile segment classified as retrograde.
Backward (%): Percent of segments in an object track that were classified as motile and retrograde.
Stop (%): This value represents the pause frequency. It is calculated as the percent of temporal segments spanning the track of an object that were classified as stationary but have a motile segment preceding it (thus marking a pause event). It is important to note that a pause event that is shorter than a single temporal segment is likely to be missed. For example, if an object pauses and restarts its movement within the same temporal segment, it will not be classified to have paused during that temporal segment as the segment containing the pause itself will be classified as motile. To avoid such missed pause events, the user should keep the temporal segmentation parameter as small as possible (see commentary section for more details).
-
Reverse (%): This value represents the rates at which an object reverses direction during the time lapse. It is calculated as the percent of temporal segments that were classified as motile and had another motile segment in an opposite direction preceding them.
Note: If two temporal segments belonging to the same object track have opposite directionality of movement while having one or more still segments in between them, they are also considered to be depicting reversal events.
Net Movement (μm): This is the net displacement of an object during the time lapse. It is calculated as the sum of distances moved in all temporal segments that were classified as motile where the retrograde motility is considered as negative distance.
Recommendation for appropriate statistical tests to analyze data obtained from Kymolyzer
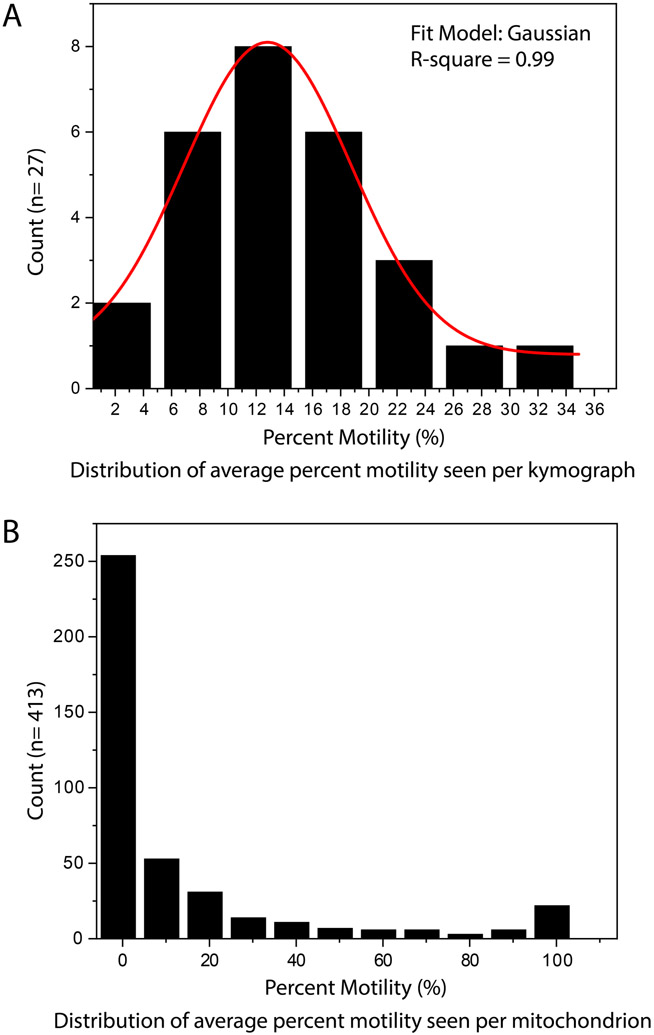
Kymolyzer allows the user to collate data object-wise or kymograph-wise. The user should choose the method that best reflects the biology. However, depending on the method of collation, the distribution of data from the same sample can be significantly different. For example, data from the same set of neurons that have been imaged for axonal mitochondrial motility shows a normal distribution when collated per axon but an atypical distribution when the data is analyzed per mitochondrion (figure 4). In such cases, the statistical tests to compare between samples and the graphical depiction of the samples should be chosen accordingly.
Figure 4. Examples of distributions of data got after tracking objects in Kymolyzer.
