Abstract
FUN14 domain-containing 1 (FUNDC1) is a receptor that has been previously reported to activate hypoxia-induced mitophagy. However, the potential role of FUNDC1 in the pathophysiology of dental pulp diseases remains unknown. Therefore, present study first collected tissue specimens from patients with pulpitis and from healthy individuals. The results of reverse transcription-quantitative PCR and immunohistochemical staining revealed markedly increased FUNDC1 and hypoxia-inducible factor-1α expression in pulpitis tissue specimens compared with those from healthy individuals. To provide a theoretical basis for the study of the occurrence, development and reparative mechanisms in the dental pulp after tissue injury, the present study then investigated the role of hypoxia-induced mitophagy in the regulation of proliferation, migration and odontoblastic differentiation in human dental pulp cells (HDPCs), in addition, to the possible involvement of FUNDC1. The surface markers and multipotent differentiation capabilities of HDPCs were performed by flow cytometry (surface markers), alizarin red (osteogenic capabilities), alcian blue (chondrogenic capabilities) and oil red O (adipogenic capabilities). Following culture under hypoxia conditions (1% O2) for varying time periods, the proliferation, migration and odontoblastic differentiation of HDPCs were measured using Cell Counting Kit-8, wound healing and Transwell migration assays, alkaline phosphatase staining and activity tests and western blotting (runt-related transcription factor 2, collagen I, osterix and osteopontin), respectively. Immunofluorescence and western blotting were performed to measure the expression levels of hypoxia-inducible factor-1α, pro-fission dynamin-related protein 1, mitochondria-related proteins translocase of inner mitochondrial membrane 23 and translocase of outer mitochondrial membrane 20, in addition to those of autophagy markers (p62, LC3II, Beclin-1 and autophagy-related 5). Transmission electron microscopy was also used to image the autophagosomes and mitochondrial morphology. In addition, to study the functional role of FUNDC1, its expression was silenced by liposome-mediated transfection with small interfering RNA into HDPCs. Compared with those in HDPCs cultured under normoxic conditions (21% O2), the ability of autophagy in HDPCs cultured under hypoxic conditions for 18 h was markedly increased, whilst the proliferation, migration and odontoblastic differentiation were also enhanced. Increased numbers of autophagosomes could also be observed in the hypoxic group. However, FUNDC1 knockdown in HDPCs reversed the aforementioned effects. Overall, data from the present study suggest that hypoxia can promote the proliferation, migration and odontoblastic differentiation of HDPCs, where the underlying mechanism may be associated with the activation of mitophagy downstream of FUNDC1.
Keywords: human dental pulp cells, hypoxia, autophagy, mitophagy, FUN14 domain-containing 1
Introduction
Autophagy is a self-digesting and auto-metabolic process that serves to stabilize intracellular metabolism under stressful conditions, such as hypoxia, infection and nutrient deficiency (1). A number of reports have suggested that autophagy is involved in a variety of physiological and pathophysiological processes in the dental pulp tissue, including regeneration (2), odontogenic differentiation (3,4), cellular aging (5) and stress adaptation (6-8). In particular, a previous study has demonstrated that lipopolysaccharide can induce autophagy in human dental pulp cells (HDPCs) (9).
Autophagy has been reported to be a selective process under certain circumstances, such that mitochondrial autophagy (mitophagy) is considered to be a specialized form of autophagy, which maintains mitochondrial quality and homeostasis (10). Mitophagy is activated by two pathways, namely the ubiquitin-dependent and receptor-dependent pathways (11). FUN14 domain-containing 1 (FUNDC1) is a receptor that has been previously found to activate hypoxia-induced mitophagy (12). FUNDC1 is an integral molecule localized to the outer mitochondrial membrane that can interact with the LC3 protein to activate mitophagy, by altering its phosphorylation states (12). Accumulating evidence has revealed that FUNDC1-related mitophagy is closely associated with the initiation, development and prognosis of various diseases, such as cardiovascular diseases (13), cancer (14) and sepsis (15). These findings suggest that mitophagy and FUNDC1 can serve as a potential prognostic marker or as a promising therapeutic target. Previously, FUNDC1 has also been demonstrated to promote the proliferation, migration and differentiation of breast cancer cells (16). However, to the best of our knowledge, the role of FUNDC1 in dental pulp diseases has not been reported previously.
Dental pulp tissue forms the soft component of the tooth that mainly consists of dental pulp cells and odontoblasts (17). HDPCs include unique mesenchymal stem cells with pluripotent potential (18). In response to injury or infection, HDPCs can differentiate into odontoblasts and migrate to the site of damage, where they mediate injury repair (19). Surrounded by the dentin and nutritionally supported by the blood vessels through the apical foramen, the dental pulp tissue is particularly susceptible to hypoxia after the occurrence of inflammation (20). Therefore, it was hypothesized that autophagy, mitophagy, proliferation, migration and odontogenic differentiation in HDPCs may all be influenced by hypoxia, a process in which FUNDC1 may also serve a potential role.
Given that injury repair and protection are of significance when the dental pulp is damaged, the present study aimed to explore the relationship between FUNDC1-related mitophagy and the regeneration of HDPCs under hypoxic conditions.
Materials and methods
Specimen collection
All samples were obtained from The Hospital of Stomatology, Guanghua School of Stomatology, Sun Yat-sen University (Guangzhou, China) between September 2019 and March 2020. Written informed consent was received from all patients, parent or guardian of the patients involved. The present study was approved by the Ethics Committee of Hospital of Stomatology and Guanghua School of Stomatology, affiliated to Sun Yat-sen University [protocol no. ERC-(2017)-27; Guangzhou, China].
Healthy human premolars or third molars were obtained from 15 orthodontic patients aged 13-29 years of either sex (sex, 8 males, 7 females; mean age, 23.9±4.3 years) from orthodontic extraction. By contrast, inflamed teeth were collected from 15 patients (sex, 8 males, 7 females; age range, 13-29 years; mean age, 23.1±4.4 years) who were diagnosed with irreversible pulpitis. The inclusion criteria were mature permanent teeth with history of spontaneous pain or severe, prolonged pain to thermal stimulation. The exclusion criteria were any teeth with periodontal diseases or radiographic evidence of internal or external resorption, inter-radicular bone loss or periapical pathology. The pulps of the healthy and inflamed groups were obtained by splitting the teeth longitudinally. In total, five tissues from each group were randomly selected for immunohistochemical staining and fixed at 25°C with 4% (w/v) paraformaldehyde (Sigma-Aldrich; Merck KGaA) for 24 h. Tissues were then embedded in paraffin and 5-µm-thick serial sections were prepared. Samples destined for reverse transcription-quantitative PCR (RT-qPCR) were preserved at −80°C.
RT-qPCR
The extraction and purification of total RNA from pulp tissues were performed according to the manufacturer's protocols of the RNA-Quick Purification kit (Shanghai Yishan Biotechnology Co., Ltd.). A NanoDrop® 2000 instrument (Thermo Fisher Scientific, Inc.) was used to measure the concentration and quality of the RNA samples. RNA (2 µg) was reverse transcribed into cDNA using an Evo M-MLV RT Premix kit (cat. no. AG11706; Hunan Aikerui Biological Engineering Co., Ltd.) by applying the following three stages: 37°C for 15 min, 85°C for 5 sec and then cooling at 4°C. Subsequently, qPCR was performed using the SYBR® Green Premix Pro Taq HS qPCR kit (cat. no. AG11701; Hunan Aikerui Biological Engineering Co., Ltd.) in a LightCycler 480 system (Roche Diagnostics), using a 20-µl reaction system. The PCR primers were synthesized by Sangon Biotech Co., Ltd. (Table I). The thermocycling conditions were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec and then 72°C for 20 sec; melting at 95°C for 5 sec, 65°C for 60 sec and 97°C for 1 sec and cooling at 40°C for 10 sec. The expression levels of each gene were normalized to β-actin using the 2−ΔΔCq method (21).
Table I.
Primer sequences.
| Gene | Primer sequence, 5′-3′ |
|---|---|
| IL-1β | Forward: GCCAGTGAAATGATGGCTTATT |
| Reverse: AGGAGCACTTCATCTGTTTAGG | |
| IL-6 | Forward: CACTGGTCTTTTGGAGTTTGAG |
| Reverse: GGACTTTTGTACTCATCTGCAC | |
| IL-8 | Forward: AACTGAGAGTGATTGAGAGTGG |
| Reverse: ATGAATTCTCAGCCCTCTTCAA | |
| TNF-α | Forward: TGGCGTGGAGCTGAGAGATAACC |
| Reverse: CGATGCGGCTGATGGTGTGG | |
| HIF-1α | Forward: ACGTTCCTTCGATCAGTTGTCACC |
| Reverse: GGCAGTGGTAGTGGTGGCATTAG | |
| FUNDC1 | Forward: GCAGTAGGTGGTGGCTTTC |
| Reverse: TGCTTTGTTCGCTCGTTT | |
| β-actin | Forward: GCATGGGTCAGAAGGATTCCT |
| Reverse: TCGTCCCAGTTGGTGACGAT |
HIF-1α, hypoxia-inducible factor-1α; FUNDC1, FUN14 domain containing 1.
