Abstract
DNA helicases of the RecD2 family are ubiquitous. Bacillus subtilis RecD2 in association with the single-stranded binding protein SsbA may contribute to replication fork progression, but its detailed action remains unknown. In this work, we explore the role of RecD2 during DNA replication and its interaction with the RecA recombinase. RecD2 inhibits replication restart, but this effect is not observed in the absence of SsbA. RecD2 slightly affects replication elongation. RecA inhibits leading and lagging strand synthesis, and RecD2, which physically interacts with RecA, counteracts this negative effect. In vivo results show that recD2 inactivation promotes RecA–ssDNA accumulation at low mitomycin C levels, and that RecA threads persist for a longer time after induction of DNA damage. In vitro, RecD2 modulates RecA-mediated DNA strand-exchange and catalyzes branch migration. These findings contribute to our understanding of how RecD2 may contribute to overcome a replicative stress, removing RecA from the ssDNA and, thus, it may act as a negative modulator of RecA filament growth.
INTRODUCTION
Accurate genome duplication is essential for life, and all organisms have multiple mechanisms to protect, repair and reactivate DNA replication forks. In addition to proteins directly involved in DNA replication, like the replicative helicase, the polymerase, primase, etc, active replication forks are pre-equipped with several accessory proteins by their interaction with replisome proteins (1–3). Our current knowledge shows that these accessory effectors may contribute to the progression of replication, and they include repair proteins, error prone polymerases and homologous recombination proteins.
The intimate links between homologous recombination and DNA replication were early noted in some lytic (e.g. Escherichia coli T4 and Bacillus subtilis SPP1) as well as temperate (Staphylococcus aureus 80α and ϕ11) dsDNA bacteriophages. These phages strictly require homologous recombination proteins for their replication (4–6). Growing evidences show that these two DNA transactions are tightly linked in all organisms (7,8). In bacteria, several recombination proteins are associated with active replication forks by its interaction with the single-stranded DNA binding protein (SSB in E. coli and SsbA in B. subtilis) (2,3). In both cases, the number of DNA helicases targeted to active replication forks in the absence of DNA damage is remarkably high, and in the case of B. subtilis these helicases are: PriA, RecG, the RecQ-like enzymes (RecQ and RecS), and RecD2 (2).
RecD enzymes, which belong to superfamily 1B (SF1B) of DNA helicases (9), are divided into two sub-groups: RecD1 (known as RecD) and RecD2 (10,11). RecD2 helicases have a longer N-terminal region prior to the first helicase motif (Supplementary Figure S1). The role of RecD2 in the cell is poorly known, whereas that of RecD is known. E. coli RecD (RecDEco) forms a part of the well-studied RecBCD complex, crucial for double-strand break repair. Bacteria that have RecD2 lack the RecBCD complex (10–12) The best biochemically characterized member of this group is Deinococcus radiodurans RecD2 (RecD2Dra). RecD2Dra is a monomeric DNA helicase with 5′→3′ polarity (13–16). RecD2 helicases also show homology with human DNA Helicase B (named HDHB, or HELB, Supplementary Figure S1), a conserved helicase present in higher eukaryotes with a poorly understood role in initiation of DNA replication and genome maintenance (17–19). Amino acid sequence identity and similarity between mouse HELB and B. subtilis RecD2 are 16.5 and 26%, respectively (20). Both proteins interact with their respective SSB (2,19,21,22).
The in vivo role of B. subtilis RecD2 is poorly understood (unless stated otherwise, indicated genes and products are of B. subtilis origin). Inactivation of recD2 confers a modest increase in the spontaneous mutation rate (21,23). A role of RecD2 in homologous recombination is inferred from its contribution to acquire DNA from related species during natural chromosomal transformation (23). recD2 mutants are sensitive to DNA damaging agents that stall or collapse replication forks such as the alkylating agent methyl methane sulfonate (MMS) and the intra- and inter-strand crosslinking agent mitomycin C (MMC) (21,24,25). RecD2 is important for replication fork progression (21). However, different results were obtained when a recD2 deletion was analyzed in D. radiodurans. The recD2Dra mutants were not sensitive to MMS and MMC, and showed greater efficiency in chromosomal transformation (26), obscuring the role of this protein in the cell.
In this work, using a combination of genetic, cytological, and biochemical techniques, we show that RecD2 is a modulator of replication restart and of the RecA recombinase. RecD2 interacts with RecA and inhibits the initiation of strand-exchange, because it inhibits the formation of the presynaptic filament. Consistent with this, RecA threads, which are formed after MMC treatment, persist for a longer period in the recD2 mutants. In the replisome context, RecD2 may inhibit unwanted recombination when replication forks stall, displacing RecA and allowing the advance of the replication fork.
MATERIALS AND METHODS
Bacterial strains and plasmids
B. subtilis BG214 is the wild type (wt) strain. TheΔrecX and ΔrecO strains were previously constructed (27). A ΔrecD2 deletion mutant was constructed replacing by homologous recombination 2193-bp of the BG214 chromosome by a six-cm-six cassette (28) and chloramphenicol selection (Cm, 5 μg/ml). Briefly, the region of the recD2 gene (781 amino acids long) has two BseRI restriction sites, one located 10-bp before the start codon and the other one overlapping codon 721. These restriction sites were used to replace this region by the six-cm-six cassette. The resulting ΔrecD2 deletion strain lacks from codon 1 to 721, so that almost the full gene is deleted. In a second step, the antibiotic resistance gene was deleted by the β site-specific recombinase, which deletes the region between the two six sites (29). The final strain (ΔrecD2) was sequenced to confirm accuracy. The construction of his-tagged recD2 on the B. subtilis chromosome was as follows. The C-ter of recD2 gene (500 bp) without the stop codon was cloned in the integration vector pSG1164-His. The resulting plasmid, pSG1164-C-ter-recD2-His, was integrated by homologous recombination into the B. subtilis BG214 chromosome at the recD2 locus by Cm selection, and the accuracy of the recombination reaction was confirmed by PCR and DNA sequencing. This allows the replacement of RecD2 by RecD2-His, a fusion protein with a linker (GPGLSG) and 6xHis, that is expressed from its natural promoter. The resulting strain was named recD2his6.
The pET-recD2-His expression vector was constructed in two steps. First, the recD2 gene with its stop codon (2397-bp) was cloned into the pET-3b vector. PCR primers to amplify the recD2 gene with flanking NdeI and BamHI restriction sites were designed. The accuracy of the resultant plasmid (pET-3b-recD2) was confirmed by sequencing. In a second step, the region in pET-3b-recD2 corresponding to the C-ter of the recD2 gene was replaced by the one of the pSG1164-Cter-recD2-His plasmid by BseRI and BamHI digestion. This resulted in the pET-recD2-His expression vector, that allows the expression in E. coli of RecD2-His, a fusion protein with linker (GPGLS) and 6xHis. The pET-recD2-K373A-His expression vector was constructed by site-directed mutagenesis using the Quickchange kit (Stratagene) with corresponding primers and pET-recD2-His as template. Plasmids to overproduce SsbA and RecO, respectively, have been described previously (30). Escherichia coli XL1 Blue was used for plasmid construction, and E. coli BL21 (DE3) pLysS was used for protein expression. B. subtilis BG214 cells bearing pBT61-recA were used to overexpress RecA as described (31). Note that the nomenclature used to denote the origin of proteins from bacteria other than B. subtills is based on the bacterial genus and species (e.g., D. radiodurans RecD2 is referred as RecD2Dra).
In vivo assays
Viability tests in the presence of MMS were performed to analyze the functionality of the RecD2-His protein. B. subtilis wt, ΔrecD2 and recD2his6 cells were grown to an OD560 = 0.5 and appropriate dilutions were spotted on LB plates containing 1.5 mM of MMS. Plates were incubated O/N at 37°C and photographed.
To measure RecD2 levels in vivo, a 250 ml culture of recD2his6 cells was grown to OD560 = 0.7 at 37°C in LB. Then, cells (0.2g wet weight) were collected and resuspended in 1 ml of buffer A (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 1 mM DTT, 10% v/v glycerol) containing lysozyme 0.2 mg/ml. After 60 min at 4°C, cells were sonicated and cell debris were separated by centrifugation (30 min, 12,000 rpm, 4°C). RecD2-His that was present in the soluble fraction was concentrated by binding to Ni-Sepharose (20 μl). RecD2 was eluted from the column with twice 100 μl of buffer A containing 200 mM imidazole. 30 μl of each fraction were loaded on a 12% SDS-polyacrylamide gel, and the amount of RecD2 was deduced from a standard curve obtained by loading increasing amounts of purified RecD2. The number of RecD2 molecules per colony forming units (CFUs) was calculated considering that for an OD560 = 0.7 there are ∼1.4 × 108 CFUs/ml.
RecA protein accumulation upon SOS induction was analyzed in wt and ΔrecD2 cells as previously described (27). Briefly, cells were grown to OD560 = 0.4 in LB, and exposed to increasing concentrations of MMC for 30 min. Then, cells were lysed, and proteins separated by 10% SDS-PAGE, followed by immunoblot transfer. Blots were developed with rabbit polyclonal anti-RecA antibodies and quantified with the ImageJ software. The amount of RecA protein in each sample was interpolated from the standard curve obtained loading different amounts of purified protein in the same gel.
