Abstract
I-motifs (iMs) are non-canonical DNA secondary structures that fold from cytosine (C)-rich genomic DNA regions termed putative i-motif forming sequences (PiMFSs). The structure of iMs is stabilized by hemiprotonated C-C base pairs, and their functions are now suspected in key cellular processes in human cells such as genome stability and regulation of gene transcription. In plants, their biological relevance is still largely unknown. Here, we characterized PiMFSs with high potential for i-motif formation in the rice genome by developing and applying a protocol hinging on an iMab antibody-based immunoprecipitation (IP) coupled with high-throughput sequencing (seq), consequently termed iM-IP-seq. We found that PiMFSs had intrinsic subgenomic distributions, cis-regulatory functions and an intricate relationship with DNA methylation. We indeed found that the coordination of PiMFSs with DNA methylation may affect dynamics of transposable elements (TEs) among different cultivated Oryza subpopulations or during evolution of wild rice species. Collectively, our study provides first and unique insights into the biology of iMs in plants, with potential applications in plant biotechnology for improving important agronomic rice traits.
INTRODUCTION
I-motifs (iMs) are non-canonical DNA quadruplexes held by hemiprotonated cytosine–cytosine (C) base pairs. iMs fold from C-rich genomic DNA regions harboring putative i-motif forming sequences (PiMFSs) under slightly acidic environment (1,2). Hemiprotonated C-C base pairs were first identified in 1962 (3) and proposed for the stabilization of hairpins (4). The structure of the first intercalated iM tetramers was elucidated by NMR in 1993 (1) and crystallographic studies in 1994 (5). Because of the acidic condition requirement, iMs were long considered as structural oddities, as their folding was supposed not to be compatible with physiological conditions (pH ∼7.4). Their biological relevance was therefore largely overlooked, and the pace of progress toward its elucidation was very slow as compared to other secondary DNA structures, notably G-quadruplexes (G4s) that fold from guanine (G)-rich sequences (6). This is, however, of utmost importance as genomic G4- and iM-forming sequences are, by definition, complementary.
Therefore, specific conditions have been discovered to favor or facilitate iM formation at physiological pH, such as DNA supercoiling (7), molecular crowding (8,9) and the presence of small molecules (ligands) (10,11), reinvigorated research on iMs (12). Recent immunodetection studies performed with the iM-specific iMab antibody have demonstrated that iM folding is cell cycle-dependent in HeLa cells, pH-dependent in MCF7 cells and occurs preferentially at the G1/S phase in U2OS cells (13). The demonstration of the relevance of iMs in a normal cellular context thus provides a key turning point in iM research.
iMs were found to be markedly overrepresented in promoters, telomeres and centromeres, which is indicative of their critical cellular roles (6,14), including maintenance of genome stability (15) and regulation of gene expression (16–24). In humans, iMs can be found in several oncogenes such as c-MYC, c-MYB, BCL-2, ILPR, Rb, RET and VEGF (18,25–29). iMs have also been involved in the regulation of the gene transcription during the metamorphosis of Bombyx mori (23). iMs frequently localize at centromeres of both humans and Drosophila (30–33), and at telomeres of both humans and Arabidopsis (34,35).
These results substantiate the putative key roles that the iMs might play in human cells (36) or other eukaryotic genomes, but much less is known about their possible roles in plants so far. Fragmentary information has been gathered over the years through low throughput-screening techniques such as circular dichroism (CD) (37), nuclear magnetic resonance (NMR) (38), native PAGE (34) and chemical (Br2) footprinting (27). Unlike G4s, high-throughput techniques, such as sequencing-based approaches, -related characterization of iMs are still missing in both mammals and plants. Here, we designed and performed iMab-based immunoprecipitation followed by sequencing (iM-IP-seq) to identify PiMFSs with high potential for i-motif formation in the rice genome. We showed the relationships between DNA methylation and iM formation, and focused on the potential impacts of iMs on TEs (transposable elements) stability and its evolution across Oryza populations.
MATERIALS AND METHODS
Plant materials
Nipponbare (Japonica) rice seeds (Oryza. sativa L.) were pre-germinated at room temperature (RT) for 3 days. Germinated seeds were grown in a greenhouse at 28–30°C and a 14 h/10 h light–dark cycle. Two-week-old rice seedlings were cut into 1–1.5 cm slices and cross-linked in HEPES buffer pH = 8.0 (20 mM HEPES, 1 mM EDTA, 100 mM NaCl and 1 mM PMSF) with a final 1% of formaldehyde under vacuum for 10 min at RT. A final concentration of 0.125 M glycine was added for an additional 5 min vacuum for quenching excess of formaldehyde. The cross-linked seedlings were ground to a fine powder using liquid nitrogen.
iM-IP-seq
The fine powder was used for nuclei preparation and genomic DNA extraction following the procedures as described previously (39). The genomic DNA extracted from wild-type or Zebularine-treated seedlings was fragmented into sizes, ranging from 100 to 500 bp, using the water-based Biorupter (Diagnode), followed by DNA extraction and purification. Total 5 μg fragmented genomic DNA was diluted in iM stabilization buffer (50 mM Tris-AcOH, pH 5.5) with (a crowding condition for mimicking the cellular and/or nucleus condition) or without (CK) 40% PEG200, then was denatured and re-associated using a PCR program as below: 95°C for 8 min, 95°C for 30 s (-0.5°C/cycles, 129 cycles), 35°C hold on. The re-associated DNA was diluted with iM-IP incubation buffer (50 mM Tris-AcOH, 1 mM MgCl2, 130 nM CaCl2, 1% BSA and Complete mini, pH 5.5), then incubated with 3 μg iMab antibody (Ab01462-23.0, Kappa) for 4 h at 4°C. The antibody incubation reaction was incubated with 30 μl of washed protein G Dynalbeads (10004D, Invitrogen) for another 4 h at 4°C. The iMab-bound DNA was finally eluted with 200 μl elution buffer (0.1 M NaHCO3 and 1% SDS) at 65°C for two times with 15 min each. Three biologically replicated PiMFS-IPed CK DNA and two biologically replicated PiMFS-IPed demethylated DNA, two biologically replicated PiMFS-IPed DNA in a crowding condition, and the corresponding Input/IgG-IPed control DNA were used for library preparation. All libraries were prepared using the NEBNext® Ultra™ II DNA Library Prep Kit (NEB, E7645S). Libraries were finally quality controlled and sequenced using the paired-end mode on Illumina NovaSeq platform.
Protoplast transient transfection
Preparation of a modified pJIT163-hGFP vector containing PiMFSs and protoplast transient transfection assay were conducted following the procedures as previously described (40). Briefly, PiMFS DNA fragments were amplified using PCR (Supplementary Table S4) and cloned into a modified pJIT163-hGFP vector. The pJIT163-hGFP vector containing mini35S promoter only (negative control), an intact 35S promoter (positive control) and a mini 35S promoter + amplified PiMFS DNA fragment were transfected into protoplasts mediated by PEG. GFP signals were captured using fluorescent microscopy.
iM-IP-qPCR assay
For iM-IP–qPCR assay, 1 μl of input and IPed DNA (2 ng/μl each) was used as DNA templates. The enrichment of IPed DNA was calculated using the 2(ΔΔCt) method and expressed as fold change over the corresponding input. Each primer set was repeated three times in each qPCR. All primer sequences are listed in Supplementary Table S4.xlsx. Significance test was performed using one-way ANOVA analysis. ***P < 0.001, **P < 0.01 and *P < 0.05.
Dot blotting assays
For the dot blotting assay, genomic DNA, synthesized oligonucleotides and M.SssI-treated genomic DNA were denatured and re-associated in iM reconstruction buffer (50 mM Tris-AcOH, pH 5.5) at 95°C for 8 min. Denatured and re-associated DNA or sonicated chromatin prepared from the purified nuclei was loaded on Amersham Hybond-N+-nylon membrane followed by pre-blocking in 5% milk for 45 min at RT. The pre-blocked membrane was incubated with the iMab antibody overnight at 4°C, then with anti-IgG (HRP) antibody for an additional 1.5 h. The procedures for immune-signal development were the same as described before (41). Each blot was repeated at least two times for quantification signal intensity.
Native polyacrylamide gel electrophoresis
Synthesized DNA oligonucleotides were denatured at 95°C for approximately 8 min in iM reconstruction buffer (50 mM Tris-AcOH, 30 mM KCl, pH 5.5), then slowly cooled down to room temperature overnight for re-association, the counterpart for each sample with denaturation but without re-association was used as control. All samples were incubated at 4°C for 10min before loading on to 15% polyacrylamide gel. The gels were run at a constant voltage of 85 V at RT for 1.5 h. Gels were stained using Stains GeneRed (Tiangen, RT211) solution and visualized under UV light.
