Abstract
CRISPR/Cas-derived base editing tools empower efficient alteration of genomic cytosines or adenines associated with essential genetic traits in plants and animals. Diversified target sequences and customized editing products call for base editors with distinct features regarding the editing window and target scope. Here we developed a toolkit of plant base editors containing AID10, an engineered human AID cytosine deaminase. When fused to the N-terminus or C-terminus of the conventional Cas9 nickase (nSpCas9), AID10 exhibited a broad or narrow activity window at the protospacer adjacent motif (PAM)-distal and -proximal protospacer, respectively, while AID10 fused to both termini conferred an additive activity window. We further replaced nSpCas9 with orthogonal or PAM-relaxed Cas9 variants to widen target scopes. Moreover, we devised dual base editors with AID10 located adjacently or distally to the adenine deaminase ABE8e, leading to juxtaposed or spaced cytosine and adenine co-editing at the same target sequence in plant cells. Furthermore, we expanded the application of this toolkit in plants for tunable knockdown of protein-coding genes via creating upstream open reading frame and for loss-of-function analysis of non-coding genes, such as microRNA sponges. Collectively, this toolkit increases the functional diversity and versatility of base editors in basic and applied plant research.
INTRODUCTION
Enormous agronomic traits of crops and genetic diseases of humans are associated with genomic single nucleotide polymorphisms (SNPs) (1,2). To ease the investigation and manipulation of these SNPs, the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas)-derived cytosine or adenine base editors (CBEs or ABEs) have been invented (3,4), which can efficiently introduce C•G to T•A and A•T to G•C mutations, respectively, to a genomic target site in both plants and humans, independently of a DNA double-stranded break (DSB) or repair template. Although the so-called prime editing technology has recently been developed to support all types of nucleotide substitutions (5), this approach exhibits an overall low editing capability in monocots and works poorly in dicot species (6–11).
Both CBEs and ABEs are chimeric enzymes containing a Cas protein and a cytosine or adenine deaminase. The prototype of CBE is known as BE3, which is composed of the rat cytosine deaminase APOBEC1 at the N-terminus, the Streptococcus pyogenes Cas9 D10A nickase (nSpCas9) in the middle, and an uracil glycosylase inhibitor (UGI) at the C-terminus (3). The nSpCas9 moiety is responsible for single-guide RNA (sgRNA)-directed DNA targeting, while the APOBEC1 moiety is able to convert a cytosine to uracil within a favorable deamination window of the target sequence (referred to as the activity window). As the recovery of U•G to C•G through the endogenous uracil-excision repair pathway is inhibited by UGI, the U•G mismatch is subsequently corrected by DNA polymerases to T•A. Similarly, ABE7.10, a canonical ABE, consists of a heterodimer between the Escherichia coli TadA adenine deaminase and TadA7.10, a laboratory-evolved TadA variant, at the N-terminus and the nSpCas9 at the C-terminus (4). Upon association with a genomic target sequence through the nSpCas9-sgRNA module, TadA7.10 deaminates an adenine within the activity window of the target sequence to inosine, which is subsequently read by DNA polymerases as guanine to induce an A•T to G•C substitution. In both CBE- and ABE-mediated base editing, nicking the non-deaminated DNA strand by nSpCas9 can facilitate its repair using the deaminated strand as a template, thus promoting the conversion of the target base pair (3,4).
In general, CBEs have been employed more extensively than ABEs in basic and applied biological research (12), especially in plants (13). Notably, in addition to the originally designed use for installing targeted C•G to T•A mutations, CBEs have demonstrated a remarkable versatility in other gene manipulation applications. In the so-called CRISPR-STOP or iSTOP application, CBEs have been repurposed as gene loss-of-function tools by converting an exonic glutamine (CAA/CAG), arginine (CGA), or tryptophan (TGG) codon of a target gene to a stop codon (14,15). CBEs have also been explored to deactivate target genes through destroying intron splice sites to cause mRNA mis-splicing (16–18). These gene loss-of-function strategies have become attractive complements to the CRISPR/Cas technology because multiplex base editing by CBEs is unlikely to induce complicated chromosomal rearrangements, as observed in the CRISPR/Cas9-mediated genome editing (19–21). Moreover, CBEs have been utilized as gene gain-of-function tools to drive directed protein evolution and functional variant screening (22,23). Furthermore, CBEs have been leveraged as molecular recorders of cellular signaling events by translating extracellular stimuli into nucleotide changes at specific genomic loci (24,25).
In CBE-mediated base editing, the requirement for a protospacer adjacent motif (PAM) in the vicinity of a target cytosine and the activity window width are two major factors determining whether the target cytosine is editable or not (26). In addition, some CBEs (e.g. BE3) work inefficiently on target cytosines within unfavorable sequence contexts (3). To address these issues, researchers have combined different Cas proteins with natural or modified cytosine deaminases to build CBEs with distinct target scopes and activity windows. For instance, the lamprey CDA1, human AID (hAID), hAPOBEC3A (hA3A), and engineered hA3B have been used to replace the rat APOBEC1 in BE3 to obtain effective CBEs in plants (27–30). Theoretically, when the base editor arsenal is filled with more CBEs with different characteristics, any single cytosine within a given genome will likely fall into the activity window of at least one CBE (26). To this end, in this study we developed a toolkit of base editors with varying activity windows and target scopes using an engineered variant of hAID cytosine deaminase (referred to as AID10) (31). Using this toolkit, we explored an unprecedented strategy to effectively down-regulate target gene activity in plants by creating upstream open reading frame (uORF). We also demonstrated that our base editor with a broad activity window was useful for probing the interactions between endogenous microRNA (miRNA) sponges and their cognate miRNAs in plants.
MATERIALS AND METHODS
Plant materials and growth conditions
The Arabidopsis thaliana ecotype Col-0 and rice (Oryza sativa) cultivar Zhonghua 11 (ZH11) plants were used as wild-type plants in this study. The seeds of bak1-4 null mutant (SALK_116202) were obtained from the Arabidopsis Biological Resource Center (ABRC). After stratification at 4°C for 2 days, Arabidopsis plants were germinated on the Jiffy soil (Jiffy Group) in a plant growth room under photoperiods of 12 h light (75 μmol/m2/s) at 23°C and 12 h dark at 21°C. For hypocotyl length comparison, Arabidopsis seeds were sterilized and sown on the 0.5 × Murashige and Skoog (MS) medium containing 1% sucrose and 0.6% agar. After stratification at 4°C for 2 days, plants were germinated in the dark at 23°C for 5 days before the hypocotyl length was measured. Rice plants were grown on the Jiffy soil in a growth chamber under photoperiods of 12 h light (200 μmol/m2/s) at 32°C and 12 h dark at 28°C. For the herbicide resistance assay, rice seeds were sterilized and grown on the 0.5 × MS medium supplemented with 0.1 mg/l imazethapyr herbicide for 9 days.
Plasmid construction
The coding sequences of AID10-XTEN and 6 × Gly-Gly-Ser (GGS)-UGI were amplified from the pGAL1-AID10-dCas9-UGI plasmid (31), and those of the nuclear localization signal (NLS) and plant codon-optimized dCas9 (D10A, H840A) were amplified from the HBT-dCas9 plasmid (32). For transient expression in rice protoplasts, the sequences of the maize Ubiquitin-1 (ZmUbi) promoter, NLS, AID10-XTEN, and dCas9 were assembled in order into the XhoI/PstI-digested HBT-FLAG vector (33) to obtain HBT-AID10-dCas9 by recombination-based cloning using the ClonExpress MultiS One Step Cloning Kit (Vazyme, China). The 6 × GGS-UGI fragment was further inserted using the ClonExpress II One Step Cloning Kit (Vazyme, China) to generate HBT-AID10-dCas9-UGI. The H840A mutation in the dCas9 coding sequence was reversed by PCR-based site-directed mutagenesis to obtain HBT-AID10-nCas9 and HBT-AID10-nCas9-UGI. The coding sequence of APOBEC1-nCas9-UGI was amplified from the pnCas9-PBE plasmid (34) and was used to replace AID10-nCas9-UGI in the HBT-AID10-nCas9-UGI plasmid through restriction digestion and ligation. The coding sequence of AtCDA1 (At2g19570) was PCR amplified from Arabidopsis cDNA and was used to replace APOBEC1 to obtain HBT-AtCDA1-nCas9-UGI by recombination-based cloning. The coding sequences of plant codon-optimized SaCas9 (35) and LbCas12a (36) harboring D10A and D832A mutations, respectively, were generated by overlapping PCR. Resultant PCR products, along with fragments of the ZmUbi promoter, NLS, AID10-XTEN and 6 × GGS-UGI, were assembled into the XhoI/PstI-digested HBT-FLAG vector to generate HBT-AID10-nSaCas9-UGI and HBT-AID10-dLbCas12a-UGI, respectively. Seven or three point mutations were introduced into nSpCas9 and nSaCas9, respectively, to obtain the HBT-AID10-nSpCas9-NG-UGI and HBT-AID10-nSaCas9-KKH-UGI plasmids encoding Cas9 derivatives with relaxed PAM requirement (37,38). For transient expression in Arabidopsis protoplasts, the ZmUbi promoter in AID10-encoding plasmids was replaced by the Arabidopsis Ubiquitin 10 (AtUBQ10) promoter through SpeI and AvrII sites. To change the fusion orientation of AID10 to nCas9, coding sequences for AtUBQ10, NLS, nCas9, XTEN, AID10, and UGI were assembled in order into the XhoI/PstI-digested HBT-FLAG vector to obtain HBT-nCas9-AID10-UGI by recombination-based cloning. The DNA fragment of XTEN-AID10-UGI was cut out from the HBT-nCas9-AID10-UGI plasmid by StuI and subsequently inserted into the HBT-AID10-nCas9 plasmid through the StuI site to generate the HBT-AID10-nCas9-AID10-UGI plasmid.