Fluorescence microscopy
A functional RecA-mVenus fusion for the visualization of fluorescent RecA in live cells was used. This C-terminal fusion is integrated at the original gene locus, so that the fusion is the sole source of RecA expressed in cells, and under the control of the original promoter (32). The fusion was placed into the ΔrecD2 background by SPP1 transduction. Epifluorescence microscopy was used to monitor filament formation and dynamics of RecA-mVenus before and after stress conditions. B. subtilis cells were grown in S750 minimal medium at 30°C under shaking conditions until exponential growth. Then, cells were treated or not with MMC (50 ng/ml) and were visualized using a Zeiss Observer Z1 (Carl Zeiss) microscope with an oil immersion objective (100× magnification, NA 1.45 alpha Plan-FLUAR) and a CCD camera (CoolSNAP EZ, Photometrics). Data were processed using Metamorph 7.5.5.0 software (Molecular Devices, Sunnyvale, CA, USA), which also allows the calibration of the fluorescence intensity and pixel size to determine the cell length and BacStalk (33). Time-lapse epifluorescence microscopy images of RecA-mVenus in the wt and ΔrecD2 background were collected every 10 min.
Purification of B. subtilis RecD2
RecD2-His (89 kDa, hereafter named RecD2), was overexpressed and purified as follows: E. coli BL21(DE3) pLysS cells carrying pET3b-recD2-His plasmid were grown in LB medium supplemented with 100 μg/ml ampicillin and 15 μg/ml chloramphenicol at 30°C to OD600= 0.2, and then at 18°C to OD600 = 0.4. Overexpression of the protein was induced by 0.25 mM IPTG addition. After overnight growth, the cell mass was recovered by centrifugation at 9,000 rpm for 10 min at 4°C. Pellets were resuspended in buffer B (50 mM Tris–HCl pH 7.5, 1 M NaCl, 1 mM DTT and 10% v/v glycerol) supplemented with 5 mM imidazole and sonicated. After centrifugation at 18,000 rpm for 45 min, the pellet was resuspended in buffer A with 5 mM imidazole and centrifugated at the same conditions. The resultant supernatant of both centrifugations was loaded onto a Ni-Sepharose column equilibrated in the same buffer. Proteins were eluted using a gradient of increasing concentrations of imidazole (5–100 mM), and analyzed by 12.5% SDS-PAGE. Fractions containing RecD2 were precipitated by the addition of 70% ammonium sulphate for 30 min. The protein pellet was resuspended in buffer B with 1 mM EDTA and 10 mM sodium phosphate pH 7.5 and then loaded onto a hydroxyapatite column. Protein elution was performed with a gradient of sodium phosphate (10–100 mM) in buffer B. Purified RecD2 was aliquoted and stored at -80°C. RecD2-K373A-His (named RecD2 K373A) was purified following the same protocol.
RecO (29.3 kDa), RecA (38 kDa) and SsbA (18.7 kDa) were purified as described (30). B. subtilis replication proteins (PriA, PolC, δ, δ’, τ, β, DnaI, DnaC, DnaG, DnaE, DnaB and DnaD) were purified as described (34,35).
Protein–protein interactions
Protein-protein interactions were analyzed by immuno dot-blot assays (36), using the Bio-Dot apparatus (Bio-Rad). Briefly, increasing amounts of RecA and SsbA were applied to a pre-wetted nitrocellulose membrane in 1× phosphate-buffered saline (PBS). Bovine serum albumin (BSA) was also applied as a negative control of interaction. After blocking with PBS and 5% (w/v) skim milk powder, the membrane was incubated for 6 h at 4°C with 200 ng/ml RecD2 in binding solution (PBS, 0.5% (w/v) skim milk powder and 0.1% (v/v) Triton X-100). The membrane was then incubated overnight at 4°C with 6× His monoclonal antibody (Clontech Ref: 631212, dilution 1:6000) and subsequently with the secondary antibody anti-mouse IgG conjugated with peroxidase (Cytiva, Ref: NXA931 dilution 1:5000) for 1 h at room temperature. The interactions were visualized by staining the membrane with the Clarity™ Western ECL Substrate kit (Bio-Rad). The images were obtained and processed with the ChemiDoc Imaging System and the Image Lab software (Bio-Rad).
DNA strand exchange
Strand exchange reactions were performed as described (37) using KpnI-linearised pGEM-3Zf (+) dsDNA (3197-bp, 20 μM in nt, 3 nM in molecules) and its homologous circular ssDNA (10 μM in nt, 3 nM in molecules) in a buffer C (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 0.05 mg/ml BSA, 5% glycerol) containing 2 mM ATP, 8 U/ml creatine phosphokinase, 8 mM phosphocreatine. RecA (1.5 μM), RecO2 (200 nM), SsbA4 (300 nM) and variable concentrations of RecD2 (1.5–50 nM) were added as indicated. Samples were deproteinized after 20 or 40 min incubation at 37°C by the addition of stop buffer to a final concentration of 25 mM EDTA, 0.5% v/v SDS, and 0.5 mg/ml proteinase K. DNA species were fractionated by 0.8% w/v agarose gel electrophoresis run in 1× TBE. Gels were stained with ethidium bromide and analyzed using a GelDoc system (Bio-Rad) and densitometric analysis was performed with the Image Lab software (Bio-Rad). The percentage of the three-stranded recombination intermediates with various lengths of heteroduplex DNA (joint molecules, jm) and full exchanged products (nicked circular dsDNA, nc) was calculated dividing each signal by the sum of the signals of the three bands (dsDNA substrate, nc and jm).
Branch migration of recombination intermediates
A large-scale standard strand exchange reaction was set up at 37°C and stopped after 10 min by addition of EDTA to 45 mM, and SDS to 0.5%. Then, the mixture was loaded into a 1 ml Sephadex-G50 spin column to remove all components except proteins and DNA, that were recovered in the Flow through fraction. DNA was further purified by ethanol precipitation. The pellet containing remaining DNA substrates and recombination intermediates was resuspended in water and incubated with increasing concentrations of RecD2 (3–24 nM) during 30 min at 37°C in buffer C containing 2 mM ATP, 8 U/ml creatine phosphokinase and 8 mM phosphocreatine, 5% glycerol. Reactions were stopped and analyzed as stated above.
ATPase assays
ATPase activity of RecD2 or RecD2 K373A was analyzed using an ATP/NADH-coupled spectrophotometric assay. PolydT80 (3 μM in nt) was preincubated 5 min at 37°C in buffer C containing 0.5 mM phosphoenolpyruvate, 0.3 mM NADH, 10 U/ml pyruvate kinase and 10 U/ml lactate dehydrogenase) with 5 nM RecD2 or RecD2 K373A. Then, 0.2 mM ATP was added and reactions were incubated for 250 s at 37°C. The ATP hydrolysis was assessed by the decrease of NADH absorbance at 340 nm due to NAD+ accumulation, using a Shimadzu CPS-20A dual-beam spectrophotometer.
DNA replication assays
Reactions were carried out using a reconstituted PriA-dependent rolling circle DNA replication system as described (34,38). Protein concentrations used were: 20 nM PriA, 20 nM PolC, 25 nM HolA (δ), 25 nM HolB (δ’), 25 nM DnaX4(τ), 25 nM DnaN2 (β), 25 nM DnaI6, 30 nM DnaC6, 30 nM DnaG, 15 nM DnaE, 50 nM DnaB4, 50 nM DnaD4, 60 nM SsbA4, RecD2 (5-40 nM), RecO2 (50 nM) and RecA (500 nM). The minicircle DNA template used allows quantification of leading and lagging strands synthesis, because the circular leading strand template is deficient in dCMP residues (G:C ratio 50:1), and the flapped lagging strand template is deficient in dGMP residues. This permits quantification of leading and lagging strand synthesis by measuring radioactive dCMP and dGMP incorporation. The B. subtilis replisome was first preassembled incubating proteins with the DNA substrate (5 nM) in buffer BsRc (40 mM Tris-acetate pH 7.8, 12 mM MgOAc, 200 mM potassium glutamate, 3 μM ZnSO4, 1% w/v polyethylene glycol MW ∼8000, 0.02% Pluronic F68 and 1 mM DTT) for 5 min at 37°C in the presence of 5 μM ATPγS. The reactions were initiated by the addition of 1 mM ATP and 24 μM dNTPs with radiolabelled dCTP or dGTP, and aliquots were stopped at certain times. Unbound nucleotides were removed by gel filtration through Sephadex G-25 mini-columns and quantification of DNA synthesis was performed by scintillation counting. For visualization of replication products, aliquots were mixed with loading dye buffer (30 mM NaOH, 2 mM EDTA, 5% v/v glycerol and 0.1% v/v bromophenol blue) and fractionated by alkaline 0.8% (w/v) agarose gel electrophoresis.
Protein-displacement assays
Streptavidin displacement assays were done according to (39). 0.5 nM of biotinylated oligo bioT-45 (5′-GTACGTATTCAAGATACCTCGTACTCTGTACTGACTCGGATCC(biodT)A-3′) was incubated with streptavidin (2.5 nM) for 5 min at room temperature in buffer D (50 mM Tris-HCl pH 7.5, 2 mM MgCl2, 2 mM DTT, 50 mM NaCl, 0.05 mg/ml BSA and 2 mM ATP). An equal volume of a solution containing increasing concentrations of RecD2 and 50 nM biotin as a trap for the displaced streptavidin was added. The reactions were incubated for 20 min at 37°C and stopped with stop buffer (25 mM EDTA and 0.5% w/v SDS) for 10 min at 37°C. The samples were separated on 10% PAGE in 1× TBE. Gels were dried and analyzed with the Personal Molecular Imager and the Image Lab software (Bio-Rad).