CD measurements
About 500 μl of 5 μM PiMFS oligonucleotide in iM stabilization buffer (50 mM Tris-AcOH, pH 5.5) was measured in an optical chamber (1 mm path length) with a JASCO J-815 spectropolarimeter (Tokyo, Japan). Dry purified nitrogen gas was used to maintain a deoxygenation atmosphere. The solution background was subtracted from the CD signal.
iM-IP-seq data analysis
Raw iM-IP-seq data were trimmed by using Fastp for removal of adapter sequence. All clean reads in three biological replicates were aligned to the MSU v7.0 reference genome(http://rice.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/all.dir/) using BWA (Burrows-Wheeler Aligner) (mem algorithm, version 0.7.17) with default parameters. SAMtools was used to exclude reads with mapping quality below 10. PCR duplicates were removed by using Picard. The Spearman rank correlation coefficient between replicates was calculated using multiBamSummary and plotCorrelation function of deepTools. Aligned reads with at least length 50 were used for calling PiMFS-IPs+ peaks, which were identified by iM-IP-seq, by using MACS2 (version 2.1.1) (42) with parameters as below: callpeak -g 3.8e + 8 -p 1e-4 -nomodel -f BAMPE. Input and IgG data were used as controls.
In silico identification of putative i-motif forming sequences
Putative i-motif forming sequences (PiMFSs) were identified through screening the MSU v7.0 reference genome using G4iMGrinder from the R package (43). Parameters were tested for obtaining regular PiMFSs with sequences as [CC2-5L1-15]3–8C2-5 followed by further selection of several subtypes of PiMFSs. To assess folding potential of each sequence, score of each individual putative sequence was calculated by considering the run size, bulges between runs and loop size. PiMFSs with score below -40 were chose for the further analyses.
To distinguish PiMFSs with long and short loop sizes, the entire regular PiMFS sequences were computationally scanned using fastaRegexFinder.py (https://github.com/dariober/bioinformaticscafe/blob/master/fastaRegexFinder.py) (44). Short loops contain sequences as (loop 1 (2-nt): loop 2 (3 to 4-nt): loop 3 (2-nt), i.e. 2:3–4:2), while long loops contain sequences as (loop1 (6–8):loop2 (2–5):loop 3 (6–7)).
Genomic distribution and calculation of GC content, GC/AT skew
ChIPseeker from the R package (45) was used to investigate genomic distribution of PiMFS-IPs+ and PiMFS-IPs–, representing PiMFSs detected and undetected by iM-IP-seq, respectively. For calculating GC content and GC/AT skew, ±1 kb regions around the center of PiMFS-IPs+/− were divided into 50 bp windows, then were calculated using the following formulas: GC content = (C + G)/(C + G + A + T); GC skew = (G-C)/(G + C) and AT skew = (A-T)/(A + T). The genomic regions lacking PiMFSs were randomly selected as controls by using bedtools shuffle.
Motif prediction
± 100 bp DNA sequences around the center of PiMFS-IPs+ peaks were used for motif identification using MEME-ChIP (http://memesuite.org/tools/meme-chip) (46) with parameters as options minimum width 5 and maximum width 20. TF database from Arabidopsis was used to match putative TF-binding sites (Tomtom tool) corresponding to all identified motifs. The motifs listed in the result represent the top three significantly enriched motifs with the highest E-values.
DNase-seq data analysis
To integrate DNase-sequencing (DNase-seq) with iM-IP-seq, the published DNase-seq data (GSE26734) (39) were used for identification of DNase I hypersensitive sites (DHSs) using F-seq (47) with 200 bp bandwidth and the FDR (false discovery rate) cutoff <0.05. The FDR represents the ratio of DHSs from DNase-seq relative to DHSs identified from 10 random data sets.
BS-seq data analysis
To examine the DNA methylation levels within PiMFS-IPs+ regions, bisulfite-sequencing (BS-seq) data (48) were analyzed by mapping clean data to the rice reference genome (MSU7.0) using Bismark (49). The methylated cytosines were counted from total uniquely mapped reads using the bismark methylation_extractor program. The DNA methylation levels were calculated using the total number of all (C + T) ≥5 in each position. The DNA methylation levels in different regions were calculated from 50 bp windows within each region.
Read count normalization
The ±1 kb upstream/downstream of center of PiMFS-IPs+/− were divided into 50 bp windows. The ±500 bp upstream/downstream of TEs and TE regions were equally divided into 25 bins for normalization. The number of reads per sliding window was divided by the window length, then by the count of all uniquely mapped reads within the genome (Mb). The midpoint of the fragment was used to determine the position in rice genome.
RESULTS
Global identification of PiMFSs with high potential for i-motif formation in the rice genome
iMab is an antibody that binds iMs with a high affinity. It was obtained by screening the Garvan-2 human single-chain variable fragment (scFv) library (13). It was reported to specifically bind to iMs present in human telomere in vitro, termed hTelo iM, in human cells (13). The use of this antibody has greatly helped gain insights into the biological relevance of iMs in eukaryotes. However, iMab-based high throughput sequencing for characterization of iMs is still missing.
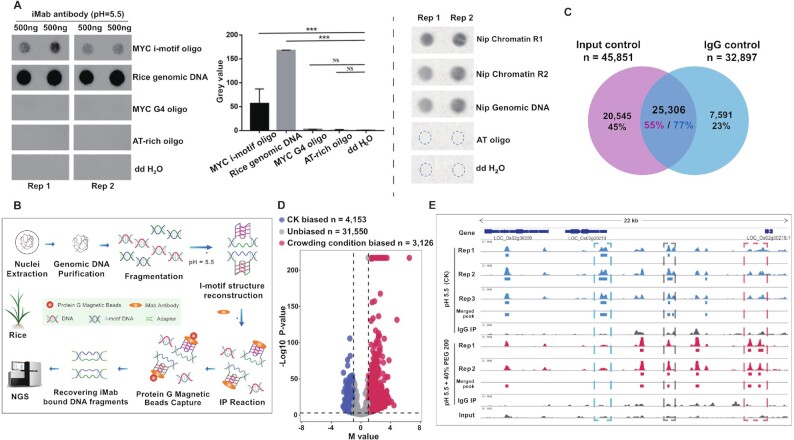
We decided to investigate this in the rice genome, using a protocol referred to as iM-IP-seq. We first validated the use of iMab by performing dot blotting assays: as seen in Figure 1A, clear dot signals were obtained using the synthetic MYC iM, the rice genomic DNA and sonicated chromatin, but almost nothing with the controls, i.e., the synthetic MYC G4 (13), an AT-rich oligonucleotide and ddH2O (Figure 1A). This first series of results confirmed the efficiency of iMab as well as its specificity for iMs due to almost non-detectable cross-reactivity of iMab with G4s in vitro.
Figure 1.
Global identification of PiMFS-IPs+ in the rice genome. (A) Dot blotting assays illustrating immuno-signals clearly detected in the synthesized MYC i-motif oligonucleotide and the rice genomic DNA, but almost undetectable in the synthesized MYC G4 oligonucleotide, the synthesized AT-rich oligonucleotide and ddH2O (left). The signal intensity was quantified from at least two replicates (middle). Immuno-signals can also be clearly detected in the sonicated chromatin (right). Significance test was determined by One-way ANOVA analysis; *** P-value < 0.001, NS: non significance. (B) Schematic diagram illustrating the main procedures for iM-IP-seq methodology. (C) Venn plot illustrating 25,306 common PiMFS-IPs+ peaks relative to the Input (n = 45 851) and IgG (n = 32 897) as controls. (D) Effects of the crowding condition on iM formation. MA plot illustrating the crowding condition biased PiMFS-IPs+ peaks (n = 3126), CK biased PiMFS-IPs+ peaks (n = 4153) and unbiased PiMFS-IPs+ peaks (n = 31 550) were identified using MAnorm with the cutoff indicated by the dotted line (x = ±1 and y = 3), P-value < 0.001 and |M value| > 1. y-axis represents -log10(P–value) and x-axis represents M value defined by MAnorm. (E) A representative Integrative Genomics Viewer (IGV) snapshot across a 22 kb window from the rice chromosome 2 illustrating reproducibility of PiMFS-IPs+ among biological replicates in normal and a crowding condition. The left, middle and right box indicating the CK biased, unbiased and the crowding condition biased PiMFS-IPs+ peaks, respectively.
