Abstract
Genes encoding carbapenemases in 15 reference strains of Chryseobacterium (Flavobacterium) meningosepticum from the United Kingdom National Collection of Type Cultures and in one recent clinical isolate were investigated. All the strains hydrolyzed imipenem, but their levels of resistance to carbapenems varied, with imipenem and meropenem MICs ranging from 2 to >32 μg/ml. The blaB gene, which encodes a molecular-class B carbapenemase, was detected in only six reference strains and in clinical isolate 97/P/5448. The gene from 97/P/5448 had 98% nucleotide identity with the published sequence of blaB (from strain NCTC 10585) and was designated blaB2. A distinct carbapenemase gene, designated blaB3, was cloned from the type strain of C. meningosepticum, NCTC 10016. blaB3 had an open reading frame of 750 bp with 82% nucleotide identity to blaB and blaB2 and encoded a β-lactamase of 249 amino acids, including the putative signal peptide. This β-lactamase showed 87.6 and 86.7% amino acid homology with BlaB and BlaB2, respectively. blaB3 was detected in one other reference strain besides NCTC 10016, but the genetic basis of the carbapenemase activity detected in the other seven reference strains was not defined. Thus, neither blaB nor blaB3 was ubiquitous in the strains of C. meningosepticum studied, indicating that the reference strains may represent more than one bacterial species, each with its own intrinsic metallo-β-lactamase. Further taxonomic studies of C. meningosepticum are necessary to resolve this topic. Chryseobacterium spp. are environmental organisms and occasional opportunist pathogens. They apparently represent a reservoir of diverse metallo-β-lactamases, which potentially spread to gram-negative bacteria of greater clinical significance.
The carbapenem antibiotics imipenem and meropenem are increasingly used for the treatment of infections caused by multidrug-resistant gram-negative pathogens. They escape hydrolysis by most β-lactamases, including AmpC and extended-spectrum TEM and SHV types, but are labile to the metallo-enzymes of molecular class B (5). An acquired metallo-β-lactamase, IMP-1, has become scattered in Pseudomonas aeruginosa and Serratia marcescens in Japan, conferring high-level resistance to carbapenems (imipenem MIC > 32 μg/ml) and all other β-lactams (22). The blaIMP gene can be associated with integrons and plasmids, and this association is likely to enable its wider dissemination in the future. Recently blaIMP has been documented in Acinetobacter baumannii from Italy (6) and Klebsiella pneumoniae from Singapore (9). Besides IMP-1, other types of metallo-β-lactamases are beginning to appear in P. aeruginosa worldwide. One, designated VIM-1, was recently reported in an isolate from Italy (10), and we have identified an enzyme distinct from both IMP-1 and VIM-1 in isolates from Canada (N. Woodford, A. P. Gibb, and D. M. Livermore, unpublished data). Determining the origins of these carbapenemases requires knowledge of the chromosomal metallo-β-lactamases that are inherent to a few bacterial species, including some flavobacteria (5, 11).
A molecular-class B carbapenemase, BlaB, was characterized from Chryseobacterium (Flavobacterium) meningosepticum NCTC 10585 (20), and was postulated to be intrinsic to this species, which is frequently resistant to carbapenems (7). Nevertheless, the distribution of blaB among isolates of C. meningosepticum has not been reported. Another carbapenem-hydrolyzing β-lactamase, Ind-1, has recently been characterized from Chryseobacterium indologenes (2), and an unsequenced metallo-β-lactamase is widespread in Myroides odoratus (previously Flavobacterium odoratum) (21). These data indicate that metallo-β-lactamases are common in this bacterial group, which may act as a potential source for dissemination of their genes to gram-negative species of greater clinical significance. We report here the distribution of blaB among reference strains of C. meningosepticum from the United Kingdom National Collection of Type Cultures (NCTC) and a recent clinical isolate; we also report the nucleotide sequence of a novel metallo-β-lactamase gene cloned from the type strain of C. meningosepticum, NCTC 10016.
MATERIALS AND METHODS
Bacterial strains.