Statistical methods
Graphs were performed with the Excel and Prism v6.0 (GraphPad) software, and data are represented as mean ± SD (standard deviation). Statistical differences between samples were assessed with the non-parametric Mann–Whitney test.
RESULTS
Purification of RecD2 and protein levels in B. subtilis
So far, all RecD2 proteins have been purified as a His-tagged protein, with the tag fused at the C-terminus (13,14,21), but if the tag affects protein activity was not analyzed. We attempted to purify a RecD2 non-tagged protein from E. coli pET-3b-recD2 cells, but the protein was poorly soluble, whereas RecD2-His was soluble. Then, we analyzed in vivo the functionality of the tagged protein. A B. subtilis strain expressing RecD2-His from its natural promoter, and as sole source of the protein in the cell was constructed. The recD2his6 cells were grown until mid-exponential phase and then dilutions were spotted on plates having 1.5 mM MMS. RecD2-His expressing cells were as resistant as wt cells to this drug (Supplementary Figure S2A) while the recD2 deletion mutant (ΔrecD2) was sensitive to this DNA damaging agent, as previously observed (25). We concluded that the C-terminal His-tag does not affect protein activity and used the purified protein without tag removal. The His-tagged protein is thereafter named RecD2.
The expression of the recD2 gene has been analyzed in global transcriptome analysis showing that gene expression is low and comparable to the level of expression of the low abundant essential replication proteins, as polC, dnaX or priA (40). RecD2 protein levels have not been determined so far. We used the recD2his6 strain that expresses RecD2 from its own promoter to quantify levels of the protein in the cell during exponential growth in LB. RecD2 was not detected when a protein extract from 1 ml of an exponential growing culture (i.e. ∼1.4 × 108 cells) was analyzed by Western blot, suggesting that RecD2 levels are low. Thus, recD2his6 cells from a 250 ml culture grown to exponential phase (∼3.5 × 1010 cells) were lysed, and the RecD2 protein, which was present in the soluble fraction, was concentrated by binding to a Ni-sepharose column. The amount of RecD2 eluted from the Ni-column was interpolated from the standard curve obtained with increasing amounts of purified RecD2 protein. These assays showed that RecD2 is low abundant in the cell, with approximate values of ∼25 RecD2 monomers/CFUs (Supplementary Figure S2B).
RecD2 may regulate replication restart
Previously it has been shown that: (i) inactivation of recD2 renders cells sensitive to DNA-damaging agents that block or impair replication fork movement, as MMS or the UV mimetic 4-nitroquinoline-1-oxide (21,25) and (ii) RecD2 is targeted to active replication forks by its interaction with SsbA (2). To test whether RecD2 affects replication restart or fork progression we performed in vitro DNA replication assays with purified B. subtilis proteins. First, we have confirmed that RecD2 physically interacts with SsbA (Figure 1A). For replication of a DNA substrate that mimics a blocked or stalled fork with a gap in the lagging strand (Figure 1B), the essential pre-primosomal PriA-DnaD-DnaB components, DnaI loader, DnaC helicase, PolC and DnaE polymerases, the clamp loader complex (τδδ’), the β-sliding clamp (DnaN), and DnaG primase are required (35). SsbA, as E. coli SSB, is not essential, but strongly stimulates in vitro DNA synthesis (38,41).
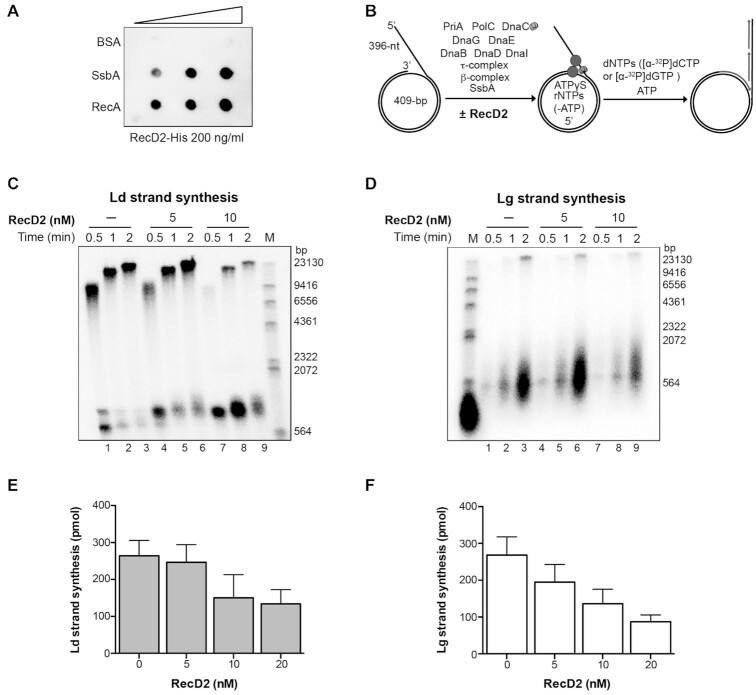
Figure 1.
RecD2 action on replication restart. (A) RecD2 interaction with SsbA and RecA is detected by immuno-dot blot. Increasing concentrations of RecA, SsbA and BSA (from 100–400 ng) were bound to a nitrocellulose membrane, which was subsequently incubated with a buffer containing 200 ng/ml of purified RecD2. After washing, the membrane was incubated with a mouse monoclonal anti-His antibody. (B) The mini-circular DNA template mimics a blocked replication fork with a 396-nt gap in the lagging strand. Diagram of the DNA template and B. subtilis replisome assembly scheme. Replisomes were assembled in the presence of RecD2 helicase incubating the minicircle DNA replication template in the presence of 5 μM ATPγS. Reactions were started by the addition of dNTPs and ATP, and stopped after the indicated times. (C) Leading strand synthesis in the presence of increasing amounts RecD2. (D) Lagging strand synthesis in the presence of increasing amounts of RecD2. Replication products were analyzed by alkaline gel electrophoresis and autoradiography. Ld, leading; Lg, lagging; M, molecular weight marker (radiolabeled λ-HindIII DNA). (E) Quantification of leading strand synthesis obtained after 2 min with increasing amounts of RecD2 (0–20 nM). (F) Quantification of lagging strand synthesis obtained after 2 min with increasing amounts of RecD2 (0–20 nM). Results are plotted as the mean ± SD of five independent experiments.
When the replisome was assembled in the presence of RecD2, and then dNTPs and ATP were added to initiate DNA synthesis, we found that a concentration of one RecD2/DNA molecule (5 nM RecD2), barely affected synthesis, but in the presence of two RecD2/DNA molecules or more, both leading and lagging strand synthesis, were significantly inhibited (P-values < 0.01; Figure 1 and Supplementary Figure S4A), suggesting that the inhibitory effect was dependent on protein concentrations.
Having observed an interaction of RecD2 with SsbA (Figure 1A), we checked if the inhibitory effect of RecD2 is due to some action on SsbA. When DNA replication was performed with all replisome components except SsbA, DNA replication was less efficient (Supplementary Figure S3), as earlier described (35,38). In the absence of SsbA, we did not observe the inhibitory effect conferred by RecD2 on leading strand synthesis (Supplementary Figures S3 and S4B). Lagging strand synthesis was inhibited at 40 nM RecD2 (Supplementary Figure S4B). It is likely that the inhibitory effect of RecD2 on DNA replication restart is conferred over SsbA, either promoting SsbA disassembly, or preventing it to properly interact with other binding partners (2,42). To test the first hypothesis, we purified the RecD2-K373A variant (21) that has no ATPase activity (Supplementary Figure S5A). No inhibition of DNA replication was observed at 40 nM RecD2-K373A, whereas at this protein concentration the wt protein strongly inhibited the reaction (Supplementary Figure S5B). This result suggests that RecD2 uses the energy from ATP hydrolysis to promote SsbA disassembly from the ssDNA.
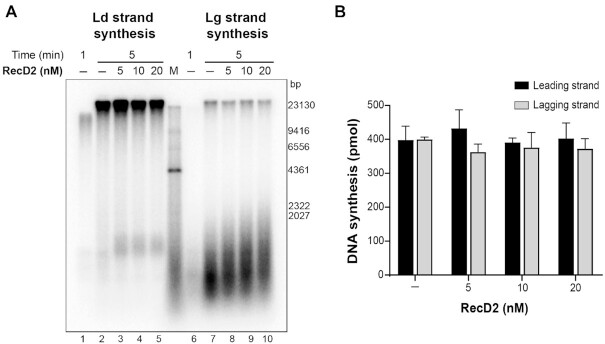
To analyze if the effect of RecD2 occurs only at the initiation step of DNA replication, RecD2 was added during ongoing DNA replication. The complete replisome was first assembled on this DNA substrate and replication was allowed to start for 1 min, then increasing concentrations of RecD2 were added and DNA replication continued for 5 min. No inhibitory effect was observed, even having an excess of RecD2 over the DNA template (Figure 2). However, we noticed that increased RecD2 concentrations led to an increase in the length of the Okazaki fragments, an effect that has been observed before in reconstituted systems lowering the concentration of DnaG primase (35,38). These results suggest that RecD2 could affect the elongation step during unperturbed replication, as well as modulate replication restart.