Next, we developed an iM-IP-seq protocol for the global identification of PiMFSs with high potential for i-motif formation, termed PiMFS-IPs+, representing the maximum possibility of i-motif formation for the rice genomic DNA. As seen in Figure 1B, the main procedures include genomic DNA purification and fragmentation, reconstruction of iM structures using iM-stabilizing conditions (50 mM Tris-AcOH, pH 5.5), iMab incubation and recovery of iMab-captured DNA fragments for library preparation and sequencing. We sequenced three biological iM-IP-seq libraries along with one input and one IgG library as controls (Supplementary Table S1). As the three biological replicates were highly correlated (r = 0.98) (Supplementary Figure S1), we merged them, which led to the identification of 45 851 and 32 897 PiMFS-IPs+ peaks relative to input and IgG control, respectively, and 25 306 common PiMFS-IPs+ peaks (Figure 1C). To mimic the cellular and/or nucleus condition, we reconstructed the i-motif structures in a crowing condition (iM stabilization buffer with 40% PEG200), then conducted iM-IP-seq with two biological replicates. After comparing PiMFS-IPs+ formed in the crowding condition relative to CK by using MAnorm (50), we identified 3126 crowding condition biased, 4153 CK biased and 31 550 unbiased PiMFS-IPs+ peaks (Figure 1D). This result indicated that a portion of iMs can be differentially formed under the normal cellular and/or nucleus condition, which is necessary to be further investigated. A representative Integrative Genomics Viewer (IGV) spanning a 22 kb region in the rice Chromosome 2 show the reproducibility of PiMFS-IPs+ detected among the replicates in normal and a crowding condition (Figure 1E). We finally determined to use 25 306 common PiMFS-IPs+ peaks for the downstream analyses.
Validation of PiMFS-IPs+
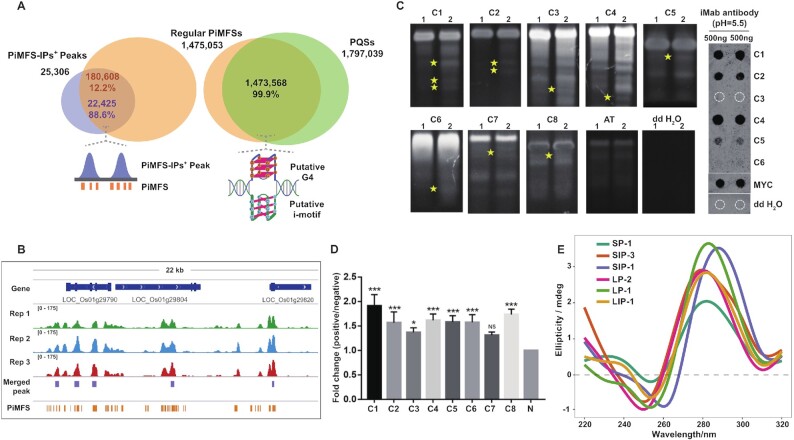
To further demonstrate that the detected C-rich sequences are iM-prone, we conducted in silico investigations with the 25 306 common PiMFS-IPs+ peaks using G4iMGrinder (43). We searched for all PiMFSs bearing at least four runs of two consecutive Cs, with loop length between 1 and 15 nt (threshold score < -40). On a genome-wide scale, we identified 1 475 053 PiMFSs of formula [CC2-5L1-15]3C2-5 (Supplementary Table S2): 99.9% of these PiMFSs were found to overlap putative G4-forming sequences (PQS) with runs of two consecutive Gs, confirming the genomic connection between G4s and iMs (Figure 2A, right). 88.6% of them belonged to PiMFS-IPs+ peaks (22 425 of 25 306) (Figure 2A left, B), implying that the remaining 2881 PiMFS-IPs+ peaks actually correspond to non-regular PiMFSs, that is, with sequences variations. This was further investigated by lowering the threshold score to < -30, which led to the identification of 25 114 (∼99.4%) PiMFS-IPs+ peaks overlapping all PiMFSs. It confirmed that the 2881 candidates correspond to non-classical iMs: a closer look to the sequences confirmed occurrence of sequence variations as compared to the regular ones (Supplementary Table S3); they included iMs in which C-runs disrupted by at least one additional nucleotide (A, T, G) and iMs with four to nine C-runs ([CC2-5L1-15]4–9C2-5). All PiMFSs were thus classified into two subtypes with PiMFS-IPs+ (n = 180 608), and PiMFS-IPs– (n = 1 294 445), representing PiMFSs detected and undetected by iM-IP-seq, respectively (Supplementary Figure S2).
Figure 2.
Validation of PiMFS-IPs+. (A) Venn plot illustrating PiMFS-IPs+ peaks overlapping regular PiMFSs (left), and regular PiMFSs overlapping G2 PQFSs (right). Schematic diagram right below the venn plot illustrating PiMFSs overlapping PiMFS-IPs+ peaks (left) or the complementary G2 PQFSs (right). (B) A representative Integrative Genomics Viewer (IGV) snapshot across a 22 kb window from the rice chromosome 1 illustrating PiMFS-IPs+ peaks overlapping PiMFSs. (C) iMab based dot blotting (right) for C1-C6 loci and Native PAGE (left) assay for C1-C8 loci corresponding to PiMFS-IPs+ peaks, synthesized MYC oligonucleotide and ddH2O only. Yellow star indicates distinct DNA bands formed in denaturing and re-associated DNA relative to denaturing DNA only, reflecting the formation of iMs after DNA re-association. (D) iM-IP-qPCR assay for 8 positive PiMFS-IPs+ peaks loci (C1-C8) and 1 non-PiMFS-IPs+ negative control (N). Fold change (P/N) indicates the enrichment levels between each positive locus over the negative locus. Significance test was determined by One-way ANOVA analysis, *** P-value < 0.001, * P-value < 0.05, NS: non-significant. (E) CD confirmation of other 6 positive loci corresponding to PiMFS-IPs+ peaks.
The iM-folding ability of randomly selected 6 PiMFS-IPs+ peaks was confirmed by dot blotting assay (Figure 2C, right panel) and native polyacrylamide gel electrophoresis (PAGE), using C1-C8 as positive controls, and a synthetic AT-rich oligonucleotide and ddH2O as negative controls (Supplementary Table S4). We observed 5 (∼83%) loci with detectable immune-signals (Figure 2C, right panel). For the native PAGE, the formation of iMs was promoted by a denaturation/renaturation cycle (lanes 2), which leads to distinct DNA bands (yellow star, as compared to lane 1) that correspond to iMs formation (Figure 2C, left panel). Moreover, eight positive loci were also significantly enriched during iM-IP-qPCR experiments (Figure 2D). In addition, for CD (circular dichroism) spectroscopy assay, we observed that randomly selected other six oligonucleotides exhibited a clear absorbance peak increased at around 290 nm but decreased at around 260 nm, confirming their ability to adopt iM structures (Figure 2E).
Collectively, all above analyses show that those PiMFS-IPs+ peaks are reliable for the downstream assays.
Genomic distribution and sequence features of PiMFS-IPs+
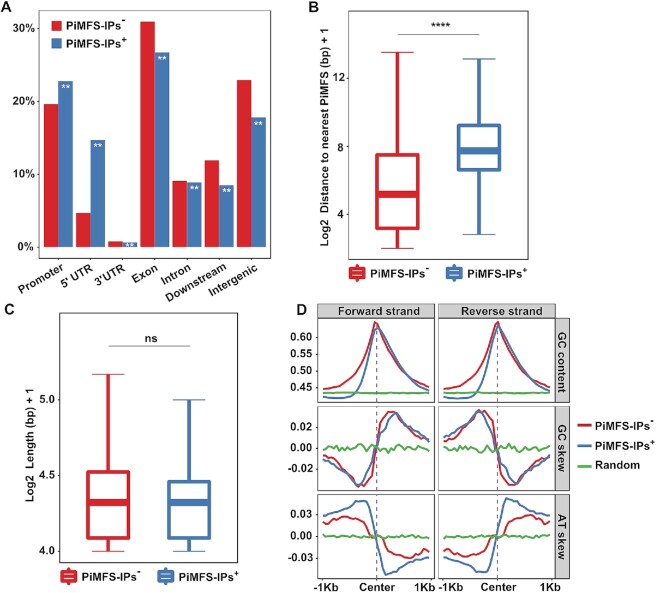
It is now established that iMs or G4s are abundantly present in promoters and untranslated regions (UTRs) of human oncogenes and tumor suppressor genes (51,52). To assess genomic distributions of iMs in the rice genome, we partitioned the whole genome into seven functionally annotated subregions, including promoters, 5′UTRs, 3′UTRs, exons, introns, downstream and distal intergenic regions. We observed that PiMFS-IPs+ were enriched in some subgenomic regions (1.2-fold in promoters, 1.3-fold in 5′UTRs) but depleted in the others (e.g., 0.8-fold in exons, 0.7-fold in distal intergenic regions) as compared to PiMFS-IPs– (Figure 3A). As shown in Figure 3B,C, we found no significant differences in the mean length of PiMFSs between PiMFS-IPs+ and PiMFS-IPs–, while the distance between PiMFSs is longer for PiMFS-IPs+ as compared to PiMFS-IPs–, indicating that the genomic density of PiMFSs may affect iM formation in vitro (Figure 3B,C).