Histograms of motility data obtained by analyzing and collating data from 27 axons with a total of 413 discrete mitochondria are shown. During data collation, when the average motility per kymograph is considered, the data generally follows a Gaussian trend (A). Data samples showing normal or close to normal distribution can be analyzed with traditional t-tests and summarized by their mean or median. However, when plotted per mitochondrion (object-wise collation), the data does not follow a normal distribution (B). Data that are not normally distributed should be analyzed with non-parametric statistical tests.
COMMENTARY
Kymographs are an elegant tool to visualize and analyze movement along a defined path, where the position of objects along the path (x’-axis) is plotted against time (Y-axis). Kymography has classically been used in molecular biology to study transport along flagella (Iomini, Babaev-Khaimov, Sassaroli, & Piperno, 2001; Piperno et al., 1998), polymerization dynamics of tubulin and actin filaments in vitro (Kuhn & Pollard, 2005; Smal, Grigoriev, Akhmanova, Niessen, & Meijering, 2010) and movement of molecular motors (Cho, Reck-Peterson, & Vale, 2008; Zhou, Brust-Mascher, & Scholey, 2001). In addition to previous uses, kymography has now become the staple method for studying intracellular cargo trafficking in a diverse array of biological samples where movement occurs along a defined axis, such as in neuronal axons and dendrites (Maday, Wallace, & Holzbaur, 2012; Pekkurnaz et al., 2014; Rangaraju, Lauterbach, & Schuman, 2019; Wang & Schwarz, 2009) and in other extended cellular processes like filopodia (Alieva et al., 2019; Kerber et al., 2009) and fungal hyphae (Bielska et al., 2014; Penalva, Zhang, Xiang, & Pantazopoulou, 2017; Salogiannis, Egan, & Reck-Peterson, 2016).
Multiple solutions to build and analyze kymographs exist. Of note, programs like the Multi Kymograph plugin in ImageJ or Kymomaker (Chiba et al., 2014) take a manual approach, wherein the user builds the kymograph and highlights straight segments in the kymographs that represent movement trajectories of objects. The algorithms then calculate the speed and directionality of the straight segments indicated by the user. While such manual approaches to analyzing kymographs are accurate, they are extremely time consuming and do not readily provide all motility parameters needed to describe long range bidirectional movement (such as pause frequency or reversal rates). On the other hand, several algorithms have tried to fully automate the analysis of kymographs through edge detection and line detection methods (Chaphalkar et al., 2016; Mangeol, Prevo, & Peterman, 2016). The automated approach to analyzing kymographs is time efficient but needs images with high signal to noise ratios and is best for measuring short unidirectional runs. When automated kymograph analysis approaches are applied to kymographs with objects displaying long range bidirectional movements, individual object tracks are often detected as broken segments, faint tracks are not detected, and tracks are wrongly assigned when two tracks crossover one another in the kymograph. More recently, computer learning driven approaches have been used to mitigate the shortfalls of automated kymography analysis (Jakobs et al., 2019), but with limited success as they require large training datasets and are computationally intensive.
In contrast to existing algorithms, Kymolyzer represents a semi-autonomous kymography software to build and analyze kymographs. Kymolyzer uses time lapse images to build kymographs, following which it relies on user assistance to track both moving and stationary objects visible on the kymographs, in a partially automated manner. The semi-autonomous approach allows Kymolyzer to track objects accurately even in images with low signal to noise ratios and high object densities with minimal user dependent variations. Kymolyzer is well suited for the analysis of complex transport events such as those encountered for organelles with bidirectional movement and that can alternate between runs and pauses. The algorithm can extract movement parameters not easily extracted by other software, such as pause rates, reversal rates, distances and speeds of movement in either direction of every object tracked, features that are particularly important to fully describe complex bidirectional motility. In addition to quantifying the time-averaged motility and flux of individual fluorescent organelles or protein complexes, Kymolyzer can be used to deconvolve the movement of an object to extract information about the active motor properties, such as instantaneous velocities.