Figure 2.
RecD2 slightly affects ongoing DNA replication. The B. subtilis replisome was assembled on the DNA template and replication was allowed to start by dNTPs and ATP addition. After 1 min, increasing concentrations of RecD2 were added and replication continued for 5 min. (A) A representative alkaline agarose gel showing leading strand and lagging strand synthesis. Ld, leading; Lg, lagging. M, molecular weight marker. (B) Quantification of DNA synthesis. Represented is the mean of three independent experiments ± SD.
RecD2 interacts with RecA and reverses RecA inhibition of replication restart
Previously it has been shown that: (i) bacterial RecD2 and human HELB interact with their cognate SSB protein (2,21,22) and (ii) GFP-tagged HELB colocalizes with RPA and also with the Rad51 recombinase (43). Since RecD2 shows homology with HELB, we analyzed whether RecD2 interacts with RecA by immuno-dot blot assays. As revealed in (Figure 1A), RecD2 physically interacted with RecA.
Measurement of replication fork progression in vivo showed that forks arrest more frequently in ΔrecD2 cells (21). We reasoned that a role for RecD2 could be to remove protein obstacles on the DNA, and thereby facilitate replisome loading or replication fork progression. In the presence of SsbA, RecA slightly affects DNA replication restart, and addition of the RecO mediator enhances RecA inhibition of DNA synthesis, facilitating the binding of RecA to the ssDNA regions, and preventing replication restart thereby (44). We tested if RecD2 could counteract RecA negative action on replication restart. The DNA template was mixed with SsbA, RecD2, RecO and RecA in buffer BsRC to allow protein assembly. Then, the replisome proteins were added, and reactions were initiated by the addition of dNTPs and ATP. We confirmed that RecO and RecA inhibit leading and lagging strand synthesis (Figure 3A, B and Supplementary Figure S6A, B) as earlier reported (44). Remarkably, a catalytic amount of RecD2 was sufficient to bypass the inhibitory effect exerted by RecO and RecA on DNA synthesis (Figure 3A, B and Supplementary Figure S6A, B). For the removal of RecA from the stalled fork substrate ATP hydrolysis was needed, because the RecD2 K373A variant could not bypass RecA inhibitory action on replication restart (Supplementary Figure S5C).
Figure 3.
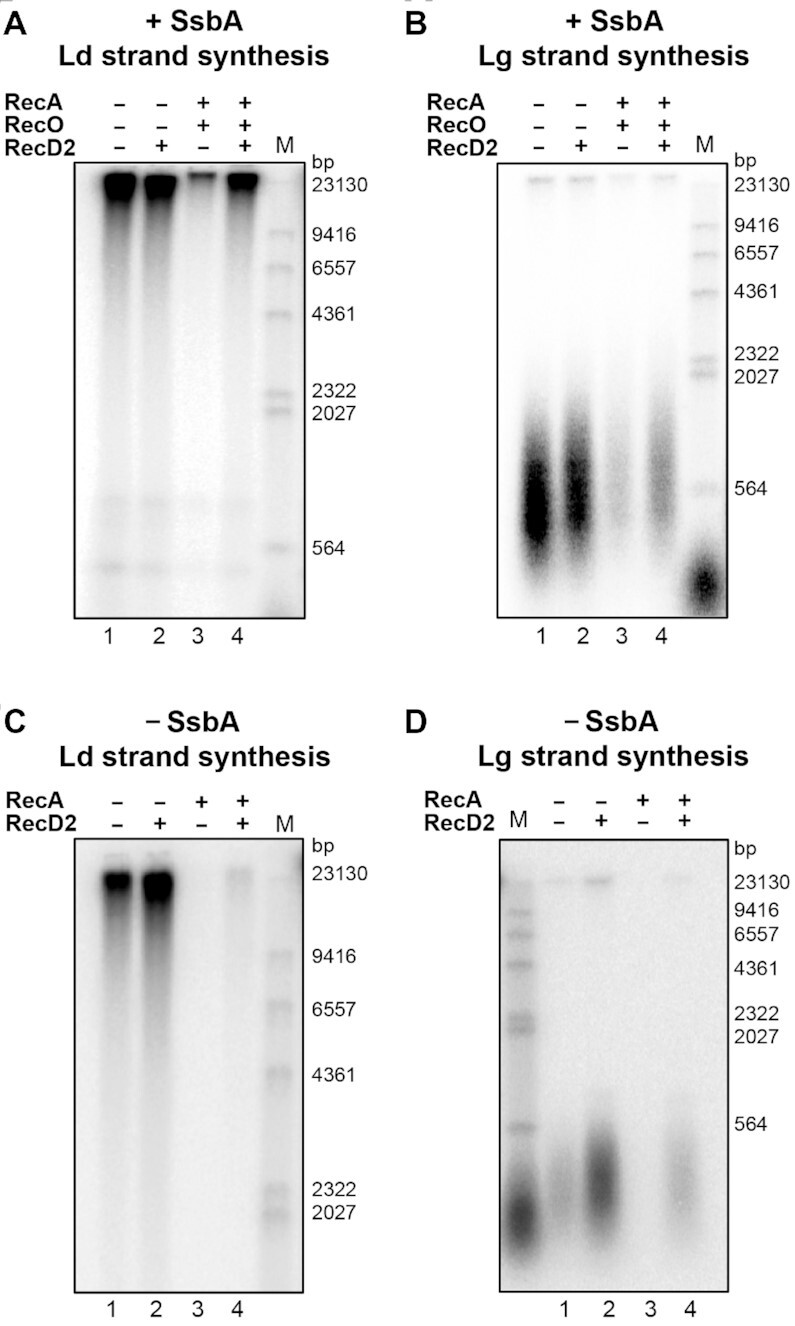
The RecD2 helicase overcomes the inhibition of replication restart by RecA. In A and B: Reactions in the presence of SsbA. An enzyme mix having B. subtilis replisome proteins was added to a substrate mix containing the DNA template, SsbA 90 nM, RecD2 5 nM, RecO 50 nM and RecA 500 nM, and DNA replication was allowed for 2 min at 37°C. (A) Leading strand synthesis. (B) Lagging strand synthesis. In C and D: Reactions without SsbA. An enzyme mix having B. subtilis replisome proteins, but no SsbA was added to a substrate mix having the DNA template, RecD2 5 nM, and RecA 500 nM, and DNA replication was allowed for 2 min at 37°C. (C) Leading strand synthesis. (D) Lagging strand synthesis. In Supplementary Figure S6, the quantifications of DNA synthesis from three independent experiments are shown.
We then tested if the interaction of RecD2 with SsbA is necessary to reverse the negative effect that RecA exerts on the resumption of DNA replication. In the absence of SsbA, RecA does not need its mediator RecO, and efficiently loads on the ssDNA (37). A RecA nucleoprotein filament assembled on the DNA template blocked both, leading and lagging strand synthesis (Figure 3C, D) as previously observed (44). RecD2 was still able to counteract the inhibitory effect exerted by RecA on DNA synthesis, but to a lesser extent (P-values < 0.06, Figure 3C, D and Supplementary Figure S5C, D). Then we tested if RecD2 can displace any other proteins from the ssDNA by its translocase activity. We detected some displacement of streptavidin from a biotinylated oligonucleotide only at high RecD2 concentrations (Supplementary Figure S7).
RecD2 modulates DNA strand exchange
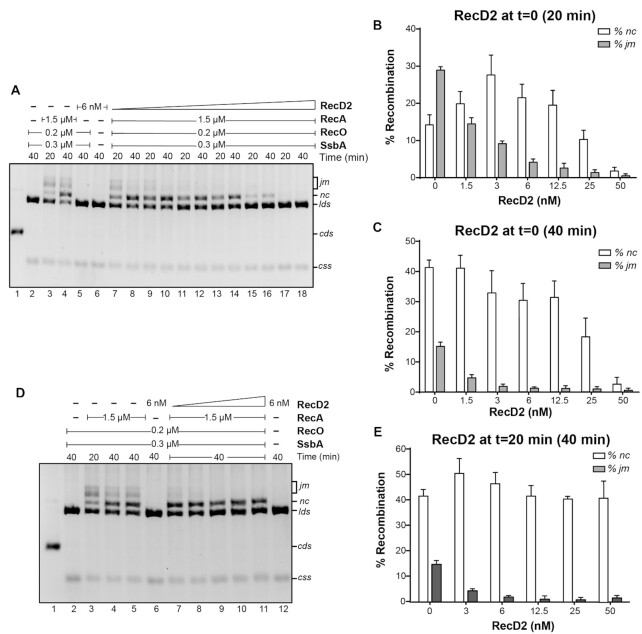
Since RecD2 seemed to displace RecA from the blocked replication fork to allow restart, we analyzed whether RecD2 also inhibits the strand exchange reaction catalyzed by RecA. We examined RecA-mediated three strand-exchange reactions with circular ssDNA (css) and its homologous linear duplex DNA (lds) in the presence of ATP and an ATP-regeneration system. In the presence of ATP, B. subtilis RecA requires the two-component RecO-SsbA mediator for efficient loading and filamentation onto the ssDNA (37). Reactions were performed in two different conditions: adding RecD2 at the beginning of the reaction, or after DNA strand-exchange has been initiated, and the outcome was monitored by agarose gel electrophoresis.