Figure 3.
Genomic distributions and sequence features of PiMFS-IPs+. (A) Subgenomic distributions of PiMFS-IPs+/−, including promoters, 5′UTRs, 3′UTRs, exons, introns, downstream and intergenic regions. Significance test was determined by hypergeometric test; ** P-value < 0.01. (B) Distance from PiMFS-IPs+ (right) or PiMFS-IPs– (left) to the nearest neighboring PiMFSs. (C) Mean length of PiMFS-IPs+ (right) and PiMFS-IPs– (left). Significance tests in (B) and (C) were determined by Wilcoxon rank-sum test; **** P-value < 0.0001, NS: non-significant. (D) Strand-specific GC content, GC and AT skews calculated around ±1 kb from the center of PiMFS-IPs+, PiMFS-IPs– and random sequences as indicated.
It is also documented that long loops facilitate more stable iMs than short loops (53). To study this, we classified PiMFS-IPs+ peaks into three subclasses according to the loop length as previously described (18), that is, with short loops (loop 1 (2-nt): loop 2 (3 to 4-nt): loop 3 (2-nt), i.e., 2:3–4:2), long loops (6–8:2–5:6–7) and both short and long loops. We then compared read intensity (normalized read counts) for each subclasses. We found that, globally, PiMFS-IPs+ peaks with both short and long loops had the highest read intensity (Supplementary Figure S3A), and that PiMFS-IPs+ with long or short loops had much higher read intensity than PiMFS-IPs– (Supplementary Figure S3B). We also found that PiMFS-IPs+ have similar GC content and skew as PiMFS-IPs– (Figure 3D, top and middle panel), while the AT skew is more pronounced in PiMFS-IPs+ than PiMFS-IPs– (Figure 3D, bottom panel).
Functional relevance of PiMFS-IPs+ in rice
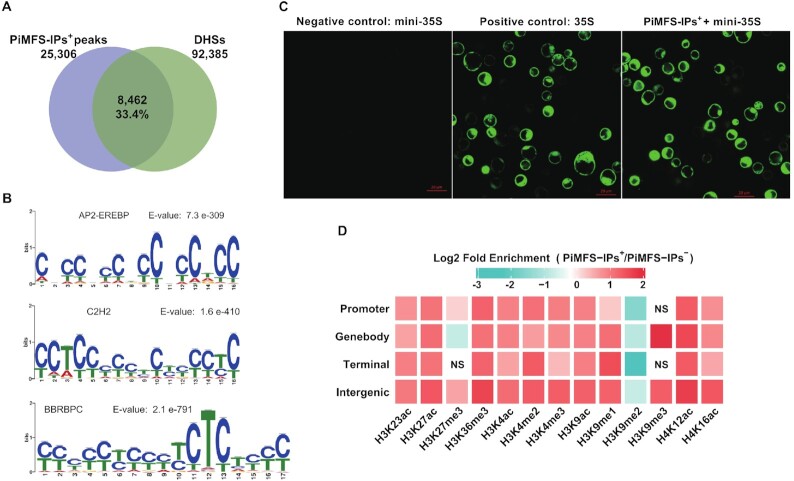
Both G4s and iMs are prevalent in promoters of human oncogenes, which suggests that they can potentially cis-regulate the expression of the corresponding genes (16). It inspired us to assess if rice iMs can serve as cis-regulatory elements or provide platforms for recruitment of trans-factors. We combined PiMFS-IPs+ peaks with DNase hypersensitive sites (DHSs) identified using DNase-seq data (GSE26610) (39). The fact that 33.4% (n = 8462) of PiMFS-IPs+ peaks overlapped DHSs (Figure 4A), confirming our hypothesis. We then conducted de novo motif identification, and found that CCTCC and CTCT motifs with potential binding of C2H2 and BBRBPC TFs, respectively, ranked highly (Figure 4B).
Figure 4.
Functional characterization of PiMFS-IPs+. (A) Venn plot illustrating PiMFS-IPs+ peaks overlapping DHSs, which were identified using published DNase-seq (39). (B) Motif discovery around sequences covering PiMFS-IPs+ peaks using MEME. The top 3 significantly enriched motifs with the highest E-values are listed. (C) Transiently expressed GFP signal detected from protoplasts transfected with a modified pJIT163-hGFP vector containing mini35S promoter only (negative control, left), an intact 35S promoter (positive control, middle) and a mini35S promoter + amplified PiMFS-IPs+ DNA fragment (right). GFP signals were captured using fluorescent microscopy. (D) Heat map showing enrichment of each mark for PiMFS-IPs+ relative to PiMFS-IPs– in each subgenomic region as indicated. The gradient representing the fold enrichment of PiMFS-IPs+ relative to PiMFS-IPs–. Chi-square test was conducted to determine the significance of the differences. NS representing non-significant and panels without NS representing significant enrichment (P-value < 0.05).
G4s or iMs are also frequently located around the transcriptional start sites (TSSs) of genes (54,55) where they can function as cis-regulatory elements for gene transcription (16). However, no direct evidence of these roles have been yet provided. To address this, we conducted protoplast-based transient expression assay. The expression of GFP gene was driven by a mini 35S promoter only (negative control), an intact 35S promoter (positive control), and a mini 35S promoter in which a DNA fragment containing PiMFSs was introduced. The GFP fluorescence signal was clearly visible with both the normal (intact 35S) and PiMFSs-containing mini 35S promoters, but was almost undetectable with the mini 35S promoter (Figure 4C), confirming that PiMFS-related DNA sequence act as enhancers to regulate gene transcription.
To examine the enrichment of histone marks around the PiMFS-IPs+ in each subgenomic region as indicated in Figure 4D, we conducted a fold enrichment assay by integrating PiMFS-IPs+ with 13 published histone marks (Supplementary Table S5) (39,40,56–58). We observed that majority of marks were enriched around all PiMFS-IPs+, except that genebody and terminal PiMFS-IPs+ exhibited less enriched H3K27me3, promoter and terminal PiMFS-IPs+ had no enrichment of H3K9me3 and all PiMFS-IPs+ had no enrichment of H3K9me2 (Figure 4D). This result indicated that epigenomic features around the PiMFS-IPs+ may be mark or genomic position dependent.
Taken together, these results indicate that PiMFS-IPs+ may have important genetic functions, notably in the regulation of gene expression.
Differential enrichment and impacts of PiMFS-IPs+ on TE activity
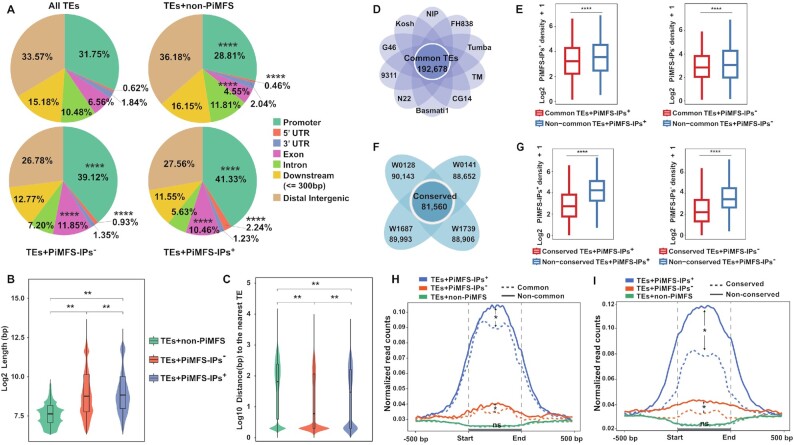
TEs serve as regulators for gene expression, genome structure variation and evolution in plants (59). It has been documented that secondary DNA structures, including G4s, can be formed in TEs and play regulatory functions in plants (60,61). We thus screened 364 741 known rice TEs to identify PiMFSs: 8131 (2.2%) TEs containing PiMFS-IPs+, 93 801 (25.7%) with PiMFS-IPs– and the rest (262 809, 72.1%) devoid of PiMFSs (Supplementary Figure S4A).
Interestingly, several parameters were found to differ within this series of data, which might be of interest in terms of mechanistic insights and evolution. First, variations in subgenomic abundance: TEs containing PiMFS-IPs+ were distributed preferentially in promoters and exons but less present in introns, downstream and distal intergenic regions as compared to TEs without PiMFSs; among three subtypes of TEs, TEs with PiMFS-IPs+ exhibited the highest and the lowest portion in promoters and distal intergenic regions/introns, respectively (Figure 5A). Second, variations in TE length: those containing PiMFS-IPs+ were the longest TEs while those without PiMFS were the shortest TEs (Figure 5B), suggesting that the former are either relatively newer insertion (62) or less active in the genome. Third, variations in distance between TEs: the distance between TEs with PiMFS-IPs+ is longer than those with PiMFS-IPs– (Figure 5C and Supplementary Figure S4B,C), again indicating that iMs may serve as an indicator of TE integrity and its insertion time in the genome.