Kymolyzer is highly versatile and can be used to process stacks of time-lapse images recorded for different lengths of time and at different temporal and spatial resolutions. As demonstrations of its versatility and robustness, the algorithm has already been used to study movement of mitochondria, synaptic vesicles, endosomes and peroxisomes in rodent hippocampal neurons and in human iPSC derived neurons (Gutnick et al., 2019; Pekkurnaz et al., 2014; Shlevkov et al., 2019) under a variety of experimental and imaging conditions.
Critical Parameters
During the analysis of multiple samples within the same experiment, the following parameters can impact the motility quantifications and should be kept constant between samples:
Duration for which the time-lapse is recorded: During motility measurement, Kymolyzer breaks down the movement of each object into small temporal segments (the duration of which is defined by the user). Most of the motility parameters are then quantified as a percent of temporal segments displaying a particular feature, such as reversals or pauses. Thus, most motility parameters calculated by Kymolyzer are intrinsically normalized for the duration of imaging. This allows Kymolyzer to compare samples that have slight differences in the duration for which they have been imaged. However, when samples with significantly different numbers of frames are compared, the quantifications by Kymolyzer can become unreliable: the number of stationary objects stays constant during the imaging, but longer movie durations allow more motile objects to traverse into the field of imaging. The fraction of motile objects will therefore likely increase as the imaging time increases. It is therefore necessary to keep the analyzed durations similar.
Number of objects imaged: During analysis and quantification by Kymolyzer, each assigned object track is quantified and saved separately. The user then has the option to collate the data from different kymographs as individual objects or as an average of all objects from individual kymographs. If the number of objects per kymograph differs by a large extent, it is advisable that the user collates the data object-wise rather than kymograph-wise. The object-wise approach is less affected by the differences in the number of objects per kymograph.
Temporal resolution of imaging: The absolute frame rate or temporal resolution needed to reliably track an object in Kymolyzer depends on the density of objects and the speed of movement. For high fidelity tracking, the frame rates should be high enough such that frame to frame displacement of the object is lower than the size of the object itself and smaller than the distance from its nearest neighbor. The user should decide upon a frame rate empirically, as movement speeds and object densities vary greatly with different experimental systems and organelles. While a frame rate that is set too high may cause phototoxicity and create data files that exceed capacity, a frame rate that is too low reduces reliability in track assignment, especially in crowded areas of a kymograph and when objects tend to cross over each other (figure 5). To compare between samples of the same experiment the rates of image acquisition should be kept constant.
-
Number of frames per temporal segment: Every object track in Kymolyzer is broken down into temporal segments and most motility parameters are quantified as a percent of temporal segments containing a desired feature of motility. This allows Kymolyzer to normalize for the duration of imaging. The temporal segmentation feature is also useful to remove small jitters (or very short non-processive movements) of an object that may not be desirable for the user to incorporate into the movement analysis (figure 6). The length of each time segment should be decided empirically with controls. This depends not only on the experimental system and the organelle being tracked but also the user’s preference in tracking a particular type of motility: long distance processive movements or short and fast movements. To decide on the temporal segment length, the user should use controls that have objects which are considered to be stationary and controls where objects can be considered to be motile. The user should then track these objects, run Step 3 of Kymolyzer multiple times on the tracked objects with different temporal segmentation lengths and monitor the motility recorded in the summary file (in the % move column of the excel file) found at “…pathTofolderWithImageName/RawPunctaFiles/Summary.xls”. The user should choose the smallest temporal segment where the control with stationary objects shows zero motility.
A temporal segmentation that is too small results in an over-estimate of motility, where even small jitters (that may be below the threshold of what is considered to be motile) are scored as movement. A segment length that is too large results in underestimation of some motility parameters, as different types of biologically significant movements can be averaged together when they occur in the same segment (figure 6). For example, including a pause together with a moving segment will decrease the measured velocity for that segment and the pause will not be scored as a pause (compare panels of figure 6B).