The lds and css DNA substrates were incubated with RecA at 37°C. In a 20 min reaction, RecA catalyzed mainly the formation of joint molecule (jm) intermediates and some nicked circular (nc) products, and these were the major species after 40 min (Figure 4A, lanes 3 and 4). When RecD2 was added at the same time as RecA, the effect observed was concentration and time dependent. After 20 min of incubation, at low RecD2 concentrations (3 and 6 nM, 1–2 RecD2/ssDNA molecule), the percentage of jm decreased and we observed an increase in nc formation (3 nM RecD2, P-value = 0.023; 6 nM, P-value = 0.026) (Figures 4A, B). However, no significant difference in the amount of nc products was detectable at these protein concentrations in a 40 min reaction (Figure 4A, lanes 10 and 12, and 4C). The presence of 25 nM RecD2, which corresponds to ∼8 RecD2/ssDNA molecule and a RecA:RecD2 molar ratio of 60:1, was sufficient to strongly inhibit the accumulation of jm intermediates and nc products at the two times of incubation tested (jm, P-value = 0.024; nc, P-value = 0.063). When RecD2 concentrations further increased (∼16 RecD2 monomers/ssDNA molecule, RecA:RecD2 30:1), the accumulation of both intermediates (jm) and products (nc) was completely blocked (Figure 4A–C). A similar result was obtained when first RecA and RecD2 were initially incubated with the ssDNA substrate and then the homologous linear dsDNA was added (Supplementary Figure S8).
Figure 4.
RecD2 modulates strand exchange. (A) Strand exchange reactions with RecD2 added at time 0. KpnI-linerarized pGEM-3Zf(+) dsDNA (lds, 3 nM) and its homologous circular ssDNA (css, 3 nM) were incubated with RecA (1.5 μM), RecO (200 nM), SsbA (300 nM) and variable concentrations of RecD2 (1.5 to 50 nM) for 5 min at 37°C with all buffer components except ATP. Then, ATP was added and reactions continued for 20 or 40 min. DNA species were separated by 0.8% w/v agarose gel electrophoresis. RecD2 concentration in lanes 7 and 8: 1.5 nM; lanes 9 and 10: 3 nM, lanes 11 and 12: 6 nM, lanes 13 and 14: 12.5 nM, lanes 15–16: 25 nM, lanes 17–18: 50 nM. In lane 1 a control with the running position of supercoiled and nicked DNA is shown. (B) Quantification of percentage of joint molecules (jm) intermediates and nicked products (nc) after 20 min. Results are the mean of three independent experiments ± SD. (C) Quantification of jm and nc after 40 min. Results are the mean of three independent experiments ± SD. (D) Strand exchange reactions with RecD2 added after strand exchange intitiates at 20 min. lds and css were incubated with RecA, RecO, and SsbA for 20 min at 37°C with all buffer components. Then, variable concentrations of RecD2 (3 to 50 nM) were added and reactions continued for another 20 min (total time 40 min). Lane 3 is a control of the extent of the strand exchange reaction when RecD2 is added (i.e. 20 min reaction). In lanes 4 and 5, two controls of 40 min reactions without RecD2. (E) Quantification of jm and nc observed when RecD2 is added after strand-exchange has been initiated. Plotted are the results from three independent experiments ± SD.
We then analyzed if this inhibition is also observed when strand exchange has initiated. RecA was pre-incubated with css and lds DNA in the presence of the two-component mediator RecO-SsbA, and strand exchange started by the addition of ATP. After 20 min, increasing concentrations of RecD2 were added and the reaction continued for another 20 min (total incubation time 40 min, Figure 4D, E). An aliquot was taken at the time of RecD2 addition to see the amount of strand exchange intermediates and products formed at this time point (Figure 4D, lane 3). Increasing concentrations of RecD2 reduced the accumulation of jm (Figure 4D, E, P-values < 0.01). We observed that the amount of nc products slightly increased at low RecD2 concentrations (P-value 0.033) and was not significantly affected (P-values >0.05) at higher RecD2 concentrations, so that most of the jm intermediates that were present at the time of RecD2 addition were converted to nc products. These results showed that RecD2 does not freeze the reaction once added, even having an excess of RecD2 over the DNA molecules (50 nM RecD2).
In summary, the strand exchange assays showed a different outcome depending on the order of protein addition. If RecD2 was added at the same time as RecA, low RecD2 concentrations (1 helicase/cssDNA molecule) slightly stimulated the production of nc product and concentrations over 50 nM blocked the reaction, whereas this effect was not observed if 50 nM RecD2 was added after strand exchange had initiated.
RecD2 catalyzes branch migration
The former results suggested that low concentrations of RecD2 can stimulate nc formation because RecD2 may branch migrate the recombination intermediate. This hypothesis is consistent with in vivo results that showed that a mutation in RecD2 is synthetically lethal when combined with mutations in RecG or RuvAB, which branch migrate recombination intermediates (25). To directly test this hypothesis, we performed standard RecA strand-exchange reactions for 10 min and purified the DNA from such reactions after EDTA/SDS treatment and ethanol precipitation to remove RecA, RecO and SsbA. This DNA, which is enriched in jm intermediates (Figure 5B, lane 8), was then incubated with RecD2 alone in the presence of ATP. The accumulation of the nc product was observed at concentrations as low as 3 nM RecD2 (Figure 5B, lane 5), and this was not observed in the reaction having the substrate incubated without RecD2 (Figure 5B, lane 4). Interestingly, under these conditions, we observed neither the accumulation of lds that could result from the disruption of the jm intermediate by the 5′→3′ translocation of RecD2 on the circular ssDNA, nor the appearance of a new jm intermediate with the displaced strand removed, that could be the product when RecD2 binds to the displaced strand and unwinds the DNA in the 5′→3′ direction.
Figure 5.
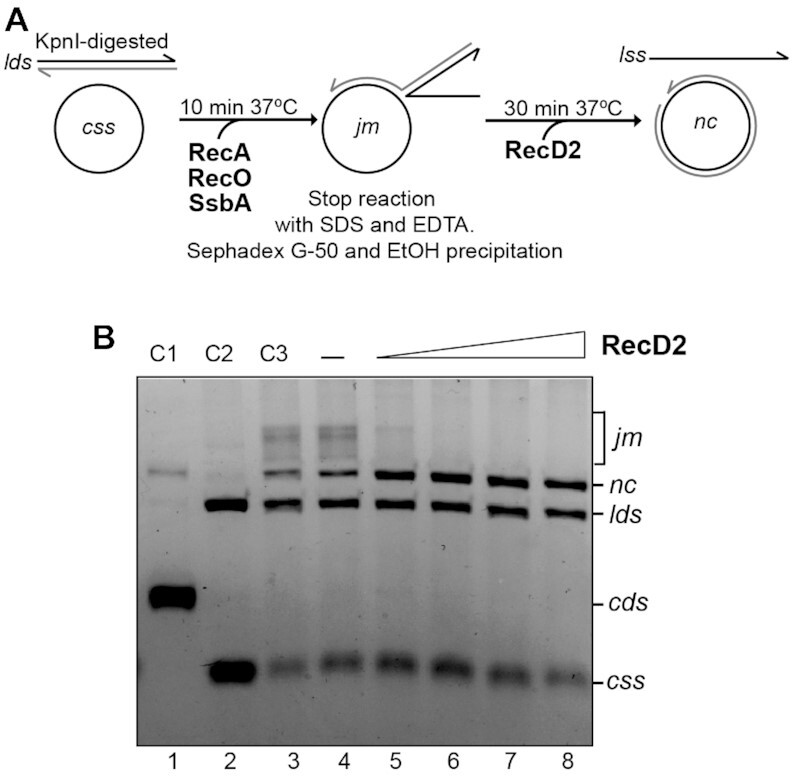
RecD2 branch migrates the recombination intermediate. (A) Scheme of the assay. Six standard strand exchange reactions were performed for 10 min at 37°C and pooled. After deproteinization, DNA was purified by gel filtration and ethanol precipitation. The DNA pellet was resuspended in MilliQ water and this DNA, that contains remaining substrates (lds and css) and recombination intermediates (jm), was used to test RecD2 branch migration activity. (B) An assay showing branch migration activity with increasing concentrations of RecD2 (from 3 to 24 nM) in a 30 min reaction. In lane 4, DNA was incubated with all buffer components without RecD2. C3 (lane 3) is a control where the DNA with all buffer components and no protein was kept on ice. Other controls are: C1, plasmid dsDNA (supercoiled, cds and nicked, nc), C2, linear dsDNA (lds) and circular ssDNA (css).
