Abstract
One of the primary clinical observations for screening the novel coronavirus is capturing a chest x-ray image. In most patients, a chest x-ray contains abnormalities, such as consolidation, resulting from COVID-19 viral pneumonia. In this study, research is conducted on efficiently detecting imaging features of this type of pneumonia using deep convolutional neural networks in a large dataset. It is demonstrated that simple models, alongside the majority of pretrained networks in the literature, focus on irrelevant features for decision-making. In this paper, numerous chest x-ray images from several sources are collected, and one of the largest publicly accessible datasets is prepared. Finally, using the transfer learning paradigm, the well-known CheXNet model is utilized to develop COVID-CXNet. This powerful model is capable of detecting the novel coronavirus pneumonia based on relevant and meaningful features with precise localization. COVID-CXNet is a step towards a fully automated and robust COVID-19 detection system.
Keywords: COVID-19, Chest X-ray, Convolutional neural networks, CheXNet, Imaging features
Introduction
Being declared as a pandemic, novel coronavirus is now a major emergency worldwide. The virus is transmitted person-to-person by respiratory droplets or close contact with a contaminated surface [52]. The most common symptoms are fever, cough, and dyspnea, which may appear 2-14 days after exposure to the virus. The standard diagnosis method, highly specific but with inconstant sensitivity [25], is based on reverse transcription polymerase chain reaction (RT-PCR) [40]. The RT-PCR test has certain shortcomings, such as availability and time-consumption. It needs special test-kits, which may not be widely available in some regions [5], and the results are generally available within hours to days [7]. A diagnostic guideline proposed by Zhongnan Hospital of Wuhan suggests that the disease could be assessed by detecting clinical symptoms as well as radiological findings of pneumonia [24]. Furthermore, Ai et al. show that chest computed tomography (CT) scans have high sensitivity for COVID-19 diagnosis and can be considered as the primary diagnostic tools in epicenters [1].
Chest x-rays (CXRs) and CT scans have been used for COVID-19 screening and disease progression evaluation in hospital admitted cases [42]. Despite offering superior sensitivity to thoracic abnormality detection [10], using CT has several challenges. CT scanners are non-portable and require sanitizing of the equipment and imaging room between patients. Besides, their radiation dose is considerably higher than x-rays [15]. On the contrary, portable x-ray units are widely available and can be easily accessed in most primary hospitals [16]. Moreover, x-ray imaging can be operated in more isolated rooms with less staff exposure to the virus [36]. In many cases, the patient’s clinical situation does not allow a CT scan; hence, CXR is a better choice for the initial assessment. Even initial reports from China and a meta-analysis performed by Korean radiologists showed a poor performance from chest CT in terms of specificity [29]. Therefore, many healthcare institutions prefer taking a CXR as the preliminary step for COVID-19 diagnosis. Radiologists in Milan also confirmed that during the peak of COVID-19 pandemic in Italy, they chose CXR instead of CT to be taken in their hospital [12]. Spanish pediatric radiologists in Madrid have also suggested that the systematic use of CT is not recommended due to the fact that children are more sensitive to radiation. As a result, CXR is considered as the main tool for clinical decision and management of children with suspected COVID-19 [44].
Since radiologists visit many patients every day and the diagnosis process takes significant time, errors may increase notably. As a result, there might be many more false negatives that will cost a lot to the patient and the medical staff. Therefore, automated computer-aided diagnostic (CAD) tools are of utmost importance. Automated deep learning-based CAD tools have previously shown promising results in medical disease classification tasks, such as early detection of arthritis [9], and specifically in detection of thoracic diseases, e.g. pulmonary nodules [55].
In this study, firstly, we collect a dataset of CXRs from COVID-19 patients from multiple publicly accessible sources. Our collected dataset is one of the largest public sources of COVID-19 CXRs, containing 1326 images. After visualizing primary dataset information, we then investigate the possibility of disease detection by an individual Convolutional Neural Network (CNN) model. On the next step, performance of prominent pretrained CNN models for fine-tuning on the dataset is investigated. Afterwards, the CheXNet pretrained model on the same type of medical images is introduced, and its efficiency is discussed. Finally, we develop our model based on the CheXNet and design a lung segmentation module to improve the model localization of lung abnormalities. Class activation map (CAM) is our main visualization leverage to compare our models. Main contributions can be summarized as:
Introducing one of the largest public datasets of COVID-19 CXR images, collected from different sources
Developing a robust pneumonia classification model by training on a large dataset of COVID-19 CXRs
Proposing a transfer learning approach to tackle the problem based on pretrained network
Precisely evaluating model performance by visualizing the results using CAMs
Related works
Identifying COVID-19 pneumonia using different types of medical images is a fast-growing topic of interest. ML-based methods, along with manual feature extraction algorithms, are used in a few articles to diagnose the disease [2, 6, 13, 18, 20]. However, most studies are utilizing DL-based techniques. Researchers have tried to tackle the problem using CT images, reaching high scoring metrics and precise abnormality localization [19, 45]. Contrarily, even though many studies have claimed to achieve excellent classification accuracy scores using CXRs, such as [32] and [28], none of them have reported visualization results. Because pneumonia diagnosis is more challenging in CXRs and the available COVID-19 pneumonia CXR datasets are small, we investigate those studies with visual interpretability used as their metric.
Zhang et al. used a dataset including 100 CXRs from COVID-19 cases and developed a ResNet-based model with pretrained weights from ImageNet as the backbone [59]. Their best model achieved an f-score of ≈ 0.72 in classifying COVID-19 pneumonia from Community-Acquired Pneumonia (CAP). Li et al. applied their multi-player model called COVID-MobileXpert on a dataset of 537 images equally divided into normal, CAP, and COVID-19 pneumonia samples [30]. Their main goal was to achieve acceptable accuracy using lightweight networks, such as SqueezeNet, for pneumonia detection on mobile devices capturing noisy snapshots. Rajaraman et al. collected a more expanded dataset containing 313 COVID-19 pneumonia CXRs from two different sources [36]. Lung segmentation was then applied using a U-Net-based model. Finally, an ensemble of different fine-tuned models was implemented and pruned iteratively to reduce parameters. Their best single pruned architecture was Inception-V3, and their best ensemble model was by weighted averaging strategy. They have achieved f-scores of 0.9841 and 0.99 detecting COVID-19 pneumonia from CAP and normal CXRs, respectively. However, their final generated visualization maps are not precisely discussed, and their model suffers some implementation drawbacks due to the significant number of parameters.
In a more advanced effort, COVID-Net was introduced by Wang et al. [53]. It was trained on COVIDx, a dataset with 358 CXR images from 266 COVID-19 patient cases. Their architecture was first trained on ImageNet and then achieved a best f-score of 0.9479 in three-class classification. Their model visualization is not properly presented nevertheless. A most recent similar research study was CovidAID conducted by Mangal et al. [31]. CovidAID is a DenseNet model built upon CheXNet weights. They compared their results with COVID-Net on the same test set. Their findings suggest that CovidAID surpassed COVID-Net with a notable margin, 0.9230 f1 score, compared with 0.3591. CovidAID image visualization shows more precise performance compared to previous studies. Consequently, developed models suffer a lack of robustness, mainly related to the insufficient number of images.
Regarding recent developments of more advanced architectures, specially ensemble methods, Kedia et al. proposed CoVNet-19 as an ensemble of VGG and DenseNet networks [26]. CoVNet-19 was trained on a relatively large dataset in terms of COVID positive images, containing 798 CXRs. Worth mentioning that the authors have collected their dataset from different public resources that some of them overlap. Hence, some of the COVID x-rays are duplicated. Their best effort led to achievement of a f1 score of 0.9891 in three-class classification. To the best of our knowledge, this study had used the dataset containing most number of COVID images to date.
Dataset overview
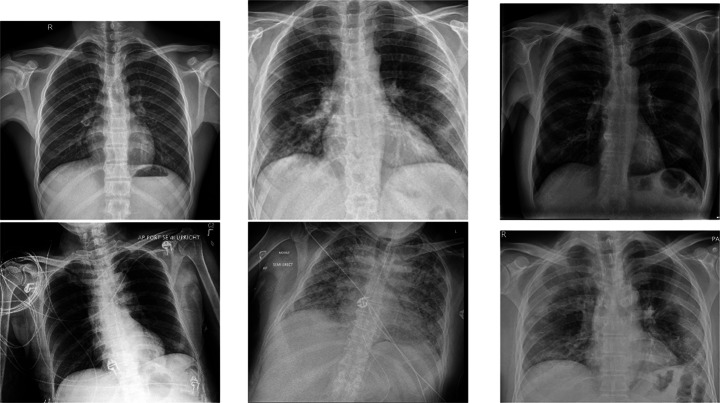
The most common imaging technique used as the first clinical step for chest-related diseases is CXR [60]. Hence, more CXRs could be collected publicly than CT images. A batch of randomly selected samples from the dataset with frontal view, also known as anteroposterior (AP) or posteroanterior (PA), is shown in Fig. 1.
Fig. 1.
Randomly selected frontal CXR images from different sources
There is another CXR imaging view called L, standing for Lateral, which is an adjunct for the main frontal view image. Lateral CXR is performed when there is diagnosis uncertainty using frontal CXR [21]. Thus it is not as common as frontal CXR, and due to its different angle, it is excluded from our data.
Dataset Sources
Since COVID-19 is a novel disease, the number of publicly available x-rays is relatively small, but constantly growing. There are different databases, regularly updated day by day, which our dataset is constructed upon them:
Radiopaedia1: open-edit radiology resource where radiologists submit their daily cases.
