Abstract
This study aimed to explore the impact of ozonated water (OW) disinfestation on soil fungal community composition in continuous ginger field. All soil samples were collected in continuous ginger field. There were two groups and 5 time points (0, 1, 3, 5, 9 day) in our study, including OW disinfestation treatment group (O3 group) and control group (CK group). Via internal transcribed spacer (ITS) sequencing and further analysis, the changes of fungal community composition were determined. As a result, at 0 and 9 days after aeration, the operational taxonomic units (OTUs) in O3 group were significantly higher than that in CK group. Compared with the CK group, in O3 group: the ACE and Chao1 index significantly increased on day 1, and the Shannon index significantly decreased while Simpson index significantly increased on day 0 after aeration. In O3 group, there were dynamic changes of top 10 abundance fungi from the genus-level and the growth of Trichoderma and Rhodotorula had been promoted while Hannaella was inhibited. In conclusion, OW disinfestation had complicated impacts on fungal communities in continuous ginger fields. The growth of Trichoderma and Rhodotorula has been promoted during disinfestation, which provided more reference information for soil OW disinfestation research.
1 Introduction
In recent decades, many countries are facing the problem of limited arable land all over the world [1]. Subsequently, intensive cultivation like continuous monocropping and overfertilization has been widely used in order to meet the increasing demands [1,2]. Long-term continuous cropping often results in plant pathogen enrichment and imbalanced microbial community in soil [3,4], which is one of the main reasons for the serious yield decline [2]. Numerous studies have demonstrated that microorganisms could play an indispensable role not only in soil healthy [2] but also in plant health and growth [1]. In addition, fungus is one of the most abundant microorganisms in soil, which plays an important part in nutrient recycling in terrestrial ecosystems [5,6]. It has been widely recognized that soil fungi can impact various soil biogchemical processes [7]. Some microorganisms could improve the quality and crop yield of the soil through organic matter degradation and transformation [8,9]. Therefore, it is quite important for planting industry to protect the balanced soil microbial community and soil ecosystem.
In order to control soil-borne diseases and support high yields, preplant soil disinfestation is usually used in most intensive cropping systems [1]. Ozone (O3) is a recognized broad-spectrum and environmentally friendly insecticide [10,11], which has been applied to several fields like plant diseases control in facility agriculture [12]. Ozone is a powerful oxidant and ozonated water (OW) usually has stronger oxidation. OW is unstable, which decomposes rapidly into oxygen and would not leave toxic residues [13]. Thus, it is an environment-friendly way leading to less influence on soil microbial community. At present, some studies have focused on the effects of OW disinfestation on crops [12,13], but few studies paid attention to it’s influence on soil microbial community as far as we know. However, the negative impact of soil disinfestation on soil microbial community is a controversial problem.
Accordingly, in this study, the continuous ginger fields treated with OW were compared with untreated ginger fields. The changes of fungal community abundance, diversity and structure were then analyzed using internal transcribed spacer (ITS) sequencing in order to explore the impact of OW disinfestation on soil fungal community composition.
2 Materials and methods
2.1 Soil samples collection
Soil was collected from farms in Shandong, China. All soil samples were collected in continuous ginger field, which had been used for many years with no disinfestation, the ginger field had serious soil diseases. Soil was randomly sampled at five points on diagonal line and samples were collected from the top 25 cm of soil, which were packed in self-sealing bags and were labeled well for future usage. Samples were collected at 0, 1, 3, 5 and 9 days after the OW treated, respectively, including the control group (CK group) samples and the OW disinfestation treatment group (O3 group) samples.
Our sequence data has been uploaded to the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) database, and the accession number is PRJNA746256.
2.2 DNA extraction
MoBio Powersoil® DNA Isolation Kit (MoBio Laboratories, USA) was used to extract the total genomic DNA from each soil sample. The DNA quality was determined by gel electrophoresis (1% agarose). The DNA concentration was determined by NanoDrop 1000 UV spectrophotometer (Thermo Scientific, USA).
2.3 High-throughput sequencing and bioinformatics analysis
The universal primers [5′-CTTGGTCATTTAGAGGAAGTAA-3′] and [5′-GCTGCGTTCTTCATCGATG-3′] were used to amplify fungal genes in ITS1 region. MiSeq sequencing was conducted on an Illumina® MiSeq sequencer (Illumina, USA) by Shanghai Tianhao Biotechnology Co., Ltd. (Shanghai, China) and paired-end sequencing (2 × 250 bp) was used. The sequencing data was quality-filtered and merged to obtain high-quality reads using TrimGalore software and FLASH software [14]. UPARSE software was used to perform operational taxonomic unit (OTU) clustering on nonrepeating sequences according to 97% similarity [15]. Moreover, the chimeras were removed during the clustering process to obtain the representative sequence of OTUs. The Mothur software was used to compare each ITS representative sequence with those deposited in the Ribosomal Database Project (RDP) ITS database (http://rdp.cme.msu.edu/) [16] to determine the classification, based on a confidence threshold of 60%. The α diversity was calculated using Mothur software, including the Coverage index, ACE index, Chao1 index, Shannon index and Simpson index. QIIME software [17] was used to calculate the unweighted UniFrac value and Principal Coordinate Analysis (PCoA) was used to visualize changes of soil microbial communities. A linear discriminant analysis effect size (LEfSe) [18] was performed to find differential biomarkers between two groups using relative abundance.