Fifteen reference strains of C. meningosepticum were obtained from the National Collection of Type Cultures (NCTC), United Kingdom (Table 1); strain NCTC 10016 is the type strain of this species. A clinical isolate of C. meningosepticum, 97/P/5448, collected from the sputum of a colonized United Kingdom hospital patient and referred to the Antibiotic Resistance Monitoring and Reference Laboratory was also investigated.
TABLE 1.
Characteristics of the strains of C. meningosepticum used in this studya
| Designation | Yr | Country of originb | MIC (μg/ml) of:
|
blaB detection by:
|
blaB3 detection by:
|
blaACME detection by PCR | blaCME-2 detection by PCR | pI(s) of β-lactamase(s) | Sp act (μmol/min/mg of protein)
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Imipenem | Meropenem | PCR | Probe | PCR | Probe | Imipenem | Cephaloridine | ||||||
| 97/P/5448 | 1997 | UK | >32 | >32 | + | + | − | − | + | + | 7.7, 8.2, 8.6, 8.9 | 0.53 | 0.17 |
| NCTC 10016T | 1958 | USA | 32 | 16 | − | − | + | + | − | − | 8.9 | 0.16 | 0.23 |
| NCTC 10585 | 1950 | USA | >32 | >32 | + | + | − | − | + | + | 7.1, 8.6 | 0.52 | 0.15 |
| NCTC 10586 | 1956 | USA | >32 | >32 | + | + | − | − | + | + | 8.6 | 0.43 | 0.14 |
| NCTC 10587 | 1961 | Ceylon | >32 | >32 | + | + | − | − | + | + | 7.1, 8.6 | 0.53 | 0.20 |
| NCTC 10588 | 1962 | USA | >32 | 16 | + | + | − | − | + | + | 7.7 | 0.39 | 0.07 |
| NCTC 10589 | 1962 | USA | 32 | 32 | − | + | − | − | + | + | 6.2 | 0.37 | 0.09 |
| NCTC 11305 | 1979 | France | >32 | >32 | − | − | − | − | + | + | 6.5, 7.7 | 0.36 | 0.09 |
| NCTC 11306 | 1979 | France | >32 | >32 | − | − | − | − | − | + | 7.1, 7.5, 7.9 | 0.23 | 0.19 |
| NCTC 11307 | 1979 | France | 4 | 2 | − | − | − | − | − | − | 8.2 | 0.66 | 0.01 |
| NCTC 11308 | 1979 | France | 4 | 8 | − | − | − | − | − | − | 7.6 | 0.18 | 0.01 |
| NCTC 11309 | 1979 | France | 32 | >32 | + | + | − | − | + | + | 8.2, 8.6 | 0.42 | 0.11 |
| NCTC 11310 | 1979 | France | 4 | 4 | − | − | − | − | − | − | 5.7, 6.2 | 0.09 | <0.01 |
| NCTC 11379 | 1980 | France | >32 | >32 | − | − | − | − | + | − | 7.1, 8.2, 8.6 | 0.04 | 0.18 |
| NCTC 11380 | 1980 | France | 2 | 4 | − | − | − | − | − | − | 5.7, 6.1 | 0.12 | 0.01 |
| NCTC 11381 | 1980 | France | >32 | 8 | − | − | + | + | − | − | 8.8, 9.1 | 0.20 | 0.01 |
PCR and probe results are presented as positive (+) or negative (−).
Abbreviations: UK, United Kingdom; USA, United States of America.
Hydrolysis of imipenem and cephaloridine.
Cultures of C. meningosepticum were grown overnight at 37°C on nutrient agar. The cells were then harvested into 2-ml amounts of 10 mM phosphate buffer, pH 7.0, and disrupted by three cycles of freezing and thawing. Debris was removed by centrifugation at 15,000 × g for 15 min, and the supernatants were retained at −20°C until needed. Hydrolysis of 0.1 mM imipenem (Merck, Hoddesdon, United Kingdom) was monitored by UV spectrophotometry at 297 nm at 37°C in 10 mM phosphate buffer, pH 7.0. Hydrolysis of 1 mM cephaloridine (Sigma, Poole, United Kingdom) was monitored at 295 nm. Specific activities of extracts were determined as described previously (12).
Antibiotic susceptibility testing.