To construct dual base editors, we synthesized the plant codon-optimized coding sequence of adenosine deaminase ABE7.10 (TadA-TadA7.10) (4) at the Genewiz company (Suzhou, China). ABE7.10 and nCas9 were fused by overlapping PCR and subsequently used to replace AID10-nCas9 in the HBT-AID10-nCas9 plasmid to obtain HBT-ABE7.10-nCas9. The fragment of XTEN-AID10 was amplified from HBT-nCas9-AID10-UGI and inserted into the StuI site of HBT-ABE7.10-nCas9 to obtain HBT-ABE7.10-nCas9-AID10. Similar cloning strategy was followed to construct HBT-TadA7.10-nCas9 and HBT-TadA7.10-nCas9-AID10. The coding sequence of XTEN-AID10-6×GGS-UGI was generated by overlapping PCR and subsequently inserted into the StuI site of HBT-ABE7.10-nCas9 to obtain HBT-ABE7.10-nCas9-AID10-UGI. To construct HBT-AID10-ABE7.10-nCas9 and HBT-AID10-TadA7.10-nCas9, the PCR amplicon of AID10-XTEN was cloned into the NcoI site of HBT-ABE7.10-nCas9 and HBT-TadA7.10-nCas9, respectively, by recombination-based cloning. The coding sequence of nCas9-6×GGS-UGI-NLS was amplified from the HBT-AID10-nCas9-UGI plasmid and was subsequently used to replace nCas9-NLS in the HBT-AID10-ABE7.10-nCas9 plasmid to obtain HBT-AID10-ABE7.10-nCas9-UGI. To replace ABE7.10 with ABE8e (39), eight mutations were introduced into HBT-ABE7.10-nCas9, HBT-ABE7.10-nCas9-AID10-UGI, and HBT-AID10-ABE7.10-nCas9-UGI through multiple rounds of PCR-based site-directed mutagenesis to obtain HBT-ABE8e-nCas9, HBT-ABE8e-nCas9-AID10-UGI, and AID10-ABE8e-nCas9-UGI, respectively. The sgRNA expression plasmids were constructed by inserting annealed oligos of guide sequences into the BsaI-digested pUC119-sgRNA plasmid, which already includes the sgRNA scaffold and an AtU6-26 promoter (32) for Arabidopsis expression or an OsU6a promoter (40) for rice expression. The sgRNA target sites are listed in Supplementary Table S1. The specificity of individual sgRNAs was evaluated using the CRISPR-GE algorithm (http://skl.scau.edu.cn) (41).
To construct binary plasmids for plant stable transformation, sequences encoding individual base editors were amplified from the corresponding HBT plasmids and used to replace APOBEC1-nCas9-UGI between AvrII and SacI sites in the pH-nCas9-PBE plasmid (34). To obtain non-chimeric T1 Arabidopsis plants, an egg cell-specific EC1.2en-EC1.1p fusion promoter was amplified from the pHEE401E plasmid (42) and used to replace the ZmUbi promoter between HindIII and AvrII sites in the binary plasmid. The sgRNA expression cassettes were amplified from the pUC119-sgRNA plasmids and inserted into the HindIII site of the binary plasmid. For multiplex editing of three OsSWEET genes in rice, the OsU6a:sgRNA-OsSWEET11:TTTTTT expression cassette was generated by inserting annealed oligos into the pUC119-sgRNA vector. Similarly, the OsU6b:sgRNA-OsSWEET13:TTTTTT and OsU6a:sgRNA-OsSWEET14:TTTTTT cassettes were sequentially inserted into the pUC119-sgRNA-OsSWEET11 plasmid through SacI/SbfI and EcoRI/EcoRV sites, respectively. The three sgRNA expression cassettes in tandem were PCR amplified and cloned into the binary plasmid at the HindIII site by recombination-based cloning.
Protoplast isolation and transfection
Protoplasts were isolated from leaves of 4-week-old Arabidopsis or leaf sheaths of 10-day-old rice plants as previously described (43,44). Briefly, plant tissues were cut into 0.5-mm strips and digested in 10 ml enzyme solution containing 1.5% Cellulase R10 (Yakult Honsha, Japan), 0.4% Macerozyme R10 (Yakult Honsha, Japan), 0.4 M mannitol, 20 mM MES, pH 5.7, 20 mM KCl, 10 mM CaCl2, 0.1% BSA for 3 h. After adding 10 ml W5 solution containing 154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7, the digestion mixture was filtered through a 75-μm cell strainer to remove tissue debris. Isolated protoplasts were collected by 2-min centrifugation at 100 g for Arabidopsis or 150 g for rice, and were resuspended in 10 ml W5 solution and rested on ice for 30 min. After centrifugation for 1 min, cells were resuspended in MMG solution containing 0.4 M mannitol, 15 mM MgCl2, 4 mM MES, pH 5.7, to a concentration of 2 × 105 cells/ml. To evaluate the editing efficiencies of base editors, 40 μg (2 μg/μl) plasmids expressing base editors and 40 μg (2 μg/μl) plasmids expressing sgRNAs were gently mixed with 400 μl protoplasts and 440 μl polyethylene glycol (PEG) solution containing 40% PEG 4000, 0.2 M mannitol, 0.1 M CaCl2 in a 2-ml round-bottom tube. The mixture was incubated at room temperature for 5 min for Arabidopsis or 15 min for rice before transfection was stopped by adding 1.6 ml W5 solution. Transfected protoplasts were collected by 3-min centrifugation at 100 g for Arabidopsis or 200 g for rice. Cells were resuspended in 200 μl W5 solution and transferred to 2 ml WI solution containing 0.5 M mannitol, 20 mM KCl, 4 mM MES, pH 5.7. After incubation at room temperature in the dark for 48 h, protoplasts were harvested by centrifugation at 200 g for 3 min for subsequent analysis.
Evaluation of base editing efficiencies in protoplasts
Genomic DNA was extracted from transfected protoplasts using the Plant Genomic DNA Kit (Tiangen, China) according to the supplier's instruction. To evaluate base editing efficiencies by restriction fragment length polymorphism (RFLP) assay, target regions were amplified by PCR using the Q5 High-Fidelity DNA Polymerase (New England Biolabs, USA) and purified using the FastPure Gel DNA Extraction Mini Kit (Vazyme, China). Equal amounts of purified amplicons were subjected to restriction digestion with XhoI (New England Biolabs, USA) at 37°C overnight and then were resolved on a 2% agarose gel by electrophoresis. To confirm mutations at the target site, XhoI-resistant PCR amplicons were cloned into the pEASY-Blunt vector (TransGen, China) and 15 clones for each sample were randomly selected for Sanger sequencing.
To evaluate base editing efficiencies by amplicon deep sequencing, genomic fragments of 220–270 bp spanning the target sites were PCR amplified using primers with a 6-nt unique barcode at the 5′ end, as listed in Supplementary Table S2. The PCR amplification was carried out with the Q5 High-Fidelity DNA Polymerase using 10 ng genomic DNA as template. The PCR cycle number was set as 28 to ensure the reaction was stopped within the linear range of amplification. Purified PCR amplicons using the FastPure Gel DNA Extraction Mini Kit were pooled at equal molar ratios and subjected to deep sequencing. Deep sequencing library construction (including dA-tailing and adaptor ligation to amplicons), sequencing, trimming of adaptor sequences, and demultiplexing based on barcodes were fulfilled by the Genewiz company (Suzhou, China). Sequencing was performed on an Illumina HiSeq platform for 150-bp paired-end reads and at least 100 000 reads were obtained for each sample. FASTQ files were analyzed using CRISPResso2 (https://crispresso.pinellolab.partners.org) (45) or BE-Analyzer (http://www.rgenome.net/be-analyzer) (46) to quantify mutation efficiencies at individual target sequences.
Generation of transgenic plants
The binary plasmid co-expressing the base editor and sgRNA was introduced into Agrobacterium tumefaciens strain GV3101 for Arabidopsis transformation or strain EHA105 for rice transformation by electroporation. Correct A. tumefaciens cells were used to transform Arabidopsis floral tissues and rice embryogenic calli according to detailed protocols described previously (47,48). Transgenic Arabidopsis T1 plants and rice T0 plants were selected on the medium containing 50 mg/l hygromycin before resistant plants were transferred to the Jiffy soil.
Genotyping of transgenic plants
For each transgenic plant, the crude extract of genomic DNA was obtained by homogenizing a total of 15 mg tissues collected from 3 different leaves in 30 μl lysis buffer containing 100 mM Tris–HCl, pH 9.5, 1 M KCl, 10 mM EDTA and then incubating at 70°C for 30 min. The 10-fold diluted lysate was used as PCR template for genotyping using the Hi-TOM method (49), which utilizes two rounds of PCR and deep sequencing to identify mutations. Briefly, genomic fragments spanning the target sites were PCR amplified using target-specific primers and barcoded primers sequentially. Resultant amplicons were pooled at equal molar ratios and sequenced with an Illumina HiSeq platform by the Novogene company (Beijing, China). Mutations in the transformants were identified by analyzing the deep sequencing data with the Hi-TOM tool (http://www.hi-tom.net/hi-tom) and further confirmed by Sanger sequencing of PCR amplicons of target regions. PCR primers used are listed in Supplementary Table S2.
RNA extraction and qRT-PCR
Leaf tissue of 20 mg from 4-week-old Arabidopsis plants were ground in liquid nitrogen and total RNA was extracted using 1 ml RNAiso Plus reagent (TAKARA, Japan) according to the manufacturer's instruction. To quantify the transcript levels of Arabidopsis miR166 target genes, the second internodes in rachis branches of 8-week-old Arabidopsis plants were sampled. Reverse transcription (RT) was performed with 1 μg RNA and oligo dT primer using the PrimeScript RT reagent Kit with gDNA Eraser (TAKARA, Japan) in a reaction volume of 20 μl. Resultant cDNA products were used as PCR template after 8-fold dilution. Quantitative RT-PCR (qRT-PCR) was performed in a LightCycler 96 Instrument (Roche, USA) using the TB Green Premix Ex Taq (TAKARA, Japan) according to the manufacturer's instruction. The relative expression levels were quantified using the 2–ΔΔCt method and the Arabidopsis Actin2 gene was used as a reference gene. Primers used for qRT-PCR are listed in Supplementary Table S3.
Immunoblot analysis
Crude protein extracts were obtained from leaf tissues of 4-week-old Arabidopsis plants by boiling homogenized tissues in the SDS-PAGE loading buffer at 95°C for 10 min. Proteins were resolved on a 10% SDS-PAGE gel and transferred to a polyvinylidene fluoride (PVDF) membrane. AtBAK1 proteins were detected by immunoblotting using the anti-BAK1 antibody, which was raised against the C-terminal peptide DSTSQIENEYPSGPR of AtBAK1 (50) by the Genscript company (Nanjing, China), as the primary antibody and the horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibody (Cell Signaling Technology, USA) as the secondary antibody.
Luciferase reporter assay
To measure the activity of the wild-type or uORF-containing AtBAK1 promoter (1,864 bp upstream of the start codon), 32 μg (2 μg/μl) promoter:LUC reporter plasmid and 8 μg (2 μg/μl) transfection control plasmid (AtUBQ10:GUS) were co-transfected into 200 μl Arabidopsis protoplasts. After incubation at room temperature for 12 h, protoplasts were harvested and homogenized in 100 μl lysis buffer containing 25 mM Tris–HCl, pH 7.8, 2 mM DTT, 2 mM trans-1,2-diaminocyclohexane-N’N’N’N’-tetraacetic acid, 10% (v/v) glycerol, 1% (v/v) Triton X-100. After incubation on ice for 5 min and centrifugation at 12,000 g for 30 sec, 10 μl supernatant was mixed with 100 μl luciferase assay reagent containing 20 mM Tricine, 1.07 mM (MgCO3)4Mg(OH)2, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 0.27 mM Coenzyme A, 0.47 mM d-luciferin potassium salt, 0.53 mM ATP, pH 7.8, in a 96-well white microplate. Luminescence was measured in a Varioskan LUX Multimode Microplate Reader (ThermoFisher Scientific, USA). In parallel, another 10 μl protoplast lysate was transferred to a 96-well black microplate with 50 μl MUG solution containing 10 mM Tris–HCl, pH 8.0, 1 mM 4-methylumbelliferyl-β-d-glucuronide (MUG), 2 mM MgCl2. The mixture was incubated at 37°C for 30 min and cooled in the ice-water bath to terminate the reaction. The GUS expression was measured by the Varioskan LUX Multimode Microplate Reader with excitation at 365 nm and emission at 455 nm. The promoter activity was calculated as the ratio between the luciferase readout and the GUS readout for each sample.