Immunohistochemical staining
Immunohistochemical staining was performed to evaluate the expression levels of hypoxia-inducible factor-1α (HIF-1α) and FUNDC1 in the healthy and inflamed groups as described previously (22). The sections were dewaxed and rehydrated with xylene and ethanol, then had their antigens retrieved in citrate buffer for 15 min in a microwave oven at 98°C. According to the manufacturer's protocol for the SP Rabbit & Mouse HRP kit (DAB; cat. no. CW2069M; CoWin Biosciences), endogenous peroxidase was blocked by using hydrogen peroxide (Solution A) for 10 min at room temperature. The sections were then blocked at room temperature for 30 min in goat serum (Solution B; contained in the kit) and incubated overnight at 4°C with the primary antibodies. The primary antibodies used were: Anti-HIF-1α (1:150 dilution; cat. no. AF1009; Affinity Biosciences) and rabbit polyclonal anti-FUNDC1 (1:150 dilution; cat. no. ab224722; Abcam). After washing with PBS (Gibco; Thermo Fisher Scientific, Inc.), the sections were incubated with biotinylated goat anti-rabbit immunoglobulin G secondary antibody (Solution C; contained in the kit) for 10 min at room temperature and stained using the streptavidin-biotin-peroxidase complex (Solution D). The chromophore used was 3,3′-diaminobenzidine (DAB). The stained sections were counterstained with hematoxylin (cat. no. C0105S; Beyotime Institute of Biotechnology) for 1 min at room temperature. For immunohistochemical staining, the slides were scanned using a light microscope (×20 and ×80 magnification; Leica Aperio AT2 instrument; Leica Microsystems, Inc.).
Isolation and culture of HDPCs
After splitting the healthy teeth, the pulp tissues were carefully separated and transferred onto a tissue culture dish, rinsed three times with sterile PBS (Gibco; Thermo Fisher Scientific, Inc.) containing 2% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.), gently cut into small pieces (1 mm2) and digested with 3 mg/ml collagenase I and 4 mg/ml dispase II (Sigma-Aldrich; Merck KGaA) at 37°C for 30 min. By the enzymatic digestion method, the resultant cell suspensions were filtered through a 70-µm cell strainer (Corning, Inc.) at room temperature after centrifugation at 300 × g for 5 min at 37°C and then cultivated in α-modified minimum essential medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc.) containing 20% heat-inactivated FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin in a 25-cm2 cell culture flask (Corning, Inc.) at 37°C in an incubator (Thermo Fisher Scientific, Inc.). The cultured cells were incubated in media mentioned above that was refreshed every 3 days and passaged with TrypLE Express (Gibco; Thermo Fisher Scientific, Inc.) into new flasks when 80-90% confluence was reached and sub-cultured with α-MEM and 10% FBS. Cells at passages 3-5 were used for experiments. HDPCs cultured with 21% O2 and 5% CO2 at 37°C were used as the normoxic group, whilst cells cultured in 1% O2, 5% CO2 and 94% N2 at 37°C were used as the hypoxic group.
Immunofluorescence staining
HPDCs were seeded into 15-mm laser confocal dishes (Biosharp Life Sciences) at a density of 1×103 cells per dish and cultured at 37°C for 24 h. Subsequently, the media were removed and replaced with 4% paraformaldehyde for fixation at room temperature. After 15 min, the cells were washed three times with PBS and permeabilized with 0.3% Triton X-100 (Sigma-Aldrich; Merck KGaA) for another 15 min, followed by blocking with 5% BSA (Biofroxx; neoFroxx GmbH) for 30 min at room temperature. Subsequently, the HPDCs were separately incubated with the rabbit anti-vimentin antibody (1:150 dilution; cat. no. AF7013; Affinity Biosciences) or rabbit anti-cytokeratin-14 antibody (1:150 dilution; cat. no. AF5370; Affinity Biosciences) overnight at 4°C. The secondary antibodies used (both 1:100 dilution; EarthOx Life Sciences) were goat anti-rabbit IgG (H + L) Dylight 594 (1:100 dilution; cat. no. E032420-02) and goat anti-rabbit IgG (H + L) Dylight 488 (1:100 dilution; cat. no. E032220-02) for 1 h at room temperature. DAPI (cat. no. C1006; Beyotime Institute of Biotechnology) was then applied for 5 min at room temperature to identify the nucleus. Finally, the cells were imaged under an LSM780 confocal microscope (×200 magnification; Carl Zeiss AG).
In addition, the subcellular locations of LC3 and translocase of outer mitochondrial membrane 20 (TOMM20) in HDPCs after being cultured in normoxia or hypoxia for 18 h at 37°C were examined by immunofluorescence staining as aforementioned. The primary antibodies used for overnight at 4°C were as follows: Rabbit anti-LC3 (1:100 dilution; cat. no. 3868S; Cell Signaling Technology, Inc.) and mouse anti-TOMM20 (1:100 dilution; cat. no. ab56783; Abcam). The corresponding secondary antibodies (both 1:100 dilution; EarthOx Life Sciences) were goat anti-rabbit IgG (H + L) Dylight 594 (cat. no. E032420-02) and goat anti-mouse IgG (H + L) Dylight 488, which were used for 1 h at room temperature. These cells were imaged using an LSM780 confocal microscope (×100 and ×400 magnification; Carl Zeiss AG).
Flow cytometry analysis of the surface markers of HDPCs
Surface markers of HDPCs were identified by flow cytometry. After collecting ~1×106 cells from the third passage, HDPCs were washed twice with PBS and then suspended in PBS, before being blocked with 5% BSA for 30 min at room temperature to remove nonspecific binding. The antibodies, including CD13-PE (1:40 dilution; cat. no. 555394; BD Biosciences), CD14-FITC (1:40 dilution; cat. no. 555397; BD Biosciences), CD29-FITC (1:40 dilution; cat. no. 11-0291-82; Thermo Fisher Scientific, Inc.), CD34-PE (1:40 dilution; cat. no. 560941; BD Biosciences), CD45-PE-Cy5 (1:40 dilution; cat. no. 560974; BD Biosciences), CD73-FITC (1:10 dilution; cat. no. 561254; BD Biosciences), CD90-PE-Cy5 (1:40 dilution; cat. no. 555597; BD Biosciences) and CD105-PE (1:40 dilution; cat. no. 560839; BD Biosciences), were added and cells were incubated for 90 min at 37°C. IgG1-PE (1:40 dilution; cat. no. 556650; BD Biosciences), IgG1-FITC (1:40 dilution; cat. no. 556649; BD Biosciences) and IgG1-PE-Cy5 (1:40 dilution; cat. no. 550618; BD Biosciences) were also used as isotype controls with incubation for 90 min at 37°C. After washing with PBS, suspended HDPCs were transferred to FACS tubes and analyzed using a MOFlo™ XDP high-performance cell sorter (Beckman Coulter, Inc.) and SUMMIT version 5.0 software (Beckman Coulter, Inc.) according to the manufacturer's instructions.
Evaluation of the osteogenic, chondrogenic and adipogenic capabilities of HDPCs
HDPCs (2×105 cells/well) in passage 3 were seeded into 6-well plates (Corning, Inc.). When the cells reached 80% confluence, the previous medium was replaced with differentiation-inducing medium and replaced every 3 days according to the type of differentiation.
Osteogenic medium [10% FBS, 10 nM dexamethasone (Beijing Solarbio Science & Technology Co., Ltd.), 10 mM β-glycerophosphate (Beijing Solarbio Science & Technology Co., Ltd.) and 50 µg/ml ascorbic acid (Beijing Solarbio Science & Technology Co., Ltd.) in α-MEM (Gibco; Thermo Fisher Scientific, Inc.)] was applied for 28 days on HDPCs at 37°C before the mineralized nodules were revealed using Alizarin Red (Cyagen Biosciences, Inc.) for 30 min staining at room temperature.
For chondrogenic differentiation, HPDCs were induced with serum-free α-MEM containing 10 nM dexamethasone, 50 µg/ml ascorbic acid, 40 ng/ml proline (Sigma-Aldrich; Merck KGaA), 0.1 ng/ml TGF-β1 (Sigma-Aldrich; Merck KGaA) and 3 µM insulin-transferrin-selenium supplement (Sigma-Aldrich; Merck KGaA) for 21 days at 37°C. Finally, the extent of chondrogenic differentiation was assessed using Alcian Blue (Cyagen Biosciences, Inc.) for 30 min staining at room temperature.
For adipogenic differentiation, HPDCs were cultured with induction medium consisting of 10% FBS, 50 nM dexamethasone, 0.1 mM indomethacin (Sigma-Aldrich; Merck KGaA), 5 µg/ml insulin (Sigma-Aldrich; Merck KGaA) and 0.25 mM 3-isobutyl-methylxanthine (Sigma-Aldrich; Merck KGaA) in α-MEM for 28 days at 37°C (23). The observation of lipid droplets was achieved by Oil Red O (Cyagen Biosciences, Inc.) for 30 min staining at room temperature.
Images of the aforementioned morphological features were captured using a light microscope (×40, ×40 and ×80 magnification; Axio Observer Z1; Carl Zeiss AG).
Cell transfection
HDPCs were first cultured in α-MEM supplemented with 10% FBS. Small interfering RNA (siRNA) targeting FUNDC1 was designed and synthesized by Guangzhou RiboBio Co., Ltd. Subsequently, cell transfection was performed with Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. After 15 min incubation at room temperature, siRNA at a final concentration of 50 nM was used to transfect cells at 70% confluency. The sequences of si-RNA1, si-RNA2 and si-RNA3 for FUNDC1 were as follows: si-RNA1 sense, 5-CAA GCA GAA CAU UGU GAU AdT dT-3 and antisense, 5-UAU CAC AAU GUU CUG CUU GdT dT-3; si-RNA2 sense, 5-GUA GCU ACC CAG AUU GUA AdT dT-3, and antisense, 5-UUA CAA UCU GGG UAG CUA CdT dT-3 and si-RNA 3 sense, 5-GCA GCA CCU GAA AUC AAC AdT dT-3 and antisense, 5-UGU UGA UUU CAG GUG CUG CdT dT-3. The corresponding non-specific control sequence was obtained from the commercial reagent (cat. no. siN0000001-1-5; Guangzhou RiboBio Co., Ltd.). After 48 h at 37°C, knockdown efficiency was evaluated by western blotting.