All antibiotic susceptibilities were determined on Iso-Sensitest agar (Oxoid, Basingstoke, United Kingdom) with E-test strips (Cambridge Diagnostics Services, Cambridge, United Kingdom), which were used in accordance with the manufacturer's directions.
Cloning and sequencing.
Genomic DNA was extracted from clinical isolate 97/P/5448 and from type strain NCTC 10016 by standard methods (18). Five-microgram amounts of this DNA were partially digested with 10 U of NdeII (Life Technologies, Paisley, United Kingdom) for 5 min at 37°C, in the buffer supplied by the manufacturer, to yield fragments predominantly in the size range of 2 to 10 kb. After purification with a Recovery DNA Purification Kit II (Hybaid, Teddington, United Kingdom), the DNA fragments were ligated into the phagemid vector pBC SK(+) (Stratagene, Cambridge, United Kingdom), which had previously been digested with BamHI (Life Technologies) and dephosphorylated with calf intestinal alkaline phosphatase (Roche, Lewes, United Kingdom). Recombinant phagemids were transformed into Escherichia coli strain XL-1 Blue MRF′ (Stratagene) by electroporation (in a Gene Pulser; Bio-Rad, Hemel Hempstead, United Kingdom) performed at 2.5 kV, 200-Ω resistance, and 25-μF capacitance. Transformants were selected on nutrient agar containing chloramphenicol (30 μg/ml) and tetracycline (12.5 μg/ml); those likely to have acquired a β-lactamase gene were selected on the same medium, but also containing ampicillin (10 μg/ml). All these antibiotics were obtained from Sigma. Selected clones were tested for the ability to hydrolyze imipenem as described above, except that they were grown on nutrient agar containing chloramphenicol (30 μg/ml), tetracycline (12.5 μg/ml), and ampicillin (10 μg/ml) prior to preparation of the crude cell extracts.
Selected recombinant phagemids were purified with the Wizard Plus SV Miniprep DNA Purification System (Promega, Southampton, United Kingdom). Parts of the inserts were sequenced with an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Warrington, United Kingdom), initially with T3 (forward) and T7 (reverse) primers, both of which correspond to sequences located in the phagemid vector, and subsequently with primers designed from the sequences thereby obtained. Once detected, the open reading frames (ORFs) encoding β-lactamases were sequenced on both strands. Samples were analyzed on an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer).
Analysis of DNA and protein sequences.
Traces from the automated sequencer were visualized with Chromas 1.45 (http://www.technelysium.com.au/chromas14x.html). The Wisconsin Genetics Computer Group package (version 9.1, UNIX) was used for primer design, manipulation, and analysis of DNA sequences. Access to this package was provided by the Human Genome Mapping Project of the Medical Research Council of the United Kingdom. CLUSTAL W (23) was used to align protein sequences, and TREEVIEW (16) was used to visualize the resulting phylogenetic tree. Molecular masses and isoelectric points of peptides were calculated using the Compute pI/Mw tool (http://www.expasy.ch/tools/pi_tool.html).
PCR and hybridization assays for blaB and blaB3.
A 205-bp fragment of blaB, corresponding to nucleotides 136 to 340 of the published sequence from C. meningosepticum NCTC 10585 (GenBank accession no. X96858) (20), was amplified with the primers 5′-GCT TGA TTC TTG CTC TTG-3′ and 5′-AAT TTG TCT TCT CCC CAC-3′. A 265-bp fragment of blaB3, corresponding to nucleotides 439 to 703 (see Results), was amplified with the primers 5′-GTA GGA AAG GAT GAG TTT CAG G-3′ and 5′-GTG TAT GCT GAA TGG CAG TC-3′. The amplification conditions were (i) one cycle of 94°C for 5 min; (ii) 30 cycles of 94°C for 25 s, 52°C for 40 s, and 72°C for 50 s; and (iii) one cycle of 72°C for 6 min.