RESULTS
AID10-nCas9-UGI confers a broad activity window at the PAM-distal protospacer
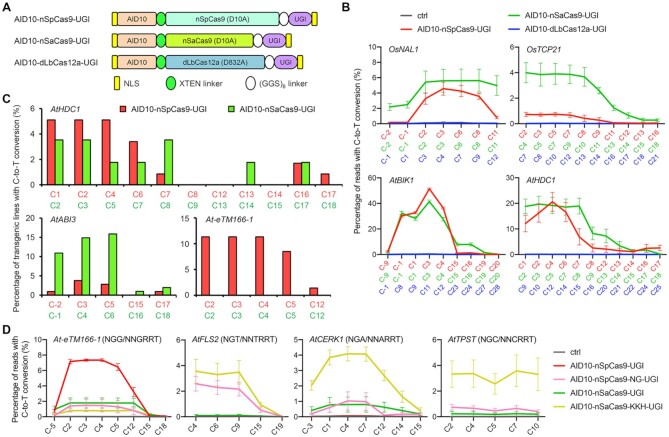
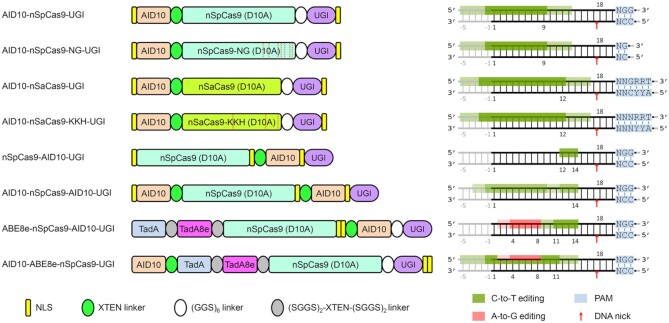
To explore CBEs with new properties in plants, we replaced the rat cytosine deaminase APOBEC1 in BE3 (i.e. APOBEC1-nCas9-UGI) with AID10 (Figure 1A and Supplementary Sequence), a newly engineered hAID variant (31). AID10 exhibits low genome-wide guide-independent off-target effects in Drosophila (31) and, as a hAID derivative, may inherently reduce guide-independent RNA off-target editing relative to APOBEC1 and hA3A (51). Unlike hAID*Δ, another hAID variant that has been successfully applied to cytosine editing in rice (28), AID10 harbors more mutations, including 9 amino acid substitutions (i.e. K10E, K34E, S38C, T82I, T110A, H130R, V152A, E156G, R174G) and a deletion of the C-terminal 18 residues relative to hAID (Supplementary Figure S1). These modifications have been shown to dramatically enhance the deamination activity of hAID (31). For comparison purpose, we also constructed AID10-nCas9, which lacked the UGI moiety, and AID10-dCas9-UGI and AID10-dCas9, both of which were based on the nuclease-dead Cas9 (dCas9) rather than the nCas9 nickase (Figure 1A). Meanwhile, an Arabidopsis cytosine deaminase, namely AtCDA1 (52), was also used to replace APOBEC1 in BE3 to obtain AtCDA1-nCas9-UGI (Figure 1A).
Figure 1.
AID10-nCas9-UGI exhibits a broader activity window than APOBEC1-nCas9-UGI. (A) Schematic diagram of CBEs with an N-terminally fused cytosine deaminase. UGI, uracil glycosylase inhibitor. NLS, nuclear localization signal. (B) Restriction fragment length polymorphism assay identifies active CBEs in rice protoplasts. The target sequence containing an XhoI site (green) is shown with the PAM in bold. bp, base pair. (C) Comparison of cytosine editing efficiencies between APOBEC1-nCas9-UGI, AID10-nCas9-UGI, and AID10-nCas9 at endogenous genomic loci in rice (left) or Arabidopsis (right) protoplasts by amplicon deep sequencing. Data are shown as mean ± s.d. of two biological replicates performed at different times. (D) Mutant allele composition of the OsNAL1 target site in protoplasts expressing APOBEC1-nCas9-UGI, AID10-nCas9-UGI, or AID10-nCas9. C-to-T conversions are highlighted in bold and the frequencies of individual alleles are shown on the right. Dashed-line box marks the protospacer and the PAM is underlined. The numbers on the top indicate the protospacer positions. Data are collected from one replicate in (C). (E, F) Comparison of cytosine editing efficiencies between APOBEC1-nCas9-UGI (top) and AID10-nCas9-UGI (bottom) at OsLAZY1 (E) and OsGL1-1 (F) target sites in transgenic T0 rice plants. Reverse complements of the protospacers and the PAMs are shadowed in blue and green, respectively. All guanines in the protospacers are in red with positions indicated by subscripted numbers. The percentages of indicated alleles among total alleles are shown and those corresponding to base substitutions are shadowed in yellow. (G) Comparison of cytosine editing efficiencies between APOBEC1-nCas9-UGI and AID10-nCas9-UGI at the AtCER10 target site in transgenic T1 Arabidopsis plants. The reverse complement of the protospacer is underlined and the PAM is in bold. The intron splice site is in lowercase letters.
The performance of these CBEs was quickly evaluated using the RFLP assay (53) in rice protoplasts co-expressing individual CBEs and an sgRNA targeting the endogenous OsER1-T1 locus that contains an XhoI restriction site (Figure 1B). Any cytosine change at the protospacer position 5 or 7 (counting the PAM as 21–23) would eliminate the XhoI site and produce an XhoI-resistant allele. As expected, we detected XhoI-resistant target amplicons in protoplasts expressing APOBEC1-nCas9-UGI (Figure 1B), which has previously been shown to enable efficient C-to-T conversions at the positions 3–9 in plant cells (34). Expression of AID10-nCas9 or AID10-nCas9-UGI, but not AID10-dCas9 and AID10-dCas9-UGI, also yielded XhoI-resistant target amplicons (Figure 1B). This observation was reminiscent of a previous finding that APOBEC1-dCas9-UGI is unable to induce C-to-T conversions in rice plants (34), implying that nicking the non-deaminated DNA strand is required for efficient cytosine editing in plant cells. Sanger sequencing of the XhoI-resistant target amplicons further validated C-to-T conversions at the OsER1-T1 locus by AID10-nCas9 and AID10-nCas9-UGI (Supplementary Figure S2). No XhoI-resistant target amplicon was detected in protoplasts expressing AtCDA1-nCas9-UGI (Figure 1B), suggesting that AtCDA1 is not suitable for building CBEs. Consistent with this finding, a recent study has shown that the ribose moiety of cytidine is required for substrate binding to AtCDA1 (54).
Next, we compared cytosine editing patterns and efficiencies between AID10-nCas9-UGI, AID10-nCas9, and APOBEC1-nCas9-UGI at four C-rich genomic target sites (Supplementary Table S1) in rice or Arabidopsis protoplasts by amplicon deep sequencing. We found that AID10-nCas9-UGI induced higher levels of C-to-T conversions than AID10-nCas9, although they exhibited similar activity windows (Figure 1C). Notably, the activity window of AID10-nCas9-UGI spanned from the protospacer positions −1 to 9, which was considerably wider than that of APOBEC1-nCas9-UGI (i.e. positions 4–8) (Figure 1C). Moreover, AID10-nCas9-UGI was able to simultaneously edit more cytosines than APOBEC1-nCas9-UGI at the same target sites (Figure 1D and Supplementary Figure S3), suggesting that AID10-nCas9-UGI has a broader activity window and higher processivity than APOBEC1-nCas9-UGI.
To verify the activity window of AID10-nCas9-UGI in planta, we generated transgenic rice plants expressing AID10-nCas9-UGI or APOBEC1-nCas9-UGI by the ZmUbi promoter and an sgRNA targeting the OsLAZY1 locus. Genotyping of T0 rice transformants identified nucleotide substitutions in 26 out of 40 (65.0%) plants expressing APOBEC1-nCas9-UGI and 24 out of 33 (72.7%) plants expressing AID10-nCas9-UGI (Supplementary Table S4). Both CBEs induced unwanted indels but at different frequencies, namely 42.5% (17 out of 40) by APOBEC1-nCas9-UGI and 21.2% (7 out of 33) by AID10-nCas9-UGI. For simplicity in editing pattern comparison, we only focused on non-chimeric (i.e. homozygous, heterozygous or biallelic) mutant plants that carried genetically homogeneous mutations. In mutant plants generated by APOBEC1-nCas9-UGI, the C-to-T conversion only occurred at the protospacer position 7 with a frequency of 31.9% (Figure 1E and Supplementary Figure S4A), while the C-to-G mutation was also detected at the position 7 with a frequency of 13.9%. By contrast, AID10-nCas9-UGI enabled cytosine editing in a broader window, including C-to-T conversions at the positions 2, 7, 13, and 15 with frequencies of 5.6–51.9% and C-to-G conversions at the positions 2 and 7 with frequencies of 1.9% (Figure 1E and Supplementary Figure S4A). Of note, only in mutant plants generated by AID10-nCas9-UGI, mutant alleles (e.g. #20 and #25) carried homogeneous C•G to T•A mutation at the protospacer position 2 (Supplementary Figure S5A). This mutation would disrupt the splicing of the first intron of OsLAZY1 and cause gene inactivation (18). Consistently, these lines displayed increased tiller angles compared to wild-type plants (Supplementary Figure S5B), resembling the previously reported rice lazy1 null alleles (55). We also compared APOBEC1-nCas9-UGI and AID10-nCas9-UGI for editing the OsGL1-1 locus in rice plants. Similarly, APOBEC1-nCas9-UGI only edited cytosines at the protospacer positions 7 and 8, whereas AID10-nCas9-UGI allowed cytosine substitutions at the positions −5, −2, 1, 7, and 8 (Figure 1F and Supplementary Figure S4B).