Cell viability assay
HPDCs were inoculated into 96-well plates (Corning, Inc.) at a density of 3×103 cells/well and incubated overnight at 37°C before treatment. Subsequently, the cells were treated under normoxic or hypoxic conditions for different durations (0, 6, 12, 18 and 24 h) at 37°C and harvested for further analysis. Cell proliferation was measured according to the manufacturer's protocols of the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.). After adding 10 µl CCK-8 solution into each well of the plate and incubation for 1 h at 37°C, the absorbance at a wavelength of 450 nm was measured by a microplate reader with BioTek Gen5 system (BioTek Instruments, Inc.).
Migration assays
The migration of HDPCs after treatment with normoxia or hypoxia was assessed by wound healing and Transwell migration assays.
For the wound healing assay, HPDCs or transfected cells were seeded into 6-well plates in α-MEM supplemented with 10% FBS and incubated to obtain a 90% cell confluence. A sterile 1-ml pipette tip was used to generate equal-width scratch wounds. After washing away the detached cells with PBS, serum-free α-MEM was added before recording the initial wounds using an EVOS M5000 microscope (Thermo Fisher Scientific, Inc.). During treatment with hypoxia at 37°C, the wound was imaged under a light microscope (×10 magnification) every 6 h for 24 h. Wound closure was defined as a mean percentage of the migrated width compared with the initial wound width. The calculation formula was: Healing rate (%)=(width of the wound at 0 h-width of the wound at 24 h)/width of the wound at 0 h ×100%.
Transwell migration assays were performed using Transwell chambers with 8-µm pores (Corning, Inc.). HPDCs or transfected HPDCs (2×105 cells/ml) suspended in 100 µl serum-free α-MEM were seeded into the upper compartment whereas 500 µl culture medium supplemented with 10% FBS was added into the lower chamber. After treatment with normoxia or hypoxia for 18 h at 37°C, migrated cells on the underside of the chamber were fixed with 4% paraformaldehyde for 15 min at room temperature, stained with 1% crystal violet (Beyotime Institute of Biotechnology) for 30 min at room temperature, imaged and counted in three random fields under an inverted light microscope (×100 magnification; Carl Zeiss AG).
Alkaline phosphatase (ALP) staining and activity assays
ALP staining was achieved using a BCIP/NBT ALP kit (cat. no. C3206; Beyotime Institute of Biotechnology). According to the manufacturer's protocols, HDPCs were seeded into 6-well plates at a density of 1×105 cells/well in α-MEM supplemented with 10% FBS. After incubation overnight at 37°C, the cells were cultured with the osteogenic medium as aforementioned and treated with normoxia or hypoxia for 3 days at 37°C. Subsequently, HDPCs were fixed with 4% paraformaldehyde for 30 min at room temperature, washed with PBS three times and stained with the aforementioned kit for 30 min at room temperature. Images were captured using an optical light microscope (×50 magnification; Carl Zeiss AG).
In addition, ALP activity was also examined using an ALP detection kit (cat. no. P0321S; Beyotime Institute of Biotechnology) according to the manufacturer's protocols.
Protein isolation and western blotting
HPDCs or transfected HPDCs were seeded into 6-well plates and treated with normoxia or hypoxia at 37°C before total protein was collected using RIPA buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) supplemented with protease inhibitor and phosphatase inhibitor (CoWin Biosciences) every 6 h for 24 h. For mineralization assay, HPDCs cultured with osteogenic medium were treated with normoxia or hypoxia at 37°C for 3 days. Subsequently, the total protein was extracted as aforementioned. The sample protein content was quantified using a BCA protein assay kit (CoWin Biosciences) before immunoblotting. Equal amounts (25 µg) of protein were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (MilliporeSigma). The membranes were blocked with 5% skimmed milk or 5% BSA for 1 h at 25°C. Subsequently, the membranes were incubated overnight at 4°C with the following primary antibodies: Rabbit anti-HIF-1α (1:1,000 dilution; cat. no. AF1009; Affinity Biosciences), rabbit anti-sequestosome 1 (SQSTM1)/p62 (1:1,000 dilution; cat. no. AF5384; Affinity Biosciences), rabbit anti-LC3B (1:1,000 dilution; cat. no. 3868S; Cell Signaling Technology, Inc.), mouse anti-dynamin-related protein 1 (DRP1; 1:1,000 dilution; cat. no. ab56788; Abcam), rabbit anti-translocase of inner mitochondrial membrane 23 (TIMM23; 1:1,000 dilution; cat. no. ab116329; Abcam), mouse anti-TOMM20 (1:1,000 dilution; cat. no. ab56783; Abcam), rabbit anti-Beclin-1 (1:1,000 dilution; cat. no. 3495S; Cell Signaling Technology, Inc.), rabbit anti-autophagy related 5 (ATG5; 1:1,000 dilution; cat. no. 12994S; Cell Signaling Technology, Inc.), rabbit anti-FUNDC1 (1:500 dilution; cat. no. ab224722; Abcam), rabbit anti-phosphorylated (p-) FUNDC1 (Ser 17; 1:500 dilution; cat. no. AF0001; Affinity Biosciences), mouse anti-β-actin (1:5,000 dilution; cat. nos. E021021-01; EarthOx Life Sciences), rabbit anti-collagen type I (Col I; 1:1,000 dilution; cat. no. AF7001; Affinity Biosciences), rabbit anti-osterix (OSX; 1:1,000 dilution; cat. no. DF7731; Affinity Biosciences), rabbit anti-osteopontin (OPN; 1:1,000 dilution; cat. no. AF0227; Affinity Biosciences) and rabbit anti-runt-related transcription factor 2 (RUNX2; 1:1,000 dilution; cat. no. AF5186; Affinity Biosciences). The membranes were washed with TBS with 0.1% Tween-20 buffer (CoWin Biosciences) for 30 min and incubated with the appropriate secondary antibodies conjugated with HRP (both 1:5,000 dilution; cat. nos. E030120-01 and E030110-01; both EarthOx Life Sciences) for 1 h at room temperature, followed by visualization using MilliporeSigma™ Immobilon™ Western Chemiluminescent HRP Substrate (ECL; MilliporeSigma). Densitometric analysis of the protein bands was performed using the ImageJ version 1.5.0 software (National Institutes of Health).
Transmission electron microscopy (TEM)
The samples were washed and harvested after normoxia or hypoxia treatment for 18 h at 37°C. Subsequently, the preparation of cells for TEM was performed as previously described (9). Briefly, cells were fixed with 2.5% glutaraldehyde (cat. no. A17876; Thermo Fisher Scientific, Inc.) for 2 h at temperature, washed six times in PBS (30 min each) and fixed in 1% osmic acid for 2 h at room temperature. The fixed samples were dehydrated through an increasing graded series of ethanol (30-100%), infiltrated and embedded in eponate 12 resin (cat. no. GP18010; Ted Pella, Inc.) at 38°C for 4 h. After the process of curing (38°C for 6 h, 60°C for 6 h and 80°C for 12 h), the embedded blocks were cut into ultrathin sections (70 nm) using a Leica Ultramicrotome EM UC7 (Leica Microsystems, Inc.). The sections were then stained with 2% uranyl acetate for 30 min at room temperature and 3.5% lead citrate for 15 min at room temperature. After that, ultrathin sections (70 nm) were viewed at 100 kV with a transmission electron microscope (magnification, ×30,000; JEM-1400 PLUS; Jeol, Ltd.).
Statistical analysis
The experiments were repeated three times independently and the data are presented as the mean ± SD. Data analysis was performed using GraphPad Prism version 9.0 software (GraphPad Software, Inc.). Two-group comparisons were assessed by unpaired Student's t-test, whilst multiple comparisons were analyzed by one-way ANOVA followed by Fisher's least significant difference post hoc test or Bonferroni's correction. P<0.05 was considered to indicate a statistically significant difference.
Results
HIF-1α and FUNDC1 expression is upregulated in inflammatory human dental pulp tissues
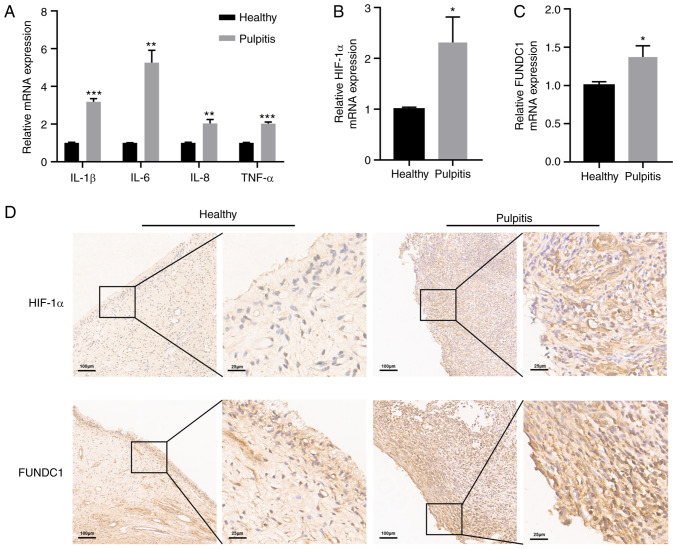
To confirm the inflammatory status of human dental pulp tissues, RT-qPCR was performed to detect the mRNA expression levels of inflammatory cytokines in healthy and pulpitis tissues separately. Pulpitis tissues exhibited statistically significantly higher levels of IL-1β, IL-6, IL-8 and TNF-α expression compared with those in the healthy group (Fig. 1A). The present study also revealed significantly higher levels of HIF-1α and FUNDC1 mRNA expression in pulpitis tissues compared with those in the healthy group (Fig. 1B and C). To measure the protein expression levels of HIF-1α and FUNDC1 further, immunohistochemistry was performed, which revealed that HIF-1α and FUNDC1 protein expression was also marked higher in pulpitis tissues compared with that in the healthy group (Fig. 1D).