The blaB and blaB3 amplicons obtained from C. meningosepticum NCTC 10585 and NCTC 10016, respectively, were used as templates in a second round of amplification, during which digoxigenin-11-dUTP (Roche) was incorporated to generate blaB-specific and blaB3-specific probes. Genomic DNA was extracted (18) from strains of C. meningosepticum and digested with EcoRI (Life Technologies). After separation on 0.8% agarose gels, the DNA fragments were transferred to Hybond-N membrane (Amersham, Amersham, United Kingdom) by vacuum blotting on a VacuGene apparatus (Pharmacia Biotech, St. Albans, United Kingdom). Hybridization with the blaB or blaB3 probes was performed overnight at 68°C under conditions of high stringency, and the hybrids were detected colorimetrically as recommended by the manufacturer (Roche).
PCR assays for blaACME and blaCME-2.
Two class A extended-spectrum β-lactamases, designated CME-1 (19) and CME-2 (3), have recently been identified in C. meningosepticum. All strains were screened for the presence of the corresponding structural genes, blaACME and blaCME-2, which show 99.2% nucleotide identity. Two pairs of primers were used. Initially, a pair of primers (forward, 5′-CCG GAA TGT CAA AGC TTC-3′, and reverse, 5′-AGC ATC CCA GAC AAT TTT C-3′) were designed from the sequence of blaACME (19), but the screening was repeated subsequently with the primers reported by Bellais and colleagues (3). The amplification conditions were as for blaB variants.
IEF.
Crude cell extracts were prepared as for the hydrolysis studies and were subjected to isoelectric focusing (IEF) at 15 W of constant power on Ampholine PAGplate gels (pH 3.5 to 9.5; Pharmacia Biotech). β-Lactamase bands were visualized with 0.5 mM nitrocefin in 10 mM phosphate buffer, pH 7.0 (12).
Nucleotide sequence accession numbers.
The sequences of blaB2 and blaB3 reported in this paper have been assigned GenBank accession no. AF126542 and AF162284, respectively.
RESULTS
Antibiotic susceptibility testing.
The strains of C. meningosepticum varied in their susceptibility to imipenem and meropenem (MIC range, 2 to >32 μg/ml [Table 1]). Most strains were resistant to both carbapenems, but three (NCTC 11307, NCTC 11310, and NCTC 11380) were susceptible to both imipenem and meropenem on NCCLS criteria (MICs, ≤4 μg/ml); strain NCTC 11308 was phenotypically susceptible to imipenem but not to meropenem.
Distribution of blaB.
The blaB gene was detected by PCR in only 5 of 15 NCTC strains of C. meningosepticum (Table 1). blaB was also detected in clinical isolate 97/P/5448. PCR with each of these positive strains gave an amplicon of ca. 200 bp. Hybridization of genomic DNA with a blaB-specific probe showed that this gene was located on a ca. 2.5-kb EcoRI fragment in these six strains. One further strain of C. meningosepticum (NCTC 10589) that was negative for blaB by PCR also hybridized with the probe (Table 1), but the hybridizing fragment was smaller (<2.0 kb). The blaB gene was not detected in NCTC 10016, which is the type strain of C. meningosepticum.
Hydrolysis of imipenem and cephaloridine.
Crude cell extracts prepared from the 16 strains of C. meningosepticum hydrolyzed 0.1 mM imipenem rapidly (i.e., faster than the spontaneous breakdown), irrespective of the MICs of imipenem and meropenem. Specific activities of the extracts for imipenem and cephaloridine are shown in Table 1; activity against imipenem varied 16-fold among the strains; that against cephaloridine varied 25-fold. There was no clear relationship between imipenem MICs and specific activity against the compound.
Cloning and sequencing of carbapenemase genes from clinical isolate 97/P/5448 and type strain NCTC 10016.
One transformant colony derived from 97/P/5448 (blaB positive) was confirmed to hydrolyze nitrocefin. A crude cell extract prepared from this clone also hydrolyzed imipenem rapidly in a spectrophotometric assay (specific activities: imipenem, 0.74 μmol/min/mg of protein; cephaloridine, 0.1 μmol/min/mg of protein). The phagemid present in this clone was designated pARL98-1. A similar imipenem-hydrolyzing clone (specific activities: imipenem, 2.58 μmol/min/mg of protein; cephaloridine, 0.33 μmol/min/mg of protein) was derived from NCTC 10016 (blaB-negative), and the phagemid was designated pARL98-2. Despite their ability to hydrolyze imipenem, neither clone showed significant increases in the MIC of imipenem or meropenem in comparison with host E. coli strain XL-1 Blue MRF′ (Table 2). In both cases the β-lactamase genes were oriented relative to the multiple cloning site of the vector such that transcription occurred from the SacI site towards the KpnI site.