2.4 Statistical analysis
Wilcoxon symbolic rank sum test (R software v3.6.2) was used to compare α diversity differences (ACE index, Chao1 index, Shannon index and Simpson index) among different groups and P < 0.05 was used as significance threshold. Wilcoxon symbolic rank sum test was used to compare the differences of microbial communities with top 10 genus abundance in soil. LEfSe analysis basing on Kruskal-Wallis sum-rank test determined the changes of the genus abundance between the OW treatment group and the control group. |LDA score|>2 and P<0.05 was used as the significance threshold.
3 Results
3.1 Changes in microbial community OTUs
Based on the genetic sequencing results of the soil microbial communities, a total of 10,724,257 effective reads were obtained and the average length of effective reads was 232.59 bp. Compared with the CK group, the shared OTUs in O3 group at 1, 3, 5 and 9 days after aeration was all more than that of shared OTUs at 0 day (S1 Fig). Additionally, at 0 and 9 days after aeration, the number of unique OTUs in O3 group was significantly higher than that in CK group (P < 0.001), which indicated that the fungal communities in soil were significantly different after the OW treatment, compared with the CK group.
3.2 Changes of fungal abundance and diversity
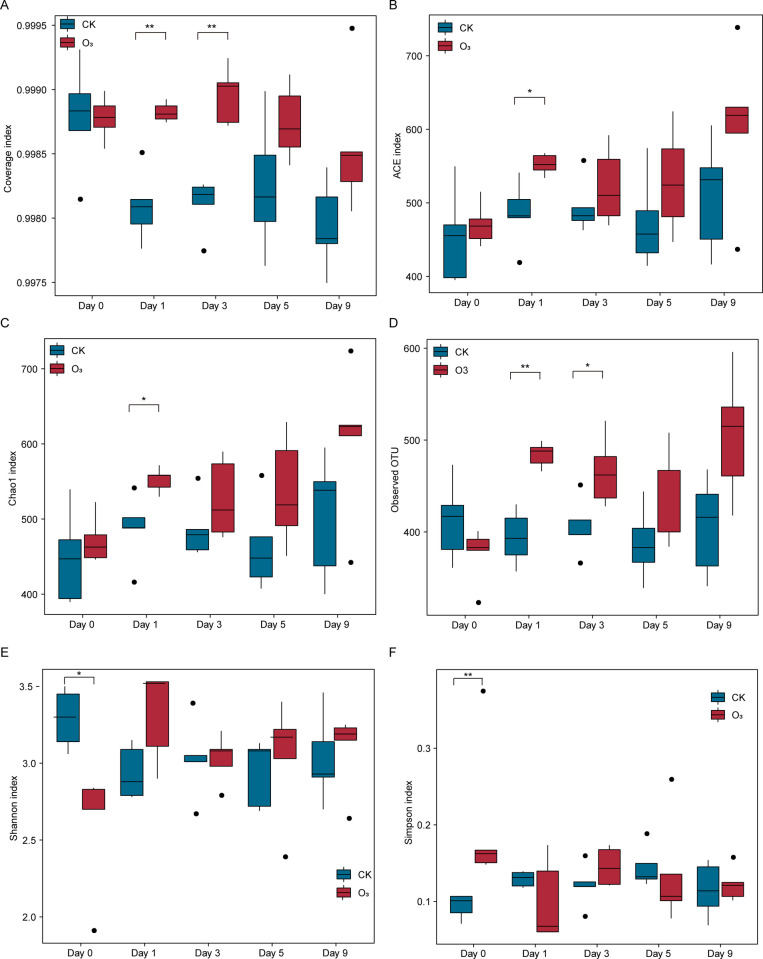
The dilution curve of samples (S2 Fig) suggested that the sampling and ITS genetic sequence data was sufficient to estimate the diversity of fungal communities. Coverage index showed that the sequencing coverage of samples was good (0.9975–0.9995) (Fig 1A). Based on 97% consistent sequence, 99% of the species presented in all samples. Compared with the CK group, the ACE and Chao1 index of the O3 group significantly increased (P < 0.05) at the first day after aeration (Fig 1B and 1C). During other time, there was no significant ACE and Chao1 index difference between the CK and O3 group. Compared with CK group, the OTU number in O3 group significantly increased at the first and third day, but there was no significant OTU number difference between the CK group and O3 group (Fig 1D). On day 0 in O3 group, the Shannon index significantly decreased while Simpson index increased significantly, but there was no significant difference between the CK group and O3 group in the following days (Fig 1E and 1F).