Figure 5.
Differential enrichment and impacts of PiMFS-IPs+ on TE activity. (A) Subgenomic distributions of all TEs, TEs containing PiMFS-IPs+/− or without PiMFSs, including promoters, 5′UTRs, 3′UTRs, exons, introns, downstream and intergenic regions. Significance test was determined by hypergeometric test; **** P-value < 0.0001. (B) Mean length of TEs containing PiMFS-IPs+/− or without PiMFSs. (C) Distance from TEs containing PiMFS-IPs+/− or without PiMFSs to the nearest neighboring TEs. Significance tests in (B) and (C) were determined by Wilcoxon rank-sum test; ** P-value < 0.01. (D) Venn plot illustrating common TEs (n = 192 678) between Nipponbare (NIP) and the other nine cultivated Oryza subpopulations. (E) Mean number per kb of PiMFS-IPs between common and non-common TEs containing PiMFS-IPs+ (left) or PiMFS-IPs– (right). (F) Venn plot illustrating 81 560 conserved TEs between Nipponbare and the four wild rice species. (G) Mean number per kb of PiMFS-IPs between conserved and non-conserved TEs containing PiMFS-IPs+ (left) or PiMFS-IPs– (right). (H) Profiling of iM-IP-seq read density across ±500 bp from the start to the end of common (dotted lines)/non-common (solid lines) TEs containing PiMFS-IPs+, PiMFS-IPs– and non-PiMFS, respectively. (I) Profiling of iM-IP-seq read density across ±500 bp from the start to the end of conserved (dotted lines)/non-conserved (solid lines) TEs overlapping PiMFS-IPs+, PiMFS-IPs– and non-PiMFS as indicated, respectively. Significance tests in (E), (G), (H) and (I) were determined by Wilcoxon rank-sum test; **** P-value < 0.0001, * P-value < 0.05, NS: non-significant.
G4s are known to be involved in the TE lifecycle by modulating their transcription, translocation and integration (60). To date, no such information is available concerning iMs. We thus compared TEs in the Nipponbare genome with those in other nine cultivated Oryza subpopulations (63) and detected 192 678 common TEs (Figure 5D). Those TEs were subsequently associated with PiMFS-IPs+/−, leading to the identification of 2825 (2.0%) with PiMFS-IPs+, 41 819 (22.0%) TEs with PiMFS-IPs– and the rest (148 034, 76%) without PiMFSs (Supplementary Figure S5A).
When analyzing the 364 741 TEs present in the Nipponbare genome, we found that 192 678 TEs (52.8%) matched well with common TEs, while the rest (172 063, 47.1%) exhibited sequence divergence, hereafter termed as non-common TEs, likely resulting from a higher activity (Supplementary Figure S5B). This was confirmed by the observation that the mean number of PiMFS-IPs+/− per kb was greater in these non-common TEs as compared to common ones (Figure 5E), by contrast, common and non-common TEs with PiMFS-IPs+/− exhibited an overall similar read intensity (normalized read counts of iMs-IP reflecting folding potential) across ±1 kb of the center of PiMFS-IPs+/− (Supplementary Figure S6A). This indicates that PiMFS number instead of folding potential of PiMFSs may be directly associated with TE activities.
We thus decided to further assess whether iMs affect TEs evolution. To this end, we aligned the 364 741 Nipponbare TEs over the genome of four wild rice species (O. rufipogon W0128, W0141, W1687 and W1739), representing distinct genetic backgrounds (64). We identified 81 560 conserved TEs, and the rest were termed as non-conserved ones (Figure 5F). Among conserved TEs, 3900 (4.8%) contained PiMFS-IPs+, 32 680 (40.0%) contained PiMFS-IPs– and the rest (44 980, 55.1%) were devoid of PiMFSs (Supplementary Figure S5C). There were 81 560 conserved TEs (∼22%) and 282 817 non-conserved TEs (∼78%) in the Nipponbare genome (Supplementary Figure S5D). As above, we observed that the mean number of PiMFS-IPs+/− per kb was greater in non-conserved TEs than that in conserved ones (Figure 5G), and also an overall similar read intensity occurred between conserved and non-conserved TEs with PiMFS-IPs+/− (Supplementary Figure S6B), again indicating that PiMFS number instead of folding potential of PiMFSs might be related to TE activity.
It was of interest to further investigate the folding ability of PiMFSs in these different TE families. To this end, we divided a region at ±500 bp around the whole TE containing similar C content (Supplementary Figure S7) into 25 bins for counting normalized PiMFS-IPs+ read counts within each bin. Higher read intensity was found in non-common and non-conserved TEs with PiMFS-IPs+/− than the corresponding common or conserved TEs; almost no difference in PiMFS-IPs+ read intensity occurred between common/conserved and non-common/non-conserved TEs without PiMFSs (Figure 5H, 5I), which suggests that higher iM formation potential in association with higher amount of PiMFSs may facilitate TE activation.
Collectively, all above analyses lend credence to the hypothesis that the amount of PiMFSs instead of iMs folding potential may affect TE dynamic among different cultivated Oryza subpopulations and/or during evolution of wild rice species.
Differential DNA methylation between TEs overlapping PiMFS-IPs+/−
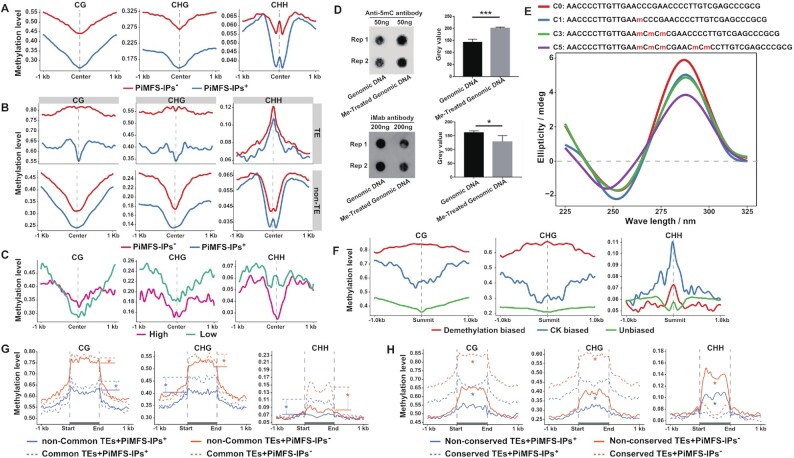
DNA methylation is essential to maintain genome stability by suppressing TE activities (65) and to control TE evolution among land plants (66). It has been shown that 5-methyl cytosine (5mC) favors iM folding while 5-hydroxymethylcytosine (5hmC) destabilizes iM structures (8,67), but no genome-wide studies on the relationship between PiMFS-IPs+ and DNA methylation have been yet reported. To explore whether DNA methylation can affect iM formation, we analyzed the DNA methylation levels at CG, CHG and CHH sites around ±1 kb of the center of PiMFS-IP+ or PiMFS-IP– with similar C content (Supplementary Figure S8A). PiMFS-IPs+ were found to be systematically hypomethylated as compared to PiMFS-IPs– (Figure 6A). PiMFS-IPs+ localized at both transposable elements (TEs, 30.3%) and non-transposable elements (non-TEs, 69.7%) in the rice genome (vide infra, Supplementary Figure S9A,B). We thus crossed the DNA methylation data of PiMFS-IPs+ and PiMFS-IPs– (with similar C-content) in TEs/non-TEs (Supplementary Figure S8B,C), and again, PiMFS-IPs+ were found to be systematically hypomethylated as compared to PiMFS-IPs– (Figure 6B), with methylation levels higher in TEs than in non-TEs, particularly at CHH sites (Supplementary Figure S10).
Figure 6.