TABLE 2.
MICs of selected β-lactams for host strain E. coli XL-1 Blue MRF′ [pBC SK(+)] and clones containing the recombinant phagemids pARL98-1 and pARL98-2
| Antibiotic | MIC (μg/ml) for:
|
||
|---|---|---|---|
| pBC SK(+) | pARL98-1 (blaB2) | pARL98-2 (blaB3) | |
| Imipenem | 0.125 | 0.125 | 0.25 |
| Meropenem | 0.012 | 0.012 | 0.016 |
| Ampicillin | 4 | 16 | >256 |
| Piperacillin | 1 | 1 | 2 |
| Aztreonam | 0.25 | 0.25 | 0.25 |
| Cefoxitin | 16 | 16 | 16 |
| Cefpirome | 0.125 | 0.125 | 0.125 |
| Ceftazidime | 0.25 | 0.25 | 0.5 |
The sequenced portion of pARL98-1 (insert of ca. 4 kb) revealed one complete ORF of 750 bp (Fig. 1). A BLAST search indicated that this had 98% homology with blaB from C. meningosepticum strain NCTC 10585. The ORF showed 10 nucleotide substitutions compared with the published blaB sequence; 8 were silent changes, but 2 resulted in conserved amino acid substitutions (the A→T change at nucleotide 93 caused a Glu→Asp substitution at amino acid 31, and the A→G change at nucleotide 698 caused a His→Arg substitution at amino acid 233). The β-lactamase gene found on phagemid pARL98-1 (derived from C. meningosepticum 97/P/5448) was therefore designated blaB2.
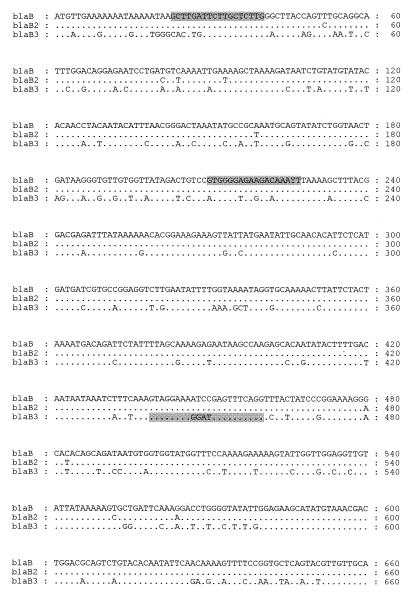
FIG. 1.
Nucleotide sequences of blaB2 (GenBank accession no. AF126542) from C. meningosepticum 97/P/5448 and blaB3 (GenBank accession no. AF162284) from the type strain, C. meningosepticum NCTC 10016, in comparison with that of blaB (20). Dots represent nucleotide identity. The positions of the blaB/blaB2-specific and blaB3-specific PCR primers used in this study are shown on respective sequences (shaded).
Sequencing of part of pARL98-2 (insert of ca. 3 kb) also revealed an ORF of 750 bp (Fig. 1), which showed 82% nucleotide identity with blaB and blaB2 and encoded a peptide of 249 amino acids. This peptide showed 87.6 and 86.7% amino acid homology with BlaB and BlaB2, respectively (Fig. 2). The β-lactamase gene found on phagemid pARL98-2 (derived from C. meningosepticum NCTC 10016) was designated blaB3.
FIG. 2.
CLUSTAL W alignment of BlaB, BlaB2, and BlaB3. Identical amino acids are indicated by asterisks, conserved substitutions are indicated by colons, and semiconserved substitutions are indicated by dots.
By comparison with the sequence of BlaB, both BlaB2 and BlaB3 were predicted to include a signal peptide of 22 amino acids (20). After cleavage of this signal peptide, the mature BlaB2 β-lactamase was calculated to have a molecular mass of 25,808 Da and a pI of 8.76; corresponding values for the mature BlaB3 enzyme were 25,849 Da and 8.65.
Comparison of BlaB3 with other metallo-β-lactamases.