Fig 1. Microbial community diversity index of the CK group and O3 group, respectively.
(A) Coverage index (B) ACE index (C) Chao1 index (D) Observed OTU (E) Shannon index (F) Simpson index. CK: Control group; O3: The OW disinfestation treatment group. Statistical significance: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
The results showed that the OW disinfestation transitorily stimulated the species abundance increasing and fungal diversity decreasing, however along with the extended time of aeration, species abundance and fungal diversity gradually returned to the untreated level.
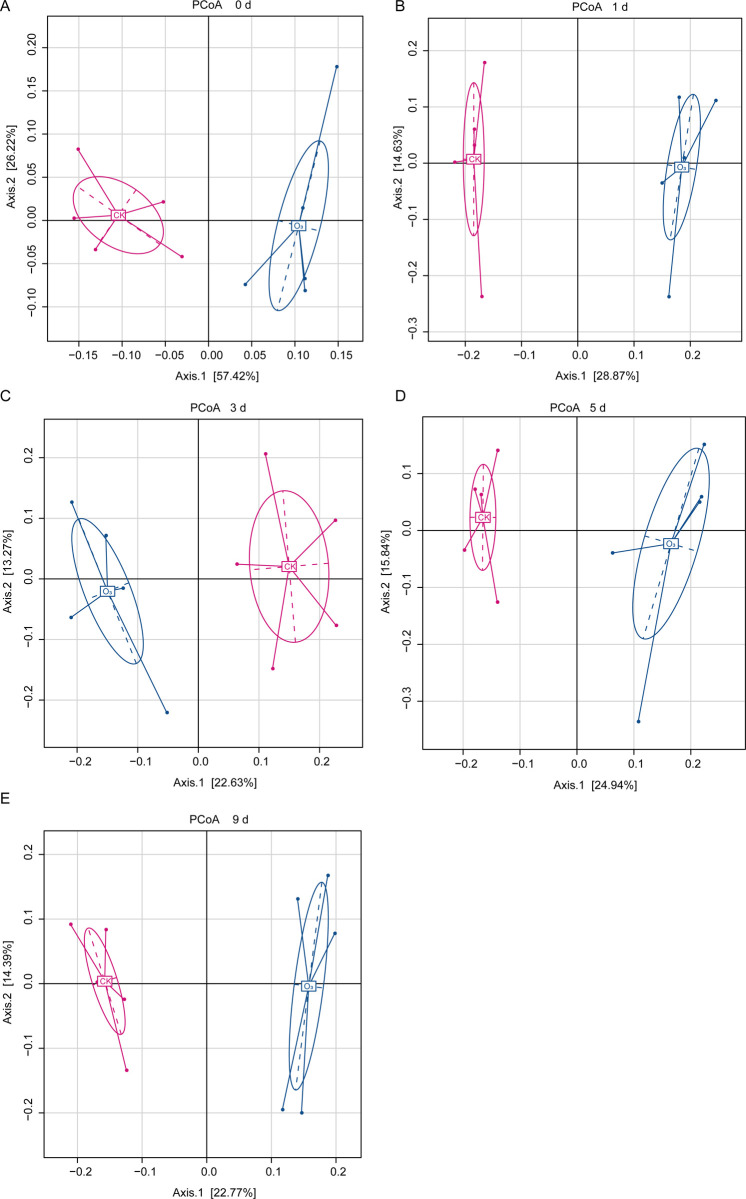
3.3 Principal coordinate analysis (PCoA)
The PCoA results showed that the soil samples of two groups (CK and O3 group) were completely separated apart at 0, 1, 3, 5 and 9 days (Fig 2), which indicated that there was difference in soil samples between CK group and O3 group.
Fig 2. Microbial community principal coordinate analysis (PCoA) of the CK group and O3 group.
(A-E) The PCoA results at 0, 1, 3, 5 and 9 days.