SIRM2: the website of the Italian Society of Medical and Interventional Radiology, which has a dedicated database of COVID-19.
EuroRad3: a peer-reviewed image resource of radiological case reports.
Figure 14: an image-based social forum that has dedicated a COVID-19 clinical cases section.
COVID-19 Image Data Collection [11]: a GitHub repository by Dr. Cohen et al. which is a combination of some of the mentioned resources and other images.
Twitter COVID-19 CXR Dataset5: a twitter thread of a cardiothoracic radiologist from Spain who has shared high-quality positive subjects.
Peer-Reviewed Papers: papers which have shared their clinical images, such as [22] and [35].
Hannover Medical School Dataset [56]: a GitHub repository containing images from the Institute for Diagnostic and Interventional Radiology in Hannover, Germany.
COVIDGR Dataset [49]: a set of CXR images from PCR-confirmed patients built under collaboration with expert radiologists in Spain.
Miscellaneous Sources: Images shared via other sources, such as RSNA cases6 and Instagram pages7.
Benefiting from several datasets with different imaging technologies and wide distribution of patient ages and locations decreases the chance of having bias. Distribution of our collected dataset among different above-mentioned sources is depicted in Fig. 2.
Fig. 2.
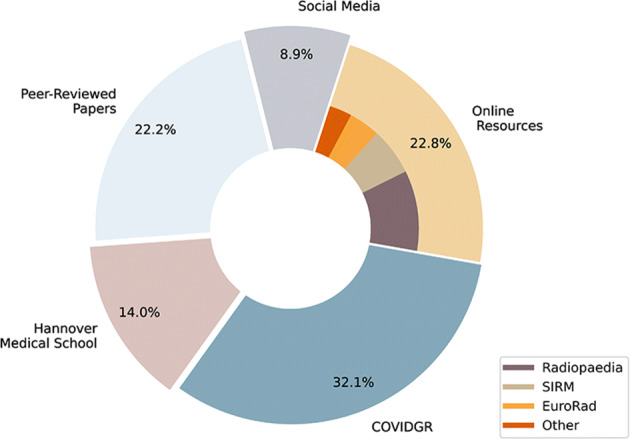
Dataset distribution. COVIDGR dataset with 426 images has the largest contribution in building our dataset
To create a more robust dataset, we aimed to have multiple sources also for images in other classes. Four sources were considered to collect normal CXRs:
Pediatric CXR dataset [27]: AP-view CXRs of children collected from Guangzhou Medical Center, including normal, bacterial pneumonia, and viral pneumonia cases.
NIH CXR-14 dataset [54]: Curated by the National Institute of Health (NIH), this large dataset has more than 100,000 images of normal chests and different abnormal lungs, including pneumonia and consolidation.
Radiopaedia: Other than infected cases, healthy CXRs taken for the purpose of medical check-ups are also available in Radiopaedia.
Tuberculosis Chest X-ray Image Datasets [23]: Provided by U.S. National Library of Medicine, it has two datasets containing 406 normal x-rays.
Currently, 1326 images of COVID-19 patients are collected in different sizes and formats. All collected images are publicly accessible in the dedicated repository8. The dataset includes 5,000 normal CXRs as well as 4,600 images of patients with CAP collected from the NIH CXR-14 dataset.
Statistical asnalysis
The dataset consists of 1326 chest x-rays labeled as COVID positive, acquired from different institutions around the world between Jan 2020 and May 2021. Due to the lack of image information availability from some of the sources, we made our best efforts to prepare the metadata of 452 chest x-rays, which is nearly 34% of the total data in COVID class. The patients had a mean of 1.62 chest x-ray studies performed at different time points, considering that each study contains one image. Current metadata consists of patient age, patient sex, x-ray view (AP/PA), PCR test result, survival result, study location, offset between imaging date and institution admission date, offset between imaging date and the onset of symptoms, a set of symptoms at the time of capturing the image, whether patient also had CT scan as the next step or not, and also notes on medical background of the patient. Since images are not stored in DICOM format, these features are extracted manually by investigating case reports one-by-one. Each metadata record stands for one image, i.e. a patient with 12 x-rays has 12 records in the metadata. Numerical variables in the metadata are patient age, number of studies per patient, admission offset, and symptom offset. Descriptive statistics related to aforementioned variables is shown in Table 1.
Table 1.
Descriptive statistics of continuous variables
| Mean ± Std | Min | Max | Most occurring value | Number of missing values | |
|---|---|---|---|---|---|
| Patient age (year) | 53.76 ± 21.20 | 0.03 | 99 | 50 and 55 (10 times) | 62 / 279 |
| Study per patient | 1.62 ± 1.28 | 1 | 12 | 1 (196 times) | - |
| Admission offset (day) | 5.93 ± 7.03 | 1 | 42 | 1 (92 times) | 196 / 452 |
| Symptom offset (day) | 10.49 ± 8.66 | 1 | 47 | 7 (21 times) | 314 / 452 |
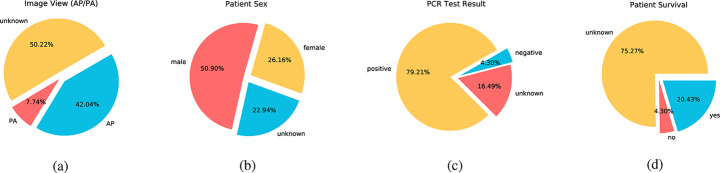
Most patients have only one chest x-ray during the study, and also most patients are adults in their 60s. Considering the first image capture performed mostly in the first day of admission to the institution, the average time for the second chest x-ray capture is 8.72 days after the admission date. Based on the symptom offsets, the average time for capturing images is 10.49 days after the onset of patient symptoms. It is also worth mentioning that all patients referred to the institutions between 0 to 14 days after noticing their symptoms. The average time between symptom onset and referral is 6.37 days. Remaining features are all categorical; x-ray view, patient sex, whether patient has survived or not, PCR test result at the time of study, and symptoms mentioned in the case report. A set of charts for each categorical variable distribution are depicted in Fig. 3.
Fig. 3.
Descriptive charts for categorical variables. a x-ray image view, b patient sex, c PCR test result, and d patient chance of survival
While image view is distinguishable by chest features in the x-ray, labels are set only when the view is directly mentioned in x-ray report or in the image itself. Firstly, the image view distribution shows a bias towards AP view, which means most images are from low quality and it makes the procedure of COVID pneumonia detection more challenging in comparison with benefiting from a dataset of mostly PA chest x-rays. Secondly, the share of male patients is approximately double the female patients. Rather than a bias, it seems like more COVID patients are males, since dataset is constructed from several sources in different institutions without a possible patient sex bias. Finally, if we consider unmentioned patient survivals as survived ones, most patients has survived. Since patient survival and the volume of manifestations in x-ray are supposed to be correlated, patient survival is possibly predictable based on the chest x-ray. This idea is not yet implementable, due to the class imbalance and lack of sufficient data.
One of the most concerning issues, specially among the community, is the relation between typical symptoms and chance of being infected by COVID-19 disease. Based on the case reports, patient symptoms are extracted and illustrated in Fig. 4 based on the frequency of happening among investigated cases.
Fig. 4.
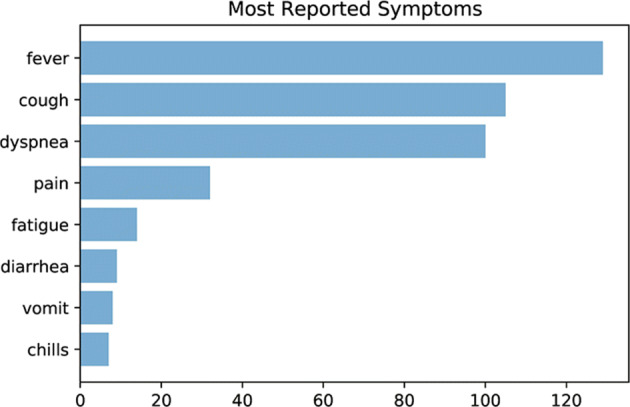
Most reported symptoms from COVID patients
Fever is the most reported symptom with a possibility of 46.24% being reported by patients as the first sign of disease. Surprisingly, pain (which includes abdominal pain, chest pain, and myalgia) and fatigue are more reported than other well-known symptoms, such as diarrhea or ageusia. Another interesting fact is that one-third of patients have complained from fever and cough at the same time, which are exact main symptoms of influenza.
Proposed method
In the following section, details about the proposed pipeline, from image preprocessing to model development, are explained in different steps. Initially, image preprocessing and contrast enhancement are mentioned. Afterwards, a base convolutional model is designed and trained on different portions of data from the dataset. Then, pretrained models based on the ImageNet dataset are discussed. Finally, a pretrained model on a similar image type is explained.
Preprocessing and enhancements
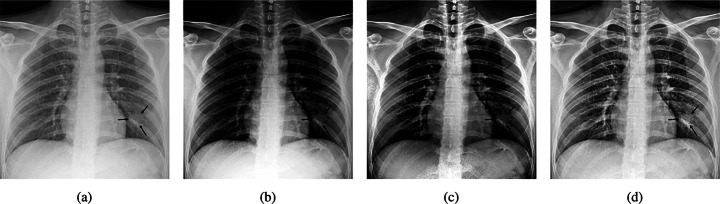
Due to the small number of images in positive class, data augmentation must be applied to prevent overfitting. For the augmentation process, Albumentations9 library is used because of its wide set of augmentation functions, from course dropout to elastic transform and different image distortions. Images are also normalized and downsized to (320,320) to prevent resource exhaustion and reduce memory utilization. There are several image enhancement methods based on histogram equalization that increase image contrast to make non-linearities more distinguishable. Radiologists also use manual contrast improvement to better diagnose mass and nodules. An example of enhancement algorithms applied on a marker-annotated CXR is shown in Fig. 5.