In general, the shorter the time segment selected, the closer the velocity parameters will be to the instantaneous velocity of the moving object. The user can select the segment duration that best addresses the user’s question and may, for example, wish to use larger segments if the goal is to understand net progress down an axon (figure 6B, left panel) or shorter segments if the goal is to describe properties of the active motor such as instantaneous velocity (figure 6B, right panel). Because temporal segmentation length influences several parameters, once decided in Step 3, it should be kept constant for all samples of the same experiment.
Lower velocity threshold: To classify an object as motile or stationary at each temporal segment, Kymolyzer utilizes a lower velocity threshold that is declared by the user in step 3. This parameter establishes the smallest distance that has to be moved by an object within a temporal segment for that segment to be considered as motile. The lower velocity threshold helps avoid counting movements due to slow drifts of the sample. The user should select the lowest possible value that causes a known stationary object to be scored with zero motility. If the lower velocity threshold is too low, it results in an over-estimate of motility by including drift. Conversely, if the lower velocity threshold is too high, the user will miss out on biologically significant movements.
During the use of Kymolyzer, the most prevalent factor that leads to user-dependent variation is the manual selection of the key points during track assignments. This variation can be minimized with imaging at higher temporal and spatial resolutions. Additionally, if the imaging parameters and thresholds (as discussed above) are kept constant and optimal, the variations are further minimized. Kymolyzer, also stands to benefit greatly with the incorporation of a line tracking algorithm (Egan, McClintock, Hollyer, Elliott, & Reck-Peterson, 2015), one that eliminates or reduces the need for manual object tracking after building the kymograph.
Supplementary Material
Movie S1. Example of mitochondrial movement in a distal axon segment.
A video of a distal axon segment expressing cytosolic GFP (top panel) to visualize axons and mitoDsRed, a mitochondrially targeted fluorescent marker (middle panel) is shown. The movement along the axon is depicted in the kymograph (bottom panel). To construct the kymograph, the path of movement has been traced from left (representing the end closer to the cell body) to right. Any movement along the x’ axis of the kymograph (in the positive direction) represents movement away from the cell body.
ACKNOWLEDGEMENTS:
We thank the members of the T. L. Schwarz laboratory for fruitful discussions, especially Dr. Jill Falk for critical reading of the manuscript and for her inputs. We also thank Dr. Shipkovenska for her help with editing of the manuscript. We thank S. Vasquez for help with hippocampal dissections from embryonic rats; we are grateful to the Harvard NeuroDiscovery Center’s Enhanced Neuroimaging Core (NINDS P30 Core Center grant no. NS072030) and the Cellular Imaging Core IDDRC at Boston Children’s Hospital (NIH U54 HD090255) for support with imaging. This research was generously supported by the National Institutes of Health grant R01 GM069808 (NIH/NIGMS) to T.L.S.