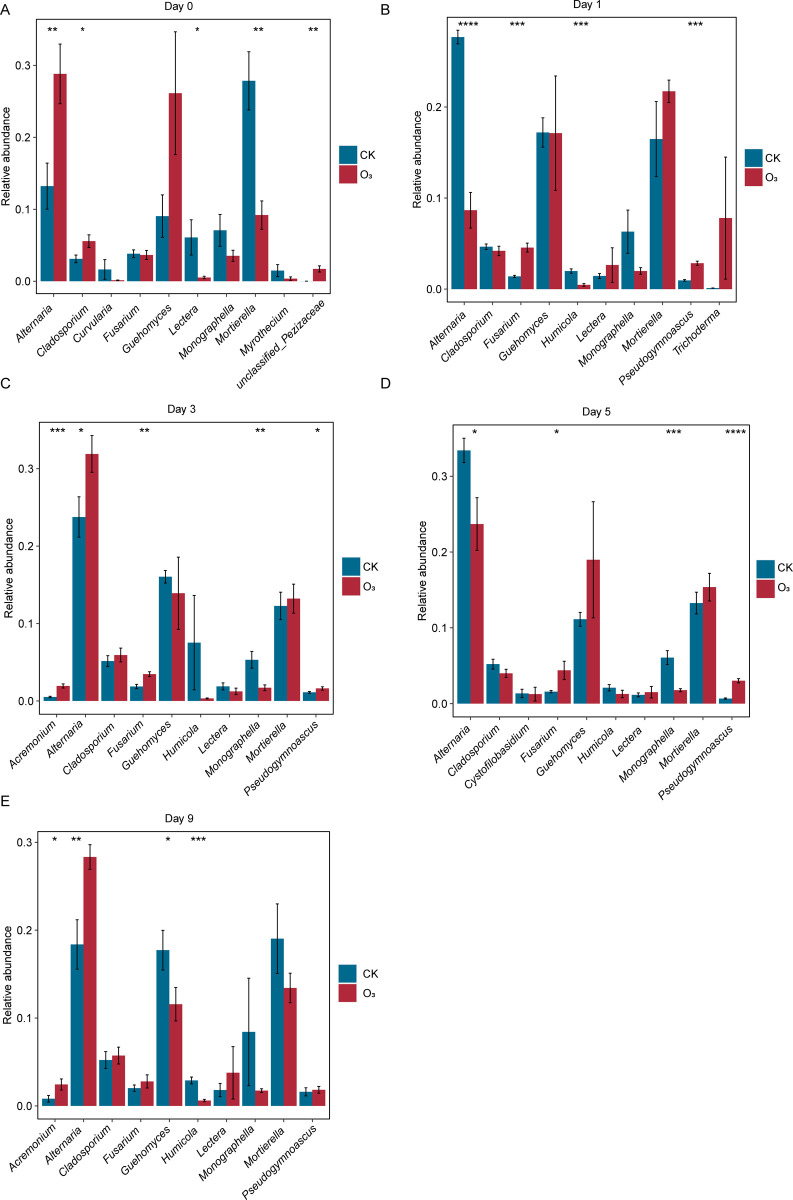
3.4 Changes of microbial communities with top 10 genus abundance in soil
In all samples, Alternaria, Mortierella, Guehomyces, Monographella and Cladosporium were dominant fungi from the genus-level (S3 Fig). The fungal community changes of top 10 abundance from the genus-level between CK group and O3 group were further compared at various days. Compared with the CK group, Alternaria in O3 group significantly increased on the 0th, 3rd and 9th day and significantly decreased on the 1st and 5th day after aeration. Cladosporium significantly increased on day 0 and then returned to untreated level. Lectera and Mortierella significantly decreased on day 0 and then returned to untreated levels. Fusarium in O3 group was significantly higher than that in CK group during 1–5 days, but it decreased gradually along with time and returned to the untreated level on the 9th day. Guehomyces was relatively stable during the first 5 days and was significantly lower than that of the CK group on the 9th day. Monographella was significantly lower than that of the CK group during 3–5 days and returned to untreated levels on the 9th day. Humicola, Pseudogymnoascus replaced Curvularia, Myrothecium and unclassified_Pezizaceae and became the dominant fungal community (among top 10) after day 1. Humicolain in O3 group was significantly higher than that in CK group on day 1 and day 9. Pseudogymnoascus in O3 group was significantly higher than that in CK group during 1–5 days, but it returned to the untreated level on the 9th day (Fig 3).
Fig 3. Changes of microbial communities with top 10 genus abundance in soil.