Figure 1.
Expressions of inflammatory cytokines, HIF-1α and FUNDC1 in healthy and pulpitis tissues. (A) mRNA expression of IL-1β, IL-6, IL-8 and TNF-α in human healthy and pulpitis tissues. mRNA expression of (B) HIF-1α and (C) FUNDC1 in human healthy and pulpitis tissues. (D) Representative immunostaining images of HIF-1α and FUNDC1 in human healthy or inflamed dental pulp tissues. Scale bars are 100 and 25 µm, respectively. Results are presented as the means ± SD from ≥ three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. healthy. HIF-1α, hypoxia-inducible factor-1α; FUNDC1, FUN14 domain-containing 1.
Identification of HDPCs
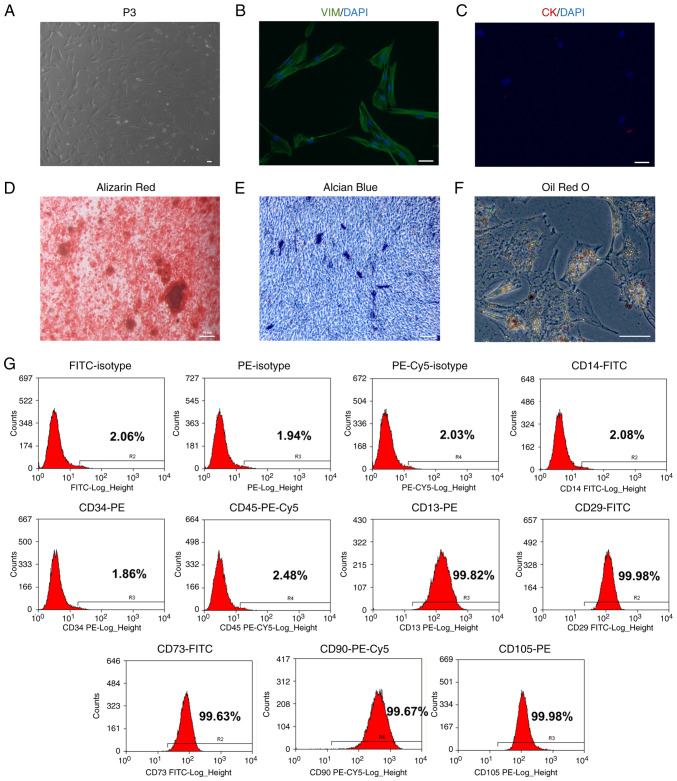
HDPCs were isolated from the human dental pulp tissues before analysis of cell morphology was performed (Fig. 2A). The morphology of cells was relatively uniform, showing a fibrous/fusiform shape. Immunofluorescence revealed that the HDPCs were positive for the expression of the mesenchymal marker vimentin (Fig. 2B) but negative for the epithelial marker cytokeratin (Fig. 2C). HDPCs were subsequently induced in osteogenic, chondrogenic and adipogenic media. The multipotent differentiation capabilities of these HDPCs were confirmed by the Alizarin Red staining of osteoblasts, Alcian Blue staining of chondroblasts and Oil Red O staining of fat droplets (Fig. 2D-F). Flow cytometry analysis of surface markers revealed that these HDPCs were positive for CD13 (99.82%), CD29 (99.98%), CD73 (99.63%), CD90 (99.67%) and CD105 (99.98%). However, they were tested negative for CD14 (2.08%), CD34 (1.86%) and CD45 (2.48%). These flow cytometry profiles are consistent with the characteristics of mesenchymal stem cells (Fig. 2G) (24).
Figure 2.
Identification of HDPCs. (A) Representative images of HDPCs at passage 3. HDPCs are (B) positive for vimentin and (C) negative for cytokeratin. (D) Representative image of mineralized nodules in HDPCs after osteogenic induction for 28 days following Alizarin Red staining. (E) Representative image of chondroblasts in HDPCs after chondrogenic induction for 21 days following Alcian Blue staining. (F) Representative image of lipid droplets in HDPCs after adipogenic induction for 28 days following Oil Red O staining. Scale bars, 50 µm. (G) Representative flow cytometry histograms showing the expression profile of markers in the p3 HDPCs. HDPCs, human dental pulp cells; VIM, vimentin; CK, cytokeratin.
Hypoxia promotes proliferation, migration and odontoblastic differentiation in HDPCs
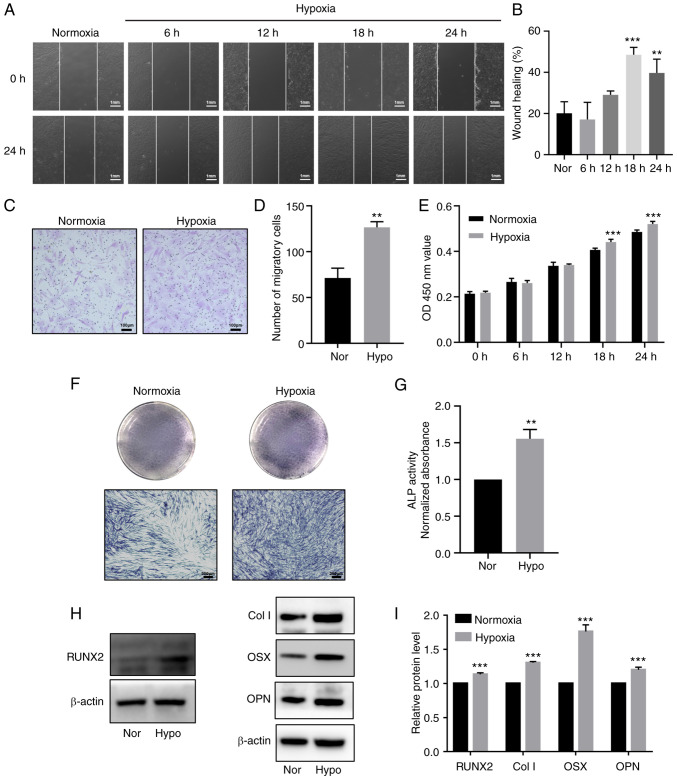
To explore the effects of hypoxia on the proliferation and differentiation of HDPCs, cells in the hypoxia group were cultured in 1% O2 in an incubator for 6, 12, 18 and 24 h, whilst cells in the normoxic group were cultured in normoxic conditions. CCK-8 assay results indicated that hypoxia significantly promoted HDPC proliferation after 18 h of stimulation compared with that in the normoxic group (Fig. 3E). Although the wound healing assay demonstrated that 24 h stimulation increased the migration, HDPCs in the 18 h hypoxia group exhibited higher horizontal and vertical migration ability compared with that in the normoxic group (Fig. 3A-D). Additionally, 3 days after odontogenic induction, ALP staining and activity were markedly enhanced in the hypoxic group (Fig. 3F-G). Western blotting also revealed significantly upregulated protein expression levels of RUNX2, Col I, OSX, OPN in the hypoxic treatment group compared with those in the normoxic group (Fig. 3H-I).
Figure 3.
Hypoxia promotes proliferation, migration and odontoblastic differentiation in HDPCs. (A) Representative images of wound healing assays in the normoxic or hypoxic group. Scale bar, 1 mm. (B) Percentages of wound closure from three independent experiments are quantified. (C) Representative images of migratory cells stained with crystal violet. (D) Statistical quantification of the number of migratory cells from three independent experiments are shown. Scale bar, 100 µm. (E) Cell viability of HDPCs was detected using Cell Counting Kit-8 analysis. (F) ALP staining and (G) activity in the normoxic or hypoxic group on day 3. Scale bar, 200 µm. (H) Representative western blotting images showing the protein expression levels of RUNX2, Col I, OSX and OPN in the normoxic or hypoxic group on day 3, (I) which were quantified. Results are presented as the means ± SD from ≥ three independent experiments. **P<0.01 and ***P<0.001 vs. normoxia. Nor, normoxia; Hypo, hypoxia; OD, optical density; ALP, alkaline phosphatase; RUNX2, runt-related transcription factor 2; Col I, collagen type I; OSX, osterix; OPN, osteopontin.
