Abstract
Aging in humans is intricately linked with alterations in circadian rhythms concomitant with physiological decline and stem cell exhaustion. However, whether the circadian machinery directly regulates stem cell aging, especially in primates, remains poorly understood. In this study, we found that deficiency of BMAL1, the only non-redundant circadian clock component, results in an accelerated aging phenotype in both human and cynomolgus monkey mesenchymal progenitor cells (MPCs). Unexpectedly, this phenotype was mainly attributed to a transcription-independent role of BMAL1 in stabilizing heterochromatin and thus preventing activation of the LINE1-cGAS-STING pathway. In senescent primate MPCs, we observed decreased capacity of BMAL1 to bind to LINE1 and synergistic activation of LINE1 expression. Likewise, in the skin and muscle tissues from the BMAL1-deficient cynomolgus monkey, we observed destabilized heterochromatin and aberrant LINE1 transcription. Altogether, these findings uncovered a noncanonical role of BMAL1 in stabilizing heterochromatin to inactivate LINE1 that drives aging in primate cells.
INTRODUCTION
Circadian rhythms regulate sleep–wake cycles, metabolism, immune function and reproduction in mammals (1–5). These processes are coordinated by the circadian clock, a biochemical oscillator that integrates physiological input signals with distinct oscillatory phases to regulate rhythms in organismal physiology, behavior and metabolism (1,6–9). Consequently, circadian dysfunction, presenting as dampened and prolonged rhythmic behavior in both aged mice and humans, is one of the aging hallmarks (7,10–14). At the cellular level, senescence is associated with extended circadian periods, decreased oscillation amplitudes and dysfunction of circadian clock proteins (15,16). At the organismal level, a dysfunctional circadian clock is a high-risk factor in multiple aging-associated diseases, including neurodegenerative diseases, osteoarthritis and metabolic disorders (6,10,17). Additionally, substantial studies in mouse models demonstrate that dysregulation of core circadian clock components results in premature aging (18,19). On aggregate, these studies reveal an intertwined relationship between the circadian clock and aging; however, the underlying mechanism remains an enigma.
Brain and muscle ARNT-like protein-1 (BMAL1), a transcription factor that initiates transcriptional-translational feedback loops (TTFLs) that drive oscillation of circadian genes, is an indispensable component of the molecular circadian clock (20–24). Loss of BMAL1 in mice leads to a complete loss of circadian rhythmicity in behavior and physiology (20,25–27). Accordingly, Bmal1–/– mice have been widely used for studies on the circadian rhythm. Notably, abnormal BMAL1 expression in mice is correlated with features of many degenerative disorders and leads to a shortened life span (19,28–32). Even though existing studies in mouse models support a correlation between BMAL1 and aging, it is still unclear how the underlying mechanism of BMAL1 deficiency leads to accelerated aging. Furthermore, the expression patterns of rhythmic genes differ between rodents and non-human primates (33,34), and given profound species-specific differences in behavior and biology between them, the knowledge we gained about BMAL1 function in mouse models is unlikely to be directly applicable to diurnal animals. Hence, the role of BMAL1 in primate aging awaits further clarification.
Stem cell exhaustion is a well-established hallmark and driving force of organismal aging (35,36). Mesenchymal progenitor cells (MPCs), also known as mesenchymal stromal cells, are multipotent stem cells residing in almost all tissues throughout our body (37). Both physiological aging and premature aging diseases are characterized by declined MPC function (38–40). Among the multiple factors contributing to MPC senescence, epigenomic instability, especially heterochromatin loss, has been verified as a prominent contributor (41–45). Normal heterochromatin organization is important for silencing transposable elements (TEs), especially long interspersed nuclear element-1 (LINE1) elements, the most abundant autonomous retrotransposons that are still active in primates (46–48). While LINE1s are usually silenced in young cells, their derepression can trigger an IFN-I response and sterile inflammation, thus leading to senescence in human mesodermal cells such as hMPCs (49,50). However, whether BMAL1 regulates human stem cell homeostasis (i.e., through LINE1-associated pathways) remains unclear.
In this study, we identified a novel role of BMAL1 in stabilizing heterochromatin and consequently repressing LINE1. Ultimately, LINE1’s activation caused by BMAL1 deficiency leads to accelerated cellular senescence. Notably, the function of BMAL1 in aging regulation is independent of its canonical role as a transcription factor regulating the expression of oscillating genes in the circadian clock machinery. Instead, it is through BMAL1’s interaction with heterochromatin-associated proteins and nuclear lamina components. In senescent hMPCs, we observed decreased BMAL1 occupancy in heterochromatin, especially in LINE1 regions, which led to destabilized heterochromatin and activation of LINE1 transcription. The resulting LINE1 transcripts then accelerated cellular senescence through the cGAS-STING signaling pathway. Likewise, heterochromatin loss and concomitant LINE1 activation were also confirmed in the MPCs or tissues (i.e., skin and muscle) derived from BMAL1-deficient cynomolgus monkey. Overall, we discovered that BMAL1 in primates, through its noncanonical role as a heterochromatin stabilizer and epigenetic LINE1 repressor, protects stem cells from senescence.
MATERIALS AND METHODS
Cell culture
BMAL1+/+ human embryonic stem cells (hESCs, line H9, WiCell Research Institute) and BMAL1–/– hESCs were seeded on feeder cells, a layer of mitomycin C-inactivated mouse embryonic fibroblasts (MEFs), and cultured in hESC culture medium containing DMEM/F12 (Thermo Fisher Scientific) supplemented with 20% Knockout Serum Replacement (Thermo Fisher Scientific), 0.1 mM nonessential amino acids (NEAAs, Thermo Fisher Scientific), 2 mM GlutaMAX (Thermo Fisher Scientific), 55 μM β-mercaptoethanol (Thermo Fisher Scientific), 1% penicillin/streptomycin (Thermo Fisher Scientific) and 10 ng/ml bFGF (Joint Protein Central, Incheon, Korea). Alternatively, hESCs were maintained on Matrigel (BD Biosciences, San Jose, CA, U.S.A.)-precoated plates with mTeSR medium (STEMCELL Technologies, Vancouver, Canada).
hESC-derived MPCs (hMPCs) were seeded on 0.1% gelatin-precoated dishes and cultured in MPC culture medium, which contains MEMα (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (Cat: 10270–106, Lot: 42F1190K, Thermo Fisher Scientific), 1% penicillin/streptomycin, 0.1 mM NEAAs and 1 ng/ml bFGF as previously described (51–54). 10 μM lamivudine (L1295, Sigma-Aldrich) was supplemented to MPC culture medium for lamivudine treatment assay in BMAL1–/– hMPCs. Primary cynomolgus monkey MPCs (cynMPCs) were also seeded on 0.1% gelatin-precoated dishes and cultured in MPC culture medium supplemented with 100 nM tenofovir (BCP02416, BiochemPartner).
Human primary fibroblasts were derived from the upper eyelid skin of a 28-year-old male individual and cultured on 0.1% gelatin-precoated dishes with fibroblast culture medium (high glucose DMEM [Hyclone] supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin and 0.1 mM NEAAs) following previous protocols (55). The skin sample was obtained from blepharoplasty of the indicated healthy donor under the ethical approval of the Peking Union Medical College Hospital Institutional Review Board.
CRISPR/Cas9-mediated gene editing in hESCs
CRISPR/Cas9-mediated BMAL1 gene editing in hESCs was performed following previously described protocols (56–58). Briefly, a guide RNA targeting exon 8 of BMAL1 was cloned into a pCAGmCherry-gRNA vector (#87110, Addgene), marking the plasmid as BMAL1-gRNA-mCherry. Next, BMAL1+/+ hESCs were resuspended in the plasmid cocktail that contained BMAL1-gRNA-mCherry and pCAG-1BPNLS-Cas9-1BPNLS-2AGFP plasmid (#87109, Addgene) in Opti-MEM (Thermo Fisher Scientific). After electroporation by a 4D-Nucleofector system (Lonza), hESCs were seeded on Matrigel precoated plates in mTeSR containing ROCK inhibitor (Y-27632, TOCRIS). Then, mCherry and GFP dual-positive cells were sorted by fluorescence-activated cell sorting (FACSAria IIIu, BD) and seeded for further expansion. hESC clones were then genotyped by genomic PCR and sequencing. The introduced single nucleic acid base in the exon 8 of BMAL1 resulted in a frameshift mutation and premature termination of translation. Successful knockout of BMAL1 in hESCs was further verified by western blot detection. Sequences of the sgRNA used for gene editing and primers used for genotyping were listed in Supplementary Table S1.
Bisulfite sequencing of the OCT4 promoter
Bisulfite sequencing of the OCT4 promoter in BMAL1+/+ and BMAL1–/– hESCs in this study was conducted as previously described (59). First, genomic DNA of BMAL1+/+ and BMAL1–/– hESCs was extracted using the DNA Extraction Kit (TIANGEN). The extracted DNA was then bisulfite modified by the CpGenome Fast DNA Modification Kit (Millipore) following the manufacturer’s protocol before amplification of the OCT4 promoter fragments using the LA Taq Hot Start Version (TAKARA). After PCR products purification by the PCR Purification Kit (Qiagen), the OCT4 promoter fragments were cloned into the pMD20 T vector (TAKARA) and sequenced. Primers used for PCR: me-OCT4 forward: ATTTGTTTTTTGGGTAGTTAAAGGT, me-OCT4 reverse: CCAACTATCTTCATCTTAATAACATCC (59).
Teratoma formation assay
Teratoma formation analysis of BMAL1+/+ and BMAL1–/– hESCs was performed as previously described (60). In Brief, approximately 3 × 106BMAL1+/+ or BMAL1–/– hESCs were resuspended in Matrigel: mTeSR (1:4) solution and injected into the inguinal region of male NOD-SCID mice (6–8 weeks old). The teratomas were collected 8–10 weeks after injection and analyzed by immunofluorescence staining.
The use and care of mice involved in this study followed the Principles for the Application Format for Ethical Approval for Research Involving Animals and were approved in advance by the Ethics Review Committee of the Institute of Biophysics, Chinese Academy of Sciences. The mice used in this study were purchased from SPF Biotechnology Co., Ltd. (Beijing) and reared at 25 ± 1°C under 12-h light–dark cycle with food and water fed ad libitum.
hMPC implantation assay
hMPC implantation assay was conducted as previously described (48,49,61,62). Briefly, approximately 1 × 106BMAL1+/+ or BMAL1–/– hMPCs were transduced with lentiviruses expressing Luc before being injected into the tibialis anterior (TA) muscle of nude mice (6–8 weeks old). Mice were further injected with D-luciferin (GoldBio) and imaged by an IVIS Spectrum imaging system (Xenogen, Caliper, Waltham, MA, USA) under ‘auto’ mode at day 0, 3 and 5 post injection of hMPCs.
Plasmid construction and lentivirus packaging
The generation of plasmids expressing BMAL1 (WT) and BMAL1 (Δ88) followed previous protocols (63). In brief, cDNA fragments amplified using primers listed in Supplementary Table S1 were cloned into the pLE4 vector (a gift from Tomoaki Hishida). cDNAs generated by reverse transcription of mRNAs from BMAL1+/+ hMPCs were used as templates for gene amplification. The sequence information of human BMAL1 (Gene ID: 406) was obtained from NCBI (https://www.ncbi.nlm.nih.gov/gene/406). The generation of plasmids expressing KAP1 and Lamin B1 followed previously described (49,64). Lentiviral CRISPR/Cas9-mediated BMAL1 knockout assay was performed as previously described (48). BMAL1-targeting sgRNA (sg-BMAL1) or non-targeting control (NTC) sgRNA (sg-NTC) was cloned into the lentiCRISPRv2 plasmid (#52961, Addgene), which carries a hSpCas9 expression cassette and a U6-driven sgRNA cassette. shRNA targeting STING or GL2 was cloned into the pLVTHM plasmid (#12247, Addgene). HEK293T cells were transfected with the plasmids and lentiviral vectors (psPAX2 (#12260, Addgene) and pMD2G (#12259, Addgene)) using Lipofectamine 3000 (Invitrogen). At 24–72 h after transfection, the culture medium of transfected HEK293T was collected and ultracentrifugated at 19,400 × g at 4°C for 2.5 h. Primers used for plasmid construction were listed in Supplementary Table S1.
hMPC generation and characterization
Directed differentiation of hESCs into hMPCs was performed following previously reported methods with some modifications (65–67). First, hESCs were individualized with 0.5 mg/ml dispase (17105041, Invitrogen) and plated on ultra-low adherent plates in the same medium for hESC culture except lower bFGF (5 ng/ml) for embryoid body (EB) formation. Next, EBs were seeded to Matrigel-precoated dishes in hMPC differentiation medium (MEMα supplemented with 10% FBS [Cat: 10270–106, Lot: 42F1190K, Thermo Fisher Scientific], 5 ng/ml TGFβ, 10 ng/ml bFGF, and 1% penicillin/streptomycin) for 10 days until the spindle-shaped cells growing out from the EBs reached 95% confluence. Then, EB-outgrowth cells were passaged by TrypLE™ Express Enzyme (1×) (Thermo Fisher Scientific) and cultured in MPC medium on Matrigel-coated plates for the first passage, and then plated on gelatin-coated dishes hereafter. Finally, the cells were purified by fluorescence-activated cell sorting (FACSAria IIIu, BD) using hMPC surface markers (CD73, CD90 and CD105). Antibodies used in flow cytometry analysis were listed in Supplementary Table S2.
The differentiation capacity of hMPCs was assessed based on adipogenic-, osteogenic- and chondrogenic-differentiation assays as previously described (61). The adipocytes, osteoblasts and chondrocytes that differentiated from hMPCs were correspondingly characterized by Oil Red O staining (O1391, Sigma) and FABP4 immunostaining (indicating adipogenesis), von Kossa staining (GMS 80045.3, GenMed Scientific Inc., U.S.A.), and Osteocalcin staining (indicating osteogenesis) and toluidine blue (T3260, Sigma) (indicating chondrogenesis) following the manufacturer’s instructions.
In vitro synchronization and circadian analysis for hMPCs and cynMPCs
BMAL1+/+ and BMAL1–/– hMPCs or cynMPCs were cultured on gelatin-coated plates in MPC culture medium until 95% confluency. Next, cells were treated with 20 μM forskolin (S2449, Selleck) for 2 h for synchronization. After forskolin treatment, cells were washed twice with PBS, and then recultured in MPC culture medium. Starting from 24 h after synchronization, cells were collected at 4-h intervals for seven time points, and then subjected for RNA extraction and RT-qPCR detection. The circadian oscillations were verified by nonparametric test JTK_CYCLE following previous described protocol (68). Both permutation-based P-values (P) and Benjamini–Hochberg adjusted Q values (Q) < 0.05 were considered as rhythmic.