LITERATURE CITED:
- Alieva NO, Efremov AK, Hu S, Oh D, Chen Z, Natarajan M, … Bershadsky AD (2019). Myosin IIA and formin dependent mechanosensitivity of filopodia adhesion. Nat Commun, 10(1), 3593. doi: 10.1038/s41467-019-10964-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloh RH, Schmidt RE, Pestronk A, & Milbrandt J (2007). Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci, 27(2), 422–430. doi: 10.1523/JNEUROSCI.4798-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska E, Higuchi Y, Schuster M, Steinberg N, Kilaru S, Talbot NJ, & Steinberg G (2014). Long-distance endosome trafficking drives fungal effector production during plant infection. Nat Commun, 5, 5097. doi: 10.1038/ncomms6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Koenderink GH, MacKintosh FC, & Weitz DA (2009). Intracellular transport by active diffusion. Trends Cell Biol, 19(9), 423–427. doi: 10.1016/j.tcb.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Chaphalkar AR, Jain K, Gangan MS, & Athale CA (2016). Automated Multi-Peak Tracking Kymography (AMTraK): A Tool to Quantify Sub-Cellular Dynamics with Sub-Pixel Accuracy. PLoS One, 11(12), e0167620. doi: 10.1371/journal.pone.0167620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetta J, & Shah SB (2011). A novel algorithm to generate kymographs from dynamic axons for the quantitative analysis of axonal transport. J Neurosci Methods, 199(2), 230–240. doi: 10.1016/j.jneumeth.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Chiba K, Shimada Y, Kinjo M, Suzuki T, & Uchida S (2014). Simple and direct assembly of kymographs from movies using KYMOMAKER. Traffic, 15(1), 1–11. doi: 10.1111/tra.12127 [DOI] [PubMed] [Google Scholar]
- Cho C, Reck-Peterson SL, & Vale RD (2008). Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem, 283(38), 25839–25845. doi: 10.1074/jbc.M802951200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard JF, Cote F, & Julien JP (1995). Defective axonal transport in a transgenic mouse model of amyotrophic lateral sclerosis. Nature, 375(6526), 61–64. doi: 10.1038/375061a0 [DOI] [PubMed] [Google Scholar]
- Dlugosz M, & Trylska J (2011). Diffusion in crowded biological environments: applications of Brownian dynamics. BMC Biophys, 4, 3. doi: 10.1186/2046-1682-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, McClintock MA, Hollyer IH, Elliott HL, & Reck-Peterson SL (2015). Cytoplasmic dynein is required for the spatial organization of protein aggregates in filamentous fungi. Cell Rep, 11(2), 201–209. doi: 10.1016/j.celrep.2015.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS, & Yang Z (2000). Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci, 23, 39–71. doi: 10.1146/annurev.neuro.23.1.39 [DOI] [PubMed] [Google Scholar]
- Guimaraes SC, Schuster M, Bielska E, Dagdas G, Kilaru S, Meadows BR, … Steinberg G (2015). Peroxisomes, lipid droplets, and endoplasmic reticulum "hitchhike" on motile early endosomes. J Cell Biol, 211(5), 945–954. doi: 10.1083/jcb.201505086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S, & Goldstein LS (2001). Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron, 32(3), 389–401. doi: 10.1016/s0896-6273(01)00496-2 [DOI] [PubMed] [Google Scholar]
- Gutnick A, Banghart MR, West ER, & Schwarz TL (2019). The light-sensitive dimerizer zapalog reveals distinct modes of immobilization for axonal mitochondria. Nat Cell Biol, 21(6), 768–777. doi: 10.1038/s41556-019-0317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafezparast M, Klocke R, Ruhrberg C, Marquardt A, Ahmad-Annuar A, Bowen S, … Fisher EM (2003). Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science, 300(5620), 808–812. doi: 10.1126/science.1083129 [DOI] [PubMed] [Google Scholar]
- Hartman MA, & Spudich JA (2012). The myosin superfamily at a glance. J Cell Sci, 125(Pt 7), 1627–1632. doi: 10.1242/jcs.094300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N (1998). Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science, 279(5350), 519–526. doi: 10.1126/science.279.5350.519 [DOI] [PubMed] [Google Scholar]
- Iomini C, Babaev-Khaimov V, Sassaroli M, & Piperno G (2001). Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J Cell Biol, 153(1), 13–24. doi: 10.1083/jcb.153.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs MA, Dimitracopoulos A, & Franze K (2019). KymoButler, a deep learning software for automated kymograph analysis. Elife, 8. doi: 10.7554/eLife.42288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, & Hoogenraad CC (2011). Which way to go? Cytoskeletal organization and polarized transport in neurons. Mol Cell Neurosci, 46(1), 9–20. doi: 10.1016/j.mcn.2010.08.015 [DOI] [PubMed] [Google Scholar]