RecD2 promotes the disassembly of RecA filaments in vivo
Given that our biochemical data are consistent with RecD2 acting as a negative regulator of RecA–ssDNA filament formation, we investigated whether this effect can be observed in vivo. Previously it has been shown that: i) in concert with mediators that act before homology search, RecA·ATP nucleates on the ssDNA produced by MMC-induced DNA damage, to form the activated RecA* (45); ii) RecA*, regulated by modulators that act during filament growth, faciltates the auto-proteolysis of the transcriptional repressor LexA to induce expression of ∼40 genuine SOS genes, being recA one of them (46,47). Previous assays have shown that in the absence of positive mediators (e.g.RecO) the maximal levels of RecA are obtained at higher MMC concentrations when compared to the wt control, whereas in the absence of negative modulators (e.g. RecX) maximal levels of RecA are obtained at lower MMC concentrations when compared to the wt strain (Supplementary Figure S9, (27,32)). Therefore, the quantification of RecA protein levels at different MMC concentrations can be an indirect measurement of RecA–ssDNA nucleoprotein extension. Exponentially grown wt (rec+) and ΔrecD2 cells were exposed to increased concentrations of MMC for 30 min. Then RecA levels were quantified by Western blot (Supplementary Figure S9). A lower MMC dose was required to achieve maximum levels of RecA in ΔrecD2 cells than in wt cells, consistent with an activity for RecD2 as negative regulator of RecA–ssDNA filament formation.
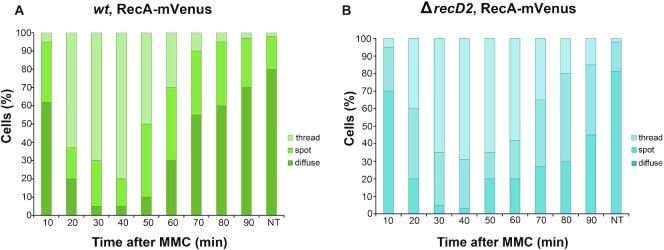
In a second test, we analyzed whether inactivation of recD2 affects the formation of RecA foci or the persisting of RecA threads. During natural competence ssDNA enters into the competent cell; fluorescent RecA forms first a focus, which is followed by a filamentous structure (termed thread or bundle) that extents from the entry channel to the nucleoid (48). In exponentially growing cells, fluorescent RecA forms spots that co-localize with the replisome (49). Upon DNA damage, the number of discrete spots decreased and were followed by the formation of RecA threads as observed previously (Figure 6A; 32). These RecA threads diminished to normal levels in the wt control after 90 min, in the time concomitant with the repair of the DNA damage (Figure 6A, movie S1). However, the threads persisted for a longer period in a ΔrecD2 strain, and about 15% of cells still contained RecA threads after 90 min (Figure 6B, movie S2). These results confirm that RecD2 in vivo acts as a negative modulator of the RecA–ssDNA filament.
Figure 6.
Epifluorescence microscopy showing that RecA assembly into threads is affected by RecD2. Quantitative analysis of RecA being diffuse, forming spots or threads in wt (A) or ΔrecD2 (B) cells after treatment with MMC at the indicated times. NT, not treated cells. The results are the average of three independent experiments (n = 400 cells). The corresponding movies are displayed in the Supplementary data (movie S1 and S2, respectively).
DISCUSSION
RecD2 helicases are proteins widely conserved in bacteria that lack the RecBCD complex. Previous in vivo studies in B. subtilis cells revealed that this DNA helicase is associated with the replisome (2), and that forks arrest more frequently in ΔrecD2 cells(21), supporting the hypothesis that RecD2 is important for normal replication fork progression. RecD2 depleted cells are synthetically lethal in the ruvAB or recG context and also show a synergistic loss of viability when combined with PcrA depletion (counterpart of UvrDEco and homologous to RepEco) (25). In contrast, the RecD2Dra enzyme in an heterologous system (E. coli) inhibited the resumption of DNA replication, and RecD2Dra expression was toxic in E. coli strains lacking Rep, but not in strains lacking RuvAB (50). All these results highlight the relevance to analyze the activity of this protein in a homologous system.
In this work we purified the B. subtilis RecD2 helicase and characterized its mode of action on replication restart, using a reconstituted in vitro DNA replication system with purified B. subtilis proteins. We found that the effect of RecD2 on PriA-dependent replication restart depends on the protein concentration used. No significant effect in replication restart was observed when one RecD2 per DNA molecule was present, but in the presence of two or more RecD2 /DNA molecule an inhibitory effect was observed. RecD2Dra also inhibited the resumption of DNA replication when a heterologous in vitro replication system and ∼50 RecD2Dra /DNA molecule were used in the assay (50). Taking into account the low levels of RecD2 in the cell, and that these protein levels are lower than the ones calculated for replisome components (51), we favour the hypothesis that the number of RecD2 helicases/replisome will be low, and suggest that RecD2 might modulate DNA replication restart in vivo. We demonstrate that this inhibition is done mostly over SsbA, because we do not observe this negative effect on DNA synthesis when SsbA is absent, unless high concentrations of RecD2 are used. The negative effect could be because during translocation, RecD2 may displace SsbA from the ssDNA. Consistent with this, the ATPase-defective mutant did not inhibit replication restart. In the absence of SsbA, several replication proteins are catalytically less efficient, such as the polymerases PolC and DnaE (38,42), and the replicative helicase (52,53). We cannot discard a direct effect of RecD2 in the activity of one of these replisome proteins, because RecD2 slightly affected the length of Okazaki fragments during replication elongation, which strictly requires PolC, DnaC and DnaE. A role in initiation of DNA replication has been proposed for the human HELB protein (18), which belongs to the same helicase superfamily.
Our results suggest that in vivo, RecD2 may favor replication restart preventing RecA from binding to the blocked fork, in order to avoid unnecessary recombination during replication restart (Figure 7, panel A). Using a DNA substrate that mimics a blocked fork with a gap in the lagging strand and in vitro replication we show that RecD2 displaces RecA from the ssDNA, and for that ATP hydrolysis is needed. Our results are consistent with a catalytic mechanism of RecA filament disassembly by RecD2 in the replisome context, and that the activity is not the result of stoichiometric binding to RecA, because RecD2:RecA at a molar ratio of 1:100, which corresponded to 1 RecD2 helicase per DNA molecule was sufficient to overcome the inhibitory effect of having RecA during replication restart.
Figure 7.
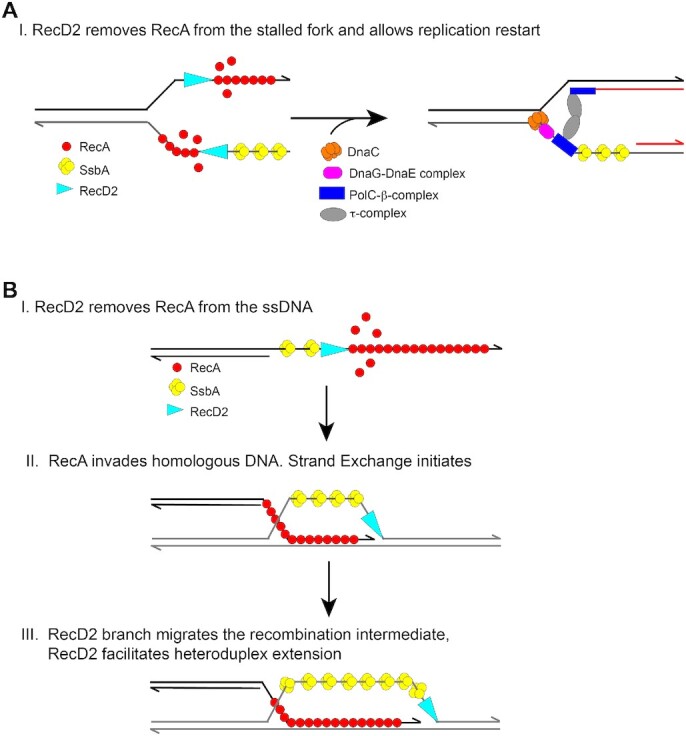
Model depicting how RecD2 helicase may modulate RecA during replication restart and strand exchange. (A) On a stalled fork, RecA may bind to the ssDNA regions inhibiting replication restart. RecD2 removes RecA from the stalled fork and thereby, facilitates replisome loading and replication restart. (B) During homologous recombination the invading strand is coated with RecA. RecD2 may translocate 5′→3′ along the ssDNA and remove RecA. However, once RecA initiates strand exchange it may accelerate heteroduplex extension.
RecA filamentation on the ssDNA is the first step in the strand exchange reaction and this is a highly regulated process (54,55). In Gram-negative bacteria, many proteins have been identified that modulate RecA activity (RecFOR, DinD, DinI, RecX, RdgC, PsiB and UvrD (54,56)), but many of these proteins are not present in B. subtilis. In B. subtilis RecX, RecU, and PcrA have been identified as negative regulators of RecA filament growth. All of them directly interact with RecA (2,57–60). PcrA, like UvrDEco that also dismantles RecA–ssDNA filaments, is a SF1A DNA helicase with a 3′→5′ polarity (9,61). In contrast, RecD2 is a SF1B DNA helicase that moves in a 5′→3′ DNA direction. Remarkably, eukaryotic Srs2, FBH1 and RecQ5 helicases also disassemble pre-synaptic filaments and interact with Rad51 (62–65). We propose that the human HELB helicase may also interact with Rad51.