Nuclear and cytoplasmic protein extraction
The extraction of nuclear and cytoplasmic proteins in hMPCs and cynMPCs was performed as previously described (69). More than 5 × 105 living cells were lysed in the Cytoplasm lysis buffer containing 10 mM KCl, 10 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 1 mM DTT, 0.4% NP-40 and 1 × protease inhibitor cocktail (Roche) and rotated for 10 min in 4°C. After centrifuged at 15,000 × g for 5 min, the supernatant was collected and marked as the cytoplasmic fraction. The remaining pellets were then resuspended in the Nucleus lysis buffer that containing 420 mM NaCl, 20 mM HEPES-KOH (pH 7.9), 0.2 mM EDTA, 0.1 mM EGTA, 25% glycerin, 1 mM DTT and 1 × protease inhibitor cocktail and vortexed for 1 h in 4°C. After centrifuged at 15,000 × g for 15 min, the supernatant was collected and marked as the nuclear fraction.
Chromatin fractionation
Chromatin fractionation analysis for hMPCs was performed as previously described with some modifications (70). At least 1 × 107 hMPCs were cultured on gelatin-coated plates with MPC culture medium until 98% confluency. hMPCs were washed with ice-cold PBS, scraped by cell scrapers, collected to a 15-ml tube, and centrifuged at 180 × g for 5 min. The cell pellets were resuspended in the Hypotonic buffer containing 10 mM Tris-HCl (pH 7.5), 10 mM KCl, 0.5 mM DTT, 1.5 mM MgCl2 and 1 × protease inhibitor cocktail (Roche) and centrifuged at 500 × g for 10 min. For cytoplasmic fractionation, the cell pellets were resuspended in the Cytoplasmic fraction lysis buffer, which contains 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM MgCl2, 0.3% NP-40 and 1 × protease inhibitor cocktail for 10-min rotation at 4°C before centrifugation at 1,000 × g for 10 min. The supernatant was then collected as the cytoplasmic fraction. The pellets were washed by the Cytoplasmic fraction washing buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM MgCl2, 0.5% NP-40 and 1 × protease inhibitor cocktail and centrifuged at 1,000 × g for 10 min. For nucleoplasm fractionation, the pellets were resuspended in the Nucleoplasmic fraction lysis buffer containing 40 μl Buffer 1 (50% glycerol, 20 mM Tris-HCl [pH 7.5], 75 mM NaCl, 0.85 mM DTT and 0.5 mM EDTA) and 360 μl Buffer 2A (20 mM HEPES [pH 7.6], 1 M urea, 300 mM NaCl, 7.5 mM MgCl2, 1 mM DTT, 0.2 mM EDTA and 1% NP-40) for 10-min vortex and rotation at 4°C followed by centrifugation at 15,000 × g for 5 min. The supernatant was then collected as the nucleoplasmic fraction. The remaining pellets were washed once in the Nucleoplasmic fraction washing buffer containing 100 μl Buffer 1 and 900 μl Buffer 2B (20 mM HEPES [pH 7.6], 1 M urea, 300 mM NaCl, 7.5 mM MgCl2, 1 mM DTT, 0.2 mM EDTA and 1.5% NP-40) and then washed twice by the Buffer 2A. Finally, the pellets were resuspended in 150 μl chromatin fraction lysis buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 0.5% sodium deoxycholate and 0.1% SDS) and marked as the chromatin fraction. All three fractions were boiled at 105°C for 10 min on a thermomixer, followed by protein quantification for western blot detection.
Western blotting
For western blotting, cells were lysed in SDS buffer (2% [wt/vol] SDS and 62.5 mM Tris-HCl [pH 6.8]) and the protein concentration was quantified by BCA quantification Kit (Dingguo Changsheng Biotechnology Co., Ltd. (Beijing, China)). Quantified lysates were subjected to SDS-PAGE gels and then electrotransferred to PVDF membranes (Millipore). Next, PVDF membranes were blocked with 5% milk for 1 h at room temperature and incubated with primary antibodies at 4°C overnight. After being washed for three times by TBST (50 mM Tris-HCl [pH 7.4], 150 mM NaCl and 0.1% Tween-20), the PVDF membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. Immunoreactive bands were then detected by a ChemiDoc XRS+ system (Bio-Rad) and statistically quantified by ImageJ software (NIH). Each image shown in the figures was representative of at least two independent experiments. Statistical significances were assessed by a two-tailed unpaired Student’s t-test. Antibodies used in western blotting were listed in Supplementary Table S2.
Transmission electron microscopy (TEM)
BMAL1+/+ and BMAL1–/– hMPCs and cynMPCs were cultured on gelatin-coated plates until 95% confluency for cell collection. Cells were digested by TrypLE™ Express Enzyme (1×) (Thermo Fisher Scientific) and then centrifuged at 1,000 rpm for 5 min at 4°C. The supernatant was removed and the pellets were fixed with 4% paraformaldehyde (PFA) in PBS (pH 7.4) at 4°C overnight. Then, cells were dehydrated in a graded series of ethanol solutions, followed by permeabilization and embedding in Lowicryl resin HM20. Finally, 200-nm sections were obtained and imaged using a 100-kV Spirit transmission electron microscope (FEI Company).
Immunofluorescence staining for cells
Immunofluorescence staining was performed as previously described (69). In brief, cells seeded on coverslips (Thermo Fisher Scientific) were washed with PBS, followed by fixation in 4% PFA for 20 min, permeabilization with 0.4% Triton X-100 in PBS for 20 min and blocking with 10% donkey serum in PBS (Jackson Immuno Research) for 1 h at room temperature. The cells were washed twice with PBS between each of the above steps. Then, cells were incubated with primary antibodies in 10% donkey serum in PBS at 4°C overnight. Subsequently, cells were washed with PBS, and incubated with secondary antibodies and Hoechst 33342 (H3570, Thermo Fisher Scientific, 1:1,000) in 10% donkey serum for 1 h at room temperature, and then washed with PBS before being mounted with antifade mounting medium (H-1000, Vector Labs). Imaging was performed with a Leica SP5 confocal system or a Zeiss LSM900 confocal microscope, followed by quantification and statistical analysis of fluorescence signals by ImageJ software (NIH). Cells were collected from three biological replicates and tissues were analyzed from at least three slides. The 3D reconstruction of H3K9me3 staining was performed by serial z-stack sectioning for up to 30 images at 100-nm intervals using a Leica SP5 confocal system, and further processed to perform 3D reconstruction by Imaris software (version 7.4.2) as previously described (71). Antibodies used in immunofluorescence staining were listed in Supplementary Table S2.
SA-β-gal staining assay
SA-β-gal staining was performed as previously described (48,72). In brief, hMPCs and cynMPCs were plated in gelatin-coated plates and grown until 70% confluency. Then, cells were washed with PBS before fixation in fixation buffer (2% formaldehyde and 0.2% glutaraldehyde) for 5 min at room temperature. After washing with PBS for two times, fixed cells were stained with staining solution at 37°C overnight. The percentage of SA-β-gal-positive cells was counted in randomly selected fields of view for each replicate using ImageJ software (NIH). Each group has three biological replicates. Statistical significances were assessed by a two-tailed unpaired Student’s t-test.
Clonal expansion assay
Clonal expansion assay was performed following previous guidance (73,74). In brief, hMPCs or cynMPCs were seeded in gelatin-coated six-well plates at a density of 6,000 cells per well and cultured for 9–12 days. Cells were then washed twice with PBS, fixed with 4% PFA for 30 min and stained with 0.2% crystal violet for 1 h at room temperature. Then, crystal violet-stained plates were scanned by a scanner (EPSON V370), and the intensity of each well was analyzed using ImageJ software (NIH). Each group has three biological replicates. Statistical significances were assessed by a two-tailed unpaired Student’s t-test.
Cell cycle analysis
The proportion of S phase cells was quantified as previously described (59,75). Briefly, 5 × 105 digested BMAL1+/+ and BMAL1–/– hMPCs were digested by TrypLE™ Express Enzyme (1×) (Thermo Fisher Scientific) and then centrifuged at 1,000 rpm for 5 min. The supernatant was removed and the pellets were fixed in 70% ethanol at −20°C overnight. On the next day, the fixed pellets were washed by PBS and then incubated in PBS containing 0.1% Triton X-100, 0.2 mg/ml RNase A (B100675-0500, Sangon biotech) and 0.02 mg/ml propidium iodide (P3566, Molecular Probes) at room temperature for 30 min. After incubation, the proportion of S phase cells was then assayed by FACS (BD FACS Calibur). Each group has three biological replicates.
LINE1 retrotransposition assay
LINE1 retrotransposition assay in hMPCs was performed following protocols as previously reported (49,76). In brief, fluorescence intensity of eGFP and mCherry in hMPCs were evaluated at 72 h after the transient transfection of plasmid DNA carrying p99-GFP-LRE3-Cherry construct (a gift from Alysson R. Muotri) using Lipofectamine™ 3000 Kit (L3000015, Thermo Fisher Scientific) (76). Cells from each group with three biological replicates were digested and washed by cold PBS before being analyzed by FACS (BD FACS Calibur). In order to calculate the adjusted retrotransposition rates of LINE1, the percentage of eGFP-positive cells out of live cells was normalized by the transfection efficiency, which was determined by the ratio of mCherry-positive cells to the total live cell number.
Enzyme-linked immunosorbent assay (ELISA)
The levels of 2’3’-cGAMP and IFN-α in the culture medium were analyzed by ELISA (CAYMAN, 501700 for 2’3’-cGAMP and Invitrogen, BMS216 for IFN-α). The ELISA was conducted following the manufacturers’ instructions and quantified by a microplate reader (Thermo Fisher Scientific, MK3).
Co-immunoprecipitation (Co-IP)
Co-immunoprecipitation (Co-IP) assay was performed as previously described (72). HEK293T cells transfected with plasmids expressing FLAG-Luc, FLAG-BMAL1 (WT) and FLAG-BMAL1 (Δ88), as well as hMPCs and cynMPCs, were lysed using CHAPS lysis buffer (120 mM NaCl, 1 mM EDTA, 40 mM HEPES [pH 7.5], 0.3% CHAPS and 1 × complete protease inhibitor cocktail [Roche]) at 4°C for 5 h and were centrifuged at 14,500 × g at 4°C for 45 min. For Co-IP with exogenous proteins in HEK293T cells, the supernatants were incubated with FLAG antibody-coupled beads (A2220, Sigma) and rotated at 4°C overnight. For Co-IP with endogenous proteins in hMPCs and cynMPCs, the supernatants were incubated with BMAL1 antibody (ab3350, Abcam) and rotated at 4°C overnight, then another 5 h incubation at 4°C with rotation after adding Protein A/G-PLUS Agarose beads (sc-2003, Santa Cruz). The beads were then washed for at least six times with CHAPS buffer, then eluted with FLAG peptides or boiled at 105°C for 10 min on a thermomixer with 1 × SDS loading buffer, followed by protein concentration quantification and western blotting.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis and protein identification
The gel bands containing the protein samples were manually excised and digested following methods as previously described (48). The solution of digested gel bands was then applied to a sample vial for LC-MS analysis, which was performed on a Q Exactive (Thermo Fisher Scientific) equipped with an Easy n-LC 1000 HPLC system (Thermo Fisher Scientific). Further MS analysis was performed with Q Exactive mass spectrometer (Thermo Fisher Scientific) in a data-dependent acquisition mode, which allowed the MS data acquired at a high resolution 70,000 (m/z 200) across the mass range of 300–1,600 m/z. Then, the raw data were transferred to analysis for protein identification with Proteome Discovery (version 1.4) using Sequest HT search engine. We searched data against UniProt human protein database (updated on 02–2016). FDR (false discovery rate) analysis by Percolator was set to < 1% for protein identification, and the peptide confidence was set as high for peptide filter.
Generation of BMAL1 knockout cynomolgus monkey
Healthy female and male cynomolgus monkeys of proven fertility were chosen as oocyte donors and sperm donors. Superovulation, sperm and oocyte collection, CRISPR injection and embryo transfer were performed as previously described (77,78). Oocyte donors were intramuscularly injected with recombinant human follicle-stimulating hormone (rhFSH, GONAL-F, Merck Serono) daily for 8 days, followed by injection of recombinant human chorionic gonadotropin (rhCG, OVIDREL, Merck Serono) on day 9. Oocytes were collected by laparoscopy 33–36 h after rhCG administration. Mature oocytes at stage Metaphase II (MII) were subjected to intracytoplasmic sperm injection (ICSI). To prepare sgRNA and SpCas9 mRNAs for embryo injection, T7-cynBMAL1 sgRNA and T7-SpCas9-expressing plasmids were linearized and in vitro transcribed with MEGAscript T7 and mMESSAGE mMACHINE T7 ULTRA Kits (Thermo Fisher Scientific). Eight to ten hours after ICSI, the zygotes were injected with SpCas9 (50 ng/μl) and cynBMAL1 sgRNA (25 ng/μl). The zygotes were transferred into surrogate females at the pronuclear stage and early pregnancy diagnosis was performed by ultrasonography around one month after embryo transfer. The sequences of sgRNA used for gene editing and primers used for genotyping were listed at Supplementary Table S1.
The use and care of cynomolgus monkeys involved in this study followed the Principles for the Application Format for Ethical Approval for Research Involving Animals and were approved in advance by the Ethics Committee of Sun Yat-sen University. The monkeys used in this study were housed at the Blooming Spring Biological Technology Development Co., Ltd, which is accredited by the Association and Accreditation of Laboratory Animal Care International (AAALAC). All monkeys were under careful surveillance and daily fed with monkey feed, drinking water and supplemented with fruits and vegetables.
Isolation and culture of primary cynMPCs
Primary cynMPCs were isolated from the bone marrow of BMAL1+/+ and BMAL1–/– cynomolgus monkeys. First, bone marrow samples from the posterior/anterior super iliac spine were extracted using puncture needles that were moistened with heparin (Sigma; H3149-500KU-9). Then, the bone marrow diluted with equal volume of PBS was slowly added to the upper layer of Ficoll-Paque PLUS (GE healthcare; 1.077 g/ml) for centrifugation at 2,000 rpm for 20 min at room temperature. The middle misty mononuclear layer was carefully aspirated and transferred into a 15-ml tube and washed with 10% FBS in PBS. After centrifuging at 1,000 rpm for 5 min, the pellets generated were incubated with red blood cell lysis buffer for 5 min at room temperature and then washed with 10% FBS in PBS. After centrifuging at 1,000 rpm for 5 min, the pellets were resuspended in cynMPC culture medium and plated onto gelatin-coated plates for further cell culture.
Hematoxylin and Eosin (H&E) staining
For histological analysis, skin and muscle tissues from BMAL1+/+ and BMAL1–/– cynomolgus monkeys were fixed with 4% PFA for 2 days, embedded in paraffin and sectioned at 5-μm intervals. The sections were deparaffinized and rehydrated by xylene and graded series of ethanol (100%, 100%, 95%, 80%, 70%, and 50%), and then washed by distilled water followed by staining with hematoxylin solution (Servicebio, China). After 5-min incubation of hematoxylin, the sections were washed with running water, differentiated with 1% acid alcohol, and washed with running tap water again until the nuclei were stained to a suitable intensity. Next, the sections were counterstained in the eosin (Solarbio), followed by dehydration in a grader series of ethanol (50%, 70%, 80%, 90%, 95%, 100%, and 100%), and immersion in xylene. Finally, the sections were mounted on the coverslip with neutral balsam for long-term preservation.
