Abstract
Twenty (8.5%) of 234 nonrepetitive clinical isolates of Klebsiella pneumoniae from southern Taiwan were found to produce extended-spectrum β-lactamases (ESBLs): 10 strains produced SHV-12, 4 produced SHV-5, 2 produced a non-TEM non-SHV ESBL with a pI of 8.3, 3 produced a novel AmpC β-lactamase designated CMY-8 with a pI of 8.25, and 1 produced SHV-12 and an unidentified AmpC enzyme with a pI of 8.2. The CMY-8 enzyme confers a resistance phenotype similar to CMY-1 and MOX-1, and sequence comparisons showed high homologies (>95%) of nucleotide and amino acid sequences among these three enzymes. Plasmid and pulse-field gel electrophoresis analyses revealed that all isolates harboring an SHV-derived ESBL were genetically unrelated, indicating that dissemination of resistance plasmids is responsible for the spread of SHV ESBLs among K. pneumoniae in this area. All three isolates carrying CMY-8 had identical genotypic patterns, suggesting the presence of an epidemic strain.
Resistance to the extended-spectrum cephalosporins among members of the family Enterobacteriaceae has become a growing worldwide problem (1, 10, 13, 16, 21, 24). Such resistance is often associated with transferable plasmid-encoded extended-spectrum β-lactamases (ESBLs) (13, 24). Most ESBLs from Escherichia coli and Klebsiella pneumoniae are derived from TEM- or SHV-type β-lactamases by one or more amino acid substitutions which confer resistance to extended-spectrum cephalosporins (13, 24). Resistance to cephamycins and broad-spectrum cephalosporins has also arisen in K. pneumoniae and E. coli via acquisition of plasmids containing the chromosomally encoded AmpC β-lactamase found in Enterobacter spp., Pseudomonas aeruginosa, and Citrobacter spp. (3, 4, 9, 11, 30). Unlike class A ESBLs, the activities of these enzymes are not inhibited by clavulanic acid or tazobactam (3, 4, 21). In the Far-Eastern countries, two closely related plasmid-mediated AmpC enzymes have been found in K. pneumoniae: MOX-1 from Japan (9) and CMY-1 from Korea (4, 14). The prevalence rates of ESBL among clinical isolates of K. pneumoniae in Taiwan have been reported to range from 10.7 to 30% (12, 15). SHV-5 has been shown to be the most-common ESBL (15). The present study was conducted to determine the prevalence and types of ESBLs among K. pneumoniae in the southern part of the country. We also describe a novel AmpC enzyme in this report.
MATERIALS AND METHODS
Bacterial isolates.
Between April and September 1998, a total of 234 nonrepetitive clinical isolates of K. pneumoniae were consecutively collected in the Department of Pathology, National Cheng Kung University Hospital, a 900-bed university hospital in southern Taiwan. All isolates were identified by using the conventional techniques (8) and/or the API 20E system (bioMérieux, Marcy l'Etoile, France).
Detection of ESBL producers and susceptibility testing.
EBSL production was detected by means of double-disk synergy tests (13) and phenotypic confirmatory tests as recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (20). ESBL producers detected by either of these two tests were investigated further. The MICs of antibiotics were determined by using E Test strips including ampicillin, amoxicillin-clavulanic acid, cefotaxime, ceftazidime, and imipenem (AB Biodisk, Solna, Sweden), and the results were interpreted by using the NCCLS criteria (20).
IEF analysis and enzyme inhibition assay.
Crude preparations of β-lactamases (5) from clinical isolates and their transconjugants were subjected to isoelectric focusing (IEF) analysis by the method of Matthew et al. (18) with an LKB Multiphor apparatus on prepared Ampholine PAG plate gels (pH 3.5 to 9.5; Pharmacia Biotech Asia Pacific, Hong Kong, China). Gels were developed with 0.5 mM nitrocefin (Oxoid, Basingstoke, United Kingdom). Inhibition assays were carried out by overlaying gels with 0.5 mM nitrocefin and 0.3 mM cloxacillin (Sigma, St. Louis, Mo.) or 0.3 mM clavulanic acid (SmithKline Beecham Pharma, Munich, Germany) (7).