Fig. 5.
Different image enhancement methods. a is the main image, b is the image with histogram equalization (HE), c is adaptive histogram equalization (AHE) applied on the image, and d is the image with contrast limited AHE
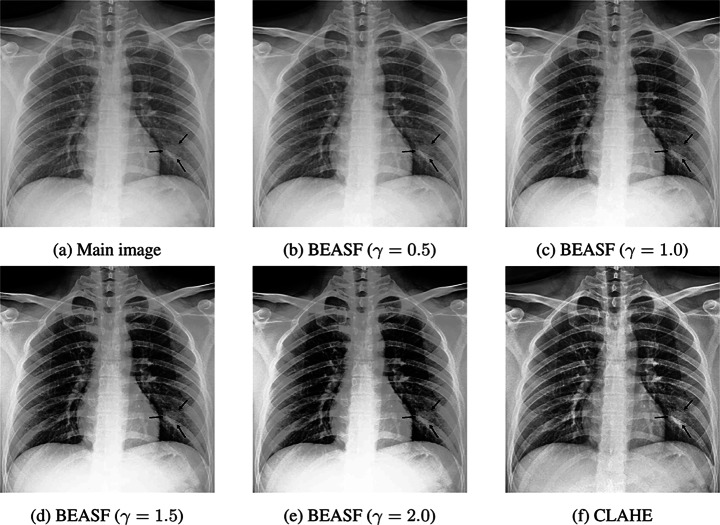
As expected, Contrast Limited Adaptive Histogram Equalization (CLAHE) has better revealed nodular-shaped opacity related to COVID-19 pneumonia. CLAHE is one of the most popular enhancement methods in different image types [34]. Another histogram equalization-based algorithm is Bi-histogram Equalization with Adaptive Sigmoid Function (BEASF) [4]. BEASF adaptively improves image contrast based on the global mean value of the pixels. It has a hyperparameter γ to define the sigmoid function slope. Figure 6 depicts the output of BEASF with different γ values.
Fig. 6.
BEASF with different hyperparameter values compared with original image and CLAHE
Although BEASF did not result in opacity detection improvement in all of the images, it could compliment the CLAHE method. Therefore, a BEASF-enhanced image with a γ = 1.5 is concatenated with CLAHE-enhanced and the main images to be fed into the model.
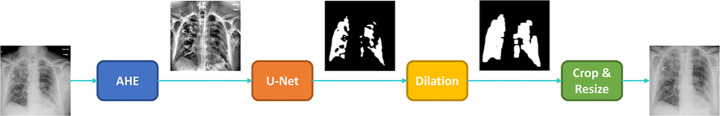
A U-Net based semantic segmentation [41] is also utilized to extract lung pixels from the body and the background. A collection of CXRs with manually segmented lung masks from Shenzhen Hospital Dataset [47] and Montgomery County Dataset [8] are used for training. The diagram of the ROI extraction block is shown in Fig. 7.
Fig. 7.
The segmentation approach based on the U-Net
Using model checkpoints, best weights are used to generate final masks. Afterwards, edge preservation is considered by applying dilation as well as adding margins to the segmented lung ROIs. Lung-segmented CXR is then used as the model input. Lung segmentation is often underestimated in similar research studies based on chest x-rays, while it can drastically improve performance by limiting search area for convolutional layes.
Base model
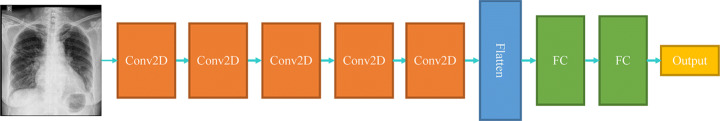
The base model consists of a couple of convolutional layers, followed by a flatten layer and some fully-connected layers. No batch normalization or pooling layers are used for this implementation stage. Figure 8 illustrates the base model architecture. This model serves as a benchmark to compare other networks with the most simple one.
Fig. 8.
A high-level illustration of the base model
Convolution layers have 32 filters, each of which has a kernel size of 3 × 3. The activation function is set as a Rectified Linear Unit (ReLU), which adds non-linearity to images helping the model with better decision making. Fully-connected layers have 10 neurons, and the last layer has one neuron, which demonstrates the probability of the input image belonging to the normal class (p = 0.0) or pneumonia class (p = 1.0).
Pretrained models
Transfer learning is to benefit from a pretrained model in a new classification task. Some pretrained models are trained on millions of images for many epochs and achieved high accuracy on a general task. We experimentally selected DenseNet, ResNet, Xception, and EfficientNet-B7 architectures pretrained on the ImageNet. Pretrained models are used for fine-tuning, i.e. training on target datasets for a small number of epochs, instead of retraining for many epochs. Since ImageNet images and labels are different from the CXR dataset, a pretrained model on the same data type should also be considered.
CheXNet is trained on CXR-14, a large publicly available CXR dataset with 14 different diseases such as pneumonia and edema [37]. CheXNet claims to have a radiologist-level diagnosis accuracy, has better performance than previous related research studies [54, 57], and has simpler architecture than later approaches [38]. CheXNet is based on DenseNet architecture and has been trained on frontal CXRs. It also could be used as a better option for the final model backbone. According to [54], pneumonia is correlated to other thoracic findings as shown in Fig. 9.
Fig. 9.
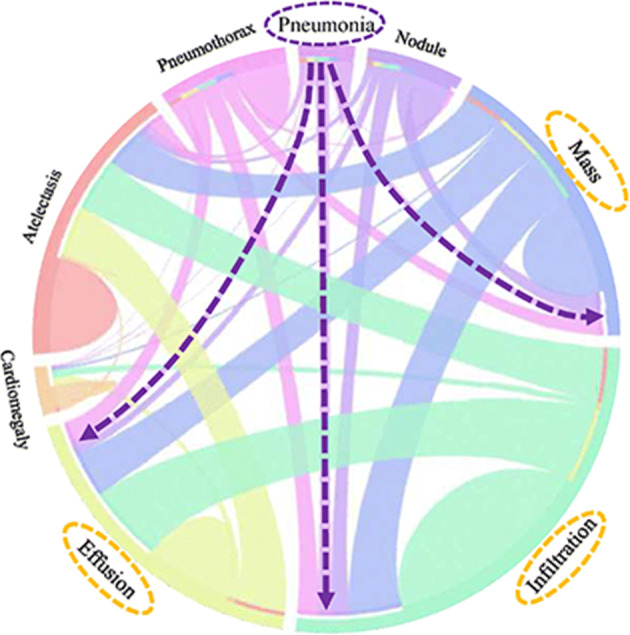
Co-occurrence of different CXR findings as a circular diagram by [54]
Considering these correlations, we can use a combination of CheXNet output neurons to use as the classifier without any fine-tuning. The co-occurrence graph suggests Mass, Effusion, Infiltration, and other labels to look into.
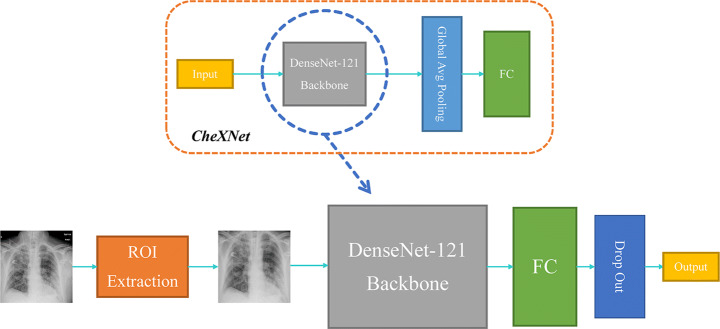
The proposed model: COVID-CXNet
The proposed COVID-CXNet is a CheXNet-based model, fine-tuned on the COVID-19 CXR dataset with 431 layers and ≈ 7M parameters. The architecture of the COVID-CXNet is presented in Fig. 10.
Fig. 10.
COVID-CXNet model architecture based on the DenseNet-121 feature extractor as the backbone
Our proposed network uses the DenseNet-121 as its backbone. There are different architectures utilized to build CheXNet, out of which the DenseNet has shown better capabilities for detecting pulmonary diseases [37]. COVID-CXNet has a fully-connected layer consisting of 10 nodes followed by a dropout layer with a 0.2 dropping rate to prevent overfitting. The activation function of the last layer is changed from SoftMax to Sigmoid because the task is a binary classification. Our proposed model’s advantage over the base model or other custom networks is the training speed as we are fine-tuning a backbone with pretrained weights. In comparison with other CheXNet-based models, COVID-CXNet is benefiting from a lung segmentation module and different image enhancements algorithms within the preprocessing section. Moreover, fine-tuning on a larger dataset along with several overfitting-prevention methods such as dropout layer and label smoothing will help our model outperform in terms of correctly localizing pneumonia in CXRs.
Experimental results
To evaluate the performance, different metrics are considered. Accuracy score is the primary metric used for statistical classification, which is required but inadequate here as we are more interested in efficiently classifying positive samples. Thus, f1-score for the positive class is also measured. The main metric here is visualization heatmaps because the small number of positive samples make the model prone to overfitting by deciding based on the wrong features. The most common manifestations of COVID-19 pneumonia in CXRs are air-space opacities in different forms, such as GGOs or consolidations. Opacities are identified as opaque regions (whiter than usual) in CXRs. They are mostly distinguished as bilateral, involving both lungs, and multifocal opacifications. Rare findings happen in the late stages of disease course, which may include pleural effusion and pneumothorax [22].