(A-E) Changes of microbial communities at 0, 1, 3, 5 and 9 days. Statistical significance: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
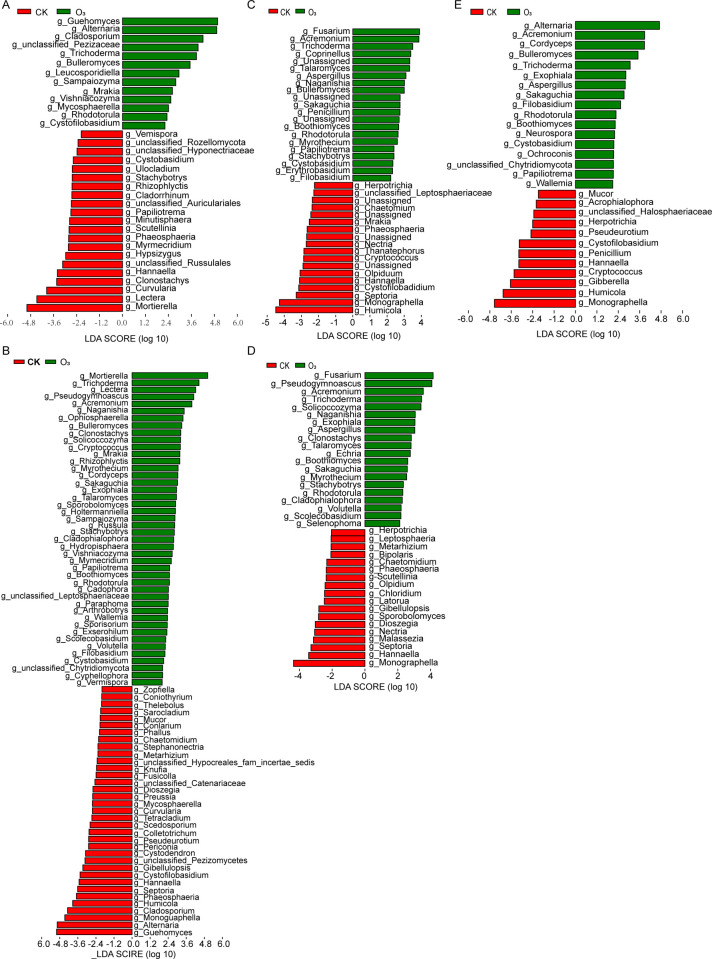
3.5 Changes of biomarkers of soil fungal communities
Significantly different biomarkers between CK group and O3 group on various days were displayed using LEFSe analysis. The number of differential biomarkers between CK group and O3 group in the initial exposure stage (0–1 days) was gradually increasing and then decreased gradually during 1–9 days. Compared with the CK group, on day 0, Guehomyces, Alternaria and other 13 genera significantly increased in O3 group, and Vermispora, Cystobasidium and other 21 genera significantly decreased (Fig 4A). Compared with the CK group, on day 1, Mortierella, Trichoderma and other 44 genera significantly increased in O3 group, and Coniothyrium, Knufia and other 35 genera significantly decreased (Fig 4B). Compared with the CK group, on day 3, Fusarium, Acremonium, Trichoderma and other 21 genera significantly increased in O3 group, and Herpotrichia, Cryptococcus and other 18 genera significantly decreased (Fig 4C). Compared with the CK group, on day 5, Fusarium, Pseudogymnoascus, Acremonium and other 20 genera significantly increased in O3 group, and Herpotrichia, Sporobolomyces and other 17 genera significantly decreased (Fig 4D). Compared with the CK group, on day 9, Alternaria, Acremonium, Cordyceps and other 17 genera significantly increased in O3 group, and Halosphaeriacea, Herpotrichia and other 12 genera significantly decreased (Fig 4E).
Fig 4. Significantly differential biomarkers between CK group and O3 group at various days were analyzed using LEFSe.
(A-E) Differential biomarkers at 0, 1, 3, 5 and 9 days. Statistical significance: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Compared with the CK group, Trichoderma and Rhodotorula significantly increased in O3 group during 0–9 days, and Boothiomyces, Sakaguchia, Acremonium and Aspergillus also significantly increased during 3–9 days (S4A Fig). Compared with the CK group, Hannaella significantly decreased in O3 group during 0–9 days, and Herpotrichia and Monographella also significantly decreased during 3–9 days (S4B Fig). The results showed that OW disinfestation had promoted the growth of Trichoderma and Rhodotorula, but it had inhibited the growth of Hannaella.
4 Discussion
In this research, via the comparison of the continuous ginger fields treated with OW and untreated ginger fields, we explored the the influence of OW disinfestation on soil fungal community composition. The OW disinfestation transitorily stimulated the fungus abundance increasing and diversity decreasing and there were dynamic changes of top 10 abundance fungi from the genus-level. The OW disinfestation had promoted the growth of Trichoderma and Rhodotorula, but it had inhibited the growth of Hannaella.