Conjugation experiments and plasmid analysis.
Conjugation experiments were performed as described previously (26) with streptomycin-resistant E. coli C600 as the recipient (2). Transconjugants were selected on tryptic soy agar plates supplemented by streptomycin (500 μg/ml; Sigma) and ceftazidime (10 μg/ml; Glaxo, Greenford, United Kingdom), cefotaxime (10 μg/ml; Hoechst-Roussel Pharmaceuticals Inc., Somerville, N.J.) or cefoxitin (64 μg/ml; Sigma). ESBL production in transconjugants was confirmed by the NCCLS confirmatory test. Plasmids from clinical isolates and transconjugants were extracted by a rapid alkaline lysis procedure (29). For the restriction enzyme analysis of transconjugant plasmids, EcoRI and PstI (Roche Molecular Biochemicals, Mannheim, Germany) were used. Digested and nondigested DNA samples were analyzed by electrophoresis on 0.8% agarose gels. The plasmid sizes of transconjugants were estimated by adding up restriction fragments.
PCR amplification and DNA sequencing.
To amplify TEM- and SHV-related genes from clinical isolates and their transconjugants, the following oligonucleotide primers were used for PCR: for blaTEM genes, forward primer (5′-CCCCTATTTGTTTATTTTTC-3′) and reverse primer (5′-GACAGTTACCAATGCTTAATCA-3′) (17), corresponding to nucleotides 112 to 130 and 1074 to 1053, respectively, of Sutcliffe (28); for blaSHV genes, forward primer (5′-GCCGGGTTATTCTTATTTGTCGC-3′) and reverse primer (5′-TCTTTCCGATGCCGCCGCCAGTCA-3′) (22), corresponding to nucleotides 55 to 77 and 1067 to 1044, respectively, of Mercier and Levesque (19). The primers used for amplification of blaCMY-1-related genes are shown in Fig. 1. PCR amplification was performed as described elsewhere (14, 17, 22) and the amplicons were totally included in these three β-lactamase structural genes. The amplicons were purified with PCR Clean Up kits (Roche), and both strands were sequenced with the same primers for PCR and sequencing primers on an ABI PRISM 377 Sequencer Analyzer (Applied Biosystems, Foster City, Calif.). The sequencing primers for blaTEM and blaSHV genes were synthesized as described elsewhere (17, 27), and for blaCMY-1-related genes are also shown in Fig. 1.
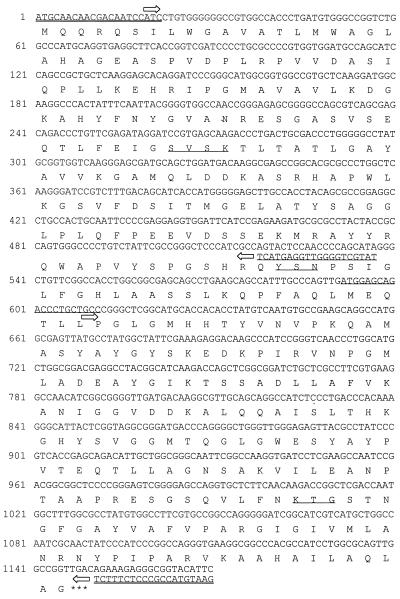
FIG. 1.
Complete nucleotide sequence and predicted amino acid sequence of the blaCMY-8 β-lactamase gene. The primers for amplification of the gene were synthesized according to the nucleotide sequence of CMY-1 (4, 14) and are double underlined. DNA sequencing was performed with a sequencing primer (underlined) and the PCR primers as well. Arrows indicate the directions of DNA sequencing. The stop codon is indicated with three asterisks. The β-lactamase active site S-V-S-K, the conserved triad K-T-G, and the class C typical motif Y-X-N are underlined.
PFGE analysis.
Genomic DNAs prepared by the procedure of Piggot et al. (25) were digested overnight with 10 U of XbaI (New England Biolabs, Beverly, Mass.) and subjected to pulsed-field gel electrophoresis (PFGE) with the Pulsaphor plus system (Pharmacia) as described previously (6). DNA fragments were separated in a 1% agarose gel in 0.5× Tris-borate-EDTA (TBE) buffer at 150 V for 30 h, with pulse times ranging from 5 to 35 s.