Base model
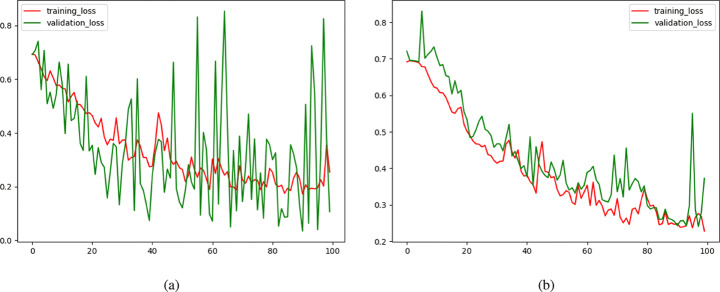
In order to train the base model, the optimizer is set to Adam with the optimal learning rate obtained using exponentially learning rate increasing method [46]. The best learning rate is where the highest decreasing slope happens (≈ 0.0001). Training the base model using 300 samples resulted in an accuracy of 96.72% on the test-set within 100 epochs. As the dataset extended, fluctuations in the curves gradually damped. With a training-set of 480 images, it reaches a reasonable accuracy on the test-set of 120 images. Model loss curves on the training-set and the validation-set (which is test-set here) on different dataset sizes are plotted in Fig. 11.
Fig. 11.
Learning Curves for base model trained on (a) 450 images, and (b) 600 images
The base model hit accuracy of 96.10%, relatively high compared to the number of images in the training-set, and complexity of pneumonia identification in CXRs. To validate the performance, CNN architecture is demystified. A popular technique for CNN visual explanation is Gradient-weighted Class Activation Mapping (Grad-CAM). Grad-CAM concept is to use the gradients of any target concept, flowing into the last convolutional layer to produce a coarse localization map with a concentration on important regions in the image for predicting the class [43]. Another visualization is Local Interpretable Model-Agnostic Explanations (LIME). LIME performs local interpretability on input images, training local surrogate models on image components to find regions with the highest impact on the decision [39]. Grad-CAM outputs of the base model for images of both normal and infected classes are illustrated in Fig. 12.
Fig. 12.
Grad-CAM heatmaps of the base model for 6 images of positive class. Important regions are wrong in most images, while classification scores are notably high
Although classification is successfully implemented with high accuracy score, extracted features are wrong. One possible reason could be the fact that normal CXRs are mostly for pediatrics. To go further about the problem of the model and to prove whether it is because of the normal CXR dataset, the model was evaluated on a small external dataset of 60 images. The confusion matrix in Table 2 shows that the model is not consistent regarding normal cases.
Table 2.
Confusion matrix of the base model on external test-set of 60 images
| Base model | Predicted | ||
|---|---|---|---|
| Normal | COVID-19 | ||
| Actual | Normal | 21 (70%) | 9 (30%) |
| COVID-19 | 3 (10%) | 27 (90%) | |
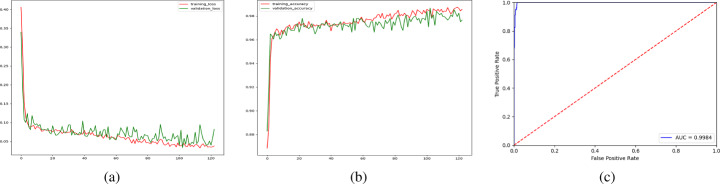
According to the results, recollecting CXRs from adult lungs is essential. The largest dataset containing normal cases is the NIH CXR-14 dataset, with almost 17,000 images. Then, we increased the number of normal CXRs in the dataset to prevent overfitting. The model is trained on a dataset of 3,400 images, 3,000 from normal and 400 from COVID-19 pneumonia classes. It is worth mentioning that classes are weighted in loss function calculation to deal with class imbalance. The results are presented in Fig. 13 and Table 3.
Fig. 13.
Results of the base model trained on 3,400 images; a training loss changes, b training accuracy score changes, and c receiver operating characteristic (ROC) plot
Table 3.
Confusion matrix of the base model on 683 samples
| Base model | Predicted | ||
|---|---|---|---|
| Normal | COVID-19 | ||
| Actual | Normal | 598 (99.17%) | 5 (0.83%) |
| COVID-19 | 4 (5%) | 76 (95%) | |
The base model has achieved a high area under the curve (AUC) of 0.9984 and an accuracy of 98.68% while reaching a reasonable f-score of 0.94. Model visualization shows better performance; however, there are still various wrong regions in image explanations illustrated in Fig. 14.
Fig. 14.
Model interpretability visualization by (a) Grad-CAM and (b) LIME image explanation
In LIME explanation, green super-pixels contribute to current predicted class, and red super-pixels are most contributing to the other class [39]. In Grad-CAM visualization, region importance decreases from red to blue areas. Due to the complexity of pneumonia manifestations in CXRs, robust detection is considered a complicated time-consuming task, even for domain experts. Recognizing complex image patterns requires deeper neural networks to extract higher level image features. As seen in Figs. 14 and 12, shallow network tends to look at simpler features to reach high classification scores, while target features, described in Section 5, are opacities observed inside the lung region with different patterns. Thus, deeper CNNs are needed. Considering the lack of sufficient amount of data, specifically for the COVID-19 class, training CNNs from scratch is likely to fall into local minima. The aforementioned hypothesis is evaluated and discussed in Section 6 with details. Consequently, a deep pretrained neural network is taken into account to overcome the complexity of resolving the problems mentioned above.
Pretrained models
Convolutional layers of the ImageNet pretrained models are kept and dense layers are added to match the number of output neurons with the number of classes for our task. Fine-tuned on the training-set images containing normal pediatric cases is done for 20 epochs. The learning curve of DenseNet-121 is plotted in Fig. 15 as an example.
Fig. 15.
DenseNet-121 fine-tuning curve over 10 epochs
Demonstrating bad results, the model is incapable of learning while fine-tuning only the last fully-connected layer and freezing feature extraction layers and overfits to the data if retrained for more epochs. As expected, ImageNet categories are everyday objects which have somewhat non-similar features as pneumonia imaging patterns in CXRs. Hence, although transfer learning techniques from ImageNet-pretrained models have remarkably improved segmentation accuracy thanks to their capability of handling complex conditions, applying them for classification is still challenging due to the limited size of annotated data and a high chance of overfitting [17]. ResNet-50, Xception, and EfficientNet-B7 have also produced almost the same results. While EfficientNet-B7 demonstrated better results in comparison with former models, it is highly over-parameterized and time-consuming to train for each epoch.
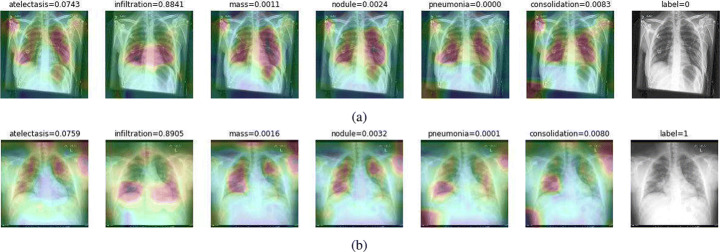
Regarding CheXNet pretrained model, we first probe to see if it is capable of correctly classifying COVID-19 pneumonia with no further improvements. Figure 16 shows results for two sample CXRs from both classes.
Fig. 16.
CheXNet probabilities of different classes for (a) a COVID-19 positive case, and (b) a normal case
Extracted heatmaps reveal that CheXNet correctly marks chest lobes to determine each class probability. The output of each class is slightly higher in positive cases for most of the diseases as well. Some of the drawbacks are extremely high predictions for infiltration in most of the dataset images, getting stuck in regions outside lung boundaries and predominantly in corners, and missing some of the opacities particularly in lower lobes.
COVID-CXNet
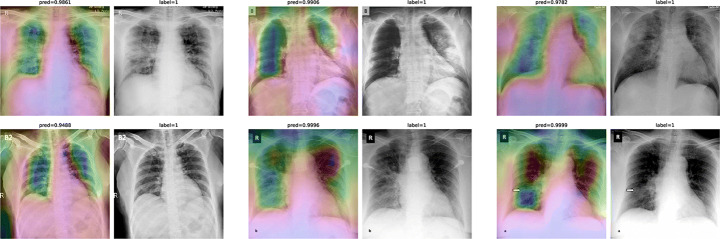
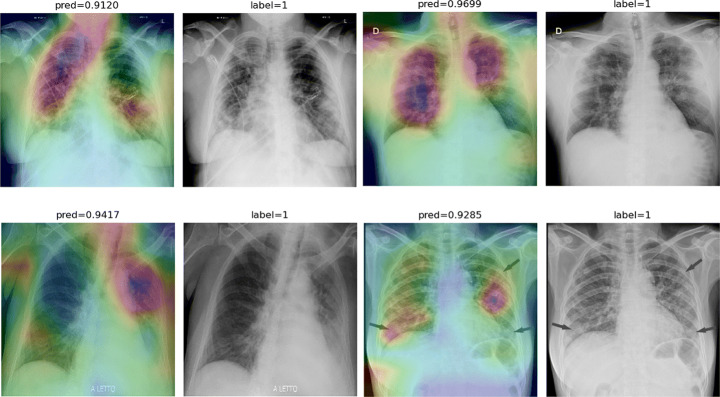
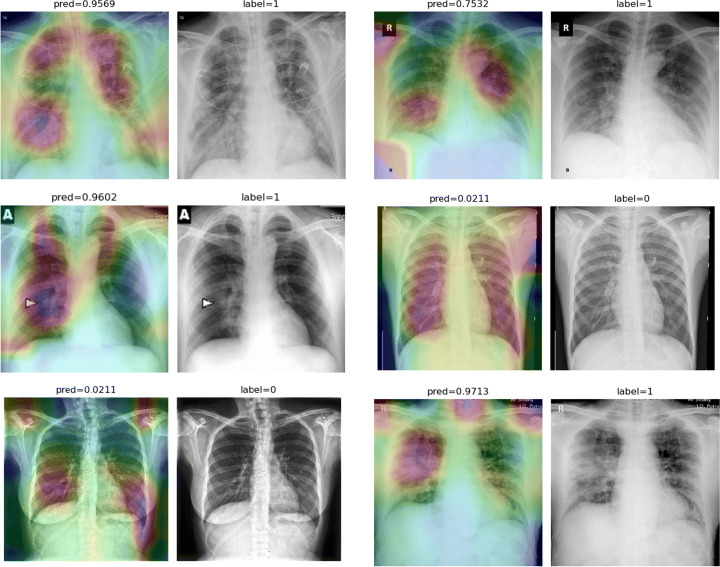
To overcome the aforementioned issues and force the model’s attention to the correct Regions of Interest (ROIs), we introduce the COVID-CXNet. Our model is initialized with the pretrained weights from CheXNet. A dataset of 3,628 images, 3,200 normal CXRs and 428 COVID-19 CXRs, are divided into 80% as training-set and 20% as test-set. Batch size is set to 16, rather than 32 in previous models, regarding memory constraints. Grad-CAMs of the COVID-CXNet for random images are plotted in Fig. 17.