Hypoxia-induces mitophagy in HDPCs
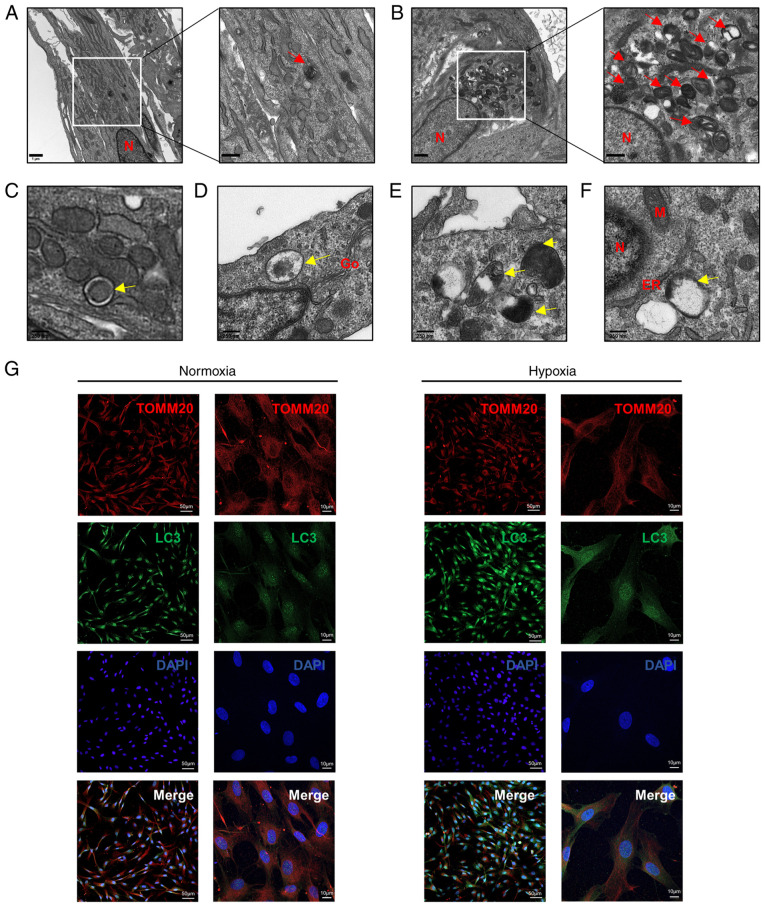
To investigate the effects of hypoxia on autophagy, TEM, western blotting and immunofluorescence staining were performed. TEM was utilized to examine the ultrastructure of HDPCs following hypoxia or normoxia treatment for 18 h. Hypoxia induced the formation of higher numbers of autophagic vacuoles in HDPCs (Fig. 4B) compared with those in the normoxic group (Fig. 4A). Furthermore, a series of processes during autophagic vesicle formation in the hypoxic group, including the early, mid and late stages, could be clearly observed (Fig. 4C-F). During the early stages (Fig. 4C), the formation of double membrane-bound vacuoles was a characteristic feature of autophagosomes (25). After fusing with lysosomes to form autolysosomes, the contents of the vacuoles were indistinct, typical of mid-stage of autophagy (Fig. 4D and E) (25). The complete degradation of organelles could be observed during late-stage autophagosome formation (Fig. 4F).
Figure 4.
Hypoxia induces mitophagy in HDPCs. (A) Cells in the normoxic group show fewer autolysosomes, which are marked by red arrows. Scale bars are 1 µm and 500 nm, respectively. (B) Cells in the hypoxic group show increased numbers of autolysosomes, which are marked by red arrows. Scale bars are 1 µm and 500 nm respectively. (C-F) The developmental process of autolysosomes. (C) Early stage of autophagosomes marked by yellow arrows in HDPCs from the hypoxic group, showing the envelope structure in the vesicles remaining intact. (D) Mid-stage autophagy, showing that the autophagosomes were fused with lysosomes to form autolysosomes and (E) the encapsulated contents were degraded to form indistinct structure. (F) Late-stage autophagy, showing the complete degradation of organelles in the vacuoles. Scale bar, 250 nm. (G) Immunofluorescence staining with the anti-TOMM20 antibody (red) and anti-LC3 antibody (green) was performed in HDPCs. DAPI (blue) staining indicated cell nuclei. Scale bars are 50 and 10 µm, respectively. TOMM20, translocase of outer mitochondrial membrane 20; LC3, microtubule-associated protein 1 light chain 3; DAPI, 4′,6-diamidino-2-phenylindole; N, nucleus; M, mitochondrion; Go, Golgi; ER, endoplasmic reticulum.
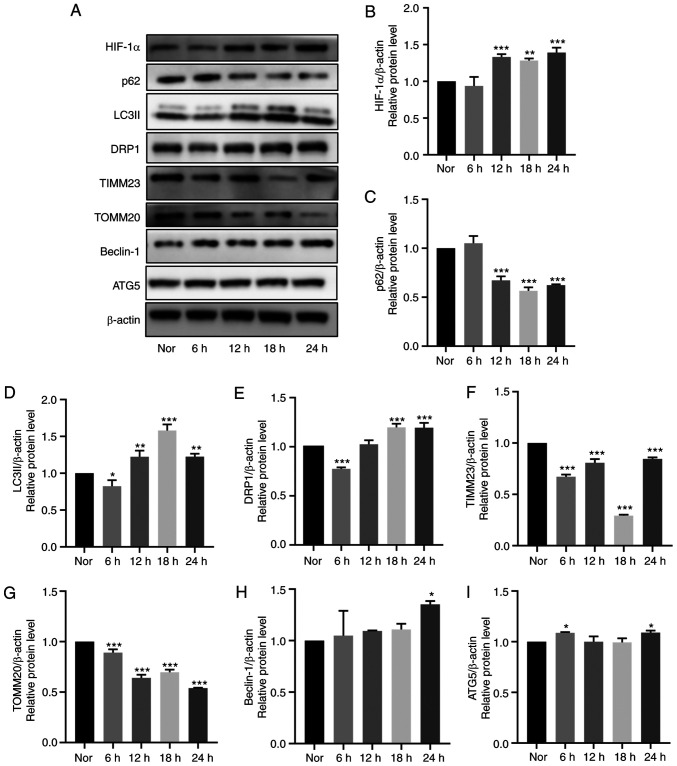
Immunofluorescence analysis demonstrated markedly higher LC3 expression and lower TOMM20 expression levels following 18-h hypoxia stimulation compared with those in the normoxic group (Fig. 4G). Additionally, the expression levels of mitochondrial membrane proteins (TOMM20 and TIMM23) and autophagy-related proteins (LC3II, SQSTM1/p62, Beclin-1 and ATG5) were detected in HDPCs after hypoxia treatment at 6-h intervals for 24 h by western blotting. Compared with those in the normoxic group, significantly increased protein expression levels of HIF-1α, LC3II and DRP1, coupled with the significantly decreased protein expression levels of SQSTM1/p62, TIMM23 and TOMM20, were observed in the hypoxia groups from 12 h onwards (Fig. 5A-I). Specifically, a plateau in LC3II expression and a trough in SQSTM1/p62 expression were reached at 18 h. Additionally, a slight by significant increase in Beclin-1 and ATG5 expression could be observed at 24 h (Fig. 5A-I). Overall, these results suggest that hypoxia markedly induced mitochondrial autophagy in HDPCs.
Figure 5.
Effects of hypoxia on mitophagy in cultured HDPCs. (A) Representative western blotting images. Protein expression of (B) HIF-1α, (C) p62, (D) LC3II, (E) DRP1, (F) TIMM23, (G) TOMM20, (H) Beclin-1 and (I) ATG5 in HDPCs cultured in hypoxia were quantified. The expression of the target proteins was measured by quantifying the intensity of the bands and normalized to that of β-actin. The results are presented as the means ± SD from ≥ three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. normoxia. Nor, normoxia; HIF-1α, hypoxia-inducible factor-1α; DRP1, dynamin-related protein 1; TIMM23, translocase of inner mitochondrial membrane 23; TOMM20, translocase of outer mitochondrial membrane 20; ATG5, autophagy related 5.
Hypoxia promotes the phosphorylation of FUNDC1 in HDPCs
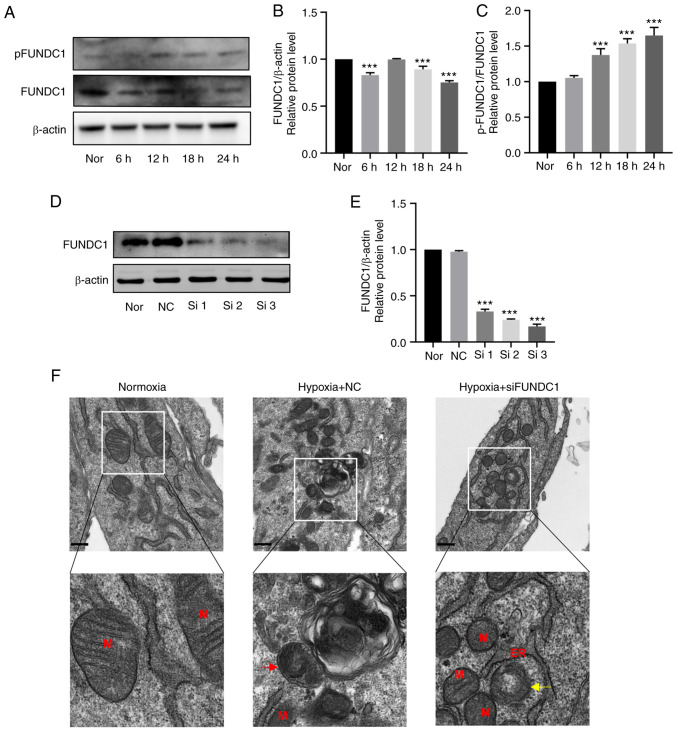
After hypoxia treatment for 24 h at 6-h intervals, the protein expression levels of FUNDC1 were slightly decreased (Fig. 6A and B). However, the ratio of p-/total FUNDC1 was significantly increased from 12 h onwards after hypoxia stimulation compared with that in the normoxic group (Fig. 6C).
Figure 6.