Nucleotide sequence accession number.
The sequence of a novel AmpC β-lactamase has been submitted to GenBank database under the accession no. AF167990.
RESULTS
Prevalence of ESBL-producing isolates in K. pneumoniae.
Among 234 nonrepetitive clinical isolates of K. pneumoniae, production of ESBL was inferred in 20 (8.5%) isolates by the NCCLS confirmatory test for ESBL. The double-disk synergy test showed clavulanic acid synergy in 15 (75%) of the 20 ESBL-producing isolates.
Resistance phenotypes.
On the basis of susceptibilities to cefotaxime, ceftazidime and cefoxitin, the ESBL producers were classified into four resistance phenotypes (Table 1). The resistance phenotype groups I and II involved high-level resistance to ceftazidime and cefotaxime, respectively. Group III involved high-level resistance to cefoxitin and cefotaxime, while group IV showed high-level resistance to cefoxitin and ceftazidime.
TABLE 1.
MICs for K. pneumoniae strains with different resistance phenotypes and their transconjugantsa
| Resistance phenotype group | No. of clinical isolates | MIC (μg/ml) of drug for clinical isolates
|
No. of transconjugants | MIC (μg/ml) of drug for transconjugants
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMC | CAZ | CTX | FOX | IPM | AMP | AMC | CAZ | CTX | FOX | IPM | |||
| I | 14 | >256 | 4–8 | ≥256 | 3–16 | 3–16 | 0.125–0.25 | 11 | >256 | 4–8 | ≥256 | 6–16 | 2–6 | 0.125–0.25 |
| II | 2 | >256 | 4–8 | 2–24 | 128–256 | 8–16 | 0.125 | 2 | >256 | 4–8 | 2–3 | 128–256 | 8–16 | 0.125 |
| III | 3 | >256 | 16 | 96 | >256 | >256 | 0.25–0.5 | 3 | >256 | 24 | 32–64 | >256 | >256 | 0.25–0.5 |
| IV | 1 | >256 | 12 | >256 | 48 | >256 | 0.5 | 1 | >256 | 12 | >256 | 8 | >256 | 0.125 |
MICs were determined by the E test. Abbreviations: AMP, ampicillin; AMC, amoxicillin-clavulanic acid combination, CAZ, ceftazidime; CTX, cefotaxime; FOX, cefoxitin; IPM, imipenem.
IEF analysis and enzyme inhibition test.
The results of IEF are summarized in Table 2. On IEF gels, the β-lactamases with a pI of 8.25 were inhibited by cloxacillin but not by clavulanic acid, suggesting that they were AmpC enzymes. The pI 8.2 β-lactamase of the isolate in group IV, O787, was not inhibited by 0.3 mM cloxacillin and 0.3 mM clavulanic acid. All other enzymes with pIs of 5.4, 7.6, 8.2, or 8.3 were inhibited by clavulanic acid but not cloxacillin.
TABLE 2.