We found that RecD2 impairs RecA nucleation and filament growth on the SsbA–ssDNA–RecO complexes, but it can also accelerate the strand exchange reaction (Figure 7, panel B). When RecD2 is added at the same time as RecA, we found that catalytic RecD2 concentrations (1 helicase/DNA substrate) stimulated the appearance of final strand-exchange products at early times of incubation, suggesting that RecD2 could branch migrate this recombination intermediate, and our experiments confirmed this activity. This has been also observed with the related Dda helicase (66). In summary, RecD2 may have both a negative and a positive action on strand exchange (Figure 7, panel B). HELB stimulated nc product formation with dsDNA with 3′-overhanging termini, but inhibited the formation of nc products when dsDNA with 5′-overhanging termini were used (43). In humans, there appear to be other helicases that also antagonize and stimulate recombination protein Rad51, as human BLM helicase (67–70). In our assays, the formation of nc products was almost unaffected when RecD2 was added after strand exchange had initiated, even in the presence of a high excess of RecD2. Similarly, ongoing DNA strand exchange was not affected by the presence of the DinI negative regulator (71). It could be a general feature that once RecA is engaged in the strand exchange reaction, it becomes more resistant to the negative action of filament regulators, but this needs to be analyzed.
As an important control, we found that RecD2 acts as a negative modulator of RecA in vivo. In response to DNA damage induced by MMC, RecA first forms visible foci, followed by filamentous structures, termed threads, which arise during repair via homologous recombination and dissipate when the DNA damage has been repaired and growth resumes. In the absence of RecD2, the threads formed by RecA-mVenus persisted for much longer time. Previous assays that analyzed the formation of RecA-GFP foci as an in vivo marker for replicative stress in B. subtilis found a nearly 2-fold elevation in the percentage of cells with RecA-GFP foci in the absence of RecD2 in untreated cells (21). Our results seem to be different, we did not observe more foci, or threads, in non-treated recD2 mutant cells, but found that filaments are longer living, as previously described in the absence of the RecX negative regulator (26). Consistent with this, we found that in ΔrecD2 mutants, a lower dose of MMC was required to achieve maximal levels of SOS induction, that requires the formation of RecA* (a RecA–ssDNA filament) to facilitate autocleavage of the LexA SOS repressor. MMC-induced replicative stress triggers the SOS response necessary for cell survival. Cells may require longer living or longer filaments to ensure proper signaling so that the SOS response is initiated. So maybe the activity of RecD2 must be overcome for the RecA–ssDNA filament to properly signal the SOS response. Taken together, our data show an important role for RecD2 in replication restart via counteracting RecA, and an additional role in controlling proper SOS induction. We suggest that these functions are widely conserved in bacteria.
Supplementary Material
ACKNOWLEDGEMENTS
CR thanks the Spanish “Ministerio de Ciencia e Innovación” for her FPI-predoctoral fellowship (BES-2017-080504).
Contributor Information
Cristina Ramos, Department of Microbial Biotechnology, Centro Nacional de Biotecnología (CNB-CSIC), Darwin 3, 28049, Madrid, Spain.
Rogelio Hernández-Tamayo, SYNMIKRO, LOEWE-Zentrum für Synthetische Mikrobiologie, Hans-Meerwein-Straße 6, 35043 Marburg, Germany; Fachbereich Chemie, Hans-Meerwein-Straße 4, 35032 Marburg, Germany.
María López-Sanz, Department of Microbial Biotechnology, Centro Nacional de Biotecnología (CNB-CSIC), Darwin 3, 28049, Madrid, Spain.
Begoña Carrasco, Department of Microbial Biotechnology, Centro Nacional de Biotecnología (CNB-CSIC), Darwin 3, 28049, Madrid, Spain.
Ester Serrano, Department of Microbial Biotechnology, Centro Nacional de Biotecnología (CNB-CSIC), Darwin 3, 28049, Madrid, Spain.
Juan C Alonso, Department of Microbial Biotechnology, Centro Nacional de Biotecnología (CNB-CSIC), Darwin 3, 28049, Madrid, Spain.
Peter L Graumann, SYNMIKRO, LOEWE-Zentrum für Synthetische Mikrobiologie, Hans-Meerwein-Straße 6, 35043 Marburg, Germany; Fachbereich Chemie, Hans-Meerwein-Straße 4, 35032 Marburg, Germany.
Silvia Ayora, Department of Microbial Biotechnology, Centro Nacional de Biotecnología (CNB-CSIC), Darwin 3, 28049, Madrid, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación MCIN/ AEI/ FEDER, EU [PGC2018-097054-B-I00 to J.C.A. and S.A.]; Deutsche Forschungsgemeinschaft (DFG-funded consortium TRR 178 to P.L.G.). Funding for open access charge: CSIC Open Access Publication Support Initiative.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lopez de Saro F.J. Regulation of interactions with sliding clamps during DNA replication and repair. Curr. Genomics. 2009; 10:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Costes A., Lecointe F., McGovern S., Quevillon-Cheruel S., Polard P.. The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS Genet. 2010; 6:e1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shereda R.D., Kozlov A.G., Lohman T.M., Cox M.M., Keck J.L.. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008; 43:289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kreuzer K.N., Brister J.R.. Initiation of bacteriophage T4 DNA replication and replication fork dynamics: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virol. J. 2010; 7:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lo Piano A., Martinez-Jimenez M.I., Zecchi L., Ayora S.. Recombination-dependent concatemeric viral DNA replication. Virus Res. 2011; 160:1–14. [DOI] [PubMed] [Google Scholar]
- 6. Neamah M.M., Mir-Sanchis I., Lopez-Sanz M., Acosta S., Baquedano I., Haag A.F., Marina A., Ayora S., Penades J.R.. Sak and Sak4 recombinases are required for bacteriophage replication in Staphylococcus aureus. Nucleic Acids Res. 2017; 45:6507–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolinjivadi A.M., Sannino V., de Antoni A., Techer H., Baldi G., Costanzo V.. Moonlighting at replication forks - a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 2017; 591:1083–1100. [DOI] [PubMed] [Google Scholar]
- 8. Ait Saada A., Lambert S.A.E., Carr A.M. Preserving replication fork integrity and competence via the homologous recombination pathway. DNA Repair (Amst). 2018; 71:135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 10. Montague M., Barnes C., Smith H.O., Chuang R.Y., Vashee S.. The evolution of RecD outside of the RecBCD complex. J. Mol. Evol. 2009; 69:360–371. [DOI] [PubMed] [Google Scholar]
- 11. Rocha E.P., Cornet E., Michel B.. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005; 1:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernheim A., Bikard D., Touchon M., Rocha E.P.C.. A matter of background: DNA repair pathways as a possible cause for the sparse distribution of CRISPR-Cas systems in bacteria. Philos Trans. R Soc. Lond B Biol. Sci. 2019; 374:20180088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J., Julin D.A.. DNA helicase activity of the RecD protein from Deinococcus radiodurans. J. Biol. Chem. 2004; 279:52024–52032. [DOI] [PubMed] [Google Scholar]
- 14. Saikrishnan K., Griffiths S.P., Cook N., Court R., Wigley D.B.. DNA binding to RecD: role of the 1B domain in SF1B helicase activity. EMBO J. 2008; 27:2222–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saikrishnan K., Powell B., Cook N.J., Webb M.R., Wigley D.B.. Mechanistic basis of 5'-3' translocation in SF1B helicases. Cell. 2009; 137:849–859. [DOI] [PubMed] [Google Scholar]
- 16. Shadrick W.R., Julin D.A.. Kinetics of DNA unwinding by the RecD2 helicase from Deinococcus radiodurans. J. Biol. Chem. 2010; 285:17292–17300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hazeslip L., Zafar M.K., Chauhan M.Z., Byrd A.K.. Genome maintenance by DNA helicase B. Genes (Basel). 2020; 11:578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerhardt J., Guler G.D., Fanning E.. Human DNA helicase B interacts with the replication initiation protein Cdc45 and facilitates Cdc45 binding onto chromatin. Exp. Cell Res. 2015; 334:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tkac J., Xu G., Adhikary H., Young J.T.F., Gallo D., Escribano-Diaz C., Krietsch J., Orthwein A., Munro M., Sol W.et al.. HELB is a feedback inhibitor of DNA end resection. Mol. Cell. 2016; 61:405–418. [DOI] [PubMed] [Google Scholar]
- 20. Tada S., Kobayashi T., Omori A., Kusa Y., Okumura N., Kodaira H., Ishimi Y., Seki M., Enomoto T.. Molecular cloning of a cDNA encoding mouse DNA helicase B, which has homology to Escherichia coli RecD protein, and identification of a mutation in the DNA helicase B from tsFT848 temperature-sensitive DNA replication mutant cells. Nucleic Acids Res. 2001; 29:3835–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walsh B.W., Bolz S.A., Wessel S.R., Schroeder J.W., Keck J.L., Simmons L.A.. RecD2 helicase limits replication fork stress in Bacillus subtilis. J. Bacteriol. 2014; 196:1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guler G.D., Liu H., Vaithiyalingam S., Arnett D.R., Kremmer E., Chazin W.J., Fanning E.. Human DNA helicase B (HDHB) binds to replication protein A and facilitates cellular recovery from replication stress. J. Biol. Chem. 2012; 287:6469–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Serrano E., Ramos C., Alonso J.C., Ayora S.. Recombination proteins differently control the acquisition of homeologous DNA during Bacillus subtilis natural chromosomal transformation. Environ. Microbiol. 2021; 23:512–524. [DOI] [PubMed] [Google Scholar]
- 24. Couve S., Ishchenko A.A., Fedorova O.S., Ramanculov E.M., Laval J., Saparbaev M.. Direct DNA lesion reversal and excision repair in Escherichia coli. EcoSal Plus. 2013; 5:2. [DOI] [PubMed] [Google Scholar]