Immunohistochemical staining
Immunohistochemical staining of BMAL1+/+ and BMAL1–/– cynomolgus monkey tissues was performed following the DAB staining method (79,80). Briefly, after deparaffinization and rehydration, sections were treated with sodium citrate buffer (pH 6.0) (ZLI-9065, ZSGB-BIO) for antigen retrieval followed by 0.4% Triton X-100 for permeabilization and 3% H2O2 for endogenous peroxidases blocking. Then, the sections were incubated with primary antibodies at 4°C overnight. After incubation with HRP-conjugated secondary antibodies, DAB application, and hematoxylin counterstain, the sections were then dehydrated and mounted on the coverslip with neutral balsam for long-term preservation. Antibodies used in immunohistochemical staining were listed in Supplementary Table S2.
Immunofluorescence staining for tissues
Immunofluorescence staining of BMAL1+/+ and BMAL1–/– cynomolgus monkey tissues was conducted as previously described (55,81–82). In short, the paraffin-embedded sections were boiled in sodium citrate buffer (pH 6.0) for antigen retrieval after deparaffination and rehydration. Then, the sections were treated with 0.4% Triton X-100 for permeabilization and 10% donkey serum for blocking. Primary antibodies were then applied at 4°C overnight. After several washes with PBS, the sections were incubated with secondary antibodies and Hoechst 33342 (Thermo Fisher Scientific). Finally, the sections were mounted with Antifade Mounting Medium (Vector Laboratories, H-1000) after several washes with PBS. The images were obtained by a ZEISS LSM900 confocal system and the mean fluorescence intensity of H3K9me3 were measured by the ImageJ software (NIH). Antibodies used in immunofluorescence staining were listed in Supplementary Table S2.
RT-qPCR and RNA-seq
TRIzol (Thermo Fisher Scientific) was used for the total RNA extraction of hMPCs, cynMPCs, and tissues derived from cynomolgus monkeys following previous protocols (63). cDNA was generated using the Go Script Reverse Transcription System (Promega) according to the manufacturer’s protocol. The resulting cDNA was then subjected to RT-qPCR amplification following MIQE guidelines (83) in ABI QuantStudio 5 Real-time system (Life Technologies). Data were evaluated on GraphPad Prism 6 and statistical significances were assessed by unpaired Student’s t-test. Primers used for RT-qPCR detection were listed in Supplementary Table S3.
For RNA-seq, a NEBNext® Ultra™ RNA Library Prep Kit for Illumina® was used for library construction and Illumina HiSeq X-Ten or NovaSeq 6000 platforms were used for paired-end sequencing with 150-bp read length. Quality control and sequencing for RNA were performed by Novogene Bioinformatics Technology Co., Ltd.
DamID-seq
DamID-seq was performed as described with some modifications (84). In brief, the Dam and Dam-EMD lentiviruses generated from HEK293T cells were concentrated by ultracentrifugation at 19,400 × g for 2.5 h. The virus pellets were resuspended in PBS. BMAL1+/+ and BMAL1–/– hMPCs were seeded in six-well dishes at 2 × 105 cells per well. Each group had three biological replicates. The next day, the culture medium was replaced with 2 ml of fresh culture medium containing either Dam or Dam-EMD lentivirus. At 72 h after transduction, cells were harvested, and genomic DNA was isolated using a TIANamp Genomic DNA Kit (Tiangen, DP304). DpnI enzyme (R0176S, New England Biolabs) digestion, adaptor ligation, DpnII enzyme (R0543S, New England Biolabs) digestion, PCR amplification and purification were performed as previously described (84). For adaptor trimming, the amplified DNA was sonicated and digested with AlwI (R0513S, New England Biolabs) enzyme. The DNA library was constructed using a NEBNext® Ultra™ DNA library prep Kit for Illumina® (E7370S, New England Biolabs). The libraries were pooled and subjected to paired-end sequencing with 150-bp read length on Illumina NovaSeq 6000 platform by Annoroad Gene Technology Company (Beijing, China).
ChIP-qPCR and ChIP-seq
ChIP-qPCR and ChIP-seq were performed as previously described with some modifications (48,85). In brief, hMPCs and cynMPCs were crosslinked in 1% (vol/vol) formaldehyde in 10% FBS in PBS for 8–12 min at room temperature, and then terminated by incubation with 125 mM glycine. Samples were then centrifuged and the resulting pellets were lysed by lysis buffer (1% SDS, 50 mM Tris-HCl [pH 8.0], 10 mM EDTA) for 5 min on ice. Sonication was performed for 10 min with a Covaris S220 Focused Ultrasonicator until chromatin were sheared to an average fragment length of 100–300 bp. After sonication, samples were centrifugated at 12,000 × g for 10 min at 4°C and the resulting supernatants were then incubated with Protein A Dynabeads (Thermo Fisher Scientific) pre-conjugated with anti-BMAL1, anti-H3K9me3 or anti-IgG antibodies. Samples were subsequently eluted and reverse cross-linked in elution buffer at 68°C for 3 h in a thermomixer. The eluted DNA was isolated using phenol-chloroform-isoamyl alcohol extraction and precipitated by isopropanol. Then, the precipitated DNA was purified by ethanol and diluted by distilled water for further RT-qPCR detection or sequencing library construction. Primers used for RT-qPCR detection were listed in Supplementary Table S3. For ChIP-seq, library construction was conducted using KAPA Hyper Prep Kit (KAPA Biosystems, KK8504) and Index Primers Set 1 for Illumina (New England Biolabs (NEB), E7335L) following the manufacturer’s instructions.
ATAC-seq
ATAC-seq was performed as previously described with minor modifications (49). In brief, a total of 30,000 cells were lysed with 50 μl of lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% [vol/vol] Nonidet P40 Substitute), followed by Tn5 transposition with 50 μl of transposition reaction mix (10 μl of 5 × TTBL buffer, 4 μl of TTE mix and 36 μl of nuclease-free H2O) from a TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme Biotech) at 37°C for 30 min. The libraries were amplified using the TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme Biotech). Finally, libraries were sequenced on Illumina NovaSeq platforms with paired-end sequencing with 150-bp read length.
RNA-seq data processing
Raw paired-end reads were trimmed by Trim Galore software (version 0.4.5) (https://github.com/FelixKrueger/TrimGalore). Then, the cleaned reads were mapped to the cynomolgus macaque (Macaca Fascicularis) MacFas5.0 genome obtained from UCSC genome browser database using hisat2 (version 2.0.4) (86). Read counts for each gene were calculated by HTSeq (version 0.11.0) with only high-quality mapped reads (mapping quality > 20) (87). Differentially expressed genes (DEGs) between tissues derived from BMAL1+/+ and BMAL1–/– cynomolgus monkeys were calculated by R package DESeq2 (version 1.22.2) with the cutoff set to ‘the adjusted P-value < 0.05 and the absolute log2(fold change) > 0.5’. Gene Ontology (GO) and pathway enrichment analysis were conducted by Metascape (88).
For calculating differentially expressed repetitive elements (REs) in cynMPCs and cynomolgus monkey tissues, trimmed reads were mapped to the cynomolgus monkey (Macaca Fascicularis) MacFas5.0 genome obtained from UCSC genome browser database using STAR software (version 2.5.2b) (89). Then, the read counts for each RepeatMasker-annotated repeats of MacFas5.0 genome were calculated by featureCounts (version 2.0.0) (90) with only high-quality mapped reads (mapping quality > 20). Differentially expressed repetitive elements were identified by R package DESeq2 (version 1.30.1) with the cutoff set to ‘the absolute log2(fold change) > 0.2 and the adjusted P-value < 0.05’ (91).
DamID-seq data processing
Raw reads of Dam and Dam-EMD data for BMAL1+/+ and BMAL1–/– hMPCs were trimmed by Trim Galore software (version 0.4.5). Trimmed reads were mapped to the UCSC human (Homo sapiens) hg19 genome using Bowtie 2 (version 2.2.9) (92). PCR duplicates were removed with the MarkDuplicates.jar program in Picard tools (version 1.119). Then, reads were sorted with SAMtools (version 1.6) (93). Replicates for each sample were merged for downstream analysis. DamID signals in BMAL1+/+ and BMAL1–/– hMPCs were calculated as the log2 ratio of the Reads Per Kilobase per Million mapped reads (RPKM) of the Dam-EMD and Dam signals [log2(Dam-EMD/Dam)]. The DamID signals were visualized for each 2,000-base pair (bp) bin using the bamCompare program in deepTools (version 2.5.4) software (94).
To define the LAD regions in BMAL1+/+ and BMAL1–/– hMPCs, we first calculated the DamID signals in BMAL1+/+ and BMAL1–/– hMPCs for each 25-kb bin. Then, LADs were identified by the R package hidden Markov models with t emissions (HMMt) (version 0.1) (https://github.com/dinovski/asDamID/blob/master/scripts/hmmt_functions.R). Genomic enrichment for LADs and iLADs (inter-LADs) was conducted by annotatePeaks.pl program in homer2 (version v4.10.3) software (95). In order to compare the DamID signals of the repetitive elements localized in LAD regions in BMAL1+/+ and BMAL1–/– hMPCs, RepeatMasker-annotated repeats located in the LAD regions of BMAL1+/+ hMPCs were identified as LAD-localized repetitive elements. LAD-localized repetitive elements with DamID signals > 0 in BMAL1+/+ hMPCs were kept for statistical analysis of DamID signals in both BMAL1+/+ and BMAL1–/– hMPCs.
ChIP-seq data processing
For the processing of H3K9me3 ChIP-seq data, raw reads were trimmed with Trim Galore software (version 0.4.5). Trimmed reads were mapped to the human (Homo sapiens) hg19 genome obtained from UCSC genome browser database using Bowtie 2 (version 2.2.9) (92). Duplicated reads were discarded by MarkDuplicates.jar program in Picard tools (version 1.119). Then, reads were sorted with SAMtools (version 1.6) (93). To minimize the effect of sequencing bias and depth, replicates for each sample were merged. Then, the same number of high-quality reads in BMAL1+/+ and BMAL1–/– hMPCs were randomly selected for downstream analysis. For H3K9me3 peak calling, SICER (version 1.1) was used with the parameters set to ‘-w 1000 -g 10’ (96). Only H3K9me3 peaks with FDR < 0.01 were retained for downstream analysis.
For the classification of H3K9me3-enriched LADs, we calculated the H3K9me3 signals for each LAD region. Then LAD regions were partitioned into ‘H3K9me3-enriched LADs’ and ‘Other LADs’ by robust k-means analysis with two clusters using R package pheatmap (version 1.0.12). Then the log2 ratios of H3K9me3 signals for each H3K9me3-enriched LAD between BMAL1+/+ and BMAL1–/– hMPCs were visualized by R package RIdeogram (version 0.2.2) (97).
ATAC-Seq data processing
For ATAC-seq data processing, raw reads were trimmed with Trim Galore software (version 0.4.5). Cleaned reads were mapped to the human (Homo sapiens) hg19 genome or the cynomolgus macaque (Macaca Fascicularis) MacFas5.0 genome obtained from UCSC genome browser database using Bowtie 2 (version 2.2.9) with the parameters ‘ -X 2000 -N 1 -L 25 –no-mixed –no-discordant -t’ (92). Duplicate reads were removed with the MarkDuplicates.jar program in Picard tools (version 1.119). To minimize the effect of sequencing bias and depth, replicates for each sample were merged. ATAC peaks were called by MACS2 (version 2.1.1.20160309) (98). Called peaks with q-value < 0.01 were retained for downstream analysis. Genomic distribution of ATAC peaks were annotated by annotatePeaks.pl program in homer2 (version 4.10.3) software (95).
Assessment of the reproducibility of the sequencing data
To evaluate the reproducibility of the H3K9me3 ChIP-seq, ATAC-seq and DamID-seq data, whole genome enrichment levels (RPKM values) or DamID signals at 2-kb resolution were calculated by bamCoverage or bamCompare program in deepTools software (version 3.5.0) and the Euclidean distance was calculated in R (version 4.0.3). To evaluate the reproducibility of RNA-seq data, Euclidean distance was calculated by DESeq2 regularized logarithm (rlog)-normalized read counts. Lower Euclidean distances mean higher correlations.
Statistical analysis
Data are presented as the means ± SEM. P-value (P) was analyzed by two-tailed Student’s t-test using GraphPad Prism 6 software.
RESULTS
BMAL1 plays a geroprotective role in hMPCs
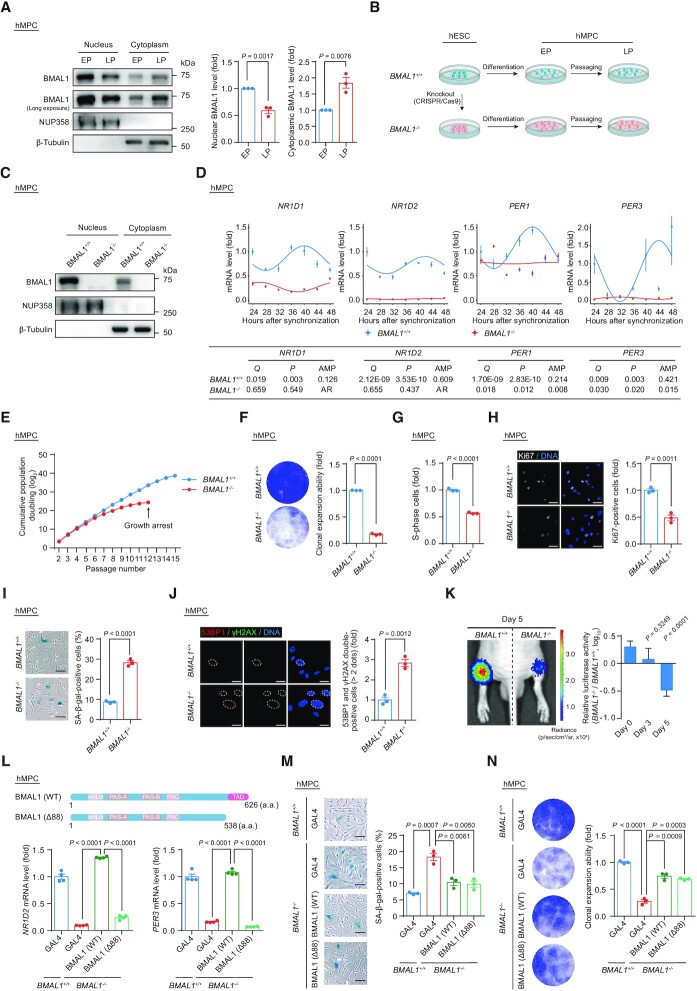
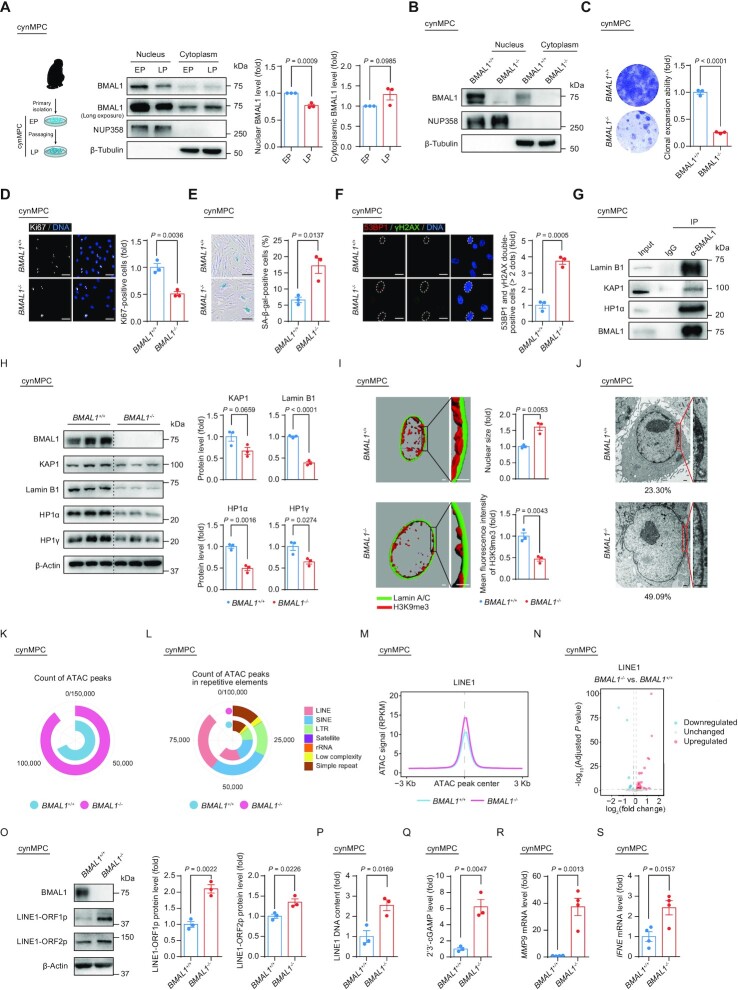
To investigate whether human MPC (hMPC) senescence was associated with a change in the cellular distribution of BMAL1, we examined its protein levels in both the cytoplasmic and nuclear fractions of hMPCs at both early passage (EP) and late passage (LP) (Figure 1A and Supplementary Figure S1A–C). In replicative senescent LP hMPCs, we found through western blot analysis that nuclear BMAL1 was reduced, concomitant with an increase of cytosolic BMAL1 (Figure 1A). These data demonstrate that BMAL1 is preferentially excluded from the nucleus in senescent hMPCs and suggest that a nucleocytoplasmic redistribution of BMAL1 might have a functional role in the process of cellular senescence.