BlaB3 was compared with BlaB, BlaB2 and several other metallo-β-lactamases using CLUSTAL W. BlaB3 shared the same phylogeny as BlaB and BlaB2. The Ind-1 enzyme from C. indologenes (2) also shared this phylogeny but appeared to have diverged from a common ancestor earlier in evolutionary history (not shown).
Distribution of blaB3.
blaB3 was detected in strains NCTC 10016 and NCTC 11381 both by PCR and by hybridization of a blaB3-specific probe to EcoRI-digested genomic DNA (Table 1). With each strain, the probe hybridized with two genomic DNA fragments, of 6.4 and <2 kb. As blaB3 has no internal EcoRI target sites, this observation may indicate the presence of two copies of blaB3 or the presence of another closely related allele in the genomes of these strains. blaB3 was not detected by PCR or hybridization in any of the other strains.
Distribution of blaACME and blaCME-2.
Isolate 97/P/5448 and seven of the 15 reference strains yielded PCR products with the primer pairs to both blaACME and blaCME-2 (Table 1). Thus, neither pair of primers was able to distinguish between these two closely related genes. One strain, NCTC 11379, gave a product only with blaACME primers, and another, NCTC 11306, gave a product only with blaCME-2 primers. These two anomalies most probably represent strains with sequence variations at positions corresponding to one or more of the primer annealing sites. All the blaB- or blaB2-containing strains also contained blaACME or blaCME-2, but these genes were detected in neither of the strains that carried blaB3 (Table 1).
IEF.
IEF of crude extracts prepared from the clone containing phagemid pARL98-1 (blaB2) produced a single nitrocefin-reactive band with a pI of 8.6. The clone containing pARL98-2 (blaB3) produced a single band with a pI of 8.3. The number of detectable bands in the C. meningosepticum strains studied varied from one to four (Table 1).
DISCUSSION
IMP-1 and VIM-1 metallo-β-lactamases have been reported in Pseudomonas, Acinetobacter, and Enterobacteriaceae from various countries (6, 9, 10, 22). Their origins remain unclear, as do those of the Sme-1, NMC-A, and IMI-1 serine carbapenemases, which have been found in a few Enterobacteriaceae in Europe and the United States (11); nevertheless the escape of intrinsic chromosomal metallo-β-lactamases has always appeared a possible route to carbapenem resistance in gram-negative opportunist pathogens. Previous examples of chromosomal β-lactamases of one species escaping to plasmids in others include SHV-1 and several AmpC types. SHV-1 is the chromosomal β-lactamase characteristic of K. pneumoniae but also occurs widely as a plasmid-mediated enzyme in both klebsiellae and other Enterobacteriaceae; chromosomal AmpC enzymes are characteristic of many Enterobacteriaceae (e.g., Citrobacter freundii and Enterobacter cloacae) but are increasingly reported as plasmid-encoded types in klebsiellae and E. coli (4, 13, 17). Knowledge of the carbapenem-hydrolyzing β-lactamases intrinsic to particular bacterial species is, therefore, invaluable for understanding the origins of the enzymes emerging in gram-negative pathogens.
In the present study, we showed that 15 NCTC strains of C. meningosepticum and a recent clinical isolate produced carbapenem-hydrolyzing β-lactamases, as demonstrated by their activity against imipenem. Surprisingly, some of these strains appeared susceptible to carbapenems in vitro (MICs, ≤4 μg/ml) (14). The blaB gene, which encodes a metallo-β-lactamase previously proposed as intrinsic to C. meningosepticum (20), or the closely related gene blaB2, was detected in only 7 of the 16 organisms, including the recent clinical isolate. A novel β-lactamase, designated BlaB3, which was phylogenetically related to BlaB, was characterized in the type strain NCTC 10016 and found in one other strain. Although blaB3 showed 82% nucleotide homology with blaB and blaB2, we were able to design two gene-specific PCR assays, and to use the products as gene-specific probes that did not cross-hybridize when used under stringent conditions because of the nucleotide divergence (Fig. 1). Seven reference strains contained neither blaB, blaB2, nor blaB3, and the basis of their carbapenem-hydrolyzing activity requires further investigation. We cannot exclude the presence of further allelic variants of blaB that were sufficiently divergent not to give products in our PCR and hybridization experiments. Neither BlaB, BlaB2, nor BlaB3 is ubiquitous in C. meningosepticum, and none could be called the sole chromosomal β-lactamase of this species.