Recently, ozone has been gradually recognized as a powerful and effective oxidant for water treatment [19]. Moreover, aqueous ozone treatments have been recently evidenced to reduce contaminant colony forming units (CFUs) [20], which makes OW disinfestation more meaningful. Thus, we expected to explore its potential influence on fungal community in field. A research in long-term continuous soybean field has reported that long-term continuous cropping led to the trends of fungal community development to antagonistic to plant health [21], whose similar situation supported our observation in continuous ginger field. Firstly, we have investigated the changes of fungal abundance and diversity in soil after treated with OW. The number of unique OTUs in O3 group (OW treated group) was significantly higher than that in CK group at 0 and 9 days aeration, which indicated that there were more fungi in soil in O3 group at 0 and 9 days after aeration. As for fungal diversity, compared with the CK group, the ACE and Chao1 index of the O3 group significantly increased at the first day after aeration. On day 0 in O3 group, the Shannon index significantly decreased while Simpson index increased significantly. Previous researches have evidenced that Simpson index, Shannon index, ACE index and Chao1 index were reliable indicators of fungal diversity [21]. Our findings suggested that the OW disinfestation transitorily stimulated the fungal abundance increasing and fungal diversity decreasing, however along with the extended time of aeration, fungal abundance and diversity gradually returned to the untreated level. Moreover, it has been demonstrated that soil fungal diversity was closely associated with the plant health and soil-borne diseases [22,23]. Collectively, the OW disinfestation would transitorily impact the fungal abundance and diversity, which might be benefit for fungal restoration after disinfestation.
Additionally, various fungal community compositions at genus level and their abundance were then analyzed. In all samples, Alternaria, Mortierella, Guehomyces, Monographella and Cladosporium were dominant fungi. Further more, the results of LEFSe analysis showed that OW disinfestation had different impacts on different fungi. The OW disinfestation had promoted the growth of Trichoderma and Rhodotorula, but it had inhibited the growth of Hannaella. A study suggested that Trichoderma and other beneficial microorganisms would help soil microbiome recover after soil fumigation disinfestation [24]. Also, it has been reported that Trichoderma could be used as the biocontrol fungi for plant disease prevention [25]. Not only that, it has also been documented that some Trichoderma strains could suppress pathogens like F. pseudograminearum, M. syringejaponicae and so on [26,27]. As for Rhodotorula, a research has suggested that one strain CAM4 of it could be used for salt and drought stress resistance biofertilizer [28]. Another study indicated that Rhodotorula strain CAH2 might be a plant growth promoting fungus in unfavourable environmental conditions [29]. Accordingly, OW disinfestation might promote the beneficial fungi growth during disinfestation. Notably, our findings indicated that the OW disinfestation could considerably relieve the soil deterioration of the continuous ginger field, whose effect was partly similar with bean dregs treatment [24]. However, many present soil disinfestation ways (i.e. reductive soil disinfestation and chemical soil disinfestation) have suppressed not only soil-borne pathogens but also fungal communities [1,24], which would alter the fungal community structure. In our study, non-pathogen fungal community has been reassembled and the growth of some non-pathogen fungi were promoted. Loganathachetti et al. have reported that there was a significant correlation between fungal community compositional changes and carbon or nitrogen availability of soil [30]. Therefore, our present study provided more insights into OW disinfestation research.
In conclusion, through a comprehensive analysis of the continuous ginger fields treated and untreated with OW, the results indicated that OW disinfestation had complicated impacts on fungal communities in continuous ginger fields. The OW disinfestation might promote the beneficial fungi growth while had a little negative influence on fungal communities in continuous ginger fields, which provided more reference information for soil disinfestation research.
Supporting information
(A-E) The OTU changes were recorded at 0, 1, 3, 5 and 9 days. CK: control group; O3: The OW disinfestation treatment group.
(TIF)
(A-E) The dilution curve at 0, 1, 3, 5 and 9 days. CK: Control group; O3: The OW disinfestation treatment group. Five replicates in each group.
(TIF)
(TIF)
(A) Venn diagram of the O3 group differential biomarkers at 0, 1, 3, 5 and 9 days. (B) Venn diagram of the CK group differential biomarkers at 0, 1, 3, 5 and 9 days. CK: Control group; O3: The OW disinfestation treatment group.
(TIF)
Data Availability
The datasets generated and analyzed during the current study are publicly available from National Center for Biotechnology Information database (accession number: PRJNA746256, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA746256).
Funding Statement
This study was funded by the National Key Research and Development Program of China (grant numbers: 2017YFD02016005, 2018YFD0201005) and the funders had role in data analysis and language editing of the manuscript.