Fig. 17.
Grad-CAM visualization of the proposed model over sample cases
More Grad-CAMs are available in Appendix A. Heatmaps are more accurate than previous models, while an accuracy of 99.04% and an f-score of 0.96 are achieved. Table 4 is the confusion matrix of the proposed model.
Table 4.
Confusion matrix of COVID-CXNet with a test-set of 726 images
| COVID-CXNet | Predicted | ||
|---|---|---|---|
| Normal | COVID-19 | ||
| Actual | Normal | 641 (99.38%) | 4 (0.62%) |
| COVID-19 | 3 (3.7%) | 78 (96.3%) | |
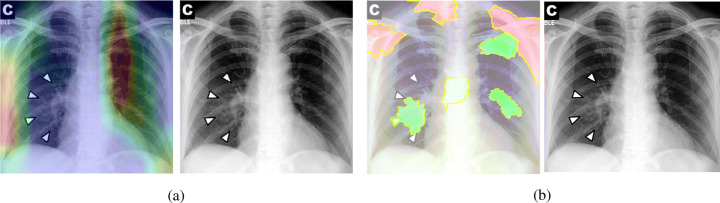
Proposed CheXNet-based model is capable of correctly classifying images. In many cases, it can localize pneumonia findings more precisely than the CheXNet. An example is illustrated in Fig. 18.
Fig. 18.
Comparison between the CheXNet and the proposed model; a is the image with patchy opacities in the upper left zone, b and c are heatmaps of the CheXNet and the proposed COVID-CXNet, respectively
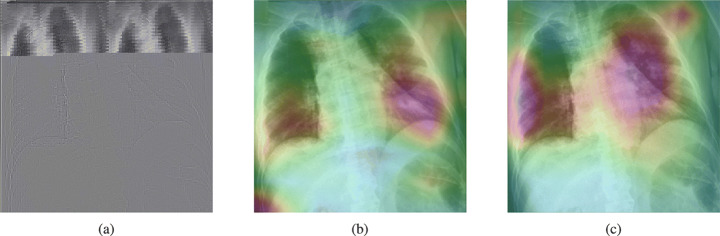
Figure 18 shows a CXR with an infiltrate in the upper lobe of the left hemithorax [33]; while CheXNet missed the region of pneumonia, the proposed model correctly uncovered the infiltration area. One concern about COVID-CXNet results is that it has pointed into other irrelevant regions, even outside the lungs. The same problem happens when there are frequently-appeared texts and signs, such as dates, present in the image. Figure 19 shows how text removal can improve model efficiency.
Fig. 19.
Text removal effect on model results. Images on the right have dates and signs, which are concealed in the images on the left
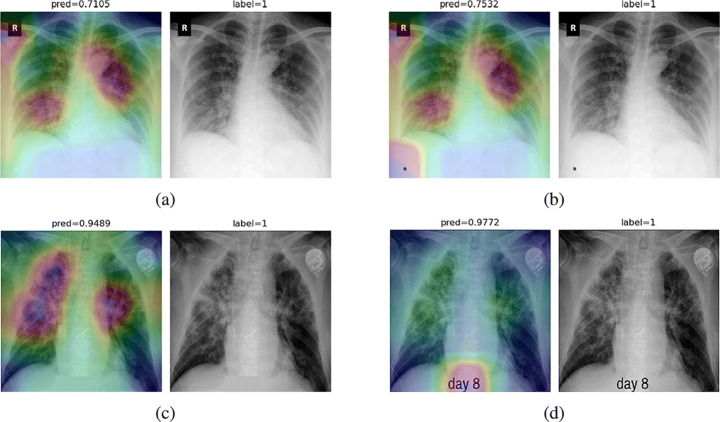
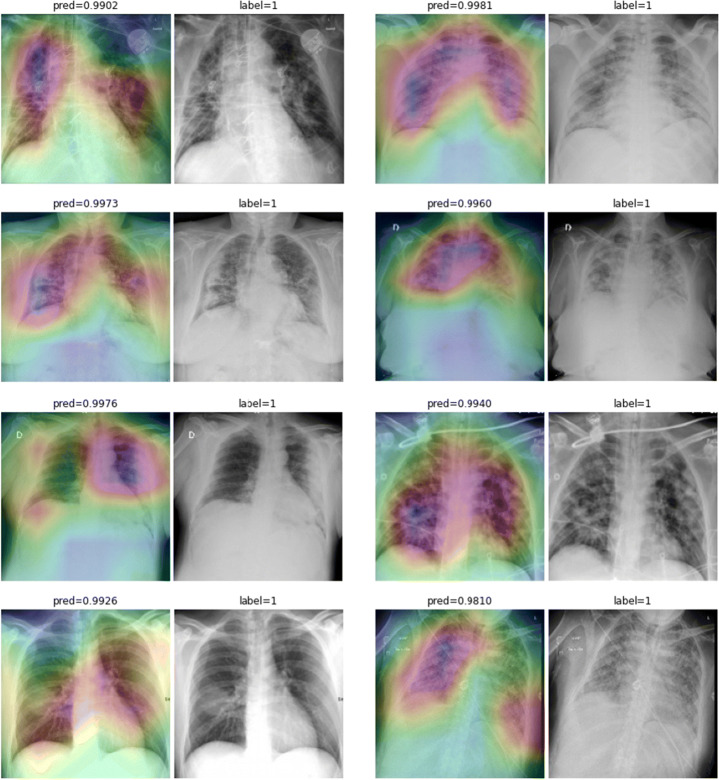
While text removal methods can prevent overfitting, we can simply force the model to look into the lungs in order to address both problems in one effort. To accomplish this task, a U-Net based segmentation illustrated in Fig. 7 is applied to the input images before enhancements. Visualization results for COVID-CXNet with the ROI-segmentation block are shown in Fig. 20.
Fig. 20.
Grad-CAM visualization of the proposed model, trained with lung-segmented CXRs, over sample cases
A figure with more Grad-CAMs is attached in Appendix A. From Fig. 20, it can be observed that COVID-CXNet with ROI-segmentation has delivered superior performance regarding the localization of pneumonia features. Worthwhile to mention that image augmentation is expanded by adding zoom-in, zoom-out, and brightness adjustment. Label smoothing is also applied to the loss function.
The proposed method has shown a negligible drop in metric scores; accuracy is decreased by 0.42%, and f-score is declined by 0.02. This decrease is a result of training with a larger dataset and accurately segmented ROIs, which means it has become more robust against unseen samples. There is a trade-off between catching good features and higher metric scores; while better features result in a more generalized model, high metric scores may indicate overfitting.
As an extra step, we expanded COVID-CXNet for multiclass classification between normal, COVID-19 pneumonia (CP), and non-COVID pneumonia to examine its performance regarding the differentiation between two types of pneumonia. Pneumonia is a common infection that inflames air sacs in one or both lungs, and is typically caused by germs like bacteria or virus. While COVID-19 is a type of virus, its pneumonia manifestations are usually different with other common viruses or bacteria that can cause pneumonia. Typical pneumonia caused by bacteria or viruses is also called Community-Acquired Pneumonia (CAP), which is the most common type. In terms of radiological findings, CP is often appeared with bilateral findings, whereas non-COVID pneumonia or CAP mostly has unilateral consolidations. Since most images are collected from the CXR-14 dataset, a histogram matching is applied to adjust histograms according to a base image. The output layer is changed to have three neurons with the SoftMax activation function. Confusion matrix is shown in Table 5.
Table 5.