As C. meningosepticum is an environmental species, the presence of diverse carbapenemases may represent acquisition from unidentified environmental sources. However, we feel that this is unlikely for two reasons: first, because the codon usage of blaB was not significantly different from that of other C. meningosepticum genes (20), and second, because BlaB, BlaB2, BlaB3, and Ind-1 (from C. indologenes) all belong to the same phylogenetic lineage. It seems more likely that the NCTC reference strains of C. meningosepticum comprise more than one species, each with its own intrinsic metallo-β-lactamase. It is known that the type strain, NCTC 10016, belongs to a distinct genomic group: thus, in DNA-DNA homology studies, NCTC 10016 shared only ca. 30% of its sequences with other C. meningosepticum strains tested, including those of the same serotype. The strains tested included NCTC 10585 and four others (NCTC 10586, 10587, 10588, and 10589) included in the present study (15). Further studies are necessary, but the solitary phylogenetic position of NCTC 10016 has caused the International Committee on Systematic Bacteriology Subcommittee on the Taxonomy of Flavobacterium and Cytophaga-like Bacteria to consider requesting the judicial commission for a change of the type strain (8).
Irrespective of their distributions, it is clear that both blaB and blaB3 were present in Chryseobacterium strains collected in the 1950s, long before the genus experienced selection pressure from human use of anti-gram-negative penicillins, let alone carbapenems. These early hosts included NCTC 10585, from which blaB was first identified (20), and NCTC 10016, from which blaB3 was characterized.
Once transferred to E. coli, BlaB2 failed to raise the MICs of imipenem and meropenem, and BlaB3 raised them by no more than twofold, with the MICs remaining well below the breakpoints of the NCCLS (14) and British Society for Antimicrobial Chemotherapy (24). Hydrolysis of imipenem nevertheless confirmed that both genes were expressed in the transformants. This behavior is identical to that seen when IMP-1 is cloned into E. coli (1) and leads to the suggestion that these enzymes require an impermeable host strain or species to confer substantial carbapenem resistance or even require the additional loss of carbapenem-specific porins, such as OprD (D2) in P. aeruginosa (22). The low levels of resistance conferred in E. coli lead to practical problems when cloning these β-lactamase genes, precluding selection with carbapenems. In this study the initial selection of transformants was with ampicillin, and carbapenemase activity was confirmed by hydrolysis assays. It also raises the worrying possibility that if these enzymes were to escape into E. coli, they might become widely disseminated before their presence were detected. Such possibilities underscore the need for cautious, appropriate use of these antibiotics and highlight the emerging need for pharmaceutical research on inhibitors of metallo-β-lactamases.
ACKNOWLEDGMENTS
We are grateful to Patricia J. Woodford (Imperial College of Science & Technology at St. Mary's, London, United Kingdom) for processing samples on the automated sequencer.
We thank the PHLS Small Scientific Initiatives Fund and Wyeth UK for financial support.