We further compared the activity windows between AID10-nCas9-UGI and APOBEC1-nCas9-UGI by targeting the AtCER10 locus in transgenic Arabidopsis plants, where both CBEs were expressed by the ZmUbi promoter. Targeted cytosine editing was identified in 27 out of 42 (64.3%) T1 plants expressing APOBEC1-nCas9-UGI and 32 out of 34 (94.1%) T1 plants expressing AID10-nCas9-UGI (Supplementary Table S4). Cytosine changes by APOBEC1-nCas9-UGI only occurred at the protospacer position 8, whereas those induced by AID10-nCas9-UGI were detected at the positions 2, 3 and 8 (Figure 1G and Supplementary Figure S6). All mutant plants created by both CBEs appeared to be chimeric (Supplementary Table S4), reminiscent of our earlier observation that Arabidopsis transgenic T1 plants expressing APOBEC1-nCas9-UGI under the constitutive promoter only produce chimeric mutations (18). Since two earlier studies reported that heterozygous base edited plants can be obtained in Arabidopsis T1 generation when CBEs are expressed by the egg cell-specific promoter (17,56), we subsequently switched to the Arabidopsis EC1.2en-EC1.1p fusion promoter (42) for AID10-nCas9-UGI expression in transgenic Arabidopsis plants. Indeed, we identified 4 out of 157 (2.5%) transgenic T1 plants carrying heterozygous C•G to T•A mutations at the AtCER10 target site, which produced homozygous mutant alleles in the T2 generation (Supplementary Figure S7A and B). In T2 lines #41-16 and #61-3, the cytosine change at the protospacer position 2 by AID10-nCas9-UGI was expected to induce mis-splicing of the first intron of AtCER10, leading to gene deactivation. Consistently, these lines exhibited severe growth defects (Supplementary Figure S7C), resembling the previously reported Arabidopsis cer10 null alleles (57). Based on these results, we chose to use the EC1.2en-EC1.1p fusion promoter for CBE expression in transgenic Arabidopsis plants throughout this study. Taken together, these data suggest that AID10-nCas9-UGI enables efficient C-to-T editing with a broader activity window than APOBEC1-nCas9-UGI in both monocot and dicot species.
Combining AID10 with orthogonal or engineered Cas9 variants expands target scopes
To expand the target scope of AID10-nCas9-UGI, we replaced nSpCas9 with other orthogonal Cas proteins, including the Staphylococcus aureus Cas9 nickase (nSaCas9) that recognizes a 3′-NNGRRT PAM (58) and the nuclease-dead Lachnospiraceae bacterium Cas12a (dLbCas12a) that recognizes a 5′-TTTV PAM (59) (Figure 2A). To compare the performance of these CBEs in protoplasts by amplicon deep sequencing, we purposely selected four genomic loci in rice or Arabidopsis (i.e. OsNAL1, OsTCP21, AtBIK1 and AtHDC1) with overlapping target sequences for SpCas9, SaCas9 and LbCas12a (Supplementary Table S1). Both AID10-nSpCas9-UGI and AID10-nSaCas9-UGI induced C-to-T conversions at all target sites, whereas AID10-dLbCas12a-UGI failed to do so (Figure 2B). We reasoned that the failure of AID10-dLbCas12a-UGI was due to its inability to nick the target loci, similar to the case of AID10-dCas9-UGI (Figure 1B). Of note, AID10-nSaCas9-UGI appeared to exhibit a comparable or even higher editing efficiency and wider activity window than AID10-nSpCas9-UGI at these target loci in protoplasts (Figure 2B).
Figure 2.
Combination of AID10 with orthogonal or engineered Cas proteins expands the target scope. (A) Schematic diagram of AID10-based CBEs derived from SpCas9, SaCas9, and LbCas12a. (B) Comparison of cytosine editing efficiencies between different CBEs at endogenous genomic loci in rice (top) or Arabidopsis (bottom) protoplasts by amplicon deep sequencing. (C) Comparison of cytosine editing efficiencies between AID10-nSpCas9-UGI and AID10-nSaCas9-UGI at three endogenous genomic loci in transgenic T1 Arabidopsis plants. (D) Comparison of cytosine editing efficiencies between CBEs derived from PAM-relaxed Cas9 variants at endogenous genomic loci with canonical or non-canonical PAMs in Arabidopsis protoplasts by amplicon deep sequencing. The PAMs are shown in the parentheses. In (B) and (C), the numbers in red, green, and blue indicate protospacer positions for SpCas9, SaCas9, and LbCas12a, respectively. Data in (B) and (D) are shown as mean ± s.d. of two biological replicates performed at different times.
We next compared the performance between AID10-nSpCas9-UGI and AID10-nSaCas9-UGI in transgenic Arabidopsis plants by targeting the AtHDC1, AtABI3 and At-eTM166-1 loci with overlapping target sequences (Supplementary Table S1). For the AtHDC1 target site, genotyping of transgenic T1 lines showed that AID10-nSaCas9-UGI induced C-to-T conversions with a similar efficiency to AID10-nSpCas9-UGI, with the former being 5.3% (3 out of 57) and the latter being 8.5% (10 out of 118) (Figure 2C and Supplementary Table S4). C-to-T conversions by AID10-nSpCas9-UGI were induced at the protospacer positions 1, 2, 4, 6, 7, 16, and 17 with frequencies of 0.9–5.1%, while those by AID10-nSaCas9-UGI occurred at the positions 2, 3, 5, 7, 8, 14 and 17 with frequencies of 1.8–3.5% (Figure 2C and Supplementary Figure S8A). Multiple cytosines could be concurrently mutated at the same target loci by both CBEs (Supplementary Figure S8A). Interestingly, AID10-nSaCas9-UGI was able to edit the cytosine at the protospacer position 14, whereas AID10-nSpCas9-UGI was unable to edit the same cytosine (i.e. at the position 13) (Figure 2C). An opposite observation was made for AID10-nSpCas9-UGI at the protospacer position 17 (corresponding to the position 18 for AID10-nSaCas9-UGI) (Figure 2C).
Further comparison between AID10-nSaCas9-UGI and AID10-nSpCas9-UGI for editing the AtABI3 and At-eTM166-1 loci revealed contrasting editing frequencies. For the AtABI3 target site, AID10-nSaCas9-UGI induced C-to-T conversions with a frequency of 17.8% (18 out of 101 T1 lines), whereas only 4 out of 107 (3.7%) T1 lines expressing AID10-nSpCas9-UGI carried C-to-T conversions (Supplementary Figure S8B and Table S4). In addition, the editing frequencies of all five cytosines at the AtABI3 target site by AID10-nSaCas9-UGI remarkably exceeded those by AID10-nSpCas9-UGI (i.e. 1.0–15.8% versus 0–3.7%) (Figure 2C). For the At-eTM166-1 site, sequencing analysis of 139 T1 lines expressing AID10-nSaCas9-UGI identified no C-to-T substitution at the target site, whereas AID10-nSpCas9-UGI mediated efficient C-to-T editing with a frequency of 11.3% (8 out of 71 T1 lines) (Figure 2C, Supplementary Figure S8C and Table S4). These results underscore that CBEs containing the same cytosine deaminase but different Cas moieties can exhibit distinct editing efficiencies at the same genomic target site.
We also utilized SpCas9-NG (37) and SaCas9-KKH (38), two engineered Cas9 variants with 3′-NG and 3′-NNNRRT PAMs, respectively, to construct AID10-based CBEs. Amplicon deep sequencing of edited targets in Arabidopsis protoplasts showed that AID10-nSaCas9-KKH-UGI enabled efficient C-to-T editing at the AtFLS2, AtCERK1, and AtTPST target sites containing non-canonical NNNRRT PAMs (Figure 2D), while AID10-nSpCas9-NG-UGI was also able to induce C-to-T editing at these target loci containing non-canonical NG PAMs, despite with lower efficiencies (Figure 2D). Notably, when compared with the PAM-stringent CBEs, these PAM-relaxed CBEs showed impaired activities at the At-eTM166-1 target site containing the canonical PAM (Figure 2D). Importantly, both AID10-nSpCas9-NG-UGI and AID10-nSaCas9-KKH-UGI maintained a wide activity window that spanned more than 10 nucleotides (Figure 2D). To evaluate the guide-dependent off-target effects that may arise from relaxed PAM requirements, we used amplicon deep sequencing to examine C-to-T editing frequencies of AID10-nSpCas9-NG-UGI and AID10-nSaCas9-KKH-UGI at potential off-target sites with high sequence homology (no more than 3 base mismatches), including some with non-canonical PAMs. We detected no off-target C-to-T editing at all of these sites (Supplementary Figure S9). Taken together, these results suggest that AID10 is compatible with diverse Cas9 nickases to confer a broad activity window at genomic target loci containing various PAMs.
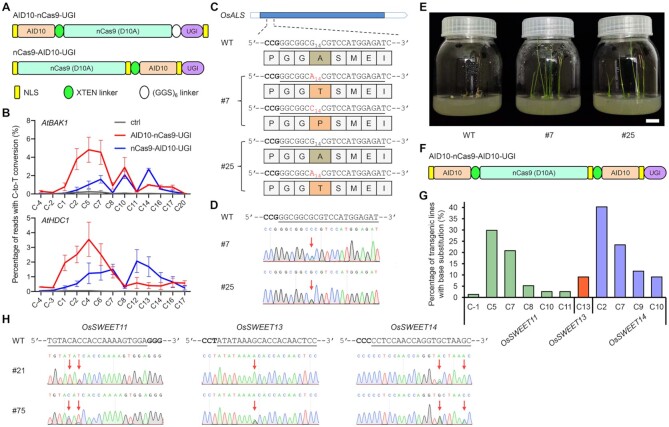
nCas9-AID10-UGI confers a narrow activity window at the PAM-proximal protospacer
Next, we wondered whether AID10 fused to the C-terminus of nSpCas9 (nCas9-AID10-UGI, Figure 3A) might give rise to a different activity window. Therefore, we employed amplicon deep sequencing to assess the performance of nCas9-AID10-UGI at the AtBAK1 and AtHDC1 target sites in Arabidopsis protoplasts. Strikingly, unlike AID10-nCas9-UGI whose activity window was located at the PAM-distal protospacer, nCas9-AID10-UGI exhibited efficient cytosine editing at the PAM-proximal protospacer (Figure 3B). Moreover, we used nCas9-AID10-UGI to edit the OsALS gene in transgenic rice plants, which encodes an acetolactate synthase targeted by many herbicides. As substitutions of the conserved Ala96 residue of OsALS can confer gain-of-function resistance to the herbicide imidazolinone (60,61), we designed an sgRNA to target the coding strand of OsALS, placing the GCG codon of Ala96 at the protospacer positions 12–14 (Figure 3C). By genotyping 39 transgenic T0 rice plants, we identified five OsALS mutant alleles with nucleotide substitution and one mutant allele with nucleotide deletion (Supplementary Table S4). Importantly, although there are nine G•C base pairs at the OsALS target site, base editing by nCas9-AID10-UGI only occurred at the position 14 with no bystander editing in all five mutant alleles (Figure 3C). We isolated two T0 alleles for further analysis, in which the line #7 carried biallelic G•C to A•T and G•C to C•G conversions and the line #25 had heterozygous G•C to A•T conversion (Figure 3C and D). The G•C to A•T or C•G conversion would generate A96T and A96P mutations, respectively, both of which have been documented to render imidazolinone resistance (60,61). Indeed, we observed that all progeny of line #7 and three quarters of the progeny of line #25 exhibited imidazolinone resistance (Figure 3E). Altogether, our data from protoplasts and transgenic plants imply that AID10 fused to the C-terminus of nSpCas9 offers a narrow activity window at the PAM-proximal protospacer.