Confusion matrix of multiclass COVID-CXNet on 1700 images
| COVID-CXNet | Predicted | |||
|---|---|---|---|---|
| Normal | CAP | CP | ||
| Actual | Normal | 629 (85.58%) | 96 (13.06%) | 10 (1.36%) |
| CAP | 110 (15.71%) | 585 (83.57%) | 5 (0.71%) | |
| CP | 8 (3.02%) | 13 (4.9%) | 244 (92.08%) | |
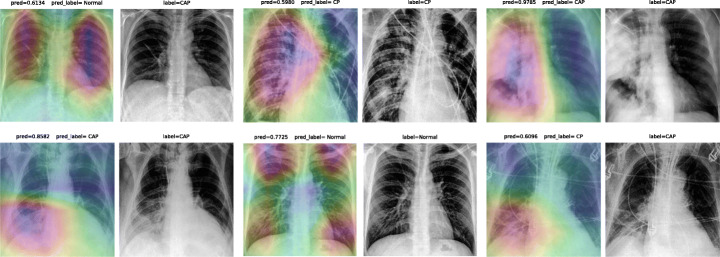
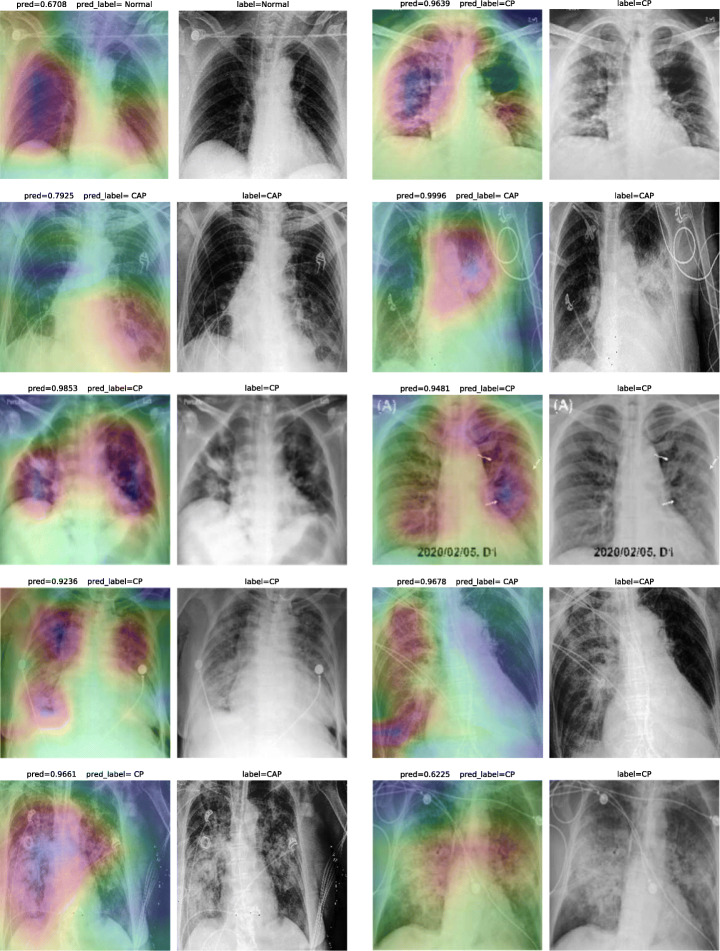
Accuracy score is 85.76%, with f-scores of 0.93 and 0.84 for CP and CAP classes, respectively. In a number of cases, especially in the first stages of virus progression, CP has unilateral findings. Also, CAP may cause bilateral consolidations. Therefore, some cases are expected to be misclassified between CP and CAP. From the confusion matrix, it could be seen that a relatively high number of images are misclassified between CAP and normal. A potential reason for this issue is considered to be related to wrong labeling. Besides, some CAP CXRs are from patients with early-stage disease development. To confirm the model performance, Grad-CAMs are plotted in Fig. 21.
Fig. 21.
COVID-CXNet multiclass classification visualization results
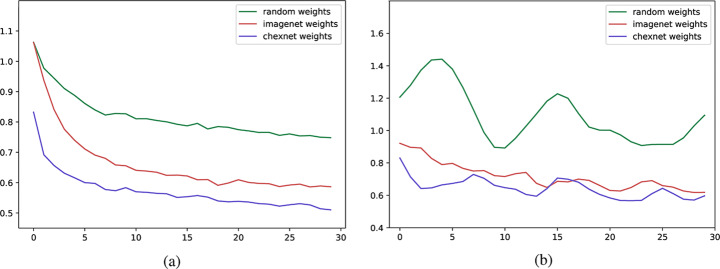
The model is properly looking at one lobe for detecting CAP and both lobes for CAP and normal images. There are some wrong labels, nevertheless. A figure containing more visualizations is found in Appendix A. In order to confirm the superiority of CheXNet weights over ImageNet weights or even using a network with random kernel initialization, several experiments are conducted with same DenseNet-121 model and hyperparameters, and the initial weights as the only difference. Statistically, models with same architecture and different initialization are expected to reach into the same results. However, using better pretrained models for fine-tuning leads to faster training and better generalization. Untrained models are generally prone to getting stuck in a local minimum during the iterative optimization procedure. To evaluate such hypothesis, three different types of weight initialization are compared in terms of learning curve, loss values, and accuracy score improvement throughout 30 epochs. Results are illustrated in Table 6 and Fig. 22.
Table 6.
Comparison of fine-tuning with different initial weights (F-score is provided for CP class only)
| Model architecture | Initial weights | Epochs | Batch Size | Validation Loss (avg) | Accuracy (avg) | F-score (avg) |
|---|---|---|---|---|---|---|
| DenseNet-121 | None | 30 | 16 | 0.792 | 68.00% | 0.76 |
| DenseNet-121 | ImageNet | 30 | 16 | 0.597 | 83.01% | 0.90 |
| DenseNet-121 | CheXNet | 30 | 16 | 0.543 | 85.76% | 0.93 |
Fig. 22.
Comparison of learning curves for (a) training loss and (b) validation loss. Note that curves of the validation loss are smoothed for better intuition
While models with different pretrained weights achieve roughly close results, untrained model is unable to properly generalize for validation data. The average elapsed time for training the models is 68.3 minutes using an NVIDIA Telsa T4 Cloud GPU provided by Google Colab. To further enhance statistical scores, a hierarchical approach is implemented. In the first level, we classify images into normal and pneumonia classes. In the second level, pneumonia images are categorized into CP and CAP. Final confusion matrix is illustrated in Table 7.
Table 7.
Confusion matrix of hierarchical multiclass COVID-CXNet on 1700 images
| COVID-CXNet | Predicted | |||
|---|---|---|---|---|
| Normal | CAP | CP | ||
| Actual | Normal | 643 (87.49%) | 83 (11.29%) | 9 (1.22%) |
| CAP | 101 (14.43%) | 597 (85.28%) | 2 (0.29%) | |
| CP | 6 (2.26%) | 5 (1.89%) | 254 (95.85%) | |
A slight improvement is observed. Overall accuracy is 87.88%, while f-scores are 0.97 for CP class and 0.86 for CAP class. Although visualizations and metrics demonstrate promising performance, the effect of dataset bias is non-negligible for wrong predictions between CAP and normal images. A dataset of 8,500 images containing 1326 CP, 3,500 CAP and 3,674 normal CXRs is used in both multiclass approaches.
Discussion
Throughout this study, several model architectures are introduced and applied to different amounts of images. Bias to pediatric CXRs and lung segmentation module are also addressed in different models. A comparison between these models is shown in Table 8.
Table 8.
Comparison between investigated models throughout this paper (F-score is provided for CP class only)
| Model | Dataset size | Pediatric Bias | Lung Seg. | Epochs | Accuracy (avg) | Confidence Intervals | F-score (avg) |
|---|---|---|---|---|---|---|---|
| Base Model v1 | 600 | Yes | No | 100 | 96.10% | 91.40 – 97.10 | 0.82 |
| Base Model v2 | 3,400 | No | No | 120 | 98.68% | 94.00 – 99.30 | 0.94 |
| COVID-CXNet v1 | 3,628 | No | No | 10 | 99.04% | 96.43 – 99.54 | 0.96 |
| COVID-CXNet v2 | 3,628 | No | Yes | 10 | 98.62% | 97.14 – 99.28 | 0.94 |
| Flat Multiclass COVID-CXNet | 8,500 | No | Yes | 30 | 85.76% | 83.74 – 87.82 | 0.93 |
| Hierarchical Multiclass COVID-CXNet | 8,500 | No | Yes | 10 + 20 | 87.88% | 86.25 – 88.98 | 0.97 |
Accuracy score ranges are achieved by running models ten different times. With the expansion of the dataset, confidence intervals shrink, and metric scores slightly decline while pneumonia symptoms localization improves. Furthermore, the proposed model is compared to other research studies discussed in Section 2 regarding several criteria, such as dataset size and f-score. The comparison is illustrated in Table 9.
Table 9.
Comparison between proposed model and related works
| Model | Num CP images | Pretrained weights | Architecture | CP class F-score | Lung segmentation | Visualization mapsα |
|---|---|---|---|---|---|---|
| Zhang et al. [59] | 100 | Yes, ImageNet | ResNet | 0.72 | No | Partially Correct |
| Khan et al. [28] | 179 | Yes, ImageNet | Xception | 0.9560 | No | Not Plotted |
| Rajaraman et al. [36] | 313 | Yes, ImageNet | Ensemble | 0.9841 | Yes, U-Net | Mostly Correct |
| Wang et al. [53] | 358 | No | COVID-Net (Custom) | 0.9480 | No | Not Plotted |
| Mangal et al. [31] | 358 | Yes, CheXNet | DenseNet | 0.9230 | No | Mostly Correct |
| Tang et al. [50] | 573 | Yes, COVID-Net | Ensemble | 0.95 | No | Not Plotted |
| Kedia et al. [26] | 798β | Yes, ImageNet | Ensemble | 0.9891 | No | Wrong |
| Proposed Model | 1326 | Yes, CheXNet | DenseNet | 0.9658 | Yes, U-Net | Mostly Correct |
α Visualization maps are confirmed by a radiologist, based on paper images, regardless of the number of provided samples. β Datasets used in this study partially overlap with each other.