Results of IEF, genotyping of β-lactamases, plasmid analysis, and PFGE
| Resistance phenotype group and strain | Clinical isolate
|
Transconjugantc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| pI(s) | SHV | TEM | AmpC | Plasmid profile | PFGE pattern | pI | SHV | AmpC | Approx plasmid size (kb) | |
| Group I | ||||||||||
| X386 | 8.2, 7.6 | 5 | A | I | 8.2 | 5 | 100 | |||
| C097 | 8.2, 7.6, 5.4 | 5 | 1 | B | II | 8.2 | 5 | 100 | ||
| Q381 | 8.2, 7.6, 5.4 | 5 | 1 | C | III | 8.2 | 5 | 70 | ||
| T923a | 8.2, 7.6, 5.4 | 5 | 1 | D | IV | |||||
| T386 | 8.2, 7.6, 5.4 | 12 | 1 | E | V | 8.2 | 12 | 70 | ||
| R436 | 8.2, 7.6, 5.4 | 12 | 1 | E | VI | 8.2 | 12 | 100 | ||
| S276 | 8.2, 7.6, 5.4 | 12 | 1 | F | VII | 8.2 | 12 | 100 | ||
| T986 | 8.2, 7.6, 5.4 | 12 | 1 | G | VIII | 8.2 | 12 | 70 | ||
| B262a | 8.2, 7.6, 5.4 | 12 | 1 | H | IX | |||||
| T255a | 8.2, 7.6, 5.4 | 12 | 1 | I | X | |||||
| V293 | 8.2, 7.6, 5.4 | 12 | 1 | G | XI | 8.2 | 12 | 70 | ||
| D155 | 8.2, 7.6 | 12 | J | XII | 8.2 | 12 | 40 | |||
| B657 | 8.2, 7.6 | 12 | K | XIII | 8.2 | 12 | 75 | |||
| C106 | 8.2, 7.6 | 12 | J | XIV | 8.2 | 12 | 40 | |||
| Group II | ||||||||||
| O574 | 8.3, 7.6, 5.4 | 11 | 1 | L | XV | 8.3 | 95 | |||
| R549 | 8.3, 7.6, 5.4 | 11 | 1 | M | XVI | 8.3 | 125 | |||
| Group III | ||||||||||
| W142 | 8.25, 7.6, 5.4 | 11 | 1 | CMY-8 | N | XVII | 8.25 | CMY-8 | 25 | |
| W580 | 8.25, 7.6, 5.4 | 11 | 1 | CMY-8 | N | XVII | 8.25 | CMY-8 | 25 | |
| T384 | 8.25, 7.6, 5.4 | 11 | 1 | CMY-8 | N | XVII | 8.25 | CMY-8 | 25 | |
| Group IV | ||||||||||
| O787 | 8.2, 7.6, 5.4 | 12 | 1 | UDb | O | XVIII | 8.2 | 12 | UD | 150 |
Conjugation experiments with strains T923, B262, and T255 were not successful.
UD, undetermined.
All transconjugates lacked TEM.
Transfer of resistance.
Resistance to extended-spectrum β-lactams was successfully transferred from 17 of 20 ESBL producers to E. coli. The transconjugants revealed similar resistance phenotypes to their donors. The susceptibility tests of the transconjugants are summarized in Table 1. It is noteworthy that the transconjugant of isolate O787 in group IV was resistant to both cefoxitin and ceftazidime, but only the pI 8.2 β-lactamase, which was not inhibited by clavulanic acid and cloxacillin, was detected on an IEF gel. The sizes of the resistance plasmids transferred to E. coli are shown in Table 2.
Sequence analysis.
The results of PCR amplification and nucleotide sequencing are summarized in Table 2. The blaSHV genes were amplified from all clinical isolates of K. pneumoniae and 12 of 17 transconjugants. Of 15 isolates producing a pI 8.2 enzyme, 4 carried an SHV-5 enzyme and 11 harbored an SHV-12 enzyme. Except for isolate O787 with group IV resistance phenotype, all isolates carrying SHV-5 or SHV-12 were distributed in group I. The SHV genotypes of the E. coli transconjugants were consistent with those of their donors. All clinical isolates in groups II and III carried SHV-11, which was consistent with the β-lactamase with a pI of 7.6, but their transconjugants gave a negative PCR result. The blaTEM genes were amplified from all 16 ESBL-producing isolates carrying a pI 5.4 β-lactamase and from none of their E. coli transconjugants. DNA sequencing of the amplicons showed that all of them were TEM-1.
A DNA fragment of approximately 1 kb was amplified with the primer pair specific for blaCMY-1-related genes from all three isolates in group III and their E. coli transconjugants, all of which expressed a pI 8.25 β-lactamase. DNA sequencing of the amplicons revealed the same nucleotide sequence among these 3 isolates. Sequence comparison with other known β-lactamases deposited in GenBank showed a close relationship to two AmpC β-lactamases, CMY-1 (accession no. X92508) and MOX-1 (accession no. D13304). The enzyme, now designated CMY-8, contains the β-lactamase active site S-V-S-K, the conserved triad K-T-G, and the class C typical motif Y-X-N on the basis of its deduced amino acid sequence (Fig. 1). Comparisons among CMY-8, CMY-1 (4) and MOX-1 (9) by using the GAP program of the Genetics Computer Group software package showed that CMY-8 shares 98.6 and 98.1% nucleotide identities, and 98.6 and 95.2% amino acid identities with CMY-1 and MOX-1, respectively. Multiple sequence alignments of the amino acid sequences of these three enzymes were performed with the PILEUP program of the Genetics Computer Group package and the result is shown in Fig. 2.