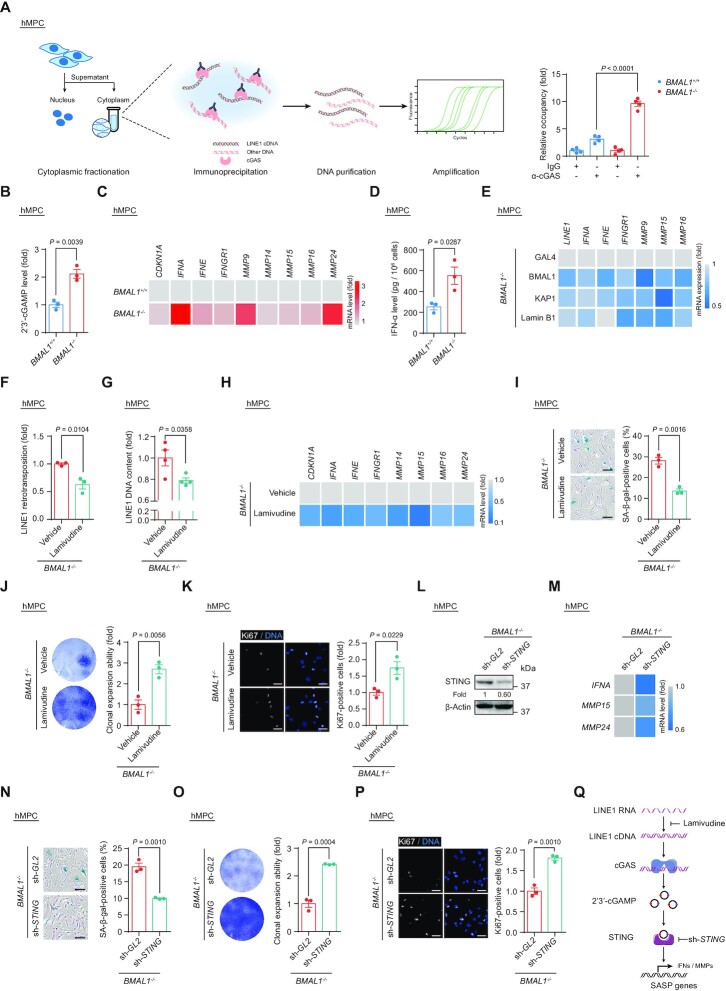
Figure 1.
(A) Western blot analysis of BMAL1 in EP (P5) and LP (P16) hMPCs. β-Tubulin and NUP358 were used as the loading control for the cytoplasmic and nuclear fractions, respectively. Western blots shown on the left are representative of three independent experiments. Quantitative data on the right are presented as the means ± SEM. Two-tailed unpaired Student’s t-test. (B) Schematic diagram for the generation of BMAL1+/+ and BMAL1–/– hESCs and hMPCs. (C) Western blot analysis of BMAL1 in EP (P4) BMAL1+/+ and BMAL1–/– hMPCs. β-Tubulin and NUP358 were used as the loading control for the cytoplasmic and nuclear fractions, respectively. (D) Relative mRNA levels of the indicated genes in EP (P4) BMAL1+/+ and BMAL1–/– hMPCs at the indicated time points after forskolin synchronization. The RT-qPCR data shown above were further analyzed by JTK_Cycle analysis to determine the rhythmicity and amplitude of the peaks (peaks with permutation-based P-values (P) and Benjamini–Hochberg adjusted Q-values (Q) < 0.05 were considered rhythmic); AR, arrhythmic. AMP, amplitude. Data are presented as the means ± SEM. n = 4 wells. Data shown are representative of two independent experiments. GAPDH was used as the reference gene. (E) Growth curve analysis of BMAL1+/+ and BMAL1–/– hMPCs. Arrowhead indicates the passage number for growth arrest of BMAL1–/– hMPCs. n = 3 biological replicates. (F) Clonal expansion analysis of LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (G) Cell cycle analysis of LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (H) Immunofluorescence staining of Ki67 in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 3 biological replicates with each replicate containing > 100 cells; scale bars: 50 μm. Two-tailed unpaired Student’s t-test. (I) SA-β-gal staining of LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 3 biological replicates; scale bars: 100 μm. Two-tailed unpaired Student’s t-test. (J) Immunofluorescence staining of 53BP1 and γH2AX in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Dashed lines indicate the nuclear boundaries of the 53BP1 and γH2AX double-positive (> 2 dots) cells. Data are presented as the means ± SEM. n = 3 biological replicates with each replicate containing > 100 cells; scale bars: 25 μm. Two-tailed unpaired Student’s t-test. (K) In vivo implantation assay of LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Photon flux from the tibialis anterior (TA) muscles of nude mice transplanted with BMAL1+/+ or BMAL1–/– hMPCs. Quantitative analysis of luciferase activity in the TA muscles was detected by an in vivo imaging system (IVIS). Data are presented as the means ± SEM. n = 8 biological replicates. Two-tailed unpaired Student’s t-test. (L) Top, schematic diagram showing the strategy for the generation of BMAL1 Δ88 mutant. Bottom, RT-qPCR detection of indicated genes in EP (P4) BMAL1+/+ hMPCs transduced with lentiviruses expressing GAL4 and EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1 (WT) or BMAL1 (Δ88) at P2 post transduction. Data are presented as the means ± SEM. n = 4 wells. GAPDH was used as the reference gene. Data shown are representative of two independent experiments. Two-tailed unpaired Student’s t-test. (M) SA-β-gal staining of EP (P4) BMAL1+/+ hMPCs transduced with lentiviruses expressing GAL4 and EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1 (WT) or BMAL1 (Δ88) at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates; scale bars: 100 μm. Two-tailed unpaired Student’s t-test. (N) Clonal expansion analysis of EP (P4) BMAL1+/+ hMPCs transduced with lentiviruses expressing GAL4 and EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1 (WT) or BMAL1 (Δ88) at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test.
To investigate the function of BMAL1 in hMPC senescence, we generated BMAL1-deficient human embryonic stem cells (hESCs) (Figure 1B). Through CRISPR/Cas9-mediated non-homologous end joining (NHEJ), we introduced a single nucleotide insertion to the exon 8 of BMAL1 in hESCs, causing a homozygous frameshift mutation and early translation termination of the BMAL1 transcript (Supplementary Figure S1D). Successful knockout of BMAL1 in both the nuclear and cytoplasmic fractions in hESCs was verified by western blotting (Supplementary Figure S1E). We found that genomic integrity and pluripotency were well maintained in BMAL1–/– hESCs, as manifested by a normal karyotype (Supplementary Figure S1F), typical expression of pluripotency markers (Supplementary Figure S1G) and DNA hypomethylation at the OCT4 promoter (Supplementary Figure S1H). In teratoma formation assay, we demonstrated that BMAL1–/– hESCs retain the pluripotency, as reflected by their capacity to differentiate into cell types of all three embryonic germ layers (Supplementary Figure S1I). Consequently, BMAL1+/+ and BMAL1–/– hMPCs derived from corresponding hESCs expressed typical hMPC surface markers, including CD73, CD90, CD105, CD166, CD29 and HLA-ABC, but did not express non-hMPC surface markers such as CD34, CD43, CD45, CD164, CD14 and CD19 (Figure 1B, C and Supplementary Figure S1J and K). Both BMAL1+/+ and BMAL1–/– hMPCs were able to further differentiate into osteoblasts, chondrocytes and adipocytes with comparable efficiency (Supplementary Figure S1L). As expected, analysis of transcript levels of classic BMAL1-targeted genes (i.e. NR1D1, NR1D2, PER1 and PER3) demonstrated dampened or disrupted oscillation in BMAL1–/– hMPCs, confirming that deficiency of BMAL1 results in the dysregulated cellular circadian activity (Figure 1D).
Notably, we found that BMAL1 deficiency in hMPCs led to an accelerated senescence phenotype, reflected as premature growth arrest (Figure 1E), compromised clonal expansion ability (Figure 1F), decreased percentage of S-phase cells (Figure 1G), and Ki67-positive cells (Figure 1H). Consistently, we observed an increased ratio of SA-β-gal positive cells (Figure 1I) and elevated levels of DNA damage response in BMAL1–/– hMPCs (Figure 1J). Moreover, when implanted into the tibialis anterior (TA) muscles of immunodeficient mice, BMAL1–/– hMPCs underwent accelerated in vivo decay relative to BMAL1+/+ hMPCs (Figure 1K). Meanwhile, accelerated cellular senescence due to BMAL1 inactivation was also observed in early passage BMAL1+/+ hMPCs and human primary fibroblasts transduced with lentiviruses encoding CRISPR/Cas9 and a sgRNA targeting BMAL1 (Supplementary Figure S2A–F). Altogether, these data revealed that BMAL1 plays a geroprotective role in various human cellular senescence models.
BMAL1 antagonizes cellular senescence independently of its canonical circadian activity
The BMAL1 protein carries a transactivation domain (TAD) in its C-terminus, which enables BMAL1 to cooperate with other circadian regulators like CRY1 or CBP (p300) to induce the expression of its circadian target genes (99–101). Accordingly, deletion of the BMAL1 TAD results in transcriptional inactivation of circadian genes and, consequently, dysregulation of the cellular circadian clock (99,102). To determine whether the senescence-antagonizing function of BMAL1 depends on its role in transcriptional regulation, we generated a BMAL1 C-terminal truncated mutant that lacks the TAD (BMAL1 (Δ88), which lacks amino acids 539–626) (Figure 1L). As expected, hMPCs harboring BMAL1 Δ88 mutant showed a compromised ability in transcriptional activation of the downstream target genes (NR1D2 and PER3) (Figure 1L). However, when we expressed the BMAL1 Δ88 mutant in BMAL1–/– hMPCs, we found that it rescued the senescence phenotypes to a similar extent as that of wild-type (WT) BMAL1 (Figure 1M, N and Supplementary Figure S2G). From these data, we concluded that BMAL1 counteracts stem cell aging in a manner independent of its canonical circadian function.
BMAL1 is required for maintaining heterochromatin in hMPCs
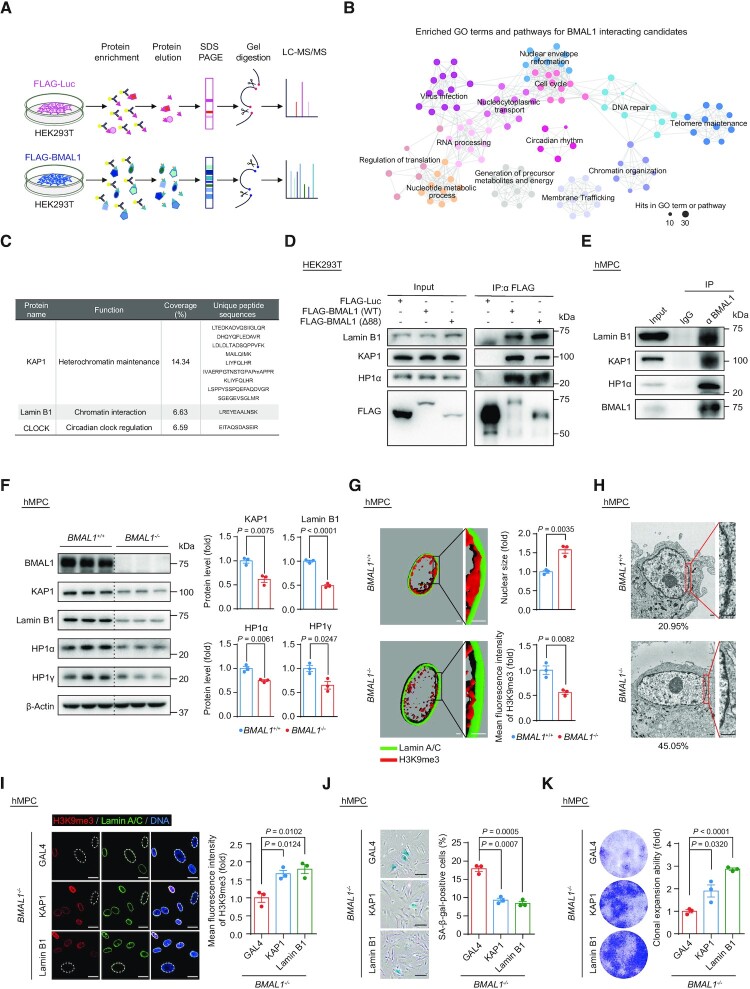
We next investigated whether BMAL1 regulates hMPC senescence through unknown interacting partners beyond those included in the circadian machinery. By immunoprecipitation followed by mass spectrometry analysis (IP-MS) (Figure 2A), we retrieved peptides corresponding to a panel of annotated human proteins as potential interacting partners with BMAL1 (Supplementary Table S4). In addition to circadian rhythm-related proteins, we identified proteins involved in nuclear envelope architecture or chromatin organization (Figure 2B and C). Through Co-IP assays in HEK293T cells and hMPCs, we verified that both exogenous and endogenous BMAL1 are associated with the nuclear lamina protein Lamin B1, as well as heterochromatin-associated proteins including HP1α and KAP1 (Figure 2D and E).