REFERENCES
- 1.Babini G S, Yuan M, Livermore D M. Interactions of β-lactamases with sanfetrinem (GV 104326) compared to those with imipenem and with oral β-lactams. Antimicrob Agents Chemother. 1998;42:1168–1175. doi: 10.1128/aac.42.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellais S, Leotard S, Poirel L, Naas T, Nordmann P. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol Lett. 1999;171:127–132. doi: 10.1111/j.1574-6968.1999.tb13422.x. [DOI] [PubMed] [Google Scholar]
- 3.Bellais S, Poirel L, Naas T, Girlich D, Nordmann P. Genetic-biochemical analysis and distribution of the Ambler class A β-lactamase CME-2, responsible for extended spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob Agents Chemother. 2000;44:1–9. doi: 10.1128/aac.44.1.1-9.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford P A, Urban C, Mariano N, Projan S J, Rahal J J, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and loss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bush K. Metallo-β-lactamases: a class apart. Clin Infect Dis. 1998;27(Suppl. 1):S48–S53. doi: 10.1086/514922. [DOI] [PubMed] [Google Scholar]
- 6.Cornaglia G, Riccio M L, Mazzariol A, Lauretti L, Fontana R, Rossolini G M. Appearance of IMP-1 metallo-β-lactamase in Europe. Lancet. 1999;353:899–900. doi: 10.1016/s0140-6736(98)05954-6. [DOI] [PubMed] [Google Scholar]
- 7.Di Pentima M C, Mason E O, Jr, Kaplan S L. In vitro antibiotic synergy against Flavobacterium meningosepticum: implications for therapeutic options. Clin Infect Dis. 1998;26:1169–1176. doi: 10.1086/520309. [DOI] [PubMed] [Google Scholar]
- 8.Holmes B. International Committee on Systematic Bacteriology Subcommittee on the Taxonomy of Flavobacterium and Cytophaga-like Bacteria. Minutes of the Meeting, 3 April 1992, Bloemfontein, Republic of South Africa. Int J Syst Bacteriol. 1993;43:623. [Google Scholar]
- 9.Koh T H, Babini G S, Woodford N, Sng L-H, Hall L M C, Livermore D M. Carbapenem-hydrolysing IMP-1 β-lactamase in Klebsiella pneumoniae from Singapore. Lancet. 1999;353:2162. doi: 10.1016/s0140-6736(05)75604-x. [DOI] [PubMed] [Google Scholar]
- 10.Lauretti L, Riccio M L, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini G M. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–1590. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livermore D M. Acquired carbapenemases. J Antimicrob Chemother. 1997;39:673–676. doi: 10.1093/jac/39.6.673. [DOI] [PubMed] [Google Scholar]
- 12.Livermore D M, Williams J D. β-Lactams: mode of action and mechanisms of bacterial resistance. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, Md: Williams & Wilkins; 1996. pp. 502–578. [Google Scholar]
- 13.Marchese A, Arlet G, Schito G C, Lagrange P H, Philippon A. Characterization of FOX-3, an AmpC-type plasmid-mediated β-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob Agents Chemother. 1998;42:464–467. doi: 10.1128/aac.42.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. 1997. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa. [Google Scholar]
- 15.Owen R J, Holmes B. Identification and classification of Flavobacterium species from clinical sources. In: Reichenbach H, Weeks O B, editors. The Flavobacterium-Cytophaga group. Proceedings of the International Symposium on Yellow-Pigmented Gram-Negative Bacteria of the Flavobacterium-Cytophaga Group. Weinheim, Germany: Verlag Chemie; 1981. pp. 39–50. [Google Scholar]
- 16.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 17.Payne D J, Woodford N, Amyes S G B. Characterization of the plasmid-mediated β-lactamase BIL-1. J Antimicrob Chemother. 1992;30:119–127. doi: 10.1093/jac/30.2.119. [DOI] [PubMed] [Google Scholar]
- 18.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 19.Rossolini G M, Franceschini N, Lauretti L, Caravelli B, Riccio M L, Galleni M, Frère J-M, Amicosante G. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaACME) encoding an extended-spectrum class A β-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER β-lactamase. Antimicrob Agents Chemother. 1999;43:2193–2199. doi: 10.1128/aac.43.9.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossolini G M, Franceschini N, Riccio M L, Mercuri P S, Perilli M, Galleni M, Frère J-M, Amicosante G. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing broad substrate profile. Biochem J. 1998;332:145–152. doi: 10.1042/bj3320145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato K, Fuji T, Okamoto R, Inoue M, Mitsuhashi S. Biochemical properties of β-lactamase produced by Flavobacterium odoratum. Antimicrob Agents Chemother. 1985;27:612–614. doi: 10.1128/aac.27.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. PCR detection of metallo-β-lactamase gene (blaimp) in gram-negative rods resistant to broad-spectrum β-lactams. J Clin Microbiol. 1996;34:2909–2913. doi: 10.1128/jcm.34.12.2909-2913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Working Party of the British Society for Antimicrobial Chemotherapy. A guide to sensitivity testing. J Antimicrob Chemother. 1991;27(Suppl. D):1–50. [PubMed] [Google Scholar]