FIG. 2.
Alignment of deduced amino acid sequences of CMY-8 with those of CMY-1 and MOX-1. Identical amino acids are marked with dots. The stop codon is indicated with an asterisk. The β-lactamase active site S-V-S-K, the conserved triad K-T-G, and the class C typical motif Y-X-N are shown in boldface type.
Plasmid analysis and PFGE.
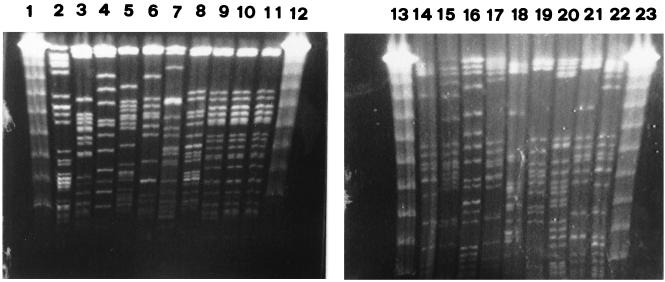
The results of plasmid and PFGE analyses are summarized in Table 2 and shown in Fig. 3. Overall, 20 clinical isolates of ESBL producers gave 15 and 18 different types of plasmid and PFGE profiles, respectively. All 14 isolates in group I showed different PFGE patterns, while all 3 isolates in group III had the same plasmid and PFGE profiles.
FIG. 3.
PFGE of XbaI-digested genomic DNAs from 19 ESBL-producing K. pneumoniae isolates. Lanes 1, 12, 13, and 23, bacteriophage lambda DNA concatemers (GibcoBRL, Gaithersburg, Md.) which served as molecular size marker; lanes 2 to 11 and 14 to 22, isolates V293, D155, B657, C106, O574, R549, O787, W142, W580, T384, X386, C097, Q381, T923, T386, R436, T986, B262, and T255, respectively. Isolate S276 is not shown here. All ESBL producers except for three isolates carrying CMY-8 (W142, W580, and T384) had different PFGE patterns.
DISCUSSION
The prevalence rate of ESBL producers among clinical isolates of K. pneumoniae in this study was 8.5%, which is similar to those reported in many other parts of the world (13, 16, 21, 24). Among 20 ESBL producers, 15 (75%) carried SHV-derived ESBLs, and 11 (55%) of them harbored SHV-12. SHV-12, identified in 1997 by Nüesch-Inderbinen et al. (23), has rarely been found in countries other than Korea (14). This report adds Taiwan to the list of countries where SHV-12 is prevalent. Another report from a district hospital in central Taiwan showed that the prevalence rate of ESBL-producing K. pneumoniae was 30% and that SHV-5 was the most common type of ESBL (18). The differences in the prevalence and types of ESBLs between different hospitals within a country may reflect variations in patient population, hospital care practices, and infection control activities.
The phenotype of group III involved high-level resistance to cefotaxime and cefoxitin (Table 1). The conjugation experiment and IEF analysis revealed a plasmid-mediated β-lactamase with a pI of 8.25 responsible for the resistance. The enzyme, designated CMY-8, could be inhibited by cloxacillin but not by clavulanic acid. Sequence analysis revealed the class C-specific amino acid motif Y-X-N and active-site serine at position 64 in the deduced amino acid sequence of the enzyme (Fig. 1). These data indicate that the enzyme is an AmpC β-lactamase. The alignments of the nucleotide and amino acid sequences showed that CMY-8 has a high degree of similarity with CMY-1 and MOX-1 (Fig. 1), two plasmid-mediated AmpC enzymes found in Korea (4) and Japan (9), respectively, suggesting that they might have a common origin. These three enzymes also have a similar resistance phenotype: high-level resistance to cefotaxime as well as cofoxitin.