References
- 1.Meng T, Ren G, Wang G, Ma Y. Impacts on soil microbial characteristics and their restorability with different soil disinfestation approaches in intensively cropped greenhouse soils. Appl Microbiol Biotechnol. 2019;103(15):6369–83. doi: 10.1007/s00253-019-09964-z . [DOI] [PubMed] [Google Scholar]
- 2.She S, Niu J, Zhang C, Xiao Y, Chen W, Dai L, et al. Significant relationship between soil bacterial community structure and incidence of bacterial wilt disease under continuous cropping system. Arch Microbiol. 2017;199(2):267–75. doi: 10.1007/s00203-016-1301-x . [DOI] [PubMed] [Google Scholar]
- 3.Yang JI, Ruegger PM, McKenry MV, Becker JO, Borneman J. Correlations between root-associated microorganisms and peach replant disease symptoms in a California soil. PLoS One. 2012;7(10):e46420. doi: 10.1371/journal.pone.0046420 ; PubMed Central PMCID: PMC3465339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Zhang J, Gu T, Zhang W, Shen Q, Yin S, et al. Microbial community diversities and taxa abundances in soils along a seven-year gradient of potato monoculture using high throughput pyrosequencing approach. PLoS One. 2014;9(1):e86610. doi: 10.1371/journal.pone.0086610 ; PubMed Central PMCID: PMC3907449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu YG, Gillings M, Simonet P, Stekel D, Banwart S, Penuelas J. Microbial mass movements. Science. 2017;357(6356):1099–100. doi: 10.1126/science.aao3007 . [DOI] [PubMed] [Google Scholar]
- 6.Kohler M, Devaux C, Grigulis K, Leitinger G, Lavorel S, Tappeiner U. Plant functional assemblages as indicators of the resilience of grassland ecosystem service provision. Ecol Indic. 2019;73:118–27. doi: 10.1016/j.ecolind.2016.09.024 ; PubMed Central PMCID: PMC6694008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551(7681):457–63. doi: 10.1038/nature24621 ; PubMed Central PMCID: PMC6192678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang W, Yan D, Wang X, Huang B, Wang X, Liu J, et al. Responses of Nitrogen-Cycling Microorganisms to Dazomet Fumigation. Front Microbiol. 2018;9:2529. doi: 10.3389/fmicb.2018.02529 ; PubMed Central PMCID: PMC6206233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crouzet O, Poly F, Bonnemoy F, Bru D, Batisson I, Bohatier J, et al. Functional and structural responses of soil N-cycling microbial communities to the herbicide mesotrione: a dose-effect microcosm approach. Environ Sci Pollut Res Int. 2016;23(5):4207–17. doi: 10.1007/s11356-015-4797-8 . [DOI] [PubMed] [Google Scholar]
- 10.Moreno JD, Rodriguez SJ, Poznyak T, Chairez I, Dorantes-Rosales HJ. Effect of the type of soil on dimethyl phthalate degradation by ozone. J Environ Manage. 2020;270:110863. doi: 10.1016/j.jenvman.2020.110863 . [DOI] [PubMed] [Google Scholar]
- 11.O’Mahony MM, Dobson AD, Barnes JD, Singleton I. The use of ozone in the remediation of polycyclic aromatic hydrocarbon contaminated soil. Chemosphere. 2006;63(2):307–14. doi: 10.1016/j.chemosphere.2005.07.018 . [DOI] [PubMed] [Google Scholar]
- 12.Prigigallo MI, Melillo MT, Bubici G, Dobrev PI, Vankova R, Cillo F, et al. Ozone treatments activate defence responses against Meloidogyne incognita and Tomato spotted wilt virus in tomato. Pest Manag Sci. 2019;75(8):2251–63. doi: 10.1002/ps.5362 . [DOI] [PubMed] [Google Scholar]
- 13.Campayo A, Serrano de la Hoz K, Garcia-Martinez MM, Sanchez-Martinez JF, Salinas MR, Alonso GL. Spraying ozonated water on Bobal grapevines: Effect on grape quality. Food Res Int. 2019;125:108540. doi: 10.1016/j.foodres.2019.108540 . [DOI] [PubMed] [Google Scholar]
- 14.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–63. doi: 10.1093/bioinformatics/btr507 ; PubMed Central PMCID: PMC3198573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–8. doi: 10.1038/nmeth.2604 . [DOI] [PubMed] [Google Scholar]
- 16.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386 ; PubMed Central PMCID: PMC2563014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303 ; PubMed Central PMCID: PMC3156573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60 ; PubMed Central PMCID: PMC3218848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ngwenya N, Ncube EJ, Parsons J. Recent advances in drinking water disinfection: successes and challenges. Rev Environ Contam Toxicol. 2013;222:111–70. Epub 2012/09/20. doi: 10.1007/978-1-4614-4717-7_4 . [DOI] [PubMed] [Google Scholar]