Figure 3.
nCas9-AID10-UGI and AID10-nCas9-AID10-UGI exhibit distinct activity windows relative to AID10-nCas9-UGI. (A) Schematic diagram of nCas9-AID10-UGI. (B) Comparison of cytosine editing efficiencies between nCas9-AID10-UGI and AID10-nCas9-UGI at endogenous genomic loci in Arabidopsis protoplasts by amplicon deep sequencing. Data are shown as mean ± s.d. of two biological replicates performed at different times. (C) Cytosine editing at the OsALS locus by nCas9-AID10-UGI in transgenic T0 rice lines #7 and #25 and resultant amino acid changes. The reverse complement of the target sequence is underlined and the PAM is in bold. The subscripted number indicates the protospacer position. (D) Sanger sequencing chromatograms show base conversions (marked by red arrows) described in (C). (E) Amino acid substitutions in OsALS confer herbicide resistance. The seedlings were germinated on the medium containing 0.1 mg/l imazethapyr for nine days. Scale bar = 2 cm. (F) Schematic diagram of AID10-nCas9-AID10-UGI. (G) Frequencies of cytosine editing at three endogenous genomic loci in transgenic T0 rice plants expressing AID10-nCas9-AID10-UGI and three sgRNAs, each targeting one indicated OsSWEET locus. (H) Sanger sequencing chromatograms show multiplex base conversions (marked by red arrows) by AID10-nCas9-AID10-UGI at the OsSWEET loci in two T0 rice lines.
AID10-nCas9-AID10-UGI confers a superimposed activity window
The observations that the N-terminal or C-terminal AID10 enabled base editing with distinct activity windows prompted us to test whether concurrent fusion of AID10 to both termini of nSpCas9 could lead to an overlaid activity window. To this end, transgenic rice plants were generated to express AID10-nCas9-AID10-UGI (Figure 3F) and three sgRNAs targeting the OsSWEET11, OsSWEET13 and OsSWEET14 loci, respectively. Cytosines at the target sequences of OsSWEET11 and OsSWEET14 are located at the PAM-distal protospacer, while the only cytosine at the OsSWEET13 target sequence lies at the position 13. In 77 transgenic T0 rice plants, AID10-nCas9-AID10-UGI successfully introduced cytosine changes to at least one OsSWEET target site (Supplementary Table S4). In particular, cytosine editing by AID10-nCas9-AID10-UGI occurred at the positions −1, 5, 7, 8, 10 and 11 of the OsSWEET11 protospacer and positions 2, 7, 9 and 10 of the OsSWEET14 protospacer, reminiscent of the activity window of AID10-nCas9-UGI (Figure 3G). Meanwhile, AID10-nCas9-AID10-UGI also edited the cytosine at the position 13 of the OsSWEET13 protospacer, showing a comparable activity window to nCas9-AID10-UGI (Figure 3G). Multiplex base editing at all three target loci was detected in two lines (i.e. #21 and #75) (Figure 3H). We also targeted AID10-nCas9-AID10-UGI to a C-rich sequence at the OsSLR1 locus in transgenic rice plants, in which the base editing efficiency appeared to be low (Supplementary Table S4). However, we still detected cytosine substitutions at either PAM-distal or -proximal protospacer in the OsSLR1-edited lines (Supplementary Figure S10). Taken together, these results indicate that AID10-nCas9-AID10-UGI possesses a highly flexible activity window at either PAM-distal or -proximal protospacer.
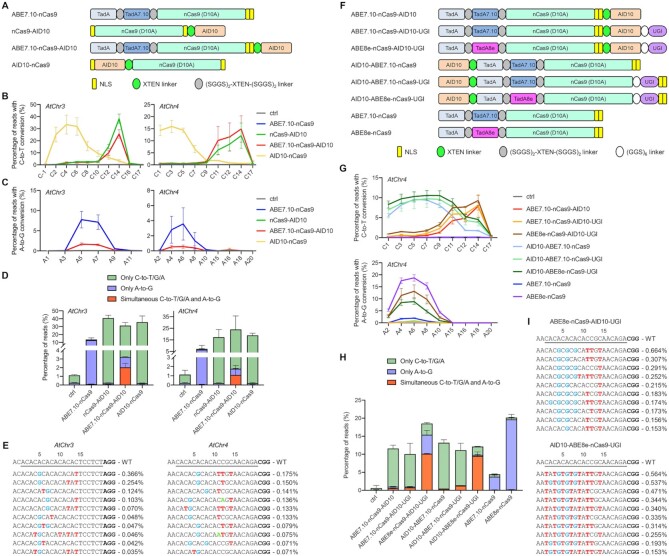
Dual base editors enable cytosine and adenine co-editing at the same target sites in plant cells
Inspired by AID10-nCas9-AID10-UGI, we reasoned that combination of AID10 and ABE7.10 (4) in a single base editor might allow flexible editing of either cytosine or adenine at a genomic target sequence. Since only the N-terminal ABE7.10 can work efficiently for A-to-G editing (4,62), we constructed ABE7.10-nCas9-AID10 (Figure 4A) and assessed its performance at the OsIPA1 or OsCKX2 locus in transgenic rice plants. However, we failed to detect any base change at the OsIPA1 site in 29 transgenic T0 lines (Supplementary Table S4), while only PAM-proximal C-to-T substitutions by AID10 were detected at the OsCKX2 site in 2 out of 23 transgenic T0 plants (Supplementary Figure S11 and Table S4).
Figure 4.
Cytosine and adenine co-editing at the same target sequences by AID10-based dual base editors. (A) Schematic diagram of the dual base editor ABE7.10-nCas9-AID10. (B, C) ABE7.10-nCas9-AID10 mediates both C-to-T (B) and A-to-G (C) conversions at two endogenous genomic loci in Arabidopsis protoplasts as revealed by amplicon deep sequencing. Data are shown as mean ± s.d. of two biological replicates performed at different times. (D) Comparison of editing product compositions between ABE7.10-nCas9-AID10 and single-deaminase base editors. The percentage of indicated sequencing reads among total reads is presented as mean ± s.d. of two replicates. (E) The ten most abundant mutant alleles with concurrent cytosine and adenine substitutions by ABE7.10-nCas9-AID10 in one replicate of (D). Frequencies of indicated sequencing reads are shown on the right. The A-to-G, C-to-T, and C-to-A substitutions are in blue, red, and green, respectively. (F) Schematic diagram of eight dual base editors with different protein architectures. (G) Comparison of cytosine (top) and adenine (bottom) editing efficiencies between these base editors at the AtChr4 locus in Arabidopsis protoplasts by amplicon deep sequencing. Data are shown as mean ± s.d. of two biological replicates performed at different times. (H) Comparison of editing product compositions at the AtChr4 locus generated by these base editors. (I) The ten most abundant mutant alleles with concurrent cytosine and adenine substitutions by ABE8e-nCas9-AID10-UGI or AID10-ABE8e-nCas9-UGI in one replicate of (H). The A-to-G and C-to-T substitutions are in blue and red, respectively.
To comprehensively evaluate the performance of ABE7.10-nCas9-AID10, we checked two more genomic loci containing AC or CA tandem repeats (referred to as AtChr3 and AtChr4, Supplementary Table S1) in Arabidopsis protoplasts by amplicon deep sequencing. ABE7.10-nCas9, nCas9-AID10, and AID10-nCas9 were included for side-by-side comparison (Figure 4A). Amplicon deep sequencing showed that ABE7.10-nCas9-AID10 was able to catalyze C-to-T (Figure 4B) or A-to-G (Figure 4C) conversions at the same target site. Although there were multiple cytosines and adenines in the two protospacers, C-to-T and A-to-G conversions by ABE7.10-nCas9-AID10 mostly occurred at the positions 11–14 and 4–8, respectively, resembling the editing pattern by nCas9-AID10 or ABE7.10-nCas9 on its own (Figure 4B and C). The C-to-T editing efficiency of ABE7.10-nCas9-AID10 was comparable to or slightly lower than that of nCas9-AID10, whereas the A-to-G editing efficiency of ABE7.10-nCas9-AID10 was dramatically reduced relative to that of ABE7.10-nCas9 (Figure 4B and C). Nevertheless, deep sequencing reads revealed synchronous cytosine and adenine conversions at the same target sequences in protoplasts expressing ABE7.10-nCas9-AID10 with frequencies of 1.09–2.04%, whereas only cytosine or adenine changes were detected in protoplasts expressing nCas9-AID10 or ABE7.10-nCas9 alone (Figure 4D and E).
The decreased A-to-G editing capability of ABE7.10-nCas9-AID10 relative to ABE7.10-nCas9 may suggest steric hindrance of the C-terminal AID10 on the activity of the N-terminal ABE7.10 (Figure 4C). Meanwhile, Hua and colleagues have recently reported that the TadA7.10 by itself can achieve higher A-to-G editing efficiency than ABE7.10 (i.e., the TadA-TadA7.10 heterodimer) in rice (63). Therefore, we constructed additional dual base editors, including TadA7.10-nCas9-AID10, AID10-ABE7.10-nCas9 and AID10-TadA7.10-nCas9 (Supplementary Figure S12A), and compared their performance with that of ABE7.10-nCas9-AID10 in Arabidopsis protoplasts by amplicon deep sequencing. Intriguingly, dual base editors containing the N-terminal or C-terminal AID10 induced C-to-T conversions at the protospacer positions 1–11 and 11–14, respectively, suggesting that the fusion orientation of AID10 is the key determinant for the cytosine editing window (Supplementary Figure S12B). By contrast, adenine editing windows of the four dual base editors were located similarly at the protospacer positions 4–8 (Supplementary Figure S12B). All dual base editors were able to mediate simultaneous cytosine and adenine conversions at the same target sequences with frequencies of 0.41–2.06% (Supplementary Figure S12C and D). AID10-ABE7.10-nCas9, despite with a different cytosine editing window from ABE7.10-nCas9-AID10 (Supplementary Figure S12B), failed to improve the adenine editing or cytosine/adenine co-editing efficiency (Supplementary Figure S12C). Using TadA7.10 to replace ABE7.10 in dual base editors retained the activity windows, but slightly impaired the adenine editing efficiency (Supplementary Figure S12B and C).