Both isolates in group II produced β-lactamases with pIs of 5.4, 7.6, and 8.3 (Table 2). PCR and sequence analysis revealed nucleotide sequences identical to those of TEM-1 and SHV-11, which were consistent with the pI 5.4 and 7.6 enzymes, respectively. The resistance phenotype was transferable and their transconjugants showed no amplification of the blaTEM and blaSHV genes. Since SHV-11 and TEM-1 are not ESBLs, the pI 8.3 lactamase must be responsible for resistance to extended-spectrum β-lactams in both isolates and should be a non-TEM and non-SHV enzyme.
The isolate in group IV had three bands reflecting pI values of 5.4, 7.6, and 8.2 on an IEF gel. The transconjugant acquiring the pI 8.2 enzyme expressed a resistance phenotype similar to that of the isolate (Table 1). The activity of the pI 8.2 β-lactamase could not be inhibited by cloxacillin or clavulanic acid. SHV-12 was detected in the isolate and its transconjugant by PCR and DNA sequencing. Since SHV-12 does not confer resistance to cefoxitin (15, 23), our findings suggest that an AmpC β-lactamase with a pI equal to or very close to 8.2 is coexistent with SHV-12 on the same plasmid in this isolate. More studies such as molecular cloning and DNA sequencing are needed to clarify the paradoxical findings.
The blaTEM genes were amplified from 80% of ESBL-producing K. pneumoniae isolates. IEF and sequence analyses showed that all TEM enzymes in these isolates were TEM-1. Our data indicate that TEM-derived β-lactamases do not play an important role in resistance to extended-spectrum β-lactams among K. pneumoniae in this area despite the prevalence of TEM-1.
All 15 isolates harboring an SHV-derived ESBL revealed different PFGE profiles (Table 2), indicating that dissemination of resistance plasmids was responsible for the spreading of SHV-derived enzymes at this hospital. All three isolates carrying CMY-8 had identical PFGE and plasmid profiles (Table 2 and Fig. 3), suggesting the presence of an epidemic strain harboring CMY-8 at this hospital. Surveillance of dissemination of this epidemic strain is important because of its high-level resistance to cephamycins as well as extended-spectrum β-lactams.
ACKNOWLEDGMENTS
We kindly thank J. Kim, Dankook University, Korea, for provision of a K. pneumoniae strain carrying CMY-1.
This work was partially supported by grants NCKUH89-054 from National Cheng Kung University Hospital and NSC89-2314-B-006-031 from the National Science Council, Republic of China.
REFERENCES
- 1.Arlet G, Rouveau M, Casin I, Bouvet P J M, Lagrange P H, Philippon A. Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV-4 β-lactamase and which were isolated in 14 French hospitals. J Clin Microbiol. 1994;32:2553–2558. doi: 10.1128/jcm.32.10.2553-2558.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B J, Low K B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980;44:1451–1456. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother. 1996;40:221–224. doi: 10.1128/aac.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Stemplinger I, Jungwirth R, Wilhelm R, Chong Y. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob Agents Chemother. 1996;40:1926–1930. doi: 10.1128/aac.40.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Grimm H, Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990;18:294–298. doi: 10.1007/BF01647010. [DOI] [PubMed] [Google Scholar]
- 6.Chu G, Vollrath D, Davis R W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 7.Danel F, Hall L M C, Gur D, Livermore D M. OXA-15, an extended-spectrum variant of OXA-2 β-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob Agents Chemother. 1997;41:785–790. doi: 10.1128/aac.41.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farmer J J., III . Enterobacteriaceae: introduction and identification. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C.: American Society for Microbiology; 1995. pp. 438–449. [Google Scholar]
- 9.Horii T, Arakawa Y, Ohta M, Sugiyama T, Wacharotayankun R, Ito H, Kato N. Characterization of a plasmid-borne and constitutively expressed blaMOX-1 gene encoding AmpC-type β-lactamase. Gene. 1994;139:93–98. doi: 10.1016/0378-1119(94)90529-0. [DOI] [PubMed] [Google Scholar]