Differential DNA methylation between TEs overlapping PiMFS-IPs+/− (A) CG, CHG and CHH methylation levels around ±1 kb of the center of PiMFS-IPs+ (n = 180 608) and PiMFS-IPs– (n = 180 608) with similar C content. (B) CG, CHG and CHH methylation levels around ±1 kb of the center of TE containing PiMFS-IPs+ (n = 54 716) and PiMFS-IPs– (n = 54 716) with similar C content (top), and non-TE containing PiMFS-IPs+ (n = 125 892) and PiMFS-IPs– (n = 125 892) with similar C content (bottom). (C) CG, CHG and CHH methylation levels between PiMFS-IPs+ peaks with high and low peak abundance (n = 8345). (D) Anti-5mC and iMab antibody based dot blotting assays for genomic DNA with or without treatment of M.SssI, a CG specific DNA methyltransferase. The dot signal in each blot was quantified from at least two replicates (right). (E) CD assay of a synthesized oligonucleotide containing 1 C methylation (C1), 3 consecutive C methylations (C3), 5 methylated Cs (C5) or non-C methylation (C0) as indicated. (F) CG, CHG and CHH methylation levels around ±1 kb of the summit of demethylated DNA biased, CK biased and unbiased PiMFS-IPs+ peaks. (G) CG, CHG and CHH methylation levels across ±1 kb from the start to the end of common (dotted lines) and non-common (solid lines) TEs (n = 19 351) containing PiMFS-IPs+/− with similar C content. (H) CG, CHG and CHH methylation levels across ±1 kb from the start to the end of conserved (dotted lines) and non-conserved (solid lines) TEs (n = 20 491) containing PiMFS-IPs+/− with similar C content. Significance tests in (G) and (H) were determined by Wilcoxon rank-sum test; * P-value < 0.05. Significance test in (D) was determined by One-way ANOVA analysis; *** P-value < 0.001, * P-value < 0.05.
Next, we grouped PiMFS-IPs+ peaks into two subtypes, i.e., with high and low peak abundance (fold change of read density between PiMFS-IPs+ peaks relative to control) with similar C-content (Supplementary Figure S8D), and conducted a similar DNA methylation analysis. We observed that PiMFS-IPs+ peaks with high peak abundance exhibited less CHG and CHH methylation (Figure 6C, middle and right), but more CG methylation around the center of PiMFS-IPs+ peaks covering from ∼200 bp upstream to 500 bp downstream of the center (Figure 6C, left). Then, we fully methylated genomic DNA in vitro using the CG specific M.SssI (M0226, NEB) DNA methyltransferase. This treatment triggered a 30% rise in total DNA methylation levels and, rather unexpectedly, a 20% decrease of iMab labeling signals (detected by anti-5mC or iMab dot blotting assay, Figure 6D). This result suggested that full CG methylation represses iM formation, in contradiction with results seen in Figure 6C, a discrepancy that might originate in the extent to which the DNA is methylated. To confirm this, we used synthetic oligonucleotides without methylated C (C0), 1 methylated C (C1) and 3 consecutive methylated Cs (C3) and 5 methylated Cs (C5). As seen by CD spectroscopy, all oligonucleotides displayed a clear iM-typical CD signature, but we found that C3 and especially C5 exhibited a weaker CD fingerprint than C1, also indicating that a higher level of methylated Cs tends to disfavor iM formation (Figure 6E). We further conducted iM-IP-seq using globally demethylated genomic DNA induced by Zebularine treatment, a chemical functioning as an inhibitor of DNA methyltransferase, as we did before (68). As shown in Supplementary Figure S11, after analyzing two biological replicates, we identified 5841 demethylated DNA biased PiMFS-IPs+ peaks with 26.5% of mean C content, 1217 CK biased PiMFS-IPs+ peaks and 67 339 unbiased PiMFS-IPs+ peaks with 31.3% and 29.1% of mean C content, respectively (Supplementary Figure S12). After plotting the DNA methylation levels at each cytosine context around ±1 kb of the summit of indicated PiMFS-IPs+ peaks, we observed that demethylated DNA biased PiMFS-IPs+ peaks had the highest CG and CHG methylation across all region examined, and the second highest CHH methylation at around ±250 bp of the summit as compared to the CK and unbiased ones (Figure 6F). This result indicated that DNA demethylation may facilitate formation of highly methylated iMs. How DNA demethylation affect iM formation in vivo is necessary to be further investigated.
Thus, all above analyses indicate that DNA methylation exhibits a complex relationship with i-motif formation in vitro, may depend on methylation extent or combined actions of all cytosine methylations.
Finally, we decided to assess whether PiMFS-IPs+ read density is related to changes in DNA methylation levels in the different TE families. To this end, we plotted DNA methylation levels of CG, CHG and CHH across ±500 bp around the whole TE region and found that common TEs with PiMF-IPs+/– had higher DNA methylation levels in all cytosine contexts relative to the corresponding non-common ones (Figure 6G). We also found that conserved TEs with PiMF-IPs+/– exhibited higher CG and CHG methylation but lower CHH methylation levels relative to the corresponding non-conserved ones (Figure 6H). An IGV showing DNA methylation distributed across a conserved/common TEs and non-conserved/non-common TEs is illustrated in Supplementary Figure S13.
Taken together, all above analyses show that DNA methylation levels are globally inversely correlated with iM formation, supporting the hypothesis that PiMFSs may coordinate with DNA methylation to control TE activity in the genome.
DISCUSSION
Our study aimed at investigating the iM landscape on a genome-wide scale in rice. To this end, we developed and applied a novel protocol, termed iM-IP-seq (Figure 1B), which allowed for the identification of rice genomic DNA capable of forming PiMFS-IPs+in vitro. The C-rich sequences prone to fold into iMs are substrates for DNA methylation. Our study showed that PiMFS-IPs+ were globally hypomethylated as compared to sequences devoid of PiMFS-IPs+-forming capability (Figure 6A,B). However, while CHG and CHH methylation exhibited an inverse correlation with PiMFS-IPs+ formation, the impact of CG methylation on PiMFS-IPs+ folding was more paradoxical (being associated with both iM formation and destabilization) (Figure 6C,D). Our study thus revealed a complex relationship between DNA methylation and iM formation in vitro, which strongly depends on methylation level and position. Such a potential impact of methylation on the stability of telomeric iM was reported in Arabidospis (34), in which a differential C-methylation frequency and a methylation position-dependent effect on iM stability were also detected (69,70). The mechanism behind these observations is not trivial: in line with the results presented here, it has been shown that one or two methylated cytosines could help stabilize telomeric iM structures, while hypermethylation results in iM destabilization in humans (67). Therefore, we can postulate that moderate levels of C-methylation could help increase C-C base-pair thermostability (71,72), while C-hypermethylation will increase more frequency of mC-mC base pairs, which could create steric hindrance, resulting in destabilization of C-C base pair. This was further evidenced in our study: iMs were overall much less enriched with repressive histone marks like H3K27me3 and H3K9me2/3 (Figure 4D), indicating that iMs are preferably present in euchromatic regions with less methylated DNA and repressive histone marks relative to heterochromatic regions; demethylated DNA biased PiMFS-IPs+ exhibited higher DNA methylation levels in CK as compared to the CK biased or unbiased PiMFS-IPs+ peaks (Figure 6F and Supplementary Figure S12). This indicates that DNA demethylation and/or the presence of euchromatic marks help(s) to open up chromatin, thereby creating more space to facilitate iM formation. Also, the opposite effects of 5mC and 5hmC on iM stability are indicative of their influences on DNA flexibility that affects iM folding, the former reducing it, the latter increasing it (73). Finally, these modifications may have distinct impacts on binding or recruitment of certain factors: as an example, 5hmC was found to reduce binding ability of proteins (nucleolin) and small-molecules (TMPyP4) to iMs (74); also, in line with what was described for G4s (75), iM formation may hamper access of DNA methyltransferase to DNA, which results in DNA hypomethylation, which in turn reduces the folding ability of PiMFSs. Moreover, we cannot exclude the possibility that effects of mC on iM stability may be sequence context-dependent. Since effects of cytosine methylation or other modifications on iM stability in vitro were mainly examined in special DNA sequences in humans like telomeric DNA and (CCG)(n)(CCG)(n) trinucleotide repeats in humans (67,71,72,76). Massive efforts must now be invested to provide evidence for these different hypotheses. In particular, it is necessary to investigate the relationship between mC and iM stability on a genome-wide scale in humans or other species. Our study casts also a new light on the intricate relationship between PiMFS-IPs+ and transposable elements (TEs). TEs, which represent 35% of the genome in rice (77), 85% in maize (78) and wheat (79), serve as a major driving force contributing to dynamics of genome size and structure, and gene/genome evolution (80). We showed here that PiMFSs were enriched in certain subtypes of TEs in rice, and that the amount of PiMFSs, rather than folding potential of PiMFSs, contributed to TE dynamics, notably modulating their methylation status. This wealth of data also highlights the possible roles of PiMFS-IPs+ in TE evolution: we showed that non-common or non-conserved TEs were highly active in different rice species likely via PiMFS-IPs+-mediated DNA hypomethylation. Collectively, this study indicates that iMs alone and in combination with epigenetic regulations play active roles in TE life cycle, thereby functioning as drivers of genome evolution in rice.
DATA AVAILABILITY
The iM-IP-seq and control data used in this study have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE184783.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Bioinformatics Center, Nanjing Agricultural University for providing computing facilities for data processing and analyses.