Figure 2.
(A) Schematic diagram of the mass spectrometry strategy for identifying BMAL1 interacting candidates in HEK293T cells. FLAG-Luc was used as a control. (B) GO terms and pathways enrichment analysis of BMAL1 interacting candidates that were identified by mass spectrometry. (C) Detailed information illustrated that KAP1 and Lamin B1 were identified as BMAL1 interacting candidates by mass spectrometry. The BMAL1 interacting candidates identified by mass spectrometry are listed in the Supplementary Table S4. (D) Co-IP analysis of Lamin B1, KAP1 and HP1α with exogenous FLAG-tagged BMAL1 (WT) and FLAG-tagged BMAL1 (Δ88) in HEK293T cells. (E) Co-IP analysis of Lamin B1, KAP1 and HP1α with endogenous BMAL1 in EP (P4) BMAL1+/+ hMPCs. (F) Western blot analysis of the protein levels of heterochromatin associated proteins and nuclear lamina protein Lamin B1 in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. β-Actin was used as a loading control. Quantitative data on the right are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (G) 3D reconstruction of z-stack immunofluorescence images of H3K9me3 (red) and Lamin A/C (green) in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Scale bars: 2 μm. Quantitative data of the mean nuclear size (top) and the mean fluorescence intensity of H3K9me3 (bottom) in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs are presented as the means ± SEM. n = 3 biological replicates with each replicate containing 100 cells. Two-tailed unpaired Student’s t-test. Related to Supplementary Figure S2H. (H) Transmission electron microscopy (TEM) analysis of the peripheral heterochromatin in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. The percentages of cells with compromised nuclear peripheral heterochromatin are presented at the bottoms of the TEM images. Scale bars: 2 μm. (I) Immunofluorescence staining of H3K9me3 in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, KAP1 or Lamin B1 at P2 post transduction. Dashed lines indicate the nuclear boundaries of the cells with decreased H3K9me3 signals. Data are presented as the means ± SEM. n = 3 biological replicates with each replicate containing 100 cells; scale bars: 25 μm. Two-tailed unpaired Student’s t-test. (J) SA-β-gal staining of EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, KAP1 or Lamin B1 at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates; scale bars: 100 μm. Two-tailed unpaired Student’s t-test. (K) Clonal expansion assay of EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, KAP1 or Lamin B1 at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test.
Given that nuclear envelope and heterochromatin-associated proteins are essential in heterochromatin maintenance, we hypothesized that BMAL1 via its interacting partners plays a role in heterochromatin modulation (103–105). We found decreased protein levels of Lamin B1 and heterochromatin-associated proteins KAP1, HP1α and HP1γ (Figure 2F), accompanied by enlarged nuclei (Figure 2G and Supplementary Figure S2H) and heterochromatin loss in BMAL1–/– hMPCs (Figure 2G, H and Supplementary Figure S2H). Overexpressing Lamin B1 and heterochromatin-associated protein KAP1 counteracted the decreased H3K9me3 intensity (Figure 2I) and rescued the senescent phenotypes in BMAL1–/– hMPCs, the latter manifested as a decrease in SA-β-gal positive cells (Figure 2J) and an increase in the cellular proliferation ability (Figure 2K and Supplementary Figure S2I). These results identified an important role of BMAL1 in stabilizing heterochromatin and regulating hMPC senescence.
Heterochromatin is destabilized at LINE1 elements in BMAL1–/– hMPCs
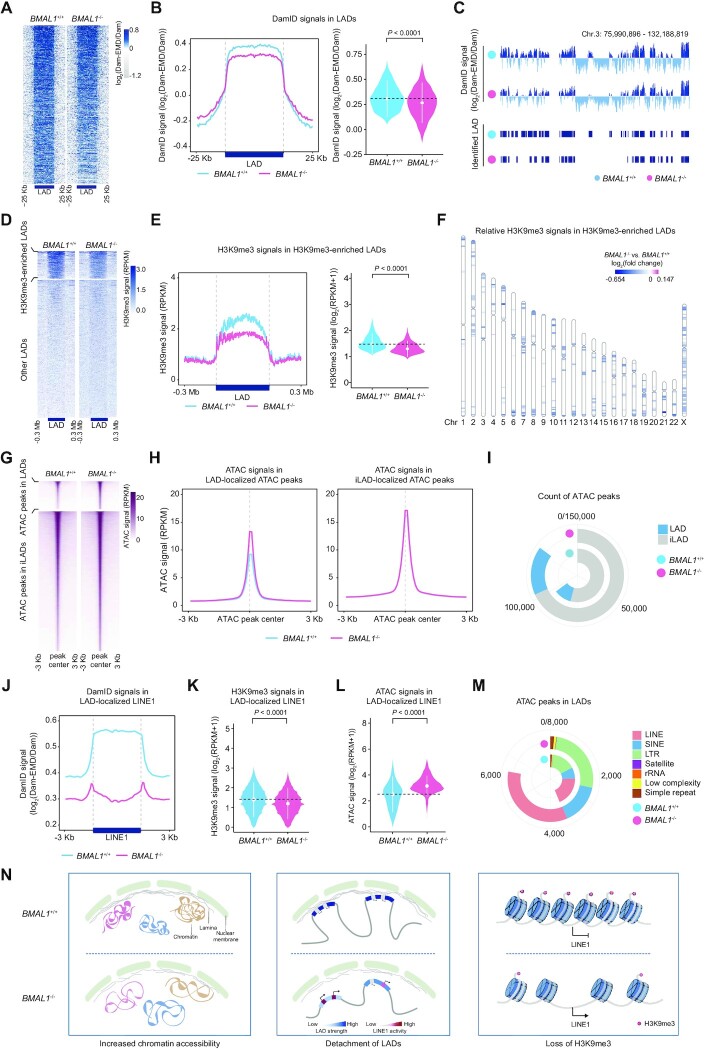
In BMAL1–/– hMPCs, we detected compromised expression levels of nuclear meshwork proteins and heterochromatin proteins as well as erosion of nuclear peripheral heterochromatin. Next, we asked if BMAL1 safeguards epigenome integrity, and especially heterochromatin. To answer this question, we first implemented DNA adenine methyltransferase identification (DamID) and H3K9me3 chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) assays to study the reorganization of peripheral heterochromatin (also known as lamina-associated domains, LADs) and the redistribution of heterochromatic marks (Supplementary Figure S3A and B). Our data demonstrated a global decrease of the lamina-association score (DamID signal) across LAD regions, indicative of a general detachment of LADs from the nuclear lamina in BMAL1–/– hMPCs (Figure 3A–C and Supplementary Figure S3C). BMAL1 depletion also led to decreased heterochromatic mark H3K9me3 in LADs (Figure 3D–F and Supplementary Figure S3D and E), consistent with the observed erosion of heterochromatin underneath the nuclear envelope assessed by immunofluorescence staining and transmission electron microscopy (Figure 2G, H and Supplementary Figure S2H). Furthermore, chromatin accessibility increased in the whole genome but more dramatically so in LADs, as evidenced by Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) (Figure 3G–I and Supplementary Figure S3F), further indicating a decompacted state for LADs upon BMAL1 deficiency. Repetitive elements (REs), such as LINEs and LTRs, are sealed in LADs and are highly decorated with the heterochromatin mark H3K9me3 (Supplementary Figure S3G) (106,107). Notably, LINE1, one of the few families of REs that are still active in primates, is highly enriched within LADs (108,109) but suffered from a dramatic loss of DamID signal strength upon BMAL1 deficiency (Figure 3J and Supplementary Figure S3H). Consistently, decreased H3K9me3 occupancy and increased chromatin accessibility were also observed in LINE1 in BMAL1–/– hMPCs (Figure 3K–M). Taken together, our data indicated that BMAL1 deficiency resulted in disorganization of LADs and derepression of the LINE1 elements harbored in LADs (Figure 3N).
Figure 3.
(A) Heatmap showing the DamID signals (log2(Dam-EMD/Dam)) ranging from 25 kb upstream to 25 kb downstream of LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. The color key from gray to blue indicates low to high DamID signal. (B) DamID signals in LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Left, metaplot showing the DamID signals (log2(Dam-EMD/Dam)) ranging from 25 kb upstream to 25 kb downstream of LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Right, violin plot showing the DamID signals (log2(Dam-EMD/Dam)) in LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. The white dots represent the median values, and the white lines represent the interquartile range (IQR) from smallest to largest. Two-sided Wilcoxon signed-rank test. (C) Representative tracks of DamID signals and identified LAD regions in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. (D) Heatmap showing the H3K9me3 signals (RPKM) ranging from 0.3 Mb upstream to 0.3 Mb downstream of H3K9me3-enriched LADs (top panel) and other LADs (bottom panel) in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. The color key from white to blue indicates low to high H3K9me3 signal. (E) H3K9me3 signals in H3K9me3-enriched LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Left, metaplot showing the H3K9me3 signals (RPKM) in H3K9me3-enriched LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Right, violin plot showing the H3K9me3 signals (log2(RPKM + 1)) in H3K9me3-enriched LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. The white dots represent the median values, and the white lines represent the interquartile range (IQR) from smallest to largest. Two-sided Wilcoxon signed-rank test. (F) Chromosome ideogram showing the relative H3K9me3 signals in H3K9me3-enriched LADs across the 23 chromosomes of LP (P9) BMAL1–/– hMPCs compared to BMAL1+/+ hMPCs. The color key from blue to amaranth indicates low to high relative H3K9me3 level. (G) Heatmap showing the ATAC signals (RPKM) ranging from 3 kb upstream to 3 kb downstream of LAD-localized ATAC peak centers (top panel) and iLAD-localized ATAC peak centers (bottom panel) in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. The color key from white to amaranth indicates low to high chromatin accessibility. (H) Metaplot showing the ATAC signals (RPKM) ranging from 3 kb upstream to 3 kb downstream of LAD-localized ATAC peak centers (left panel) and iLAD-localized ATAC peak centers (right panel) in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. (I) Ring plot showing the count of ATAC peaks in LADs and iLADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. (J) Metaplot showing the DamID signals (log2(Dam-EMD/Dam)) ranging from 3 kb upstream to 3 kb downstream of LAD-localized LINE1 regions in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. (K) Violin plot showing the H3K9me3 signals (log2(RPKM + 1)) in LINE1s localized in H3K9me3-enriched LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. The white dots represent the median values, and the white lines represent the interquartile range (IQR) from smallest to largest. Two-sided Wilcoxon signed-rank test. (L) Violin plot showing the ATAC signals (log2(RPKM + 1)) for ATAC peaks in LAD-localized LINE1 regions in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. The white dots represent the median values, and the white lines represent the interquartile range (IQR) from smallest to largest. Two-sided Wilcoxon signed-rank test. (M) Ring plot showing the count of ATAC peaks in indicated repetitive elements in LADs in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. (N) Diagrams summarizing epigenomic alterations in BMAL1–/– hMPCs detected by DamID-seq, H3K9me3 ChIP-seq and ATAC-seq.
LINE1 elements are repressed via the heterochromatin stabilization maintained by BMAL1
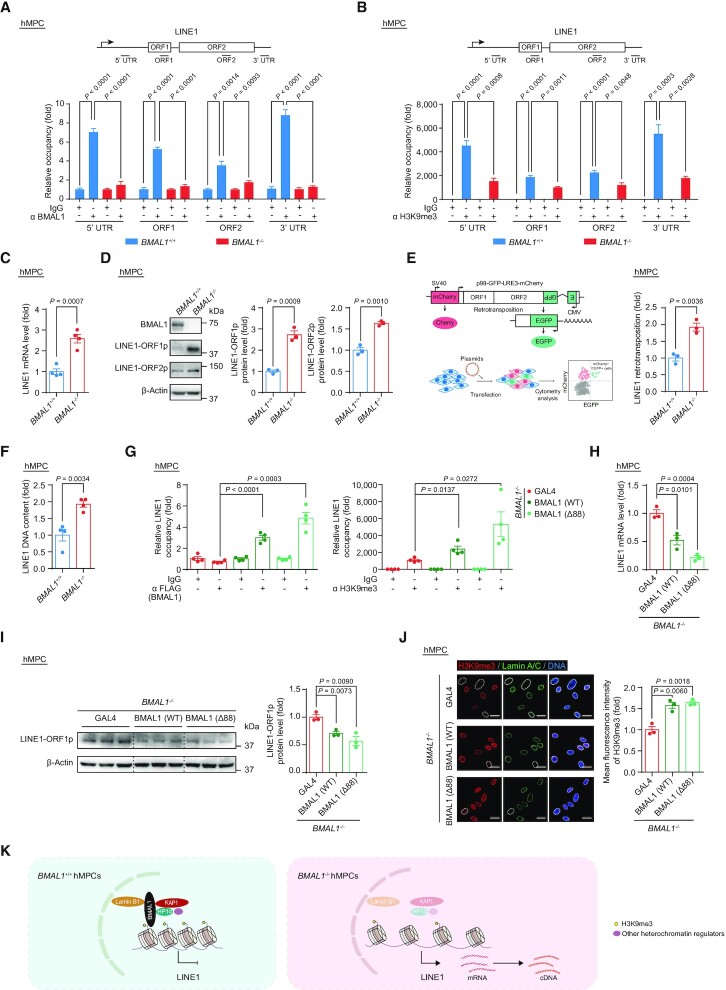
Next, we sought to investigate whether BMAL1 maintains the repressive state at LINE1 elements by binding to the genomic regions of LINE1. Using ChIP-qPCR assays, we confirmed that BMAL1 was associated with LINE1 regions (Figure 4A). Consistently, loss of BMAL1 occupancy led to weakened H3K9me3 intensity at LINE1 regions (Figure 4B). Given that all evidence points to a decompaction of LINE1 in BMAL1–/– hMPCs, we evaluated the activation of LINE1 elements at both RNA and protein levels. As expected, the levels of LINE1 transcripts and LINE1 ORF-encoded proteins were markedly upregulated in BMAL1–/– hMPCs (Figure 4C and D). Because aberrant LINE1 expression is linked to an elevated retrotransposon activity (49,50), we next set out to detect the de novo LINE1 retrotransposition events in both BMAL1+/+ and BMAL1–/– hMPCs using a previously reported LINE1 reporter system (49,50,76). As expected, BMAL1-deficient hMPCs showed a higher frequency of LINE1 retrotransposition (Figure 4E). Consistently, DNA contents of LINE1 were increased in BMAL1–/– hMPCs compared to that of the wild-type counterparts (Figure 4F).
Figure 4.