- 10.Jacoby G A, Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34:908–911. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacoby G A, Tran J. Sequence of the MIR-1 β-lactamase gene. Antimicrob Agents Chemother. 1999;43:1759–1760. doi: 10.1128/aac.43.7.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jan I S, Hsueh P R, Teng L J, Ho S W, Luh K T. Antimicrobial susceptibility testing for Klebsiella pneumoniae isolates resistant to extended-spectrum β-lactam antibiotics. J Formos Med Assoc. 1998;97:661–666. [PubMed] [Google Scholar]
- 13.Jarlier V, Nicolas M-H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Kwon Y, Pai H, Kim J W, Cho D T. Survey of Klebsiella pneumoniae strains producing extended-spectrum β-lactamase: prevalence of SHV-12 and SHV-2a in Korea. J Clin Microbiol. 1998;36:1446–1449. doi: 10.1128/jcm.36.5.1446-1449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu P Y F, Tung J C, Ke S C, Chen S L. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates in a district hospital in Taiwan. J Clin Microbiol. 1998;36:2759–2762. doi: 10.1128/jcm.36.9.2759-2762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore D M, Yuan M. Antibiotic resistance and production of extended-spectrum β-lactamases amongst Klebsiella spp. from intensive care units in Europe. J Antimicrob Chemother. 1996;38:409–424. doi: 10.1093/jac/38.3.409. [DOI] [PubMed] [Google Scholar]
- 17.Mabilat C, Goussard S. PCR detection and identification of genes for extended-spectrum β-lactamases. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 553–559. [Google Scholar]
- 18.Matthew M, Harris M, Marshall M J, Rose G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 19.Mercier J, Levesque R C. Cloning of SHV-2, OHIO-1, and OXA-6 β-lactamase. Antimicrob Agents Chemother. 1990;34:1577–1583. doi: 10.1128/aac.34.8.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; ninth informational supplement. M100-S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 21.Nordmann P. Trends in β-lactam resistance among Enterobacteriaceae. Clin Infect Dis. 1998;27(Suppl. 1):S100–S106. doi: 10.1086/514905. [DOI] [PubMed] [Google Scholar]
- 22.Nüesch-Inderbinen M T, Hächler H, Kayser F H. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur J Clin Microbiol Infect Dis. 1996;15:398–402. doi: 10.1007/BF01690097. [DOI] [PubMed] [Google Scholar]
- 23.Nüesch-Inderbinen M T, Kayser F H, Hächler H. Survey and molecular genetics of SHV β-lactamase in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob Agents Chemother. 1997;41:943–949. doi: 10.1128/aac.41.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philippon A, Arlet G, Lagrange P H. Origin and impact of plasmid-mediated extended-spectrum beta-lactamases. Eur J Clin Microbiol Infect Dis. 1994;13(Suppl. 1):S17–S29. doi: 10.1007/BF02390681. [DOI] [PubMed] [Google Scholar]
- 25.Piggot P J, Amjad M, Wu J J, Sandoval H, Castro J. Genetic and physical maps of Bacillus subtilis 168. In: Harwood C R, Cutting S M, editors. Molecular biology methods for bacillus. West Sussex, England: John Wiley & Sons Ltd.; 1990. pp. 493–543. [Google Scholar]
- 26.Provence D L, Curtiss R., III . Gene transfer in gram-negative bacteria. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 319–347. [Google Scholar]
- 27.Rasheed J K, Metchock B, Berkowitz F, Weigel L, Crellin J, Steward C, Hill B, Medeiros A A, Tenover F C. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother. 1997;41:647–653. doi: 10.1128/aac.41.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi S, Nagano Y. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J Clin Microbiol. 1984;20:608–613. doi: 10.1128/jcm.20.4.608-613.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzouvelekis L S, Tzelepi E, Mentis A F. Nucleotide sequence of a plasmid-mediated cephalosporinase gene (blaLAT-1) found in Klebsiella pneumoniae. Antimicrob Agents Chemother. 1994;38:2207–2209. doi: 10.1128/aac.38.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]