Next, we sought to test whether adding a UGI moiety to the dual base editors could promote the cytosine and adenine co-editing efficiency. Therefore, we constructed ABE7.10-nCas9-AID10-UGI and AID10-ABE7.10-nCas9-UGI (Figure 4F) and compared their performance with that of ABE7.10-nCas9-AID10 and AID10-ABE7.10-nCas9 in protoplasts. Amplicon deep sequencing of the AtChr4 locus revealed that the addition of UGI led to a slightly enhanced C-to-T editing efficiency for both dual base editors (Figure 4G). Notably, ABE7.10-nCas9-AID10-UGI also exhibited a 1.4-fold increase in the cytosine and adenine co-editing efficiency relative to ABE7.10-nCas9-AID10, while there was a 3.4-fold increase in the cytosine and adenine co-editing efficiency for AID10-ABE7.10-nCas9-UGI relative to AID10-ABE7.10-nCas9 (Figure 4H). On the other hand, we assumed that enhancing the adenine editing activity by using ABE8e (i.e., the TadA-TadA8e heterodimer), a hyperactive adenine deaminase (39,64–66), in our dual base editors may further boost the cytosine and adenine co-editing efficiency. Indeed, amplicon deep sequencing revealed that the newly constructed ABE8e-nCas9-AID10-UGI and AID10-ABE8e-nCas9-UGI (Figure 4F) exhibited a dramatically increased A-to-G editing efficiency when compared to the corresponding ABE7.10-containing versions (Figure 4G). Accordingly, ABE8e-nCas9-AID10-UGI and AID10-ABE8e-nCas9-UGI induced simultaneous cytosine and adenine conversions at the same target sequences with a frequency of 10.1% and 9.5%, respectively, which represented a 13.4- or 7.5-fold increase relative to the co-editing frequencies by the corresponding ABE7.10-containing versions (Figure 4H and I). Importantly, C-to-T and A-to-G conversions by ABE8e-nCas9-AID10-UGI mostly occurred at the protospacer positions 11–14 and 4–8, respectively, while those by AID10-ABE8e-nCas9-UGI were mainly induced at the positions 1–11 and 4–8, respectively (Figure 4G and I). Taken together, these results indicate that ABE8e-nCas9-AID10-UGI and AID10-ABE8e-nCas9-UGI enable efficient generation of different cytosine and adenine co-editing products at the same target sites in plant cells.
Base editing can be repurposed for tunable gene knockdown via creating uORFs
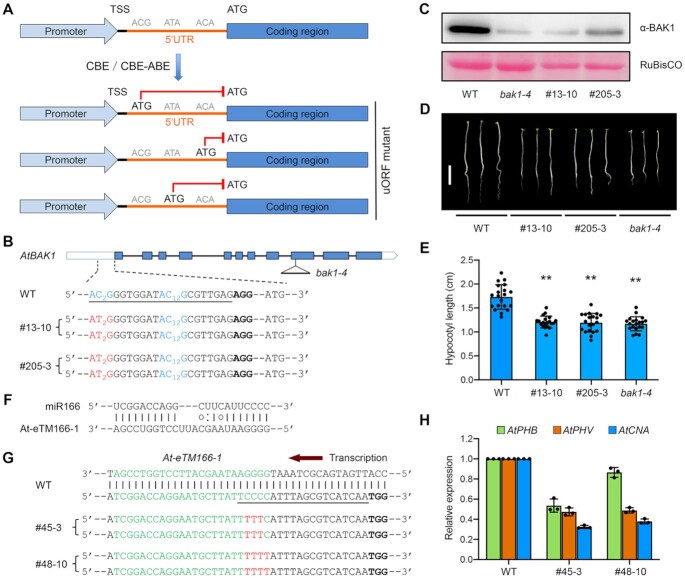
To expand the usefulness of our base editors in plant research, we explored whether tunable gene knockdown could be achieved by base editors via creating uORFs in the 5′ untranslated region (5′ UTR) of the target gene. As the uORFs have been shown to inhibit the translation initiation of downstream primary ORFs (pORFs) through ribosome stalling (67), we reasoned that creation of a uORF by changing the ACG, ATA or ACA trinucleotide in the 5′ UTR to ATG using our base editor toolkit would provide an effective strategy to down-regulate target gene activity (Figure 5A).
Figure 5.
Repurposing base editors as loss-of-function tools for studying protein-coding or non-coding genes. (A) Schematic diagram illustrating the gene knockdown strategy through creating a uORF in the 5′ UTR of the target gene by base editing. TSS, transcription start site. (B) Two homozygous mutant alleles of Arabidopsis BAK1 containing AID10-nCas9-UGI-created uORF in the 5′ UTR. The target sequence is underlined and the PAM is in bold. The ACG trinucleotides are in blue with subscripted numbers indicating the protospacer positions of target cytosines. (C) Immunoblot analysis indicates significantly decreased AtBAK1 abundances in bak1 mutant alleles relative to wild-type (WT) plants. The T-DNA insertion null mutant bak1-4 was used for reference. Note that the basal immunoblot signals were due to the cross-reactivity of the anti-BAK1 antibody to AtBKK1, a AtBAK1 paralog. RuBisCO, loading control. (D, E) Bak1 5′ UTR mutant seedlings resemble T-DNA insertion null seedlings. The photograph in (D) was taken at five days post germination. Scale bar = 0.5 cm. Quantification of hypocotyl lengths in (E) is presented as means ± s.d. of 20 seedlings. ** P< 0.01 (two-tailed Student's t test). (F) Predicted base pairing between miR166 and the miRNA binding site of At-eTM166-1 in Arabidopsis. (G) Two homozygous mutant alleles of the At-eTM166-1 locus generated by AID10-nCas9-UGI. The miR166 binding site is in green. The target sequence is underlined and the PAM is in bold. Cytosine conversions in mutant lines are in red. (H) qRT-PCR analysis reveals reduced transcript levels of miR166 target genes in At-eTM166-1 mutant lines. Data are presented as means ± s.d. of three biological replicates.
To provide a proof of concept, we intended to generate uORFs in the 5′ UTR of the AtBAK1 gene in Arabidopsis, which encodes the co-receptor for the plant hormone brassinosteroid (68). As there are three ACG trinucleotides in the 5′ UTR of AtBAK1, we evaluated the effects of changing them to ATG, singly or combinatorially, on the AtBAK1 expression in protoplasts using a luciferase reporter as a proxy (Supplementary Figure S13A). Creation of different uORFs inhibited the luciferase expression to variable levels, among which the uORF-2 was able to reduce the luciferase expression by 84% (Supplementary Figure S13B). Therefore, we employed AID10-nCas9-UGI to generate an endogenous uORF-2 in the 5′ UTR of AtBAK1 by inducing a C-to-T change at the protospacer position 2 (Figure 5B). Among 325 transgenic T1 plants, five heterozygous mutant lines carrying the intended C-to-T conversion were identified (Supplementary Figure S13C and Table S4), from which we harvested two T2 alleles (#13-10 and #205-3) with homozygous C-to-T mutation (Figure 5B). Strikingly, immunoblot analysis using the anti-AtBAK1 antibody indicated that the accumulation of endogenous AtBAK1 in these two mutant alleles was almost abolished using the bak1-4 T-DNA insertion null allele (68) as a reference (Figure 5C). The basal immunoblot signals in these bak1 mutant plants were attributed to the cross-reactivity of the anti-AtBAK1 antibody to AtBKK1 (50), a paralog of AtBAK1. Consistently, #13–10 and #205–3 seedlings exhibited shortened hypocotyls in the dark resembling the bak1-4 null allele (Figure 5D and E), indicative of functional knockout of AtBAK1 in these mutant alleles. These results suggest that creation of a suitable uORF in the 5′ UTR of the target gene through base editing can quantitatively knockdown or even knockout target gene expression.
Base editing is useful for loss-of-function analysis of endogenous target mimics of miRNAs
Plant endogenous target mimics (eTMs) and animal competing endogenous RNAs (ceRNAs) are analogous eukaryotic machineries regulating miRNA activities through sequestrating and preventing miRNAs from interacting with their authentic target mRNAs (69–71). While a large number of eTMs and ceRNAs have been computationally predicted, only a few of them were experimentally validated (71). In addition, several studies in animals have challenged the biological relevance of miRNA-ceRNA interactions (72). Therefore, an efficient tool for dissecting the interactions between miRNAs and predicted eTMs or ceRNAs is highly desirable.
We conceived an approach for loss-of-function analysis of plant eTMs by introducing a cluster of nucleotide changes to disrupt the putative miRNA binding site in eTMs through base editing. To demonstrate a proof of concept, we employed AID10-nCas9-UGI to destroy the miR166 binding site in At-eTM166-1 (Figure 5F), an eTM that has been reported to negatively regulate the activity of miR166 in Arabidopsis (71). An sgRNA was designed to target the At-eTM166-1 locus, placing the four consecutive C•G base pairs at the protospacer positions 2–5 (Figure 5G). Among 71 transgenic T1 plants, eight lines harboring C•G to T•A mutations were identified (Supplementary Figure S8C and Table S4). We further isolated two T2 lines (i.e. #45–3 and #48–10) carrying homozygous C•G to T•A mutations at the positions 2–4 and 2–5, respectively (Figure 5G). If the interaction between At-eTM166-1 and miR166 was biologically relevant, we expected that these mutations would unleash miR166 to further down-regulate its target genes, including AtPHB, AtPHV and AtCNA (73). Indeed, we observed that the transcript levels of AtPHV and AtCNA were comparably decreased in these two lines (Figure 5H), implying that disruption of three internal cytosines has been sufficient to fully abolish the interaction between At-eTM166-1 and miR166. The transcript abundances of AtPHB were also diminished in both lines, whereas the line #48–10 showed a less dramatic reduction than the line #45–3 (Figure 5H), which may be ascribed to the counteracting effect of unknown environmental stimuli on the miR166-mediated AtPHB transcriptional repression in the line #48–10. Overall, our results indicate that the proposed strategy can ease the functional validation of plant eTMs.
DISCUSSION
The CRISPR-derived base editing technology has continuously evolved since its emergence to gain increased efficiency, flexibility, and versatility in both plant and animal research. In contrast to limited forms of laboratory-evolved adenine deaminases for building ABEs, the great divergence of natural or engineered cytosine deaminases has facilitated the birth of a handful of CBEs with different editing properties regarding the width and position of the activity window within the protospacer as well as the sequence context preference (26,74). These CBEs in turn helped satisfy different mutagenesis needs of cytosines in highly variable target sequences in a given genome (26).