Author contributions: W.L.Z. conceived and designed the study. X.M. analyzed the data. Y.L.F. performed iM-DNA-IP, qPCR and dot blotting experiments. X.J.C. helped with material preparation. Y.N.S. helped with the genomic DNA preparation. Y.Y. prepared DNA plasmids for transient transformation. Y.L. and C.Y.C. assisted with rice protoplast transformation. X.E.W. supervised the experiments. S.T.T. helped with data analyses. J.H. assisted with CD experiment. X.M., Y.L.F., D.M. and W.L.Z. interpreted the results. D.M. and W.L.Z. wrote the manuscript with contributions from all other authors.
Contributor Information
Xing Ma, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), Nanjing Agricultural University, No.1 Weigang, Nanjing, Jiangsu 210095, P.R. China.
Yilong Feng, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), Nanjing Agricultural University, No.1 Weigang, Nanjing, Jiangsu 210095, P.R. China.
Ying Yang, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), Nanjing Agricultural University, No.1 Weigang, Nanjing, Jiangsu 210095, P.R. China.
Xin Li, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, Hunan, 410125, P.R. China.
Yining Shi, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), Nanjing Agricultural University, No.1 Weigang, Nanjing, Jiangsu 210095, P.R. China.
Shentong Tao, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), Nanjing Agricultural University, No.1 Weigang, Nanjing, Jiangsu 210095, P.R. China.
Xuejiao Cheng, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), Nanjing Agricultural University, No.1 Weigang, Nanjing, Jiangsu 210095, P.R. China.
Jian Huang, School of Biology & Basic Medical Science, Soochow University, Suzhou, Jiangsu 215123, P.R. China.
Xiu-e Wang, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), Nanjing Agricultural University, No.1 Weigang, Nanjing, Jiangsu 210095, P.R. China.
Caiyan Chen, Institute of Subtropical Agriculture, Chinese Academy of Sciences, Changsha, Hunan, 410125, P.R. China.
David Monchaud, Institut de Chimie Moleculaire, ICMUB CNRS UMR 6302, UBFC Dijon, France.
Wenli Zhang, State Key Laboratory for Crop Genetics and Germplasm Enhancement, Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), Nanjing Agricultural University, No.1 Weigang, Nanjing, Jiangsu 210095, P.R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [32070561, U20A2030]. Funding for open access charge: National Natural Science Foundation of China [32070561, U20A2030].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gehring K., Leroy J.L., Gueron M.. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993; 363:561–565. [DOI] [PubMed] [Google Scholar]
- 2. Leroy J.L., Gehring K., Kettani A., Gueron M.. Acid multimers of oligodeoxycytidine strands: stoichiometry, base-pair characterization, and proton exchange properties. Biochemistry. 1993; 32:6019–6031. [DOI] [PubMed] [Google Scholar]
- 3. Marsh R.E., Bierstedt R., Eichhorn E.L.. The crystal structure of cytosinecetic acid. Acta Cryst. 1962; 15:310. [Google Scholar]
- 4. Ahmed S., Henderson E.. Formation of novel hairpin structures by telomeric C-strand oligonucleotides. Nucleic Acids Res. 1992; 20:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kang C.H., Berger I., Lockshin C., Ratliff R., Moyzis R., Rich A.. Crystal structure of intercalated four-stranded d(C3T) at 1.4 a resolution. Proc. Natl. Acad. Sci. USA. 1994; 91:11636–11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benabou S., Aviñó A., Eritja R., González C., Gargallo R.. Fundamental aspects of the nucleic acid i-motif structures. RSC Adv. 2014; 4:26956–26980. [Google Scholar]
- 7. Sun D., Hurley L.H.. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009; 52:2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhavsar-Jog Y.P., Van Dornshuld E., Brooks T.A., Tschumper G.S., Wadkins R.M.. Epigenetic modification, dehydration, and molecular crowding effects on the thermodynamics of i-motif structure formation from C-rich DNA. Biochemistry. 2014; 53:1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cui J., Waltman P., Le V.H., Lewis E.A.. The effect of molecular crowding on the stability of human c-MYC promoter sequence I-motif at neutral pH. Molecules. 2013; 18:12751–12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li X., Peng Y., Ren J., Qu X.. Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Natl. Acad. Sci. USA. 2006; 103:19658–19663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen X., Zhou X., Han T., Wu J., Zhang J., Guo S.. Stabilization and induction of oligonucleotide i-motif structure via graphene quantum dots. ACS Nano. 2013; 7:531–537. [DOI] [PubMed] [Google Scholar]
- 12. Wright E.P., Huppert J.L., Waller Z.A.E.. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017; 45:2951–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeraati M., Langley D.B., Schofield P., Moye A.L., Rouet R., Hughes W.E., Bryan T.M., Dinger M.E., Christ D.. I-motif DNA structures are formed in the nuclei of human cells. Nat. Chem. 2018; 10:631–637. [DOI] [PubMed] [Google Scholar]
- 14. Abou Assi H., Garavis M., Gonzalez C., Damha M.J.. i-Motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 2018; 46:8038–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bacolla A., Wells R.D.. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004; 279:47411–47414. [DOI] [PubMed] [Google Scholar]
- 16. Kendrick S., Hurley L.H.. The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl. Chem. 2010; 82:1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kang H.J., Kendrick S., Hecht S.M., Hurley L.H.. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014; 136:4172–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brooks T.A., Kendrick S., Hurley L.. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010; 277:3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ruggiero E., Lago S., Sket P., Nadai M., Frasson I., Plavec J., Richter S.N.. A dynamic i-motif with a duplex stem-loop in the long terminal repeat promoter of the HIV-1 proviral genome modulates viral transcription. Nucleic Acids Res. 2019; 47:11057–11068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saha P., Panda D., Muller D., Maity A., Schwalbe H., Dash J.. In situ formation of transcriptional modulators using non-canonical DNA i-motifs. Chem. Sci. 2020; 11:2058–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaiser C.E., Van Ert N.A., Agrawal P., Chawla R., Yang D., Hurley L.H.. Insight into the complexity of the i-Motif and G-Quadruplex DNA structures formed in the KRAS promoter and subsequent drug-induced gene repression. J. Am. Chem. Soc. 2017; 139:8522–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kendrick S., Kang H.J., Alam M.P., Madathil M.M., Agrawal P., Gokhale V., Yang D., Hecht S.M., Hurley L.H.. The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 2014; 136:4161–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niu K., Zhang X., Deng H., Wu F., Ren Y., Xiang H., Zheng S., Liu L., Huang L., Zeng B.et al.. BmILF and i-motif structure are involved in transcriptional regulation of bmpoum2 in bombyx mori. Nucleic Acids Res. 2018; 46:1710–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dzatko S., Krafcikova M., Hansel-Hertsch R., Fessl T., Fiala R., Loja T., Krafcik D., Mergny J.L., Foldynova-Trantirkova S., Trantirek L.. Evaluation of the stability of DNA i-Motifs in the nuclei of living mammalian cells. Angew. Chem. Int. Ed. Engl. 2018; 57:2165–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simonsson T., Pribylova M., Vorlickova M.. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 2000; 278:158–166. [DOI] [PubMed] [Google Scholar]
- 26. Xu Y., Sugiyama H.. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb). Nucleic Acids Res. 2006; 34:949–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guo K., Gokhale V., Hurley L.H., Sun D.. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008; 36:4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cogoi S., Xodo L.E.. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006; 34:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H., Hai J., Zhou J., Yuan G.. The formation and characteristics of the i-motif structure within the promoter of the c-myb proto-oncogene. J. Photochem. Photobiol. B. 2016; 162:625–632. [DOI] [PubMed] [Google Scholar]
- 30. Garavis M., Mendez-Lago M., Gabelica V., Whitehead S.L., Gonzalez C., Villasante A.. The structure of an endogenous drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep. 2015; 5:13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garavis M., Escaja N., Gabelica V., Villasante A., Gonzalez C.. Centromeric alpha-satellite DNA adopts dimeric i-Motif structures capped by AT hoogsteen base pairs. Chemistry. 2015; 21:9816–9824. [DOI] [PubMed] [Google Scholar]
- 32. Gallego J., Chou S.H., Reid B.R.. Centromeric pyrimidine strands fold into an intercalated motif by forming a double hairpin with a novel T:G:G:T tetrad: solution structure of the d(TCCCGTTTCCA) dimer. J. Mol. Biol. 1997; 273:840–856. [DOI] [PubMed] [Google Scholar]
- 33. Nonin-Lecomte S., Leroy J.L.. Structure of a C-rich strand fragment of the human centromeric satellite III: a pH-dependent intercalation topology. J. Mol. Biol. 2001; 309:491–506. [DOI] [PubMed] [Google Scholar]