(A) ChIP-qPCR detection of BMAL1 enrichment at indicated regions of LINE1 in EP (P4) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 4 wells. Data shown are representative of two independent experiments. Two-tailed unpaired Student’s t-test. (B) ChIP-qPCR detection of H3K9me3 enrichment at indicated regions of LINE1 in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 4 wells. Data shown are representative of two independent experiments. Two-tailed unpaired Student’s t-test. (C) RT-qPCR detection of LINE1 mRNA levels in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 4 biological replicates. ACTB was used as the reference gene. Two-tailed unpaired Student’s t-test. (D) Western blot detection of LINE1-ORF1p and LINE1-ORF2p in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. β-Actin was used as the loading control. Quantitative data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (E) Quantification of the de novo LINE1 retrotransposition events in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Left, schematic diagram of the de novo LINE1 retrotransposition assay. Right, quantification of the de novo LINE1 retrotransposition events in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (F) qPCR detection of LINE1 DNA contents in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 4 biological replicates. 5S rDNA was used as the reference gene. Two-tailed unpaired Student’s t-test. (G) ChIP-qPCR detection of BMAL1 and H3K9me3 at 3’UTR region of LINE1 sequences. Left, ChIP-qPCR detection of enrichment of FLAG-tagged BMAL1 at 3’UTR region of LINE1 sequences in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1(WT) or BMAL1 (Δ88) at P2 post transduction. Right, ChIP-qPCR detection of H3K9me3 enrichment at 3’UTR region of LINE1 sequences in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1(WT) or BMAL1 (Δ88) at P2 post transduction. Data are presented as the means ± SEM. n = 4 wells. Data shown are representative of two independent experiments. Two-tailed unpaired Student’s t-test. (H) RT-qPCR detection of LINE1 mRNA levels in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1 (WT) or BMAL1 (Δ88) at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates. ACTB was used as the reference gene. Two-tailed unpaired Student’s t-test. (I) Western blot detection of LINE1-ORF1p levels in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1 (WT) or BMAL1 (Δ88) at P2 post transduction. β-Actin was used as the loading control. Data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (J) Immunofluorescence staining of H3K9me3 and Lamin A/C in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1 (WT) or BMAL1 (Δ88) at P2 post transduction. Dashed lines indicate the nuclear boundaries of the cells with decreased H3K9me3 signals. Quantification of mean fluorescence intensity of H3K9me3 are presented as the means ± SEM. n = 3 biological replicates with each replicate containing 100 cells; scale bars: 25 μm. Two-tailed unpaired Student’s t-test. (K) Diagram showing that BMAL1, together with KAP1, HP1α, Lamin B1 and other heterochromatin regulators, acts as a complex that binds to LINE1 regions and represses its activation.
Although the BMAL1 Δ88 mutant failed to initiate transcription of its target genes (Figure 1L), it retained the ability to form complexes with heterochromatin-associated and nuclear lamina proteins (Figure 2D). Ectopically expressed BMAL1 (WT) and BMAL1 (Δ88) in BMAL1–/– hMPCs restored its occupancy in LINE1 regions and effectively reestablished H3K9me3 domains (Figure 4G), thereby repressing LINE1 activation (Figure 4H and I). Furthermore, by introducing BMAL1 or BMAL1 Δ88 mutant, heterochromatin was reestablished (Figure 4J), and cellular senescence was alleviated in BMAL1–/– hMPCs (Figure 1M, N and Supplementary Figure S2G). Taken together, these results supported a noncanonical role of BMAL1 in stabilizing heterochromatin and silencing LINE1 (Figure 4K).
Derepression of LINE1 induces interferon response and SASP in BMAL1–/– hMPCs
Mounting evidence suggests that intermediates of activated LINE1 transposition, i.e. cytosolic LINE1 cDNA, can be recognized by cGAS, thus triggering a type I interferon response and senescence-associated secretory phenotype (SASP) (49,110,111). We observed that BMAL1 deficiency led to an elevated level of cytosolic LINE1 cDNA that can be immunoprecipitated with cGAS (Figure 5A). Also, upregulation of 2'3'-cGAMP, a cGAS downstream nucleotide messenger, and activation of the interferon response and SASP were all features of BMAL1–/– hMPCs (Figure 5B–D); whereas either expression of BMAL1 or overexpression of heterochromatin modulators, i.e. KAP1 and Lamin B1, restrained the aberrant transcription of LINE1 and thus downregulated the activated interferon response and SASP-associated genes caused by BMAL1 deficiency (Figure 5E). In parallel, treatment with the reverse-transcriptase inhibitor lamivudine, which has been reported to inhibit the synthesis of LINE1 cDNA (49,112), effectively attenuated the LINE1 retrotransposition (Figure 5F and G), interferon response and SASP (Figure 5H) caused by BMAL1 deficiency. It is worth noting that the restraint of LINE1 by lamivudine treatment can also counteract cellular senescence in BMAL1–/– hMPCs, as evidenced by decreased SA-β-gal positive cells (Figure 5I) and increased cellular proliferative ability (Figure 5J and K). Moreover, ablation of cGAS signaling in BMAL1–/– hMPCs by silencing STING (Figure 5L) contributed to decreased transcriptional levels of interferon response and SASP-associated factors, such as IFNA, MMP15 and MMP24 (Figure 5M) and alleviated hMPC senescence phenotypes (Figure 5N–P). These data substantiated that BMAL1 deficiency promotes cellular senescence via an intensified interferon response and SASP caused by derepressed LINE1 elements (Figure 5Q).
Figure 5.
(A) Co-immunoprecipitation of cGAS with cytoplasmic LINE1 cDNA in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Left, schematic diagram of the ChIP-qPCR strategy for measuring the level of cGAS-immunoprecipitated cytoplasmic LINE1 cDNA. Right, quantitative data of the levels of cytoplasmic LINE1 cDNA immunoprecipitated by cGAS in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 4 wells. Data shown are representative of two independent experiments. Two-tailed unpaired Student’s t-test. (B) ELISA detection of 2’3’-cGAMP levels in culture medium of LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (C) Heatmap showing the mRNA levels of indicated genes in LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the mean values of three biological replicates. The color key from gray to red indicates low to high of the relative expression level. ACTB was used as the reference gene. (D) ELISA detection of IFN-α levels in culture medium of LP (P9) BMAL1+/+ and BMAL1–/– hMPCs. Data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (E) Heatmap showing the mRNA levels of indicated genes in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing GAL4, BMAL1, KAP1 and Lamin B1 at P2 post transduction. Data are presented as the mean values of three biological replicates. The color key from gray to blue indicates high to low of the relative expression level. ACTB was used as the reference gene. (F) Quantification of the de novo LINE1 retrotransposition events in BMAL1–/– hMPCs (P7) treated with vehicle or lamivudine at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates. Two-tailed unpaired Student’s t-test. (G) qPCR detection of LINE1 DNA contents in BMAL1–/– hMPCs (P7) treated with vehicle or lamivudine at P2 post treatment. Data are presented as the means ± SEM. n = 4 wells. Data shown are representative of two independent experiments. 5S rDNA was used as the reference gene. Two-tailed unpaired Student’s t-test. (H) Heatmap showing the mRNA levels of indicated genes in BMAL1–/– hMPCs (P7) treated with vehicle or lamivudine at P2 post treatment. Data are presented as the mean values of four wells. The color key from gray to blue indicates high to low of the relative expression level. ACTB was used as the reference gene. (I) SA-β-gal staining in BMAL1–/– hMPCs (P7) treated with vehicle or lamivudine at P2 post treatment. Data are presented as the means ± SEM. n = 3 biological replicates; scale bars: 100 μm. Two-tailed unpaired Student’s t-test. (J) Clonal expansion assay of BMAL1–/– hMPCs (P7) treated with vehicle or lamivudine at P2 post treatment. Data are presented as the means ± SEM. n = 3 biological replicates; Two-tailed unpaired Student’s t-test. (K) Immunofluorescence staining of Ki67 in BMAL1–/– hMPCs (P7) treated with vehicle or lamivudine at P2 post treatment. Data are presented as the means ± SEM. n = 3 biological replicates with each replicate containing > 100 cells; scale bars: 50 μm. Two-tailed unpaired Student’s t-test. (L) Western blot analysis of STING levels in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing sh-GL2 and sh-STING at P2 post transduction. β-Actin was used as the loading control. (M) Heatmap showing the mRNA levels of indicated genes in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing sh-GL2 and sh-STING at P2 post transduction. Data are presented as the mean values of 3 biological replicates. The color key from gray to blue indicates high to low of the relative expression level. ACTB was used as the reference gene. (N) SA-β-gal staining in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing sh-GL2 and sh-STING at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates; scale bars: 100 μm. Two-tailed unpaired Student’s t-test. (O) Clonal expansion assay of EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing sh-GL2 and sh-STING at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates; two-tailed unpaired Student’s t-test. (P) Immunofluorescence staining of Ki67 in EP (P4) BMAL1–/– hMPCs transduced with lentiviruses expressing sh-GL2 and sh-STING at P2 post transduction. Data are presented as the means ± SEM. n = 3 biological replicates with each replicate containing > 100 cells; scale bars: 50 μm. Two-tailed unpaired Student’s t-test. (Q) Schematic diagram showing how LINE1 triggers the activation of cGAS-STING pathway and interferon and SASP response in BMAL1–/– hMPCs. Lamivudine treatment or STING knockdown in BMAL1–/– hMPCs blocks the activation of cGAS-STING pathway and SASP response induced by LINE1.
BMAL1 association with LINE1 is impaired during cellular senescence
To further dissect BMAL1 involvement in regulating hMPC senescence, we performed chromatin fractionation assay in EP and LP hMPCs. We detected a decreased BMAL1 occupancy in the chromatin fraction (Supplementary Figure S4A), which suggested that BMAL1 is less capable of binding to chromatin in LP hMPCs. ChIP-qPCR further confirmed the compromised binding of BMAL1 at LINE1 regions, accompanied by decreased H3K9me3 intensity during replicative senescence (Supplementary Figure S4B and C). In line with the data showing a loss of heterochromatin or BMAL1 occupancy at LINE1s, we observed increased expression levels of both LINE1 mRNA and ORF1p and ORF2p proteins in LP hMPCs (Supplementary Figure S4D and E). In addition, LINE1 DNA contents were upregulated in replicatively senescent hMPCs (Supplementary Figure S4F), indicating increased retrotransposition activity.
LINE1 is activated in BMAL1–/– cynomolgus monkey stem cells and tissues
We next evaluated whether the non-canonical role we had discovered for BMAL1 might be conserved in non-human primate models. As we saw in senescent hMPCs, the nuclear content of BMAL1 was also decreased with replicative senescence in primary MPCs derived from cynomolgus monkey (cynMPCs) (Figure 6A and Supplementary Figure S4G–I). Senescent cynMPCs also showed a decrease in BMAL1 and H3K9me3 occupancy at LINE1 regions (Supplementary Figure S4J), accompanied by decreased H3K9me3 intensity (Supplementary Figure S4K). Concurrently, LINE1 transcription and translation levels were elevated (Supplementary Figure S4L and M), accompanied by an increase in LINE1 DNA contents in LP cynMPCs (Supplementary Figure S4N). Altogether, these data indicated that BMAL1 occupancy is reduced at LINE1 regions during senescence in hMPCs and cynMPCs, suggesting a conserved role of BMAL1 in counteracting excessive LINE1 activation in primate aging.
Figure 6.
(A) Schematic diagram illustrating the isolation and culture of EP (P5) and LP (P13) BMAL1+/+ cynMPCs and western blot analysis of BMAL1 in EP (P5) and LP (P13) BMAL1+/+ cynMPCs. β-Tubulin and NUP358 were used as the loading control for the cytoplasmic and nuclear fractions, respectively. Quantitative data are presented as the means ± SEM. n = 3 independent experiments. Two-tailed unpaired Student’s t-test. (B) Western blot analysis of BMAL1 in BMAL1+/+ and BMAL1–/– cynMPCs (P3). β-Tubulin and NUP358 were used as the loading control for the cytoplasmic and nuclear fractions, respectively. (C) Clonal expansion assay of BMAL1+/+ and BMAL1–/– cynMPCs (P5). Data are presented as the means ± SEM. n = 3 independently cultured wells. Two-tailed unpaired Student’s t-test. (D) Immunofluorescence staining of Ki67 in BMAL1+/+ and BMAL1–/– cynMPCs (P5). Data are presented as the means ± SEM. n = 3 replicates from independently cultured wells with each replicate containing > 100 cells; scale bars: 50 μm. Two-tailed unpaired Student’s t-test. (E) SA-β-gal staining in BMAL1+/+ and BMAL1–/– cynMPCs (P5). Data are presented as the means ± SEM. n = 3 independently cultured wells; scale bars: 100 μm. Two-tailed unpaired Student’s t-test. (F) Immunofluorescence staining of 53BP1 and γH2AX in BMAL1+/+ and BMAL1–/– cynMPCs (P5). Dashed lines indicate the nuclear boundaries of the 53BP1 and γH2AX double-positive (> 2 dots) cells. Data are presented as the means ± SEM. n = 3 replicates from independently cultured wells with each replicate containing > 100 cells; scale bars: 25 μm. Two-tailed unpaired Student’s t-test. (G) Co-IP analysis of KAP1, Lamin B1 and HP1α with endogenous BMAL1 protein in BMAL1+/+ cynMPCs (P5). (H) Western blot analysis of the protein levels of heterochromatin associated proteins and nuclear lamina protein Lamin B1 in BMAL1+/+ and BMAL1–/– cynMPCs (P5). β-Actin was used as the loading control. Quantification of protein levels are presented on the right as the means ± SEM. n = 3 independently cultured wells. Two-tailed unpaired Student’s t-test. (I) 3D reconstruction of z-stack immunofluorescence images of H3K9me3 (red) and Lamin A/C (green) in BMAL1+/+ and BMAL1–/– cynMPCs (P5); scale bars: 2 μm. Quantitative data of the mean nuclear size (top) and the mean fluorescence intensity of H3K9me3 (bottom) of BMAL1+/+ and BMAL1–/– cynMPCs are presented as the means ± SEM. n = 3 replicates from independently cultured wells with each replicate containing 100 cells. Two-tailed unpaired Student’s t-test. Related to Supplementary Figure S5E. (J) Transmission Electron microscopy (TEM) analysis of the heterochromatin architecture at the nuclear periphery in BMAL1+/+ and BMAL1–/– cynMPCs (P5). The percentages of cells with compromised nuclear peripheral heterochromatin are presented at the bottom of each TEM image; scale bars: 2 μm. (K) Ring plot showing the count of ATAC peaks in BMAL1+/+ and BMAL1–/– cynMPCs (P5). (L) Ring plot showing the count of ATAC peaks in indicated repetitive elements in BMAL1+/+ and BMAL1–/– cynMPCs (P5). (M) Metaplot showing the ATAC signals (RPKM) ranging from 3 kb upstream to 3 kb downstream of LINE1-localized ATAC peak centers in BMAL1+/+ and BMAL1–/– cynMPCs (P5). (N) Volcano plot showing the differentially expressed LINE1 elements in BMAL1–/– cynMPCs compared to BMAL1+/+ cynMPCs (P5). (O) Western blot detection of LINE1-ORF1p and LINE1-ORF2p in BMAL1+/+ and BMAL1–/– cynMPCs (P5). β-Actin was used as the loading control. Quantitative data are presented as the means ± SEM. n = 3 independently cultured wells. Two-tailed unpaired Student’s t-test. (P) qPCR detection of LINE1 DNA contents in BMAL1+/+ and BMAL1–/– cynMPCs (P5). Data are presented as the means ± SEM. n = 3 independently cultured wells. 5S rDNA was used as the reference gene. Two-tailed unpaired Student’s t-test. (Q) ELISA detection of 2’3’-cGAMP levels in culture medium of BMAL1+/+ and BMAL1–/– cynMPCs (P5). Data are presented as the means ± SEM. n = 3 independently cultured wells. Two-tailed unpaired Student’s t-test. (R) RT-qPCR detection of MMP9 mRNA levels in BMAL1+/+ and BMAL1–/– cynMPCs (P5). Data are presented as the means ± SEM. n = 4 wells. ACTB was used as the reference gene. Data shown are representative of two independent experiments. Two-tailed unpaired Student’s t-test. (S) RT-qPCR detection of IFNE mRNA levels in BMAL1+/+ and BMAL1–/– cynMPCs (P5). Data are presented as the means ± SEM. n = 4 wells. ACTB was used as the reference gene. Data shown are representative of two independent experiments. Two-tailed unpaired Student’s t-test.