Hypoxia promotes the phosphorylation of FUNDC1 but the knockdown of FUNDC1 influences mitochondria morphology. (A) Representative western blotting images. (B) Protein expression of FUNDC1 and (C) phosphorylation of FUNDC1 (Ser 17) in HDPCs cultured in hypoxia were quantified. (D) Protein expression of FUNDC1 in HDPCs cultured in normoxia after transfection with siNC or three different siRNAs, (E) which was quantified. The results are presented as the means ± SD from ≥ three independent experiments. (F) Transmission electron microscopy images of normal mitochondria in the normoxic group, single-membrane autolysosomes containing partially degraded mitochondria in the hypoxia + NC group (red arrow) and blurred mitochondrial cristae (yellow arrow) in the hypoxia + siFUNDC1 group. Scale bars, 500 nm. ***P<0.001 vs. normoxia. Nor, normoxia; N, nucleus; M, mitochondrion; Go, Golgi; ER, endoplasmic reticulum; NC, negative control; si, small-interfering; p, phosphorylated; FUNDC1, FUN14 domain-containing.
FUNDC1 knockdown suppresses hypoxia-induced mitophagy, weakening the promotion of hypoxia-induced proliferation, migration and odontoblastic differentiation
To identify the effects of FUNDC1 on hypoxia-induced mitophagy, cell proliferation, migration and differentiation, FUNDC1 expression was silenced by siRNA transfection. FUNDC1 expression in HDPCs was significantly knocked down by siRNA number three compared with that in the NC group (Fig. 6D and E). This siRNA was therefore used in the subsequent 18-h hypoxia stimulation experiments. TEM was first used to assess mitochondria morphology. The ultrastructure of mitochondria in the normoxic group was clear, including intact membranes, dense mitochondrial cristae and clear mitochondrial matrices (Fig. 6F). Under hypoxia treatment, there appeared to be autophagosomes containing partially degraded mitochondria (Fig. 6F), whilst the FUNDC1-knockdown group exhibited damaged mitochondria with blurred, disordered or even vacuolized mitochondrial cristae (Fig. 6F).
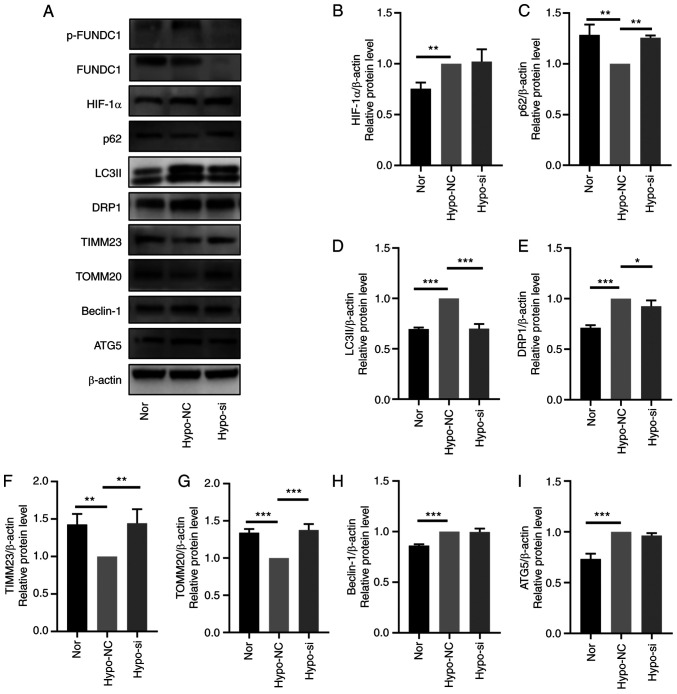
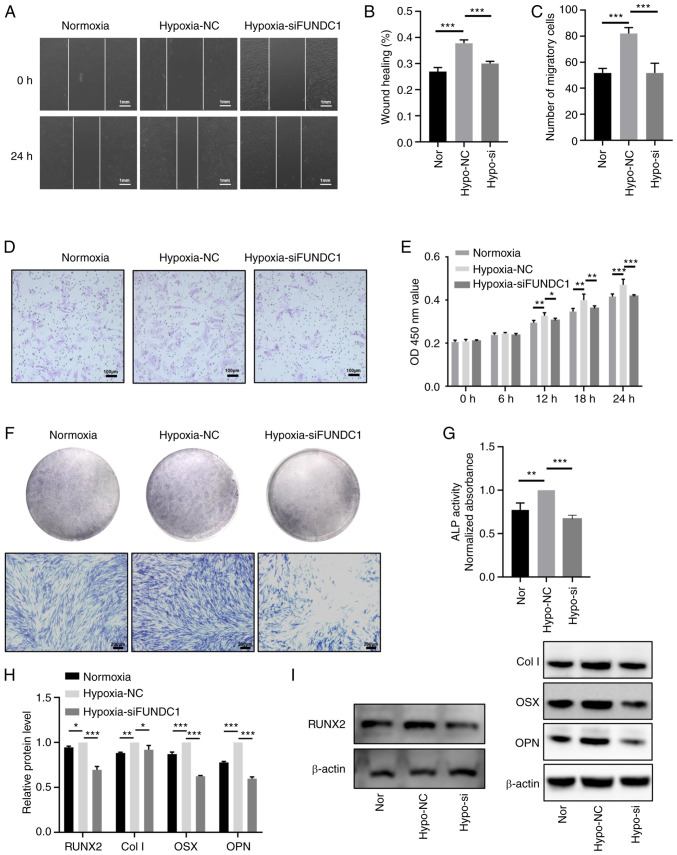
After FUNDC1 expression was knocked down, hypoxia-induced mitophagy was inactivated, which was demonstrated by western blotting with decreased protein expression levels of LC3II and significantly increased protein expression levels of SQSTM1/p62, TIMM23 and TOMM20 (Fig. 7A-I). Based on these findings, FUNDC1 knockdown appeared to have reversed hypoxia-induced mitophagy. CCK-8 assay (Fig. 8E), migration assays (Fig. 8A-D), ALP staining (Fig. 8F) and activity assays (Fig. 8G) and western blotting (Fig. 8H and I) were utilized to detect the proliferation, migration and odontoblastic differentiation in the FUNDC1-knockdown group. The western blotting results showed that the protein levels of RUNX2, Col I, OSX, OPN in hypoxia-siFUNDC1 group were significantly downregulated. Compared with those in the hypoxia-NC group, FUNDC1 knockdown markedly reversed hypoxia-induced proliferation, migration and odontoblastic differentiation.
Figure 7.
FUNDC1 knockdown prevents hypoxia-induced mitophagy. (A) Representative western blotting images. The protein expression of (B) HIF-1α, (C) p62, (D) LC3II, (E) DRP1, (F) TIMM23, (G) TOMM20, (H) Beclin-1 and (I) ATG5 in HDPCs after hypoxic culture and transfection with NC or siRNA was all quantified. The results are presented as the means ± SD from ≥ three independent experiments. *P<0.05, **P<0.01 and ***P<0.001. FUNDC1, FUN14 domain-containing; p-, phosphorylated; Nor, normoxia; Hypo, hypoxia; NC, negative siRNA; si, small interfering; HIF-1α, hypoxia-inducible factor-1α; DRP1, dynamin-related protein 1; TIMM23, translocase of inner mitochondrial membrane 23; TOMM20, translocase of outer mitochondrial membrane 20; ATG5, autophagy related 5.
Figure 8.
Knocking down FUNDC1 expression weakens the promotion of hypoxia-induced proliferation, migration and odontoblastic differentiation. (A) Representative images of the wound healing assay (scale bar, 1 mm) and (B) percentages of wound closure from three independent experiments are quantified. (C) The number of migratory cells stained with crystal violet solution from three independent experiments was quantified. (D) Representative images of migratory cells are shown. Scale bar, 100 µm. (E) Viability of HDPCs was detected by Cell Counting Kit-8 analysis. (F) ALP staining and (G) activity in the normoxic, hypoxia + NC and hypoxia + siFUNDC1 groups on days 3. Scale bar, 200 µm. (H) Protein expression levels of RUNX2, Col I, OSX and OPN in the normoxic or hypoxic group on day 3 were quantified, (I) the corresponding representative western blotting images of which were shown. Results are presented as the means ± SD from ≥ three independent experiments. *P<0.05, **P<0.01 and ***P<0.001. FUNDC1, FUN14 domain-containing 1; Nor, normoxia; Hypo, hypoxia; NC, negative control; si, small interfering; ALP, alkaline phosphatase; RUNX2, runt-related transcription factor 2; Col I, collagen type I; OSX, osterix; OPN, osteopontin.
Discussion
Hypoxia may be caused by inflammation resulting from bacterial infection or trauma, which frequently occurs in injured dental pulp tissues (20). Using RT-qPCR and immunohistochemistry, the present study verified the increased expression levels of inflammatory cytokines IL-1β, IL-6, IL-8 and TNF-α in addition to HIF-1α in pulpitis tissues, suggesting the presence of a hypoxic environment after the inflammation occurred. In some tissues, hypoxia induces cell proliferation, migration or angiogenesis to promote cell survival and tissue repair (22). HDPCs can differentiate into odontoblasts and migrate to the site of damage, where they then activate their stimulatory effects after injury or infection (19). A marked increase in the proliferation of HDPCs under hypoxic conditions was previously observed by Sakdee et al (26). In addition, Du et al (27) revealed that the migration and odontoblastic differentiation of HDPCs were enhanced by treatment with 10 µM deferoxamine, a hypoxia-mimetic agent. HIF-1α knockdown in stem cells isolated from human exfoliated deciduous teeth (SHEDs) resulted in reduced proliferative and migratory capabilities, which in turn impaired the paracrine angiogenic effects (28). Therefore, the present study aimed to investigate the influence of 1% O2 hypoxia on the proliferation, migration and odontoblastic differentiation of HDPCs. The results suggest that 1% O2 hypoxia could promote all of these aforementioned processes. However, other studies have also revealed opposite effects. Chen et al (29) found that cobalt chloride, a hypoxia-mimetic agent, inhibited the osteogenic differentiation of SHEDs. Hu et al (30) revealed that hypoxia (5% O2) diminished cell survival whilst suppressing the osteogenic differentiation of dental pulp stem cells. These discrepancies are likely to be due to the use of different cell lines, oxygen deprivation protocols and/or the hypoxia treatment time. Zhou et al (31) previously demonstrated that hypoxia serves a key role in maintaining the stemness and differentiation capacity of periodontal ligament and dental pulp cells. In addition, another study reported that hypoxia can induce the differentiation of stem cells from the apical papilla into cells of osteogenic and neurogenic lineages (32). HDPCs are capable for multilineage differentiation, but the effects of hypoxia on chondrogenesis or differentiation into other lineages warrant further study, which is a limitation of the present study.