In this study, we developed a toolkit of plant base editors containing AID10, an improved variant of hAID (31). AID10 exhibits no bias across TC, AC, GC and CC contexts in plant cells (Supplementary Figure S14), outperforming APOBEC1 that disfavors the GC context (3). Notably, AID10 confers varying activity windows when fused to different termini of nSpCas9. AID10-nCas9-UGI enables a broad activity window at the PAM-distal protospacer (i.e. positions -1–9), whereas nCas9-AID10-UGI exhibits a narrow and PAM-proximal activity window (i.e. positions 12–14) (Figure 6). By contrast, AID10-nCas9-AID10-UGI displays an additive activity window of AID10-nCas9-UGI and nCas9-AID10-UGI (Figure 6). In particular, the narrow and PAM-proximal editing window of nCas9-AID10-UGI (Figure 3B–D) has not been observed in other CBEs containing a C-terminal fusion of hAID in mammalian cells (22,75), thus representing a novel editing feature. The target scope of this toolkit was further expanded using orthogonal or engineered Cas9 variants, including nSpCas9-NG, nSaCas9 and nSaCas9-KKH, which have dissimilar or relaxed PAM requirements (Figure 6). Of particular note, the CBEs derived from SaCas9 or SpCas9 could outperform each other at different target sequences or for different target cytosines at the same target sequences (Figure 2B and C). Moreover, our toolkit can generate amino acid substitutions that cannot be fulfilled by existing CBEs. For example, we used nCas9-AID10-UGI to generate the herbicide-resistant OsALS allele containing an A96T or A96P substitution (Figure 3C), whereas an earlier report obtained the herbicide-resistant OsALS allele with an A96V substitution using Target-AID, a CBE with a PAM-distal activity window, in combination with a different sgRNA (27). Very recently, dual base editors have been invented to confer cytosine and adenine co-editing at the same target sequences in mammalian and plant cells (76–80). We further devised and evaluated eight AID10-based dual base editors, all of which enabled concurrent C-to-T and A-to-G conversions at the same protospacers in plant cells (Figure 4). Among them, ABE8e-nCas9-AID10-UGI and AID10-ABE8e-nCas9-UGI exhibited the highest cytosine and adenine co-editing efficiency, although their performance in planta still needs to be validated. Noteworthily, ABE8e-nCas9-AID10-UGI allowed simultaneous editing of cytosines and adenines at separate windows within the same protospacers (Figure 4F and I). To our knowledge, such an editing pattern has not been reported for other dual base editors (76–80) and will be particularly useful for tailoring new dual base editing products.
Figure 6.
Summary of editing windows and scopes of the eight representative base editors developed in this study. Dark colors indicate optimal editing windows, while light colors indicate possible editing windows. Dashed lines in nSpCas9-NG and nSaCas9-KKH denote extra mutations.
In addition to developing useful plant base editors, we also explored unprecedented applications of base editing in plant research. On the one hand, we invented a strategy to down-regulate target gene activity by creating uORFs in the 5′ UTR of the target gene through base editing. When compared with the CRISPR/Cas9 technology or other base editor-mediated gene loss-of-function strategies, such as CRISPR-STOP (14) and mRNA mis-splicing (18), this new strategy for gene inactivation has a particular advantage in terms of tunability. By creating uORFs at different positions relative to the pORF, quantitative gene knockdown to varying degrees or even functional knockout can be selectively achieved for the target gene, making it a flexible and versatile tool for manipulating both lethal and non-lethal genes. When compared with other gene knockdown strategies, such as RNA inference (RNAi) and CRISPR inference (CRISPRi) (81), this strategy can mediate stable and permanent gene knockdown free of any transgene. On the other hand, we utilized base editing to effectively perturb the putative miRNA binding site in a plant eTM to study its biological function in miRNA regulation, which fills in the blank of loss-of-function tools for experimental validation of predicted plant eTMs. It is conceivable that such a strategy is immediately transferrable to studying regulatory mechanisms of predicted animal ceRNAs, which are functionally analogous to plant eTMs (70).
The PAM requirement and the width of deamination window of a CBE combinatorially determine the total number of editable cytosines in a given genome. When compared to CBEs with a narrow activity window, those with a broad activity window, such as AID10-nCas9-UGI, not only make more genomic cytosines accessible but also increase mutations in the protospacers, which is advantageous for applications such as directed protein evolution (22,23,76,82) and loss-of-function analysis of eTMs/ceRNAs (Figure 5G and H). The downside of broad-window CBEs is the accompanied risk of bystander editing. Although the bystander editing can be circumvented by rational selection of an optimal base editor and sgRNA in some cases, it is inevitable in others. However, it should be emphasized that bystander mutations can be tolerated when they are synonymous in the exons or occur in the non-coding regions. Moreover, for the base editor-mediated gene inactivation, such as inducing intron mis-splicing (Supplementary Figures S5 and S7) (18,83,84) or creating premature stop codons (14,15) or uORFs (Figure 5A–E), the bystander editing is inconsequential. Also, it is possible in practice to screen a large quantity of edited plant alleles to identify those without or with acceptable bystander mutations.
In summary, here we report a suite of plant base editors with different activity windows and target scopes (Figure 6 and Supplementary Table S5). This toolkit will be a valuable addition to the current base editor arsenal in plants by expanding editable cytosines in a given plant genome or creating diverse base editing products at the same target sequences. We also open up novel routes for manipulating the function of protein-coding or non-coding genes through base editing.
DATA AVAILABILITY
Raw data of amplicon deep sequencing have been deposited in the NCBI Sequence Read Archive under BioProject accession code PRJNA762247 (http://www.ncbi.nlm.nih.gov/sra/PRJNA762247). Amino acid sequences of all base editors tested in this study are provided in the Supplementary Sequence.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Xionglei He for providing the pGAL1-AID10-dCas9-UGI plasmid, Caixia Gao for the nCas9-PBE plasmid, Qi-Jun Chen for the pHEE401E plasmid, Kejian Wang for the LbCas12a construct, and Holger Puchta for the SaCas9 construct.
Author contributions: J.-F.L. and Z.L. conceived and designed the study, and wrote the manuscript. X.X., Z.L., J.L., K.L. and C.L. performed the experiments and analyzed the data.
Contributor Information
Xiangyu Xiong, MOE Key Laboratory of Gene Function and Regulation, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China.
Zhenxiang Li, MOE Key Laboratory of Gene Function and Regulation, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China.
Jieping Liang, MOE Key Laboratory of Gene Function and Regulation, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China.
Kehui Liu, MOE Key Laboratory of Gene Function and Regulation, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China.
Chenlong Li, MOE Key Laboratory of Gene Function and Regulation, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China.
Jian-Feng Li, MOE Key Laboratory of Gene Function and Regulation, State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou 510275, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Transgenic Science and Technology Major Program of China [2019ZX08010003-001-009]; Foundation of Guangzhou Science and Technology Key Project [201904020041]; National Natural Science Foundation of China [32125004]; China Postdoctoral Science Foundation [2020TQ0387]. Funding for open access charge: Foundation of Guangzhou Science and Technology Key Project.
Conflict of interest statement. The authors have filed a China invention patent (application number: ZL202011444311.9) about the gene knockdown approach through creating upstream open reading frames using base editors.
REFERENCES
- 1. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J.et al.. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu J., Zeng Q., Wang Q., Liu S., Yu S., Mu J., Huang S., Sela H., Distelfeld A., Huang L.et al.. SNP-based pool genotyping and haplotype analysis accelerate fine-mapping of the wheat genomic region containing stripe rust resistance gene Yr26. Theor. Appl. Genet. 2018; 131:1481–1496. [DOI] [PubMed] [Google Scholar]
- 3. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R.. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017; 551:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A.et al.. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019; 576:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., Wang Y., Anzalone A.V., Raguram A., Doman J.L.et al.. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020; 38:582–585. [DOI] [PubMed] [Google Scholar]
- 7. Tang X., Sretenovic S., Ren Q., Jia X., Li M., Fan T., Yin D., Xiang S., Guo Y., Liu L.et al.. Plant prime editors enable precise gene editing in rice cells. Mol. Plant. 2020; 13:667–670. [DOI] [PubMed] [Google Scholar]
- 8. Li H., Li J., Chen J., Yan L., Xia L.. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant. 2020; 13:671–674. [DOI] [PubMed] [Google Scholar]
- 9. Xu W., Zhang C., Yang Y., Zhao S., Kang G., He X., Song J., Yang J.. Versatile nucleotides substitution in plant using an improved prime editing system. Mol. Plant. 2020; 13:675–678. [DOI] [PubMed] [Google Scholar]
- 10. Hua K., Jiang Y., Tao X., Zhu J.K.. Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 2020; 18:2167–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Butt H., Rao G.S., Sedeek K., Aman R., Kamel R., Mahfouz M.. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 2020; 18:2370–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Molla K.A., Yang Y.. CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol. 2019; 37:1121–1142. [DOI] [PubMed] [Google Scholar]
- 13. Mishra R., Joshi R.K., Zhao K.. Base editing in crops: current advances, limitations and future implications. Plant Biotechnol. J. 2019; 18:20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuscu C., Parlak M., Tufan T., Yang J., Szlachta K., Wei X., Mammadov R., Adli M.. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat. Methods. 2017; 14:710–712. [DOI] [PubMed] [Google Scholar]
- 15. Billon P., Bryant E.E., Joseph S.A., Nambiar T.S., Hayward S.B., Rothstein R., Ciccia A.. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell. 2017; 67:1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gapinske M., Luu A., Winter J., Woods W.S., Kostan K.A., Shiva N., Song J.S., Perez-Pinera P.. CRISPR-SKIP: programmable gene splicing with single base editors. Genome Biol. 2018; 19:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xue C., Zhang H., Lin Q., Fan R., Gao C.. Manipulating mRNA splicing by base editing in plants. Sci. China Life Sci. 2018; 61:1293–1300. [DOI] [PubMed] [Google Scholar]
- 18. Li Z., Xiong X., Wang F., Liang J., Li J.F.. Gene disruption through base editing-induced messenger RNA missplicing in plants. New Phytol. 2019; 222:1139–1148. [DOI] [PubMed] [Google Scholar]
- 19. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cullot G., Boutin J., Toutain J., Prat F., Pennamen P., Rooryck C., Teichmann M., Rousseau E., Lamrissi-Garcia I., Guyonnet-Duperat V.et al.. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019; 10:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt C., Pacher M., Puchta H.. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 2019; 98:577–589. [DOI] [PubMed] [Google Scholar]
- 22. Ma Y., Zhang J., Yin W., Zhang Z., Song Y., Chang X.. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods. 2016; 13:1029–1035. [DOI] [PubMed] [Google Scholar]