To further investigate the mechanism of BMAL1’s geroprotective role in primates, we used CRISPR/Cas9-mediated gene editing to generate BMAL1–/– cynomolgus monkey, from which we derived primary cynMPCs lacking BMAL1 (Figure 6B and Supplementary Figure S5A–C). As we had detected in hMPCs, NR1D2 oscillations were compromised, and NR1D1, PER1 and PER3 amplitudes dampened in BMAL1–/– cynMPCs relative to WT cynMPCs, confirming the disrupted classic function of BMAL1 in the non-human primate model (Supplementary Figure S5D). Further analysis of cynMPCs derived from BMAL1–/– monkey revealed phenotypes congruent with those we had observed in BMAL1–/– hMPCs, such as impaired clonal expansion ability (Figure 6C), decreased percentage of Ki67-positive cells (Figure 6D), increased percentage of SA-β-gal-positive cells (Figure 6E), and an elevated DNA damage response (Figure 6F). Hence, BMAL1 deficiency led to accelerated senescence in cynMPCs.
Next, we asked whether BMAL1’s geroprotective function in monkey MPCs relied on its ability to maintain heterochromatin and to repress LINE1, as we demonstrated in hMPCs. Co-IP analysis verified that BMAL1 forms a similar complex with KAP1, HP1α and Lamin B1 in cynMPCs (Figure 6G). Consequently, BMAL1–/– cynMPCs phenocopied BMAL1–/– hMPCs, demonstrating downregulation of heterochromatin-associated proteins and nuclear lamina protein (Figure 6H), concomitant with enlarged nuclear size (Figure 6I and Supplementary Figure S5E), disorganized heterochromatin (Figure 6I, J and Supplementary Figure S5E) and increased chromatin accessibility (Figure 6K, L and Supplementary Figure S5F), especially in LINE1 regions (Figure 6M). Most importantly, LINE1 activation manifests as dramatically upregulated transcription (Figure 6N and Supplementary Figure S5G), protein levels (Figure 6O) and elevated DNA contents (Figure 6P) in BMAL1–/– cynMPCs, similar to the observations in LP cynMPCs. Furthermore, activation of the cGAS-STING pathway and transcription induction of interferon response and SASP factors were observed in BMAL1–/– cynMPCs (Figure 6Q–S). In summary, these results indicated that BMAL1 regulates monkey MPC senescence by stabilizing heterochromatin architecture in LINE1 regions, analogous to its function in human stem cells.
Ultimately, we sought to understand whether the BMAL1-deficiency induced senescence we had characterized in cell models also extended to affect tissues. To this end, we collected skin and muscle biopsies from BMAL1+/+ and BMAL1–/– cynomolgus monkeys to evaluate their heterochromatin states, LINE1 expression levels and aging defects. In both skin and muscle tissues from BMAL1–/– cynomolgus monkey, we found decreased H3K9me3 intensity (Supplementary Figure S6A and B). Moreover, through RNA-seq analysis, we discovered that LINE1 elements are concordantly upregulated in both monkey tissues and MPCs (Figure 6N and Supplementary Figure S6C and D). Similar to the activated interferon response and SASP observed in aged hMPCs, genes highly correlated with an activated inflammatory response and aging were dramatically elevated in monkey MPCs, skin, and muscle tissues with BMAL1 deficiency (Supplementary Figure S6E). Correspondingly, p21, the molecular marker for aging, and 53BP1, the DNA damage marker, were both increased (Supplementary Figure S6F–I). Furthermore, skin thickness (Supplementary Figure S6J) and cross-sectional area (CSA) of muscle fibers (Supplementary Figure S6K) were decreased, indicating tissue degeneration of BMAL1–/– cynomolgus monkey (113–116). Taken together, using human and cynomolgus monkey mesenchymal progenitor cell models and tissues, we document a cell-autonomous role of BMAL1 in repressing LINE1 by stabilizing heterochromatin, thereby modulating senescence.
DISCUSSION
Aging is known to be correlated with dampened oscillation of clock gene expression (32,117). However, the impact of circadian protein deregulation in cellular senescence in primates is virtually unknown. In the past few decades, BMAL1 has become well-known as a transcription factor regulating metabolism, mitochondrial biogenesis, bone development and so on (29,118,119). Here, we discovered a transcription-independent role for BMAL1 in stabilizing heterochromatin and protecting hMPCs against senescence by repressing LINE1. Using CRISPR/Cas9-mediated gene editing in combination with directed differentiation, we generated BMAL1-deficient hESCs and their hMPC derivatives, providing valuable isogenic models to investigate the roles of BMAL1 in regulating human stem cell homeostasis. BMAL1 deficiencies in both hMPCs and cynMPCs led to the loss of perinuclear heterochromatin and simultaneously accelerated cellular senescence. Further mechanical analysis revealed that BMAL1 forms complexes with Lamin B1 and heterochromatin-associated proteins KAP1 and HP1α to stabilize heterochromatin. Using the DamID assay in BMAL1-deficient hMPCs to track the maintenance of LADs, the gene-poor regions of chromosomes that interact with the nuclear lamina (107), combined with H3K9me3 ChIP-seq and ATAC-seq, we further demonstrated a profound heterochromatin dysregulation, especially the derepression of LINE1. By evaluating a BMAL1 mutant lacking the transactivation domain (TAD), we demonstrated that the ability of BMAL1 to stabilize heterochromatin did not reside with its transcriptional activity. The exclusion of BMAL1 from the nucleus during hMPC aging decreased the capacity of BMAL1 binding to LINE1 regions, which led to the activation of LINE1 and ultimately cellular senescence. More importantly, we showed that BMAL1’s moonlighting role as a gatekeeper of LINE1 derepression is conserved in primates, emphasizing the BMAL1-heterochromatin axis as a potential mechanism for attenuating primate aging.
From an epigenomic angle, we found decreased interaction between the nuclear lamina and LADs, loss of the constitutive heterochromatic mark H3K9me3 and increased chromatin accessibility in BMAL1–/– hMPCs, which coincided with previous observations in other senescent cell models (44,45,104,120–123). Mechanistically, we demonstrated that BMAL1 forms complexes with nuclear lamina components and heterochromatin-associated proteins to maintain heterochromatin stability. Mutation or reduction of heterochromatin components such as HP1 or defection in nuclear lamina have been identified as the major risk for aging-associated heterochromatin structure loss (42,44,104,124–128). Consistently, decreased protein levels of heterochromatin components KAP1, HP1α and nuclear lamina protein Lamin B1 accompanied by heterochromatin loss were observed in BMAL1 deficient hMPCs and cynMPCs. Moreover, ectopically expressed heterochromatin modulators KAP1 and Lamin B1 counteracted the H3K9me3 loss and rescued cellular senescence phenotypes in BMAL1–/– hMPCs, further confirming that compromised heterochromatin is the mediator of BMAL1-deficiency induced cellular senescence. Meanwhile, previous studies have also identified several important regulators of heterochromatin in senescence, such as Rb, SIRT6, SIRT7 and ZKSCAN3 (16,48,49,62,64,125,129). Whether BMAL1 stabilizes heterochromatin through collaboration with other regulators remains to be further investigated.
Repetitive elements, especially LINE1 elements, are enriched at and safeguarded by heterochromatic regions (107,129). In this study, we delineated a cell-autonomous role of BMAL1 in stabilizing heterochromatin, repressing LINE1 therein, and thus counteracting senescence. This is in line with previous reports that > 10% of BMAL1 binding sites were mapped within TEs by ChIP-seq in mouse liver (130,131), while its overall binding to chromatin in the same tissue was dampened with age (117). Similarly, we found decreased BMAL1 occupancy in the chromatin fraction, especially at LINE1 regions, leading to the activation of LINE1 during senescence in various cells in primates. The role of BMAL1 in regulating repetitive elements was also supported by a recent study that suggested the involvement of BMAL1 in safeguarding telomeric heterochromatin (132). Collectively, these studies demonstrated the noncanonical and cell-autonomous function of core circadian proteins in regulating chromatin homeostasis.
As a result of heterochromatin loss, activated LINE1 elements have been proven to be, to a considerable extent, the cause of genomic instability and inflammation, thereby accelerating senescence (50,129,133). Consistently, we found that derepression of LINE1 contributes to the activation of the cGAS-STING pathway and SASP in BMAL1–/– hMPCs. In support of this mechanism, blocking the LINE1-cGAS-STING-SASP cascade, either through treatment with a reverse-transcriptase inhibitor or by silencing the expression of STING, rejuvenated senescent hMPCs with BMAL1 deficiency. We revealed that BMAL1 antagonizes primate stem cell senescence by stabilizing heterochromatin to suppress the LINE1-cGAS-STING-SASP axis. Our study, along with previous research, emphasizes that stabilizing heterochromatin-associated proteins and nuclear lamina proteins or inhibiting the cGAS-STING pathway are mechanisms that counteract cellular senescence (48,49,64,129,133). In conclusion, for the first time, our findings provide clear evidence and underlying mechanism for the role of BMAL1 in regulating cellular senescence in primates. Although our conclusion is to a certain extent limited by the number of non-human primate individuals available in this study, we found additional evidence in a recent work with another BMAL1 deficient cynomolgus monkey model generated by different sgRNAs that also exhibited an upregulation of inflammation- and aging-associated genes in the blood sample (33). Yet future investigations with a larger cohort of BMAL1 deficient individuals are needed to further confirm the role of BMAL1 in regulating primate aging. The moonlighting function of BMAL1, independent of the canonical circadian machinery, may reflect the cell-autonomous regulation during cellular aging. Moreover, lamivudine supplementation prevented cellular senescence triggered by BMAL1 deficiency, which indicates the possibility of identifying other compounds for the treatment of aging-associated disorders in the future.
DATA AVAILABILITY
The sequencing data obtained in this study have been deposited in the Genome Sequence Archive (134) in the National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation) of the Chinese Academy of Sciences, under accession number HRA001460 and CRA005216 that are publicly accessible at http://bigd.big.ac.cn/gsa-human or http://bigd.big.ac.cn/gsa. Data of BMAL1 interacting candidates that identified by LC-MS/MS data have been uploaded in the ProteomeXchange under the dataset identifier PXD030570. The differentially expressed genes identified in tissues derived from BMAL1–/– cynomolgus monkey were uploaded to the Aging Atlas (AA, https://ngdc.cncb.ac.cn/aging/index) database (135).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Anqi Hu for kind help in BMAL1 deficiency cynomolgus monkey associated experiments; Jingyi Jia for management of laboratory animals; Lei Bai, Ruijun Bai, Jing Lu, Luyang Tian, Ying Yang, Qun Chu, Shikun Ma and Xiangmei Jin for administrative assistance; Yandong Zheng for assisting image organization. Junying Jia from the Institute of Biophysics, Chinese Academy of Sciences for performing the fluorescence-activated cell sorting; Jifeng Wang from the Institute of Biophysics, Chinese Academy of Sciences for the help in liquid chromatography-tandem mass spectrometry (LC-MS/MS) experiment. We thank all members from the Liu lab for their helpful discussion and technical assistance.
Contributor Information
Chuqian Liang, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Qiong Ke, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, Guangdong 510080, China; Department of Genetics and Cell Biology, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, Guangdong 510080, China.
Zunpeng Liu, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jie Ren, Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China.
Weiqi Zhang, Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China.
Jianli Hu, CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Zehua Wang, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Hong Chen, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, Guangdong 510080, China.
Kai Xia, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, Guangdong 510080, China.
Xingqiang Lai, Cardiovascular Department, The Eighth Affiliated Hospital, Sun Yat-sen University, Shenzhen, Guangdong 518033, China.
Qiaoran Wang, CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Kuan Yang, CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China.
Wei Li, Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; Aging Translational Medicine Center, International Center for Aging and Cancer, Xuanwu Hospital, Capital Medical University, Beijing 100053, China.
Zeming Wu, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Chao Wang, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, Guangdong 510080, China; Department of Genetics and Cell Biology, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, Guangdong 510080, China.
Haoteng Yan, Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; Aging Translational Medicine Center, International Center for Aging and Cancer, Xuanwu Hospital, Capital Medical University, Beijing 100053, China.
Xiaoyu Jiang, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Zhejun Ji, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Miyang Ma, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Xiao Long, Division of Plastic Surgery, Peking Union Medical College Hospital, Beijing 100032, China.
Si Wang, Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; Aging Translational Medicine Center, International Center for Aging and Cancer, Xuanwu Hospital, Capital Medical University, Beijing 100053, China; Chongqing Renji Hospital, University of Chinese Academy of Sciences, Chongqing 400062, China.
Huating Wang, Department of Orthopaedics and Traumatology, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong 999077, China.
Hao Sun, Department of Chemical Pathology, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong 999077, China.
Juan Carlos Izpisua Belmonte, Gene Expression Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, USA.
Jing Qu, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Andy Peng Xiang, Center for Stem Cell Biology and Tissue Engineering, Key Laboratory for Stem Cells and Tissue Engineering, Ministry of Education, Sun Yat-Sen University, Guangzhou, Guangdong 510080, China; Department of Biochemistry, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, Guangdong 510080, China.
Guang-Hui Liu, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China; Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; Aging Translational Medicine Center, International Center for Aging and Cancer, Xuanwu Hospital, Capital Medical University, Beijing 100053, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2020YFA0804000, 2018YFA0107203]; Strategic Priority Research Program ofthe Chinese Academy of Sciences [XDA16000000]; National Natural Science Foundation of China [81861168034, 81921006, 81625009, 91949209, 92049304, 81822018, 82071588, 92049116, 82125011, 82122024, 32100937, 92149301, 92168201]; National Key Research and Development Program of China [2018YFC2000100, 2020YFA0112200]; Key Research Program of the Chinese Academy of Sciences [KFZD-SW-221]; Program of the Beijing Natural Science Foundation [Z190019]; K.C. Wong Education Foundation [GJTD-2019-06, GJTD-2019-08]; Key-Area Research and Development Program of Guangdong Province [2019B020235002]; Youth Innovation Promotion Association of CAS [2021078, E1CAZW0401]; Informatization Plan of Chinese Academy of Sciences [WX145XQ07-18; CAS-WX2021SF-0301]; State Key Laboratory of Stem Cell and Reproductive Biology; State Key Laboratory of Membrane Biology; Tencent Foundation [2021-1045]; Milky Way Research Foundation (MWRF). Funding for open access charge: National Key Research and Development Program of China [2020YFA0804000].