While other models have higher f-scores, they have different issues. For example, CoroNet [28] has a small dataset in which most images are from pediatrics. The pruned ensemble method [36] also suffers from over-parameterization and a lack of proper visualization discussion in the paper. Considering the significant number of parameters of the model and their small dataset, Grad-CAM visualizations must be investigated to a certain extent. Our proposed model is the only study to use a lung segmentation module with a CheXNet-based fine-tuning on a relatively large dataset of CP CXRs. Besides, our model benefits from different approaches, such as label smoothing and hierarchical strategy, to enhance its performance and prevent overfitting.
Dataset
Data: While DL-based methods have demonstrated promising results in medical image classification, a significant challenge is data shortage. Since COVID-19 diagnosis using CXR images is recently becoming an interesting topic, accessible data is limited. Data augmentation is an essential method of coping with data shortcomings. However, designing a pneumonia detection model still needs much more CXR data. Most COVID-19 pneumonia detection articles have very small datasets. Although [53] has claimed to introduce the largest open-access dataset of 385 CXRs, to the best of our knowledge, our dataset of 1326 COVID-19 CXRs has the most number of images.
Pediatric Bias: Many research papers have benefited from databases built upon the proposed dataset by [27]. Children have different pulmonary anatomy. Hence, developed models based on normal pediatric and adult pneumonia images are highly vulnerable to the “right decision with wrong reason” problem. Besides, previous studies have proved that using various datasets containing images from different hospitals improves the pneumonia-detection results [51]. To prevent pediatric bias, we not only collected normal CXRs from different sources, but also meticulously filtered images of [27]. Furthermore, COVID-19 pneumonia CXRs were collected from 9 different sources to improve cross-dataset robustness.
In the future, other information regarding patient status can be used alongside x-rays. Clinical symptoms can remarkably help radiologists in COVID-19 differential diagnosis. Metadata could be concatenated with the input CXR and be fed into the model to help increase its decision certainty. Providing the metadata, it is possible to have more detailed predictions, e.g. the chance of patient survival, based on clinical symptoms and the severity of pneumonia features presented by CXR.
Architecture
Metrics: In Section 4.2 we introduced a simple base model. The purpose was to show how it can achieve very high accuracy scores. Digging into explainability, the model revealed wrong features responsible for its excellent metric scores. Therefore, high accuracy scores of sophisticated models from a small number of CXRs, which have high texture complexity, are tricky. Investigation of model performance based upon confusion matrices and accuracy scores could not be usually validated unless demonstrating appropriate localization of imaging features.
Transfer Learning: Using pretrained models with ImageNet weights, some studies such as [36] showed acceptable heatmaps, but only a few images were visualized. While these pretrained models may help, having small dataset sizes suggested us to fine-tune models previously trained on similar data. Our CheXNet-based model shows better performance over ImageNet-pretrained models, while not hindered by problems like overparameterization. Besides, lung segmentation was also performed by a U-Net based architecture previously trained on similar frontal CXRs. Among studies conducted, there was only one article to use CheXNet as its backbone [31], which applied the model on a fewer number of images and without lung segmentation as its image preprocessing procedure.
CheXNet: CheXNet is trained on a very large dataset of CXRs and has been used for transfer learning by some other thoracic disease identification studies [3, 48]. However, it has its own deficiencies, such as individual sample variability as a result of data ordering changes [58] and vulnerability to adversarial attacks [14]. Enhancing thoracic abnormality detection in CXRs using CheXNet requires the development of ensemble models, which is currently prone to overfitting due to the number of images from COVID-19 positive patients.
Conclusion
In this paper, we firstly collected a dataset of CXR images from normal lungs and COVID-19 infected patients. The constructed dataset is made from images of different datasets from multiple hospitals and radiologists and is the one of the largest public datasets to the best of our knowledge. Next, we designed and trained an individual CNN and also investigated the results of ImageNet-pretrained models. Then, a DenseNet-based model is designed and fine-tuned with weights initially set from the CheXNet model. Comparing model visualization over a batch of samples as well as accuracy scores, we denoted the significance of Grad-CAM heatmaps and its priority to be considered as the primary model validation metric. Finally, we discussed several points like data shortage and the importance of transfer learning for tackling similar tasks. In three-class classification, COVID-CXNet achieved final f-scores of 0.8621 and 0.9658 for community-acquired pneumonia and COVID pneumonia classes, respectively, while the total accuracy was 87.88%. The proposed model development procedure is visualization-oriented as it is the best method to confirm its generalization as a medical decision support system.
Appendix:
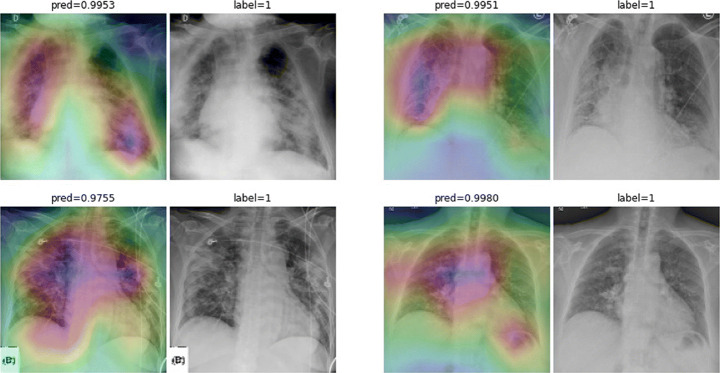
A More Grad-CAMs of the COVID-CXNet Model
Fig. 23.
Grad-CAMs from COVID-CXNet
B More Grad-CAMs of the COVID-CXNet Model with Lung Segmentation Preprocessing
Fig. 24.
Grad-CAMs from COVID-CXNet with lung segmentation module
C More Grad-CAMs of the Multiclass COVID-CXNet from Different Classes
Fig. 25.
Grad-CAMs from multiclass COVID-CXNet
Data and Code Availability
A dataset of COVID-19 positive CXR images, used in this study, as well as source codes and the pretrained network weights are hosted on a public repository on GitHub to help accelerate further research studies. https://github.com/armiro/COVID-CXNet
Footnotes
COVID-19 Chest X-ray Image Data Collection. Available on: https://github.com/armiro/COVID-CXNet
This manuscript is the updated version of our arXiv preprint (available here: https://arxiv.org/abs/2006.13807) which has been cited 34 times as of Sep 15, 2021 since June 16, 2020.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Arman Haghanifar, Email: arman.haghanifar@usask.ca.
Mahdiyar Molahasani Majdabadi, Email: m.molahasani@usask.ca.
Younhee Choi, Email: younhee.choi@irdinc.com.
S. Deivalakshmi, Email: deiva@nitt.edu
Seokbum Ko, Email: seokbum.ko@usask.ca.
References
- 1.Ai T et al (2020) Correlation of chest ct and rt-pcr testing in coronavirus disease 2019 (covid-19) in china: a report of 1014 cases. Radiology:200642 [DOI] [PMC free article] [PubMed]
- 2.Al-Karawi D, Al-Zaidi S, Polus N, Jassim S (2020) Ai based chest x-ray (cxr) scan texture analysis algorithm for digital test of covid-19 patients. medRxiv
- 3.Almuhayar M, Lu HH-S, Iriawan N (2019) Classification of abnormality in chest x-ray images by transfer learning of chexnet. In: 2019 3Rd international conference on informatics and computational sciences (ICICos). IEEE, pp 1–6
- 4.Arriaga-Garcia EF, Sanchez-Yanez RE, Garcia-Hernandez M (2014) Image enhancement using bi-histogram equalization with adaptive sigmoid functions. In: 2014 International conference on electronics, communications and computers (CONIELECOMP). IEEE, pp 28–34
- 5.Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran TML, Pan I, Shi L-B, Wang D-C, Mei J et al (2020) Performance of radiologists in differentiating covid-19 from viral pneumonia on chest ct. Radiology:200823 [DOI] [PMC free article] [PubMed]
- 6.Barstugan M, Ozkaya U, Ozturk S (2020) Coronavirus (covid-19) classification using ct images by machine learning methods, arXiv:2003.09424
- 7.Brueck H (2020) There is only one way to know if you have the coronavirus, and it involves machines full of spit and mucus. [Online]. Available: https://www.businessinsider.com/how-to-know-if-you-have-the-coronavirus-pcr-test-2020-1https://www.businessinsider.com/how-to-know-if-you-have-the-coronavirus-pcr-test-2020-1
- 8.Candemir S, Jaeger S, Palaniappan K, Musco JP, Singh RK, Xue Z, Karargyris A, Antani S, Thoma G, McDonald CJ. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans Med Imaging. 2013;33(2):577–590. doi: 10.1109/TMI.2013.2290491. [DOI] [PubMed] [Google Scholar]
- 9.Castro-Zunti R, Park EH, Choi Y, Jin GY, Ko S-B (2020) Early detection of ankylosing spondylitis using texture features and statistical machine learning, and deep learning, with some patient age analysis. Comput Med Imaging Graph 82:101718 [DOI] [PubMed]
- 10.Cellina M, Orsi M, Toluian T, Pittino C, Oliva G (2020) False negative chest x-rays in patients affected by covid-19 pneumonia and corresponding chest ct findings. Radiography [DOI] [PMC free article] [PubMed]
- 11.Cohen JP, Morrison P, Dao L (2020) Covid-19 image data collection, arXiv:2003.11597. [Online] Available: https://github.com/ieee8023/covid-chestxray-dataset