- 23. Hess G.T., Frésard L., Han K., Lee C.H., Li A., Cimprich K.A., Montgomery S.B., Bassik M.C.. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods. 2016; 13:1036–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang W., Liu D.R.. Rewritable multi-event analog recording in bacterial and mammalian cells. Science. 2018; 360:eaap8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Farzadfard F., Gharaei N., Higashikuni Y., Jung G., Cao J., Lu T.K.. Single-nucleotide-resolution computing and memory in living cells. Mol. Cell. 2019; 75:769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anzalone A.V., Koblan L.W., Liu D.R.. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020; 38:824–844. [DOI] [PubMed] [Google Scholar]
- 27. Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H., Teramura H., Yamamoto T., Komatsu H., Miura K.et al.. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017; 35:441–443. [DOI] [PubMed] [Google Scholar]
- 28. Ren B., Yan F., Kuang Y., Li N., Zhang D., Zhou X., Lin H., Zhou H.. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. Plant. 2018; 11:623–626. [DOI] [PubMed] [Google Scholar]
- 29. Zong Y., Song Q., Li C., Jin S., Zhang D., Wang Y., Qiu J., Gao C.. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018; 36:950–953. [DOI] [PubMed] [Google Scholar]
- 30. Jin S., Fei H., Zhu Z., Luo Y., Liu J., Gao S., Zhang F., Chen Y.H., Wang Y., Gao C.. Rationally designed APOBEC3B cytosine base editors with improved specificity. Mol. Cell. 2020; 79:728–740. [DOI] [PubMed] [Google Scholar]
- 31. Liu K., Deng S., Ye C., Yao Z., Wang J., Liu L., He X.. Mapping single-cell-resolution cell phylogeny reveals cell population dynamics during organ development. Nat. Methods. 2021; 18:1506–1514. [DOI] [PubMed] [Google Scholar]
- 32. Li Z., Zhang D., Xiong X., Yan B., Xie W., Sheen J., Li J.F.. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants. 2017; 3:930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J.F., Norville J.E., Aach J., McCormack M., Zhang D., Bush J., Church G.M., Sheen J.. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotianabenthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013; 31:688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.L., Wang D., Gao C.. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017; 35:438–440. [DOI] [PubMed] [Google Scholar]
- 35. Steinert J., Schiml S., Fauser F., Puchta H.. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcusthermophilus and Staphylococcusaureus. Plant J. 2015; 84:1295–1305. [DOI] [PubMed] [Google Scholar]
- 36. Hu X., Wang C., Liu Q., Fu Y., Wang K.. Targeted mutagenesis in rice using CRISPR-Cpf1 system. J. Genet. Genomics. 2017; 44:71–73. [DOI] [PubMed] [Google Scholar]
- 37. Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., Noda T., Abudayyeh O.O., Gootenberg J.S., Mori H.et al.. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018; 361:1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K.. Broadening the targeting range of Staphylococcusaureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015; 33:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Richter M.F., Zhao K.T., Eton E., Lapinaite A., Newby G.A., Thuronyi B.W., Wilson C., Koblan L.W., Zeng J., Bauer D.E.et al.. Phage-assisted evolution of an adenine base editor with improved cas domain compatibility and activity. Nat. Biotechnol. 2020; 38:883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiong X., Liang J., Li Z., Gong B.Q., Li J.F.. Multiplex and optimization of dCas9-TV-mediated gene activation in plants. J. Integr. Plant Biol. 2021; 63:634–645. [DOI] [PubMed] [Google Scholar]
- 41. Xie X., Ma X., Zhu Q., Zeng D., Li G., Liu Y.G.. CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol. Plant. 2017; 10:1246–1249. [DOI] [PubMed] [Google Scholar]
- 42. Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J.. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015; 16:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoo S.D., Cho Y.H., Sheen J.. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007; 2:1565–1572. [DOI] [PubMed] [Google Scholar]
- 44. Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D.et al.. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011; 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E.et al.. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019; 37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E., Kim S.T., Eils R., Kim J.S., Bae S.. Web-based design and analysis tools for CRISPR base editing. BMC Bioinf. 2018; 19:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clough S.J., Bent A.F.. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsisthaliana. Plant J. 1998; 16:735–743. [DOI] [PubMed] [Google Scholar]
- 48. Nishimura A., Aichi I., Matsuoka M.. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 2006; 1:2796–2802. [DOI] [PubMed] [Google Scholar]
- 49. Liu Q., Wang C., Jiao X., Zhang H., Song L., Li Y., Gao C., Wang K.. Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019; 62:1–7. [DOI] [PubMed] [Google Scholar]
- 50. Gong B.Q., Guo J., Zhang N., Yao X., Wang H.B., Li J.F.. Cross-microbial protection via priming a conserved immune co-receptor through juxtamembrane phosphorylation in plants. Cell Host Microbe. 2019; 26:810–822. [DOI] [PubMed] [Google Scholar]
- 51. Grunewald J., Zhou R., Iyer S., Lareau C.A., Garcia S.P., Aryee M.J., Joung J.K.. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 2019; 37:1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen M., Herde M., Witte C.P.. Of the nine cytidine deaminase-like genes in Arabidopsis, eight are pseudogenes and only one is required to maintain pyrimidine homeostasis in vivo. Plant Physiol. 2016; 171:799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bennett E.P., Petersen B.L., Johansen I.E., Niu Y., Yang Z., Chamberlain C.A., Met O., Wandall H.H., Frodin M.. INDEL detection, the ‘Achilles heel’ of precise genome editing: a survey of methods for accurate profiling of gene editing induced indels. Nucleic Acids Res. 2020; 48:11958–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang J., Guo Q., Liu L., Wang X.. Crystal structure of Arabidopsisthaliana cytidine deaminase. Biochem. Biophys. Res. Commun. 2020; 529:659–665. [DOI] [PubMed] [Google Scholar]
- 55. Li P., Wang Y., Qian Q., Fu Z., Wang M., Zeng D., Li B., Wang X., Li J.. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007; 17:402–410. [DOI] [PubMed] [Google Scholar]
- 56. Chen Y., Wang Z., Ni H., Xu Y., Chen Q., Jiang L.. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci. China Life Sci. 2017; 60:520–523. [DOI] [PubMed] [Google Scholar]
- 57. Zheng H., Rowland O., Kunst L.. Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell. 2005; 17:1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S.et al.. In vivo genome editing using Staphylococcusaureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A.et al.. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015; 163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Powles S.B., Yu Q.. Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 2010; 61:317–347. [DOI] [PubMed] [Google Scholar]
- 61. Yu Q., Powles S.B.. Resistance to AHAS inhibitor herbicides: current understanding. Pest Manag. Sci. 2014; 70:1340–1350. [DOI] [PubMed] [Google Scholar]
- 62. Li C., Zong Y., Wang Y., Jin S., Zhang D., Song Q., Zhang R., Gao C.. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018; 19:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hua K., Tao X., Liang W., Zhang Z., Gou R., Zhu J.K.. Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol. J. 2020; 18:770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lapinaite A., Knott G.J., Palumbo C.M., Lin-Shiao E., Richter M.F., Zhao K.T., Beal P.A., Liu D.R., Doudna J.A.. DNA capture by a CRISPR-Cas9-guided adenine base editor. Science. 2020; 369:566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ren Q., Sretenovic S., Liu S., Tang X., Huang L., He Y., Liu L., Guo Y., Zhong Z., Liu G.et al.. PAM-less plant genome editing using a CRISPR-SpRY toolbox. Nat Plants. 2021; 7:25–33. [DOI] [PubMed] [Google Scholar]
- 66. Xu Z., Kuang Y., Ren B., Yan D., Yan F., Spetz C., Sun W., Wang G., Zhou X., Zhou H.. SpRY greatly expands the genome editing scope in rice with highly flexible PAM recognition. Genome Biol. 2021; 22:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang H., Wang Y., Lu J.. Function and evolution of upstream ORFs in eukaryotes. Trends Biochem. Sci. 2019; 44:782–794. [DOI] [PubMed] [Google Scholar]
- 68. Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C.. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002; 110:213–222. [DOI] [PubMed] [Google Scholar]
- 69. Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., Garcia J.A., Paz-Ares J.. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007; 39:1033–1037. [DOI] [PubMed] [Google Scholar]
- 70. Rubio-Somoza I., Weigel D., Franco-Zorilla J.M., Garcia J.A., Paz-Ares J.. ceRNAs: miRNA target mimic mimics. Cell. 2011; 147:1431–1432. [DOI] [PubMed] [Google Scholar]
- 71. Wu H.J., Wang Z.M., Wang M., Wang X.J.. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013; 161:1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Thomson D.W., Dinger M.E.. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016; 17:272–283. [DOI] [PubMed] [Google Scholar]
- 73. Williams L., Grigg S.P., Xie M., Christensen S., Fletcher J.C.. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development. 2005; 132:3657–3668. [DOI] [PubMed] [Google Scholar]
- 74. Arbab M., Shen M.W., Mok B., Wilson C., Matuszek Z., Cassa C.A., Liu D.R.. Determinants of base editing outcomes from target library analysis and machine learning. Cell. 2020; 182:463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cheng T.L., Li S., Yuan B., Wang X., Zhou W., Qiu Z.. Expanding C-T base editing toolkit with diversified cytidine deaminases. Nat. Commun. 2019; 10:3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li C., Zhang R., Meng X., Chen S., Zong Y., Lu C., Qiu J.L., Chen Y.H., Li J., Gao C.. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020; 38:875–882. [DOI] [PubMed] [Google Scholar]
- 77. Zhang X., Zhu B., Chen L., Xie L., Yu W., Wang Y., Li L., Yin S., Yang L., Hu H.et al.. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 2020; 38:856–860. [DOI] [PubMed] [Google Scholar]
- 78. Grunewald J., Zhou R., Lareau C.A., Garcia S.P., Iyer S., Miller B.R., Langner L.M., Hsu J.Y., Aryee M.J., Joung J.K.. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 2020; 38:861–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sakata R.C., Ishiguro S., Mori H., Tanaka M., Tatsuno K., Ueda H., Yamamoto S., Seki M., Masuyama N., Nishida K.et al.. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020; 38:865–869. [DOI] [PubMed] [Google Scholar]
- 80. Wang Y., Zhou L., Tao R., Liu N., Long J., Qin F., Tang W., Yang Y., Chen Q., Yao S.. sgBE: a structure-guided design of sgRNA architecture specifies base editing window and enables simultaneous conversion of cytosine and adenosine. Genome Biol. 2020; 21:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li Z., Xiong X., Li J.F.. The working dead: repurposing inactive CRISPR-associated nucleases as programmable transcriptional regulators in plants. aBIOTECH. 2019; 1:32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li Q., Li Y., Yang S., Huang S., Yan M., Ding Y., Tang W., Lou X., Yin Q., Sun Z.et al.. CRISPR–Cas9-mediated base-editing screening in mice identifies DND1 amino acids that are critical for primordial germ cell development. Nat. Cell Biol. 2018; 20:1315–1325. [DOI] [PubMed] [Google Scholar]
- 83. Kang B.C., Yun J.Y., Kim S.T., Shin Y., Ryu J., Choi M., Woo J.W., Kim J.S.. Precision genome engineering through adenine base editing in plants. Nat. Plants. 2018; 4:427–431. [DOI] [PubMed] [Google Scholar]
- 84. Webber B.R., Lonetree C.L., Kluesner M.G., Johnson M.J., Pomeroy E.J., Diers M.D., Lahr W.S., Draper G.M., Slipek N.J., Smeester B.A.et al.. Highly efficient multiplex human t cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 2019; 10:5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of amplicon deep sequencing have been deposited in the NCBI Sequence Read Archive under BioProject accession code PRJNA762247 (http://www.ncbi.nlm.nih.gov/sra/PRJNA762247). Amino acid sequences of all base editors tested in this study are provided in the Supplementary Sequence.