- 25. Torres R., Romero H., Rodriguez-Cerrato V., Alonso J.C.. Interplay between Bacillus subtilis RecD2 and the RecG or RuvAB helicase in recombinational repair. DNA Repair (Amst). 2017; 55:40–46. [DOI] [PubMed] [Google Scholar]
- 26. Servinsky M.D., Julin D.A.. Effect of a recD mutation on DNA damage resistance and transformation in Deinococcus radiodurans. J. Bacteriol. 2007; 189:5101–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cardenas P.P., Carrasco B., Defeu Soufo C., Cesar C.E., Herr K., Kaufenstein M., Graumann P.L., Alonso J.C.. RecX facilitates homologous recombination by modulating RecA activities. PLoS Genet. 2012; 8:e1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanchez H., Cozar M.C., Martinez-Jimenez M.I.. Targeting the Bacillus subtilis genome: an efficient and clean method for gene disruption. J. Microbiol. Methods. 2007; 70:389–394. [DOI] [PubMed] [Google Scholar]
- 29. Canosa I., Rojo F., Alonso J.C.. Site-specific recombination by the beta protein from the streptococcal plasmid pSM19035: minimal recombination sequences and crossing over site. Nucleic Acids Res. 1996; 24:2712–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manfredi C., Carrasco B., Ayora S., Alonso J.C.. Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J. Biol. Chem. 2008; 283:24837–24847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carrasco B., Manfredi C., Ayora S., Alonso J.C.. Bacillus subtilis SsbA and dATP regulate RecA nucleation onto single-stranded DNA. DNA Repair. 2008; 7:990–996. [DOI] [PubMed] [Google Scholar]
- 32. Romero H., Serrano E., Hernandez-Tamayo R., Carrasco B., Cardenas P.P., Ayora S., Graumann P.L., Alonso J.C.. Bacillus subtilis RarA Acts as a Positive RecA Accessory Protein. Front. Microbiol. 2020; 11:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hartmann R., van Teeseling M.C.F., Thanbichler M., Drescher K.. BacStalk: a comprehensive and interactive image analysis software tool for bacterial cell biology. Mol. Microbiol. 2020; 114:140–150. [DOI] [PubMed] [Google Scholar]
- 34. Dallmann H.G., Fackelmayer O.J., Tomer G., Chen J., Wiktor-Becker A., Ferrara T., Pope C., Oliveira M.T., Burgers P.M., Kaguni L.S.et al.. Parallel multiplicative target screening against divergent bacterial replicases: identification of specific inhibitors with broad spectrum potential. Biochemistry. 2010; 49:2551–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sanders G.M., Dallmann H.G., McHenry C.S.. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol. Cell. 2010; 37:273–281. [DOI] [PubMed] [Google Scholar]
- 36. Walsh B.W., Lenhart J.S., Schroeder J.W., Simmons L.A.. Far Western blotting as a rapid and efficient method for detecting interactions between DNA replication and DNA repair proteins. Methods Mol. Biol. 2012; 922:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carrasco B., Yadav T., Serrano E., Alonso J.C.. Bacillus subtilis RecO and SsbA are crucial for RecA-mediated recombinational DNA repair. Nucleic Acids Res. 2015; 43:5984–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Seco E.M., Ayora S.. Bacillus subtilis DNA polymerases, PolC and DnaE, are required for both leading and lagging strand synthesis in SPP1 origin-dependent DNA replication. Nucleic Acids Res. 2017; 45:8302–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bruning J.G., Howard J.A., McGlynn P.. Use of streptavidin bound to biotinylated DNA structures as model substrates for analysis of nucleoprotein complex disruption by helicases. Methods. 2016; 108:48–55. [DOI] [PubMed] [Google Scholar]
- 40. Nicolas P., Mader U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., Bidnenko E., Marchadier E., Hoebeke M., Aymerich S.et al.. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012; 335:1103–1106. [DOI] [PubMed] [Google Scholar]
- 41. Antony E., Weiland E., Yuan Q., Manhart C.M., Nguyen B., Kozlov A.G., McHenry C.S., Lohman T.M.. Multiple C-terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J. Mol. Biol. 2013; 425:4802–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paschalis V., Le Chatelier E., Green M., Nouri H., Kepes F., Soultanas P., Janniere L.. Interactions of the Bacillus subtilis DnaE polymerase with replisomal proteins modulate its activity and fidelity. Open Biol. 2017; 7:170146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu H., Yan P., Fanning E.. Human DNA helicase B functions in cellular homologous recombination and stimulates Rad51-mediated 5'-3' heteroduplex extension in vitro. PLoS One. 2015; 10:e0116852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vlasic I., Mertens R., Seco E.M., Carrasco B., Ayora S., Reitz G., Commichau F.M., Alonso J.C., Moeller R.. Bacillus subtilis RecA and its accessory factors, RecF, RecO, RecR and RecX, are required for spore resistance to DNA double-strand break. Nucleic Acids Res. 2014; 42:2295–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sassanfar M., Roberts J.W.. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 1990; 212:79–96. [DOI] [PubMed] [Google Scholar]
- 46. Goranov A.I., Kuester-Schoeck E., Wang J.D., Grossman A.D.. Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. J. Bacteriol. 2006; 188:5595–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cardenas P.P., Gandara C., Alonso J.C.. DNA double strand break end-processing and RecA induce RecN expression levels in Bacillus subtilis. DNA Repair (Amst). 2014; 14:1–8. [DOI] [PubMed] [Google Scholar]
- 48. Kidane D., Graumann P.L.. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell. 2005; 122:73–84. [DOI] [PubMed] [Google Scholar]
- 49. Lenhart J.S., Brandes E.R., Schroeder J.W., Sorenson R.J., Showalter H.D., Simmons L.A.. RecO and RecR are necessary for RecA loading in response to DNA damage and replication fork stress. J. Bacteriol. 2014; 196:2851–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gupta M.K., Guy C.P., Yeeles J.T., Atkinson J., Bell H., Lloyd R.G., Marians K.J., McGlynn P.. Protein-DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7252–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li G.W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Biswas E.E., Chen P.H., Biswas S.B.. Modulation of enzymatic activities of Escherichia coli DnaB helicase by single-stranded DNA-binding proteins. Nucleic Acids Res. 2002; 30:2809–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang H., Zhang Z., Yang J., He Z.G.. Functional characterization of DnaB helicase and its modulation by single-stranded DNA binding protein in Mycobacterium tuberculosis. FEBS J. 2014; 281:1256–1266. [DOI] [PubMed] [Google Scholar]
- 54. Cox M.M. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007; 8:127–138. [DOI] [PubMed] [Google Scholar]
- 55. Bell J.C., Kowalczykowski S.C.. RecA: regulation and mechanism of a molecular search engine. Trends Biochem. Sci. 2016; 41:491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Uranga L.A., Balise V.D., Benally C.V., Grey A., Lusetti S.L.. The Escherichia coli DinD protein modulates RecA activity by inhibiting postsynaptic RecA filaments. J. Biol. Chem. 2011; 286:29480–29491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cañas C., Carrasco B., Ayora S., Alonso J.C.. The RecU Holliday junction resolvase acts at early stages of homologous recombination. Nucleic Acids Res. 2008; 36:5242–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Le S., Serrano E., Kawamura R., Carrasco B., Yan J., Alonso J.C.. Bacillus subtilis RecA with DprA-SsbA antagonizes RecX function during natural transformation. Nucleic Acids Res. 2017; 45:8873–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Serrano E., Carrasco B., Gilmore J.L., Takeyasu K., Alonso J.C.. RecA regulation by RecU and DprA during Bacillus subtilis natural plasmid transformation. Front. Microbiol. 2018; 9:1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carrasco B., M M.-d.A., Torres R., Alonso J.C. PcrA dissociates RecA filaments and the SsbA and RecO mediators counterbalance such activity. Front. Mol. Biosci. 2022; 9:836211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F.. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003; 423:309–312. [DOI] [PubMed] [Google Scholar]
- 63. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P.. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003; 423:305–309. [DOI] [PubMed] [Google Scholar]
- 64. Simandlova J., Zagelbaum J., Payne M.J., Chu W.K., Shevelev I., Hanada K., Chatterjee S., Reid D.A., Liu Y., Janscak P.et al.. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 2013; 288:34168–34180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schwendener S., Raynard S., Paliwal S., Cheng A., Kanagaraj R., Shevelev I., Stark J.M., Sung P., Janscak P.. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 2010; 285:15739–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kodadek T., Alberts B.M.. Stimulation of protein-directed strand exchange by a DNA helicase. Nature. 1987; 326:312–314. [DOI] [PubMed] [Google Scholar]
- 67. Branzei D., Szakal B.. DNA helicases in homologous recombination repair. Curr. Opin. Genet. Dev. 2021; 71:27–33. [DOI] [PubMed] [Google Scholar]
- 68. Huselid E., Bunting S.F.. The regulation of homologous recombination by helicases. Genes (Basel). 2020; 11:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bugreev D.V., Mazina O.M., Mazin A.V.. Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J. Biol. Chem. 2009; 284:26349–26359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bugreev D.V., Yu X., Egelman E.H., Mazin A.V.. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007; 21:3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lusetti S.L., Voloshin O.N., Inman R.B., Camerini-Otero R.D., Cox M.M.. The DinI protein stabilizes RecA protein filaments. J. Biol. Chem. 2004; 279:30037–30046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.