- 12.Cozzi A, Schiaffino S, Arpaia F, Della Pepa G, Tritella S, Bertolotti P, Menicagli L, Monaco CG, Carbonaro LA, Spairani R et al (2020) Chest x-ray in the covid-19 pandemic: Radiologists’ real-world reader performance. Eur J Radiol 132:109272 [DOI] [PMC free article] [PubMed]
- 13.Dey N, Rajinikanth V, Fong SJ, Kaiser MS, Mahmud M. Social group optimization–assisted kapur’s entropy and morphological segmentation for automated detection of covid-19 infection from computed tomography images. Cogn Comput. 2020;12(5):1011–1023. doi: 10.1007/s12559-020-09751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finlayson SG, Chung HW, Kohane IS, Beam AL (2018) Adversarial attacks against medical deep learning systems, arXiv:1804.05296
- 15.Furlow B. Radiation dose in computed tomography. Radiol Technol. 2010;81(5):437–450. [PubMed] [Google Scholar]
- 16.Geffen D (2020) Covid-19 chest x-ray guideline. [Online] Available: https://www.uclahealth.org/radiology/covid-19-chest-x-ray-guideline
- 17.Godasu R, Zeng D, Sutrave K (2020) Transfer learning in medical image classification: Challenges and opportunities. In: Proceedings of the 15th Annual Conference of the Midwest Association for Information Systems, pp 28–2020
- 18.Gomes JC, Barbosa V A d F, Santana MA, Bandeira J, Valenċa MJS, de Souza RE, Ismael AM, dos Santos WP (2020) Ikonos: an intelligent tool to support diagnosis of covid-19 by texture analysis of x-ray images. Res Biomed Eng:1–14
- 19.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, Bernheim A, Siegel E (2020) Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis, arXiv:2003.05037
- 20.Hassanien AE, Mahdy LN, Ezzat KA, Elmousalami HH, Ella HA (2020) Automatic x-ray covid-19 lung image classification system based on multi-level thresholding and support vector machine. medRxiv
- 21.Ittyachen AM, Vijayan A, Isac M. The forgotten view: Chest x-ray-lateral view. Respiratory Med Case Rep. 2017;22:257–259. doi: 10.1016/j.rmcr.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobi A, Chung M, Bernheim A, Eber C (2020) Portable chest x-ray in coronavirus disease-19 (covid-19): A pictorial review. Clinical Imaging [DOI] [PMC free article] [PubMed]
- 23.Jaeger S, Candemir S, Antani S, Wáng Y-XJ, Lu P-X, Thoma G. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg. 2014;4(6):475. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin YH, et al. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-ncov) infected pneumonia (standard version) Military Med Res. 2020;7(1):4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH (2020) Essentials for radiologists on covid-19: an update—radiology scientific expert panel [DOI] [PMC free article] [PubMed]
- 26.Kedia P, Katarya R et al (2021) Covnet-19: A deep learning model for the detection and analysis of covid-19 patients. Appl Soft Comput 104:107184 [DOI] [PMC free article] [PubMed]
- 27.Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Khan AI, Shah JL, Bhat MM (2020) Coronet: A deep neural network for detection and diagnosis of covid-19 from chest x-ray images. Comput Methods Programs Biomed:105581 [DOI] [PMC free article] [PubMed]
- 29.Kim H, Hong H, Yoon SH. Diagnostic performance of ct and reverse transcriptase polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology. 2020;296(3):E145–E155. doi: 10.1148/radiol.2020201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Li C, Zhu D (2020) Covid-mobilexpert: On-device covid-19 screening using snapshots of chest x-ray
- 31.Mangal A, Kalia S, Rajgopal H, Rangarajan K, Namboodiri V, Banerjee S, Arora C (2020) Covidaid: Covid-19 detection using chest x-ray, arXiv:2004.09803
- 32.Pereira RM, Bertolini D, Teixeira LO, Silla Jr CN, Costa YM (2020) Covid-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed:105532 [DOI] [PMC free article] [PubMed]
- 33.Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen HT, Le HQ, Nguyen TT, Cao TM, Pham QD (2020) Importation and human-to-human transmission of a novel coronavirus in vietnam. N Engl J Med 382(9):872–874 [DOI] [PMC free article] [PubMed]
- 34.Pizer SM, Amburn EP, Austin JD, Cromartie R, Geselowitz A, Greer T, ter Haar Romeny B, Zimmerman JB, Zuiderveld K. Adaptive histogram equalization and its variations. Comput Vis Graph Image Process. 1987;39(3):355–368. doi: 10.1016/S0734-189X(87)80186-X. [DOI] [Google Scholar]
- 35.Qian L, Yu J, Shi H (2020) Severe acute respiratory disease in a huanan seafood market worker: Images of an early casualty. Radiol Cardiothoracic Imaging 2(1):e200033 [DOI] [PMC free article] [PubMed]
- 36.Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR, Antani S. Iteratively pruned deep learning ensembles for covid-19 detection in chest x-rays. IEEE Access. 2020;8:115 041–115 050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K et al (2017) Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning, arXiv:1711.05225
- 38.Ranjan E, Paul S, Kapoor S, Kar A, Sethuraman R, Sheet D (2018) Jointly learning convolutional representations to compress radiological images and classify thoracic diseases in the compressed domain. In: 11th indian conference on computer vision, graphics and image processing (ICVGIP)
- 39.Ribeiro MT, Singh S, Guestrin C (2016) “why should i trust you?” explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp 1135–1144
- 40.Rio DC. Reverse transcription-polymerase chain reaction. Cold Spring Harbor Protocols. 2014;2014(11):pdb–prot080, 887. doi: 10.1101/pdb.prot080887. [DOI] [PubMed] [Google Scholar]
- 41.Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
- 42.Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, Schluger NW, Volpi A, Yim J. -J., Martin IB et al (2020) The role of chest imaging in patient management during the covid-19 pandemic: a multinational consensus statement from the fleischner society. CHEST [DOI] [PMC free article] [PubMed]
- 43.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp 618–626
- 44.Serrano CO, Alonso E, Andrés M, Buitrago N, Vigara AP, Pajares MP, López EC, Moll GG, Espin IM, Barriocanal MB et al (2020) Pediatric chest x-ray in covid-19 infection. Eur J Radiol 131:109236 [DOI] [PMC free article] [PubMed]
- 45.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shen D, Shi Y (2020) Lung infection quantification of covid-19 in ct images with deep learning, arXiv:2003.04655
- 46.Smith LN (2017) Cyclical learning rates for training neural networks. In: 2017 IEEE Winter conference on applications of computer vision (WACV). IEEE, pp 464–472
- 47.Stirenko S, Kochura Y, Alienin O, Rokovyi O, Gordienko Y, Gang P, Zeng W (2018) Chest x-ray analysis of tuberculosis by deep learning with segmentation and augmentation. In: 2018 IEEE 38Th international conference on electronics and nanotechnology (ELNANO). IEEE, pp 422–428
- 48.Sze-To A, Wang Z (2019) Tchexnet Detecting pneumothorax on chest x-ray images using deep transfer learning. In: International conference on image analysis and recognition. Springer, pp 325–332
- 49.Tabik S, Gómez-ríos A, Martín-rodríguez JL, Sevillano-garcía I, Rey-area M, Charte D, Guirado E, Suárez JL, Luengo J, Valero-González M, et al. Covidgr dataset and covid-sdnet methodology for predicting covid-19 based on chest x-ray images. IEEE J Biomed Health Inf. 2020;24(12):3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang S, Wang C, Nie J, Kumar N, Zhang Y, Xiong Z, Barnawi A (2021) Edl-covid: Ensemble deep learning for covid-19 cases detection from chest x-ray images. IEEE Transactions on Industrial Informatics [DOI] [PMC free article] [PubMed]
- 51.Tilve A, Nayak S, Vernekar S, Turi D, Shetgaonkar PR, Aswale S (2020) Pneumonia detection using deep learning approaches. In: 2020 International conference on emerging trends in information technology and engineering (ic-ETITE). IEEE, pp 1–8
- 52.WHO (2020) Coronavirus disease 2019 (covid-19): situation report, 51. [Online]. Available: https://apps.who.int/iris/bitstream/handle/10665/331475/nCoVsitrep11Mar2020-eng.pdf
- 53.Wang L, Lin ZQ, Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 2097–2106
- 55.Wang Y, Zhang H, Chae KJ, Choi Y, Jin GY, Ko S-B. Novel convolutional neural network architecture for improved pulmonary nodule classification on computed tomography. Multidim Syst Sign Process. 2020;31(3):1163–1183. doi: 10.1007/s11045-020-00703-6. [DOI] [Google Scholar]
- 56.Winther HB, Laser H, Gerbel S, Maschke SK, Hinrichs JB, Vogel-Claussen J, Wacker FK, Höper MM, Meyer BC (2020) Covid-19 image repository. [Online] Available: https://github.com/ml-workgroup/covid-19-image-repository
- 57.Yao L, Poblenz E, Dagunts D, Covington B, Bernard D, Lyman K (2017) Learning to diagnose from scratch by exploiting dependencies among labels, arXiv:1710.10501
- 58.Zech JR, Forde JZ, Littman ML (2019) Individual predictions matter: Assessing the effect of data ordering in training fine-tuned cnns for medical imaging, arXiv:1912.03606
- 59.Zhang J, Xie Y, Li Y, Shen C, Xia Y (2020) Covid-19 screening on chest x-ray images using deep learning based anomaly detection, arXiv:2003.12338
- 60.Zompatori M, Ciccarese F, Fasano L (2014) Overview of current lung imaging in acute respiratory distress syndrome [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A dataset of COVID-19 positive CXR images, used in this study, as well as source codes and the pretrained network weights are hosted on a public repository on GitHub to help accelerate further research studies. https://github.com/armiro/COVID-CXNet