- 20.Martinelli M, Giovannangeli F, Rotunno S, Trombetta CM, Montomoli E. Water and air ozone treatment as an alternative sanitizing technology. J Prev Med Hyg. 2017;58(1):E48–E52. Epub 2017/05/19. ; PubMed Central PMCID: PMC5432778. [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Pan F, Han X, Song F, Zhang Z, Yan J, et al. Response of Soil Fungal Community Structure to Long-Term Continuous Soybean Cropping. Front Microbiol. 2018;9:3316. doi: 10.3389/fmicb.2018.03316 ; PubMed Central PMCID: PMC6333693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rime D, Nazaret S, Gourbiere F, Cadet P, Moenne-Loccoz Y. Comparison of sandy soils suppressive or conducive to ectoparasitic nematode damage on sugarcane. Phytopathology. 2003;93(11):1437–44. doi: 10.1094/PHYTO.2003.93.11.1437 . [DOI] [PubMed] [Google Scholar]
- 23.Liang J, Tang S, Gong J, Zeng G, Tang W, Song B, et al. Responses of enzymatic activity and microbial communities to biochar/compost amendment in sulfamethoxazole polluted wetland soil. J Hazard Mater. 2020;385:121533. doi: 10.1016/j.jhazmat.2019.121533 . [DOI] [PubMed] [Google Scholar]
- 24.Zhao J, Zhou X, Jiang A, Fan J, Lan T, Zhang J, et al. Distinct impacts of reductive soil disinfestation and chemical soil disinfestation on soil fungal communities and memberships. Appl Microbiol Biotechnol. 2018;102(17):7623–34. Epub 2018/06/23. doi: 10.1007/s00253-018-9107-1 . [DOI] [PubMed] [Google Scholar]
- 25.Xian HQ, Liu L, Li YH, Yang YN, Yang S. Molecular tagging of biocontrol fungus Trichoderma asperellum and its colonization in soil. J Appl Microbiol. 2020;128(1):255–64. doi: 10.1111/jam.14457 . [DOI] [PubMed] [Google Scholar]
- 26.Stummer BE, Zhang Q, Zhang X, Warren RA, Harvey PR. Quantification of Trichoderma afroharzianum, Trichoderma harzianum and Trichoderma gamsii inoculants in soil, the wheat rhizosphere and in planta suppression of the crown rot pathogen Fusarium pseudograminearum. J Appl Microbiol. 2020;129(4):971–90. doi: 10.1111/jam.14670 . [DOI] [PubMed] [Google Scholar]
- 27.Liu B, Ji S, Zhang H, Wang Y, Liu Z. Isolation of Trichoderma in the rhizosphere soil of Syringa oblata from Harbin and their biocontrol and growth promotion function. Microbiol Res. 2020;235:126445. doi: 10.1016/j.micres.2020.126445 . [DOI] [PubMed] [Google Scholar]
- 28.Silambarasan S, Logeswari P, Cornejo P, Abraham J, Valentine A. Simultaneous mitigation of aluminum, salinity and drought stress in Lactuca sativa growth via formulated plant growth promoting Rhodotorula mucilaginosa CAM4. Ecotoxicol Environ Saf. 2019;180:63–72. doi: 10.1016/j.ecoenv.2019.05.006 . [DOI] [PubMed] [Google Scholar]
- 29.Silambarasan S, Logeswari P, Cornejo P, Kannan VR. Evaluation of the production of exopolysaccharide by plant growth promoting yeast Rhodotorula sp. strain CAH2 under abiotic stress conditions. Int J Biol Macromol. 2019;121:55–62. doi: 10.1016/j.ijbiomac.2018.10.016 . [DOI] [PubMed] [Google Scholar]
- 30.Sanka Loganathachetti D, Poosakkannu A, Muthuraman S. Fungal community assemblage of different soil compartments in mangrove ecosystem. Sci Rep. 2017;7(1):8560. doi: 10.1038/s41598-017-09281-3 ; PubMed Central PMCID: PMC5561109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-E) The OTU changes were recorded at 0, 1, 3, 5 and 9 days. CK: control group; O3: The OW disinfestation treatment group.
(TIF)
(A-E) The dilution curve at 0, 1, 3, 5 and 9 days. CK: Control group; O3: The OW disinfestation treatment group. Five replicates in each group.
(TIF)
(TIF)
(A) Venn diagram of the O3 group differential biomarkers at 0, 1, 3, 5 and 9 days. (B) Venn diagram of the CK group differential biomarkers at 0, 1, 3, 5 and 9 days. CK: Control group; O3: The OW disinfestation treatment group.
(TIF)
Data Availability Statement
The datasets generated and analyzed during the current study are publicly available from National Center for Biotechnology Information database (accession number: PRJNA746256, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA746256).