The intracellular mechanism underlying the stimulation by hypoxia in HDPCs remain poorly understood. Autophagy is a degradative pathway that can regulate various physiological processes, including cell proliferation, migration, angiogenesis and odontoblastic differentiation (33-35). Gong et al (36) reported that HIF-1α overexpression can trigger mitochondrial autophagy, which promotes neuron survival, whereas Song et al (37) demonstrated that overexpression of HIF-1α promoted papillary thyroid carcinoma progression by inducing autophagy during hypoxia stress. Zhang et al (38) revealed that hypoxia increased the migration of epidermal keratinocytes during wound healing through BCL2 interacting protein 3-induced autophagy, highlighting the reactive oxygen species-mediated activation of p38 and JNK MAPK signaling in the process. Additionally, a previous study reported that typical ultrastructure of autophagosomes could be observed in radicular cysts and periapical granulomas, suggesting that autophagy is activated in inflamed and hypoxic periapical lesions (39). As observed using electron microscopy in the present study, higher numbers of autophagic vacuoles were present in hypoxic HDPCs. Several markers of autophagy are involved in this process, including LC3, Beclin-1, p62 and ATG5 (40). LC3-II, Beclin-1 and ATG5 are directly associated with autophagic activity at various stages, whereas p62 expression levels are inversely associated with this (41). Since TOMM20 and TIMM23 are markers of the mitochondrial membrane, their protein expression levels can be used to directly reflect the number of mitochondria. As a pro-fission protein, DRP1 is involved in the modulation of the dynamic mitochondrial cycle, which can coordinate with the autophagic machinery to mediate mitophagy (42). The present study demonstrated that the protein expression levels of LC3-II, Beclin-1 and DRP1 were increased, whereas those of p62, TOMM20 and TIMM23 were decreased, in HDPCs cultured under hypoxic conditions. Subsequently, double immunofluorescence staining of LC3 and TOMM20 supported these western blotting findings. These results suggest that hypoxia can induce mitophagy in HPDCs.
The involvement of mitochondrial dysfunction and mitophagy in diseases has been previously reported (43) As a receptor for activating hypoxia-induced mitophagy, FUNDC1 expression has been demonstrated to be upregulated in patients with chronic obstructive pulmonary disease (44) and breast cancer (16). The present study first reported the elevated mRNA and protein levels of FUNDC1 in pulpitis tissues whereas the in vitro experiments revealed that hypoxia upregulated phosphorylation of Ser 17 of FUNDC1 in HDPCs. Increasing the phosphorylation of Ser 17 on FUNDC1 enhances the binding of FUNDC1 to LC3, leading to increased mitophagy (41). In addition, silencing FUNDC1 expression reversed hypoxia-induced mitophagy in HDPCs. These results suggest that FUNDC1-mediated mitophagy is therefore activated by hypoxia, where FUNDC1 served a key role in the process. Furthermore, it was revealed that not only was mitophagy inhibited, silencing FUNDC1 expression also reversed hypoxia-induced proliferation, migration and odontoblastic differentiation. FUNDC1-mediated mitophagy is associated with various cell activities (45,46). Wu et al (16) previously showed that upregulating the expression of FUNDC1 can stimulate human breast cancer cell proliferation and migration by increasing Ca2+ influx. By contrast, Wu et al (47) previously demonstrated that knocking down FUNDC1 or DRP1 expression suppressed mitophagy under hypoxic conditions. In cervical cancer cell lines, FUNDC1 knockdown inhibited cell proliferation (48). Lampert et al (49) previously revealed that FUNDC1-mediated mitophagy was required for mitochondrial network remodeling during cardiac progenitor cell differentiation. To initiate protective effects following injury or infection, HDPCs can differentiate into odontoblasts and migrate to the site of damage (19). Based on the aforementioned results, FUNDC1-mediated mitophagy may be associated with the proliferation, migration and odontoblastic differentiation in HDPCs, which may be a mechanism underlying the activities of HDPCs.
In conclusion, the present results suggested that FUNDC1-mediated mitophagy can serve as a key factor for the physiology of HDPCs. However, further studies are necessary to explore the pathways underlying this association and to investigate how these activities can be modulated during the responses of HDPCs to hypoxia.
Acknowledgments
Not applicable.
Funding Statement
The present study was supported by funds from the National Natural Science Foundation of China (grant nos. 81700957 and 81870750) and Natural Science Foundation of Guangdong Province (grant no. 2016A030310197).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL and LC performed the experiments, collected and visualized the data. YL, QG and YH analyzed the data. HJ and YH conceptualized the study. YL and YH confirm the authenticity of all the raw data. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Written informed consent was received from all patients, parent or guardian of the patients involved. The present study was approved by the Ethics Committee of Hospital of Stomatology and Guanghua School of Stomatology, affiliated to Sun Yat-sen University [protocol no. ERC-(2017)-27; Guangzhou, China].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yin Z, Pascual C, Klionsky DJ. Autophagy: Machinery and regulation. Microb Cell. 2016;3:588–596. doi: 10.15698/mic2016.12.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang JW, Zhang YF, Wan CY, Sun ZY, Nie S, Jian SJ, Zhang L, Song GT, Chen Z. Autophagy in SDF-1a-mediated DPSC migration and pulp regeneration. Biomaterials. 2015;44:11–23. doi: 10.1016/j.biomaterials.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Pantovic A, Krstic A, Janjetovic K, Kocic J, Harhaji-Trajkovic L, Bugarski D, Trajkovic V. Coordinated time-dependent modulation of AMPK/Akt/mTOR signaling and autophagy controls osteogenic differentiation of human mesenchymal stem cells. Bone. 2013;52:524–531. doi: 10.1016/j.bone.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Pan Y, Li Z, Wang Y, Yan M, Wu J, Beharee RG, Yu J. Sodium fluoride regulates the osteo/odontogenic differentiation of stem cells from apical papilla by modulating autophagy. J Cell Physiol. 2019 Feb 14; doi: 10.1002/jcp.28269. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 5.Couve E, Osorio R, Schmachtenberg O. Mitochondrial autophagy and lipofuscin accumulation in aging odontoblasts. J Dent Res. 2012;91:696–701. doi: 10.1177/0022034512449347. [DOI] [PubMed] [Google Scholar]
- 6.Park SY, Sun EG, Lee Y, Kim MS, Kim JH, Kim WJ, Jung JY. Autophagy induction plays a protective role against hypoxic stress in human dental pulp cells. J Cell Biochem. 2018;119:1992–2002. doi: 10.1002/jcb.26360. [DOI] [PubMed] [Google Scholar]
- 7.Roa-Mansergas X, Fado R, Atari M, Mir JF, Muley H, Serra D, Casals N. CPT1C promotes human mesenchymal stem cells survival under glucose deprivation through the modulation of autophagy. Sci Rep. 2018;8:6997. doi: 10.1038/s41598-018-25485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuang H, Hu D, Singer D, Walker JV, Nisr RB, Tieu K, Ali K, Tredwin C, Luo S, Ardu S, Hu B. Local anesthetics induce autophagy in young permanent tooth pulp cells. Cell Death Discov. 2015;1:15024. doi: 10.1038/cddiscovery.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Li X, Liu Y, Gong Q, Tian J, Jiang H. LPS-induced autophagy in human dental pulp cells is associated with p38. J Mol Histol. 2021;52:919–928. doi: 10.1007/s10735-021-10004-2. [DOI] [PubMed] [Google Scholar]
- 10.Yang F, Li Y, Duan H, Wang H, Pei F, Chen Z, Zhang L. Activation of mitophagy in inflamed odontoblasts. Oral Dis. 2019;25:1581–1588. doi: 10.1111/odi.13110. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Liu N, Lu B. Mechanisms and roles of mitophagy in neurodegenerative diseases. CNS Neurosci Ther. 2019;25:859–875. doi: 10.1111/cns.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Siraj S, Zhang R, Chen Q. Mitophagy receptor FUNDC1 regulates mitochondrial homeostasis and protects the heart from I/R injury. Autophagy. 2017;13:1080–1081. doi: 10.1080/15548627.2017.1300224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Q, Sun N, Zheng J, Wang Y, Yan X, Mai W, Liao Y, Chen X. Prognostic and Immunological role of FUN14 domain containing 1 in pan-cancer: Friend or foe? Front Oncol. 2019;9:1502. doi: 10.3389/fonc.2019.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan M, Yu Y, Mao X, Feng J, Wang Y, Chen H, Xie K, Yu Y. Hydrogen gas inhalation attenuates sepsis-induced liver injury in a FUNDC1-dependent manner. Int Immunopharmacol. 2019;71:61–67. doi: 10.1016/j.intimp.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Wu L, Zhang D, Zhou L, Pei Y, Zhuang Y, Cui W, Chen J. FUN14 domain-containing 1 promotes breast cancer proliferation and migration by activating calcium-NFATC1-BMI1 axis. EBioMedicine. 2019;41:384–394. doi: 10.1016/j.ebiom.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veis A. The role of dental pulp-thoughts on the session on pulp repair processes. J Dent Res. 1985;64:552–554. doi: 10.1177/002203458506400409. Spec No. [DOI] [PubMed] [Google Scholar]