- Kerber ML, Jacobs DT, Campagnola L, Dunn BD, Yin T, Sousa AD, … Cheney RE (2009). A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr Biol, 19(11), 967–973. doi: 10.1016/j.cub.2009.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman E, & Holzbaur EL (2016). Comparative analysis of axonal transport markers in primary mammalian neurons. Methods Cell Biol, 131, 409–424. doi: 10.1016/bs.mcb.2015.06.011 [DOI] [PubMed] [Google Scholar]
- Kuhn JR, & Pollard TD (2005). Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J, 88(2), 1387–1402. doi: 10.1529/biophysj.104.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth EG, Peremyslov VV, Turner HL, Makarova KS, Iranzo J, Mekhedov SL, … Dolja VV (2017). Myosin-driven transport network in plants. Proc Natl Acad Sci U S A, 114(8), E1385–E1394. doi: 10.1073/pnas.1620577114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli G, Gschmeissner S, & Schiavo G (2003). Myosin Va and microtubule-based motors are required for fast axonal retrograde transport of tetanus toxin in motor neurons. J Cell Sci, 116(Pt 22), 4639–4650. doi: 10.1242/jcs.00727 [DOI] [PubMed] [Google Scholar]
- Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, … Rohrer B (2015). Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochim Biophys Acta, 1853(2), 348–360. doi: 10.1016/j.bbamcr.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo AT, Nelson SR, Kennedy GG, Trybus KM, Walcott S, & Warshaw DM (2019). Myosin Va transport of liposomes in three-dimensional actin networks is modulated by actin filament density, position, and polarity. Proc Natl Acad Sci U S A, 116(17), 8326–8335. doi: 10.1073/pnas.1901176116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, & Holzbaur EL (2014). Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron, 84(2), 292–309. doi: 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE, & Holzbaur EL (2012). Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol, 196(4), 407–417. doi: 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeol P, Prevo B, & Peterman EJ (2016). KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol Biol Cell, 27(12), 1948–1957. doi: 10.1091/mbc.E15-06-0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Okada Y, & Hirokawa N (2005). Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol, 15(9), 467–476. doi: 10.1016/j.tcb.2005.07.006 [DOI] [PubMed] [Google Scholar]
- Misgeld T, & Schwarz TL (2017). Mitostasis in Neurons: Maintaining Mitochondria in an Extended Cellular Architecture. Neuron, 96(3), 651–666. doi: 10.1016/j.neuron.2017.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourao MA, Hakim JB, & Schnell S (2014). Connecting the dots: the effects of macromolecular crowding on cell physiology. Biophys J, 107(12), 2761–2766. doi: 10.1016/j.bpj.2014.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux CW (2017). How bacteria keep proteins moving. Elife, 6. doi: 10.7554/eLife.33590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Chassefeyre R, Campbell GE, & Encalada SE (2017). KymoAnalyzer: a software tool for the quantitative analysis of intracellular transport in neurons. Traffic, 18(1), 71–88. doi: 10.1111/tra.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS, … Lippincott-Schwartz J (2016). Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science, 354(6311). doi: 10.1126/science.aaf3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkurnaz G, Trinidad JC, Wang X, Kong D, & Schwarz TL (2014). Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell, 158(1), 54–68. doi: 10.1016/j.cell.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva MA, Zhang J, Xiang X, & Pantazopoulou A (2017). Transport of fungal RAB11 secretory vesicles involves myosin-5, dynein/dynactin/p25, and kinesin-1 and is independent of kinesin-3. Mol Biol Cell, 28(7), 947–961. doi: 10.1091/mbc.E16-08-0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Morfini G, Pelsman A, Mattson MP, Brady ST, & Busciglio J (2003). Alzheimer's presenilin 1 mutations impair kinesin-based axonal transport. J Neurosci, 23(11), 4499–4508. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/12805290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, & Saxton WM (2006). Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell, 17(4), 2057–2068. doi: 10.1091/mbc.e05-06-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Siuda E, Henderson S, Segil M, Vaananen H, & Sassaroli M (1998). Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol, 143(6), 1591–1601. doi: 10.1083/jcb.143.6.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V, Lauterbach M, & Schuman EM (2019). Spatially Stable Mitochondrial Compartments Fuel Local Translation during Plasticity. Cell, 176(1-2), 73–84 e15. doi: 10.1016/j.cell.2018.12.013 [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Redwine WB, Vale RD, & Carter AP (2018). The cytoplasmic dynein transport machinery and its many cargoes. Nat Rev Mol Cell Biol, 19(6), 382–398. doi: 10.1038/s41580-018-0004-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Goodman BS, & Reck-Peterson SL (2014). Reconstitution of dynein transport to the microtubule plus end by kinesin. Elife, 3, e02641. doi: 10.7554/eLife.02641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salogiannis J, Egan MJ, & Reck-Peterson SL (2016). Peroxisomes move by hitchhiking on early endosomes using the novel linker protein PxdA. J Cell Biol, 212(3), 289–296. doi: 10.1083/jcb.201512020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp BJ, & Reese TS (1989). Dynein is the motor for retrograde axonal transport of organelles. Proc Natl Acad Sci U S A, 86(5), 1548–1552. doi: 10.1073/pnas.86.5.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz TL (2013). Mitochondrial trafficking in neurons. Cold Spring Harb Perspect Biol, 5(6). doi: 10.1101/cshperspect.a011304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim SH, Xia C, Zhong G, Babcock HP, Vaughan JC, Huang B, … Zhuang X (2012). Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc Natl Acad Sci U S A, 109(35), 13978–13983. doi: 10.1073/pnas.1201882109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov E, Basu H, Bray MA, Sun Z, Wei W, Apaydin K, … Schwarz TL (2019). A High-Content Screen Identifies TPP1 and Aurora B as Regulators of Axonal Mitochondrial Transport. Cell Rep, 28(12), 3224–3237 e3225. doi: 10.1016/j.celrep.2019.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smal I, Grigoriev I, Akhmanova A, Niessen WJ, & Meijering E (2010). Microtubule dynamics analysis using kymographs and variable-rate particle filters. IEEE Trans Image Process, 19(7), 1861–1876. doi: 10.1109/TIP.2010.2045031 [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Morfini GA, Babcock A, Gould M, Selkoe K, Stenoien DL, … Brady ST (2003). Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron, 40(1), 41–52. doi: 10.1016/s0896-6273(03)00569-5 [DOI] [PubMed] [Google Scholar]
- Titus MA (2018). Myosin-Driven Intracellular Transport. Cold Spring Harb Perspect Biol, 10(3). doi: 10.1101/cshperspect.a021972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Kaul N, & Soppina V (2011). Kinesin assembly and movement in cells. Annu Rev Biophys, 40, 267–288. doi: 10.1146/annurev-biophys-042910-155310 [DOI] [PubMed] [Google Scholar]
- Wang X, & Schwarz TL (2009). The mechanism of Ca2+ -dependent regulation of kinesin-mediated mitochondrial motility. Cell, 136(1), 163–174. doi: 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson TL, & Cleveland DW (1999). Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat Neurosci, 2(1), 50–56. doi: 10.1038/4553 [DOI] [PubMed] [Google Scholar]
- Wu X, Bowers B, Rao K, Wei Q, & Hammer JA 3rd. (1998). Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function In vivo. J Cell Biol, 143(7), 1899–1918. doi: 10.1083/jcb.143.7.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HM, Brust-Mascher I, & Scholey JM (2001). Direct visualization of the movement of the monomeric axonal transport motor UNC-104 along neuronal processes in living Caenorhabditis elegans. J Neurosci, 21(11), 3749–3755. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11356862 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Example of mitochondrial movement in a distal axon segment.
A video of a distal axon segment expressing cytosolic GFP (top panel) to visualize axons and mitoDsRed, a mitochondrially targeted fluorescent marker (middle panel) is shown. The movement along the axon is depicted in the kymograph (bottom panel). To construct the kymograph, the path of movement has been traced from left (representing the end closer to the cell body) to right. Any movement along the x’ axis of the kymograph (in the positive direction) represents movement away from the cell body.