- 34. Skolakova P., Badri Z., Foldynova-Trantirkova S., Rynes J., Sponer J., Fojtova M., Fajkus J., Marek R., Vorlickova M., Mergny J.L.et al.. Composite 5-methylations of cytosines modulate i-motif stability in a sequence-specific manner: implications for DNA nanotechnology and epigenetic regulation of plant telomeric DNA. Biochim. Biophys. Acta Gen. Subj. 2020; 1864:129651. [DOI] [PubMed] [Google Scholar]
- 35. Megalathan A., Cox B.D., Wilkerson P.D., Kaur A., Sapkota K., Reiner J.E., Dhakal S.. Single-molecule analysis of i-motif within self-assembled DNA duplexes and nanocircles. Nucleic Acids Res. 2019; 47:7199–7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amato J., Iaccarino N., Randazzo A., Novellino E., Pagano B.. Noncanonical DNA secondary structures as drug targets: the prospect of the i-motif. Chem. Med. Chem. 2014; 9:2026–2030. [DOI] [PubMed] [Google Scholar]
- 37. Manzini G., Yathindra N., Xodo L.E.. Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res. 1994; 22:4634–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dai J., Ambrus A., Hurley L.H., Yang D.. A direct and nondestructive approach to determine the folding structure of the I-motif DNA secondary structure by NMR. J. Am. Chem. Soc. 2009; 131:6102–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang W., Wu Y., Schnable J.C., Zeng Z., Freeling M., Crawford G.E., Jiang J.. High-resolution mapping of open chromatin in the rice genome. Genome Res. 2012; 22:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fang Y., Wang X., Wang L., Pan X., Xiao J., Wang X.E., Wu Y., Zhang W.. Functional characterization of open chromatin in bidirectional promoters of rice. Sci. Rep. 2016; 6:32088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang Y., Chen L., Lin K., Feng Y., Zhang P., Pan X., Sanders J., Wu Y., Wang X.E., Su Z.et al.. Characterization of functional relationships of R-loops with gene transcription and epigenetic modifications in rice. Genome Res. 2019; 29:1287–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of chip-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Belmonte-Reche E., Morales J.C.. G4-iM grinder: when size and frequency matter. G-Quadruplex, i-Motif and higher order structure search and analysis tool. NAR. Genome Bioinform. 2020; 2:lqz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fujimoto A., Fujita M., Hasegawa T., Wong J.H., Maejima K., Oku-Sasaki A., Nakano K., Shiraishi Y., Miyano S., Yamamoto G.et al.. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Res. 2020; 30:334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu G., Wang L.G., He Q.Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 46. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E.. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008; 132:311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu L., Li N., Xu C., Zhong S., Lin X., Yang J., Zhou T., Yuliang A., Wu Y., Chen Y.R.et al.. Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA. 2014; 111:10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shao Z., Zhang Y., Yuan G.C., Orkin S.H., Waxman D.J.. MAnorm: a robust model for quantitative comparison of chip-Seq data sets. Genome Biol. 2012; 13:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Verma A., Halder K., Halder R., Yadav V.K., Rawal P., Thakur R.K., Mohd F., Sharma A., Chowdhury S.. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008; 51:5641–5649. [DOI] [PubMed] [Google Scholar]
- 52. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 53. Kendrick S., Akiyama Y., Hecht S.M., Hurley L.H.. The i-motif in the bcl-2 P1 promoter forms an unexpectedly stable structure with a unique 8:5:7 loop folding pattern. J. Am. Chem. Soc. 2009; 131:17667–17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eddy J., Maizels N.. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008; 36:1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu L., Chen X., Sanders D., Qian S., Zhong X.. High-resolution mapping of H4K16 and H3K23 acetylation reveals conserved and unique distribution patterns in arabidopsis and rice. Epigenetics. 2015; 10:1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tan F., Zhou C., Zhou Q., Zhou S., Yang W., Zhao Y., Li G., Zhou D.X.. Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol. 2016; 171:2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. He G., Zhu X., Elling A.A., Chen L., Wang X., Guo L., Liang M., He H., Zhang H., Chen F.et al.. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010; 22:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kejnovsky E., Lexa M.. Quadruplex-forming DNA sequences spread by retrotransposons may serve as genome regulators. Mob. Genet. Elements. 2014; 4:e28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kejnovsky E., Tokan V., Lexa M.. Transposable elements and G-quadruplexes. Chromosome Res. 2015; 23:615–623. [DOI] [PubMed] [Google Scholar]
- 61. Tokan V., Puterova J., Lexa M., Kejnovsky E.. Quadruplex DNA in long terminal repeats in maize LTR retrotransposons inhibits the expression of a reporter gene in yeast. BMC Genomics. 2018; 19:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Le N.T., Harukawa Y., Miura S., Boer D., Kawabe A., Saze H.. Epigenetic regulation of spurious transcription initiation in arabidopsis. Nat. Commun. 2020; 11:3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qin P., Lu H., Du H., Wang H., Chen W., Chen Z., He Q., Ou S., Zhang H., Li X.et al.. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell. 2021; 184:3542–3558. [DOI] [PubMed] [Google Scholar]
- 64. Zhao Q., Feng Q.. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018; 50:278–284. [DOI] [PubMed] [Google Scholar]
- 65. Deniz O., Frost J.M., Branco M.R.. Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 2019; 20:417–431. [DOI] [PubMed] [Google Scholar]
- 66. Brautigam K., Cronk Q.. DNA methylation and the evolution of developmental complexity in plants. Front. Plant Sci. 2018; 9:1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu B., Devi G., Shao F.. Regulation of telomeric i-motif stability by 5-methylcytosine and 5-hydroxymethylcytosine modification. Org. Biomol. Chem. 2015; 13:5646–5651. [DOI] [PubMed] [Google Scholar]
- 68. Feng Y., Tao S., Zhang P., Sperti F.R., Liu G., Cheng X., Zhang T., Yu H., Wang X.E., Chen C.et al.. Epigenomic features of DNA G-quadruplexes and their roles in regulating rice gene transcription. Plant Physiol. 2021; 10.1093/plphys/kiab566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ogrocka A., Polanska P., Majerova E., Janeba Z., Fajkus J., Fojtova M.. Compromised telomere maintenance in hypomethylated arabidopsis thaliana plants. Nucleic Acids Res. 2014; 42:2919–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vrbsky J., Akimcheva S., Watson J.M., Turner T.L., Daxinger L., Vyskot B., Aufsatz W., Riha K.. siRNA-mediated methylation of Arabidopsis telomeres. PLos Genet. 2010; 6:e1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang B., Wu R.R., Rodgers M.T.. Base-pairing energies of proton-bound homodimers determined by guided ion beam tandem mass spectrometry: application to cytosine and 5-substituted cytosines. Anal. Chem. 2013; 85:11000–11006. [DOI] [PubMed] [Google Scholar]
- 72. Yang B., Rodgers M.T.. Base-pairing energies of proton-bound heterodimers of cytosine and modified cytosines: implications for the stability of DNA i-motif conformations. J. Am. Chem. Soc. 2014; 136:282–290. [DOI] [PubMed] [Google Scholar]
- 73. Ngo T.T., Yoo J., Dai Q., Zhang Q., He C., Aksimentiev A., Ha T.. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat. Commun. 2016; 7:10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Morgan R.K., Molnar M.M., Batra H., Summerford B., Wadkins R.M., Brooks T.A.. Effects of 5-hydroxymethylcytosine epigenetic modification on the stability and molecular recognition of VEGF i-Motif and G-Quadruplex structures. J. Nucleic Acids. 2018; 2018:9281286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mao S.Q., Ghanbarian A.T., Spiegel J., Martinez Cuesta S., Beraldi D., Di Antonio M., Marsico G., Hansel-Hertsch R., Tannahill D., Balasubramanian S.. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018; 25:951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang B., Moehlig A.R., Frieler C.E., Rodgers M.T.. Base-pairing energies of protonated nucleobase pairs and proton affinities of 1-methylated cytosines: model systems for the effects of the sugar moiety on the stability of DNA i-motif conformations. J. Phys. Chem. B. 2015; 119:1857–1868. [DOI] [PubMed] [Google Scholar]
- 77. Takata M., Kiyohara A., Takasu A., Kishima Y., Ohtsubo H., Sano Y.. Rice transposable elements are characterized by various methylation environments in the genome. BMC Genomics. 2007; 8:469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schnable P.S., Ware D., Fulton R.S., Stein J.C., Wei F., Pasternak S., Liang C., Zhang J., Fulton L., Graves T.A.et al.. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009; 326:1112–1115. [DOI] [PubMed] [Google Scholar]
- 79. Wicker T., Gundlach H., Spannagl M., Uauy C., Borrill P., Ramirez-Gonzalez R.H., De Oliveira R.International Wheat Genome Sequencing, C. International Wheat Genome Sequencing, C. Mayer K.F.X., Paux E.et al.. Impact of transposable elements on genome structure and evolution in bread wheat. Genome Biol. 2018; 19:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bennetzen J.L., Wang H.. The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 2014; 65:505–530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The iM-IP-seq and control data used in this study have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE184783.