- 18.Kim SA, Choi HS, Ahn SG. Pin1 induces the ADP-induced migration of human dental pulp cells through P2Y1 stabilization. Oncotarget. 2016;7:85381–85392. doi: 10.18632/oncotarget.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Jiang H. Altered expression of circular RNA in human dental pulp cells during odontogenic differentiation. Mol Med Rep. 2019;20:871–878. doi: 10.3892/mmr.2019.10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyeraas KJ, Berggreen E. Interstitial fluid pressure in normal and inflamed pulp. Crit Rev Oral Biol Med. 1999;10:328–336. doi: 10.1177/10454411990100030501. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Gong Q, Jiang H, Wei X, Ling J, Wang J. Expression of erythropoietin and erythropoietin receptor in human dental pulp. J Endod. 2010;36:1972–1977. doi: 10.1016/j.joen.2010.08.041. [DOI] [PubMed] [Google Scholar]
- 23.Luke AM, Patnaik R, Kuriadom S, Abu-Fanas S, Mathew S, Shetty KP. Human dental pulp stem cells differentiation to neural cells, osteocytes and adipocytes-An in vitro study. Heliyon. 2020;6:e03054. doi: 10.1016/j.heliyon.2019.e03054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al Madhoun A, Sindhu S, Haddad D, Atari M, Ahmad R, Al-Mulla F. Dental pulp stem cells derived from adult human third molar tooth: A brief review. Front Cell Dev Biol. 2021;9:717624. doi: 10.3389/fcell.2021.717624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakraborty J, Caicci F, Roy M, Ziviani E. Investigating mitochondrial autophagy by routine transmission electron microscopy: Seeing is believing? Pharmacol Res. 2020;160:105097. doi: 10.1016/j.phrs.2020.105097. [DOI] [PubMed] [Google Scholar]
- 26.Sakdee JB, White RR, Pagonis TC, Hauschka PV. Hypoxia-amplified proliferation of human dental pulp cells. J Endod. 2009;35:818–823. doi: 10.1016/j.joen.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Du R, Zhao J, Wen Y, Zhu Y, Jiang L. Deferoxamine enhances the migration of dental pulp cells via hypoxia-inducible factor 1alpha. Am J Transl Res. 2021;13:4780–4787. [PMC free article] [PubMed] [Google Scholar]
- 28.Han Y, Chen Q, Zhang L, Dissanayaka WL. Indispensable role of HIF-1a signaling in post-implantation survival and angio-/vasculogenic properties of SHED. Front Cell Dev Biol. 2021;9:655073. doi: 10.3389/fcell.2021.655073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Zhao Q, Yang X, Yu X, Yu D, Zhao W. Effects of cobalt chloride on the stem cell marker expression and osteogenic differentiation of stem cells from human exfoliated deciduous teeth. Cell Stress Chaperones. 2019;24:527–538. doi: 10.1007/s12192-019-00981-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu HM, Mao MH, Hu YH, Zhou XC, Li S, Chen CF, Li CN, Yuan QL, Li W. Artemisinin protects DPSC from hypoxia and TNF-alpha mediated osteogenesis impairments through CA9 and Wnt signaling pathway. Life Sci. 2021;277:119471. doi: 10.1016/j.lfs.2021.119471. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Fan W, Xiao Y. The effect of hypoxia on the stemness and differentiation capacity of PDLC and DPC. Biomed Res Int. 2014;2014:890675. doi: 10.1155/2014/890675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanacker J, Viswanath A, De Berdt P, Everard A, Cani PD, Bouzin C, Feron O, Diogenes A, Leprince JG, des Rieux A. Hypoxia modulates the differentiation potential of stem cells of the apical papilla. J Endod. 2014;40:1410–1418. doi: 10.1016/j.joen.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Xu H, Xu F, Zhao J, Zhou C, Liu J. Platelet-rich plasma induces autophagy and promotes regeneration in human dental pulp cells. Front Bioeng Biotechnol. 2021;9:659742. doi: 10.3389/fbioe.2021.659742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Zhao X, He B, Wu W, Zhang H, Yang X, Cheng W. Autophagy activation by hypoxia regulates angiogenesis and apoptosis in oxidized low-density lipoprotein-induced preeclampsia. Front Mol Biosci. 2021;8:709751. doi: 10.3389/fmolb.2021.709751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pei F, Wang HS, Chen Z, Zhang L. Autophagy regulates odontoblast differentiation by suppressing NF-kappaB activation in an inflammatory environment. Cell Death Dis. 2016;7:e2122. doi: 10.1038/cddis.2015.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong G, Hu L, Liu Y, Bai S, Dai X, Yin L, Sun Y, Wang X, Hou L. Upregulation of HIF-1alpha protein induces mitochondrial autophagy in primary cortical cell cultures through the inhibition of the mTOR pathway. Int J Mol Med. 2014;34:1133–1140. doi: 10.3892/ijmm.2014.1850. [DOI] [PubMed] [Google Scholar]
- 37.Song H, Chen X, Jiao Q, Qiu Z, Shen C, Zhang G, Sun Z, Zhang H, Luo QY. HIF-1alpha-mediated telomerase reverse transcriptase activation inducing autophagy through mammalian target of rapamycin promotes papillary thyroid carcinoma progression during hypoxia stress. Thyroid. 2021;31:233–246. doi: 10.1089/thy.2020.0023. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Zhang C, Jiang X, Li L, Zhang D, Tang D, Yan T, Zhang Q, Yuan H, Jia J, et al. Involvement of autophagy in hypoxia-BNIP3 signaling to promote epidermal keratinocyte migration. Cell Death Dis. 2019;10:234. doi: 10.1038/s41419-019-1473-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Huang HY, Wang WC, Lin PY, Huang CP, Chen CY, Chen YK. The roles of autophagy and hypoxia in human inflammatory periapical lesions. Int Endod J. 2018;51(Suppl 2):e125–e145. doi: 10.1111/iej.12782. [DOI] [PubMed] [Google Scholar]
- 40.Ren C, Xu Y, Liu H, Wang Z, Ma T, Li Z, Sun L, Huang Q, Zhang K, Zhang C, et al. Effects of runt-related transcription factor 2 (RUNX2) on the autophagy of rapamycin-treated osteoblasts. Bioengineered. 2022;13:5262–5276. doi: 10.1080/21655979.2022.2037881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Wang P, Dong H, Wang S, Chu H, Yan W, Zhang X. Ulk1/FUNDC1 prevents nerve cells from hypoxia-induced apoptosis by promoting cell autophagy. Neurochem Res. 2018;43:1539–1548. doi: 10.1007/s11064-018-2568-x. [DOI] [PubMed] [Google Scholar]
- 42.Wu W, Li W, Chen H, Jiang L, Zhu R, Feng D. FUNDC1 is a novel mitochondrial-associated-membrane (MAM) protein required for hypoxia-induced mitochondrial fission and mitophagy. Autophagy. 2016;12:1675–1676. doi: 10.1080/15548627.2016.1193656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De R, Mazumder S, Bandyopadhyay U. Mediators of mitophagy that regulate mitochondrial quality control play crucial role in diverse pathophysiology. Cell Biol Toxicol. 2021;37:333–366. doi: 10.1007/s10565-020-09561-1. [DOI] [PubMed] [Google Scholar]
- 44.Wen W, Yu G, Liu W, Gu L, Chu J, Zhou X, Liu Y, Lai G. Silencing FUNDC1 alleviates chronic obstructive pulmonary disease by inhibiting mitochondrial autophagy and bronchial epithelium cell apoptosis under hypoxic environment. J Cell Biochem. 2019;120:17602–17615. doi: 10.1002/jcb.29028. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, Zang C, Yuan F, Ju C, Shang M, Ning J, Yang Y, Ma J, Li G, Bao X, Zhang D. The role of FUNDC1 in mitophagy, mitochondrial dynamics and human diseases. Biochem Pharmacol. 2022;197:114891. doi: 10.1016/j.bcp.2021.114891. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W. The mitophagy receptor FUN14 domain-containing 1 (FUNDC1): A promising biomarker and potential therapeutic target of human diseases. Genes Dis. 2021;8:640–654. doi: 10.1016/j.gendis.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu W, Lin C, Wu K, Jiang L, Wang X, Li W, Zhuang H, Zhang X, Chen H, Li S, et al. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J. 2016;35:1368–1384. doi: 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou H, Er P, Cheng J, Chen X, Ding X, Wang Y, Chen X, Yuan Z, Pang Q, Wang P, Qian D. High expression of FUNDC1 predicts poor prognostic outcomes and is a promising target to improve chemoradiotherapy effects in patients with cervical cancer. Cancer Med. 2017;6:1871–1881. doi: 10.1002/cam4.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lampert MA, Orogo AM, Najor RH, Hammerling BC, Leon LJ, Wang BJ, Kim T, Sussman MA, Gustafsson ÅB. BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy. 2019;15:1182–1198. doi: 10.1080/15548627.2019.1580095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.