Conflict of Interest. The authors declare no competing interests.
REFERENCES
- 1. Reinke H., Asher G.. Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 2019; 20:227–241. [DOI] [PubMed] [Google Scholar]
- 2. Dierickx P., Van Laake L.W., Geijsen N.. Circadian clocks: from stem cells to tissue homeostasis and regeneration. EMBO Rep. 2018; 19:18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kinouchi K., Mikami Y., Kanai T., Itoh H.. Circadian rhythms in the tissue-specificity from metabolism to immunity; insights from omics studies. Mol. Aspects Med. 2021; 80:100984. [DOI] [PubMed] [Google Scholar]
- 4. Shao S., Zhao H., Lu Z., Lei X., Zhang Y.. Circadian rhythms within the female HPG axis: from physiology to etiology. Endocrinology. 2021; 162:bqab117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Musiek E.S., Holtzman D.M.. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science. 2016; 354:1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bechtold D.A., Gibbs J.E., Loudon A.S.. Circadian dysfunction in disease. Trends Pharmacol. Sci. 2010; 31:191–198. [DOI] [PubMed] [Google Scholar]
- 7. Hood S., Amir S.. The aging clock: circadian rhythms and later life. J. Clin. Invest. 2017; 127:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dang F., Wu R., Wang P., Wu Y., Azam M.S., Xu Q., Chen Y., Liu Y.. Fasting and feeding signals control the oscillatory expression of angptl8 to modulate lipid metabolism. Sci. Rep. 2016; 6:36926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu R., Dang F., Li P., Wang P., Xu Q., Liu Z., Li Y., Wu Y., Chen Y., Liu Y.. The circadian protein period2 suppresses mTORC1 activity via recruiting tsc1 to mTORC1 complex. Cell Metab. 2019; 29:653–667. [DOI] [PubMed] [Google Scholar]
- 10. Mattis J., Sehgal A.. Circadian rhythms, sleep, and disorders of aging. Trends Endocrinol. Metab. 2016; 27:192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofman M.A., Swaab D.F.. Living by the clock: the circadian pacemaker in older people. Ageing Res. Rev. 2006; 5:33–51. [DOI] [PubMed] [Google Scholar]
- 12. Chen C.Y., Logan R.W., Ma T., Lewis D.A., Tseng G.C., Sibille E., McClung C.A.. Effects of aging on circadian patterns of gene expression in the human prefrontal cortex. Proc. Natl. Acad Sci. USA. 2016; 113:206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakamura T.J., Nakamura W., Tokuda I.T., Ishikawa T., Kudo T., Colwell C.S., Block G.D.. Age-related changes in the circadian system unmasked by constant conditions. Eneuro. 2015; 2:ENEURO.0064-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakamura T.J., Nakamura W., Yamazaki S., Kudo T., Cutler T., Colwell C.S., Block G.D.. Age-related decline in circadian output. J. Neurosci. 2011; 31:10201–10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmed R., Ashimori A., Iwamoto S., Matsui T., Nakahata Y., Bessho Y.. Replicative senescent human cells possess altered circadian clocks with a prolonged period and delayed peak-time. Aging. 2019; 11:950–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liang C., Liu Z., Song M., Li W., Wu Z., Wang Z., Wang Q., Wang S., Yan K., Sun L.et al.. Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration. Cell Res. 2021; 31:187–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benitah S.A., Welz P.S.. Circadian regulation of adult stem cell homeostasis and aging. Cell Stem Cell. 2020; 26:817–831. [DOI] [PubMed] [Google Scholar]
- 18. Dubrovsky Y.V., Samsa W.E., Kondratov R.V.. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging. 2010; 2:936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kondratov R.V., Kondratova A.A., Gorbacheva V.Y., Vykhovanets O.V., Antoch M.P.. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006; 20:1868–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bunger M.K., Wilsbacher L.D., Moran S.M., Clendenin C., Radcliffe L.A., Hogenesch J.B., Simon M.C., Takahashi J.S., Bradfield C.A.. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000; 103:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hogenesch J.B., Gu Y.Z., Jain S., Bradfield C.A.. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA. 1998; 95:5474–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin X., Shearman L.P., Weaver D.R., Zylka M.J., de Vries G.J., Reppert S.M.. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999; 96:57–68. [DOI] [PubMed] [Google Scholar]
- 23. Patke A., Young M.W., Axelrod S.. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020; 21:67–84. [DOI] [PubMed] [Google Scholar]
- 24. Ripperger J.A., Schibler U.. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian dbp transcription and chromatin transitions. Nat. Genet. 2006; 38:369–374. [DOI] [PubMed] [Google Scholar]
- 25. Laposky A., Easton A., Dugovic C., Walisser J., Bradfield C., Turek F.. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005; 28:395–409. [DOI] [PubMed] [Google Scholar]
- 26. Hemmeryckx B., Frederix L., Lijnen H.R.. Deficiency of bmal1 disrupts the diurnal rhythm of haemostasis. Exp. Gerontol. 2019; 118:1–8. [DOI] [PubMed] [Google Scholar]
- 27. Engeland W.C., Massman L., Mishra S., Yoder J.M., Leng S., Pignatti E., Piper M.E., Carlone D.L., Breault D.T., Kofuji P.. The adrenal clock prevents aberrant light-induced alterations in circadian glucocorticoid rhythms. Endocrinology. 2018; 159:3950–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma Z., Jin X., Qian Z., Li F., Xu M., Zhang Y., Kang X., Li H., Gao X., Zhao L.et al.. Deletion of clock gene bmal1 impaired the chondrocyte function due to disruption of the HIF1alpha-VEGF signaling pathway. Cell Cycle. 2019; 18:1473–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dudek M., Gossan N., Yang N., Im H.J., Ruckshanthi J.P., Yoshitane H., Li X., Jin D., Wang P., Boudiffa M.et al.. The chondrocyte clock gene bmal1 controls cartilage homeostasis and integrity. J. Clin. Invest. 2016; 126:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gossan N., Boot-Handford R., Meng Q.J.. Ageing and osteoarthritis: a circadian rhythm connection. Biogerontology. 2015; 16:209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakazato R., Kawabe K., Yamada D., Ikeno S., Mieda M., Shimba S., Hinoi E., Yoneda Y., Takarada T.. Disruption of bmal1 impairs blood-brain barrier integrity via pericyte dysfunction. J. Neurosci. 2017; 37:10052–10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chhunchha B., Kubo E., Singh D.P.. Clock protein bmal1 and nrf2 cooperatively control aging or oxidative response and redox homeostasis by regulating rhythmic expression of prdx6. Cells. 2020; 9:1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qiu P., Jiang J., Liu Z., Cai Y., Huang T., Wang Y., Liu Q., Nie Y., Liu F., Cheng J.et al.. BMAL1 knockout macaque monkeys display reduced sleep and psychiatric disorders. Natl. Sci. Rev. 2019; 6:87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mure L.S., Le H.D., Benegiamo G., Chang M.W., Rios L., Jillani N., Ngotho M., Kariuki T., Dkhissi-Benyahya O., Cooper H.M.et al.. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018; 359:eaao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G.. The hallmarks of aging. Cell. 2013; 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee B.C., Yu K.R.. Impact of mesenchymal stem cell senescence on inflammaging. BMB Rep. 2020; 53:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uccelli A., Moretta L., Pistoia V.. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008; 8:726–736. [DOI] [PubMed] [Google Scholar]
- 38. Zhang W., Li J., Suzuki K., Qu J., Wang P., Zhou J., Liu X., Ren R., Xu X., Ocampo A.et al.. Aging stem cells. A werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015; 348:1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu Z., Zhang W., Song M., Wang W., Wei G., Li W., Lei J., Huang Y., Sang Y., Chan P.et al.. Differential stem cell aging kinetics in hutchinson-gilford progeria syndrome and werner syndrome. Protein Cell. 2018; 9:333–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chojnowski A., Ong P.F., Foo M.X.R., Liebl D., Hor L.P., Stewart C.L., Dreesen O.. Heterochromatin loss as a determinant of progerin-induced DNA damage in hutchinson-gilford progeria. Aging Cell. 2020; 19:e13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J.H., Kim E.W., Croteau D.L., Bohr V.A.. Heterochromatin: an epigenetic point of view in aging. Exp. Mol. Med. 2020; 52:1466–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Janssen A., Colmenares S.U., Karpen G.H.. Heterochromatin: guardian of the genome. Annu. Rev. Cell Dev. Biol. 2018; 34:265–288. [DOI] [PubMed] [Google Scholar]
- 43. Chen Y., Geng A., Zhang W., Qian Z., Wan X., Jiang Y., Mao Z.. 2020) Fight to the bitter end: DNA repair and aging. Ageing Res. Rev. 64:101154. [DOI] [PubMed] [Google Scholar]
- 44. Chandra T., Ewels PhilipA., Schoenfelder S., Furlan-Magaril M., Wingett StevenW., Kirschner K., Thuret J.-Y., Andrews S., Fraser P., Reik W.. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015; 10:471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sati S., Bonev B., Szabo Q., Jost D., Bensadoun P., Serra F., Loubiere V., Papadopoulos G.L., Rivera-Mulia J.-C., Fritsch L.et al.. 4D Genome rewiring during oncogene-induced and replicative senescence. Mol. Cell. 2020; 78:522–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. De Cecco M., Criscione S.W., Peckham E.J., Hillenmeyer S., Hamm E.A., Manivannan J., Peterson A.L., Kreiling J.A., Neretti N., Sedivy J.M.. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013; 12:247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peng J.C., Karpen G.H.. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 2007; 9:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deng L., Ren R., Liu Z., Song M., Li J., Wu Z., Ren X., Fu L., Li W., Zhang W.et al.. Stabilizing heterochromatin by DGCR8 alleviates senescence and osteoarthritis. Nat. Commun. 2019; 10:3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bi S., Liu Z., Wu Z., Wang Z., Liu X., Wang S., Ren J., Yao Y., Zhang W., Song M.et al.. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 2020; 11:483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Cecco M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., Caligiana A., Brocculi G., Adney E.M., Boeke J.D.et al.. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019; 566:73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shan H., Geng L., Jiang X., Song M., Wang J., Liu Z., Zhuo X., Wu Z., Hu J., Ji Z.et al.. Large-scale chemical screen identifies gallic acid as a geroprotector for human stem cells. Protein Cell. 2021; 10.1007/s13238-021-00872-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang S., Cheng F., Ji Q., Song M., Wu Z., Zhang Y., Ji Z., Feng H., Belmonte J.C.I., Zhou Q.et al.. Hyperthermia differentially affects specific human stem cells and their differentiated derivatives. Protein Cell. 2021; 10.1007/s13238-021-00887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lei J., Wang S., Kang W., Chu Q., Liu Z., Sun L., Ji Y., Esteban C.R., Yao Y., Belmonte J.C.I.et al.. FOXO3-engineered human mesenchymal progenitor cells efficiently promote cardiac repair after myocardial infarction. Protein Cell. 2021; 12:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li W., Zou Z., Cai Y., Yang K., Wang S., Liu Z., Geng L., Chu Q., Ji Z., Chan P.et al.. Low-dose chloroquine treatment extends the lifespan of aged rats. Protein Cell. 2022; 10.1007/s13238-021-00903-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zou Z., Long X., Zhao Q., Zheng Y., Song M., Ma S., Jing Y., Wang S., He Y., Esteban C.R.et al.. A single-cell transcriptomic atlas of human skin aging. Development. Cell. 2021; 56:383–397. [DOI] [PubMed] [Google Scholar]
- 56. Li J., Hong S., Chen W., Zuo E., Yang H.. Advances in detecting and reducing off-target effects generated by CRISPR-mediated genome editing. J. Genet. Genom. = Yi Chuan Xue Bao. 2019; 46:513–521. [DOI] [PubMed] [Google Scholar]
- 57. Li H., Wu Z., Liu X., Zhang S., Ji Q., Jiang X., Liu Z., Wang S., Qu J., Zhang W.et al.. ALKBH1 deficiency leads to loss of homeostasis in human diploid somatic cells. Protein Cell. 2020; 11:688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chu Q., Liu F., He Y., Jiang X., Cai Y., Wu Z., Yan K., Geng L., Zhang Y., Feng H.et al.. mTORC2/RICTOR exerts differential levels of metabolic control in human embryonic, mesenchymal and neural stem cells. Protein Cell. 2022; 10.1007/s13238-021-00898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang X., Liu Z., Liu X., Wang S., Zhang Y., He X., Sun S., Ma S., Shyh-Chang N., Liu F.et al.. Telomere-dependent and telomere-independent roles of RAP1 in regulating human stem cell homeostasis. Protein Cell. 2019; 10:649–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ling C., Liu Z., Song M., Zhang W., Wang S., Liu X., Ma S., Sun S., Fu L., Chu Q.et al.. Modeling CADASIL vascular pathologies with patient-derived induced pluripotent stem cells. Protein Cell. 2019; 10:249–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fu L., Hu Y., Song M., Liu Z., Zhang W., Yu F.X., Wu J., Wang S., Izpisua Belmonte J.C., Chan P.et al.. Up-regulation of FOXD1 by YAP alleviates senescence and osteoarthritis. PLoS Biol. 2019; 17:e3000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Diao Z., Ji Q., Wu Z., Zhang W., Cai Y., Wang Z., Hu J., Liu Z., Wang Q., Bi S.et al.. 2021) SIRT3 consolidates heterochromatin and counteracts senescence. Nucleic Acids Res. 49:4203–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu Z., Shi Y., Lu M., Song M., Yu Z., Wang J., Wang S., Ren J., Yang Y.G., Liu G.H.et al.. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 2020; 48:11083–11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hu H., Ji Q., Song M., Ren J., Liu Z., Wang Z., Liu X., Yan K., Hu J., Jing Y.et al.. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res. 2020; 48:6001–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pan H., Guan D., Liu X., Li J., Wang L., Wu J., Zhou J., Zhang W., Ren R., Zhang W.et al.. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 2016; 26:190–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang S., Hu B., Ding Z., Dang Y., Wu J., Li D., Liu X., Xiao B., Zhang W., Ren R.et al.. ATF6 safeguards organelle homeostasis and cellular aging in human mesenchymal stem cells. Cell Discov. 2018; 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang J., Li J., Suzuki K., Liu X., Wu J., Zhang W., Ren R., Zhang W., Chan P., Izpisua Belmonte J.C.et al.. Genetic enhancement in cultured human adult stem cells conferred by a single nucleotide recoding. Cell Res. 2017; 27:1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hughes M.E., Hogenesch J.B., Kornacker K.. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhyth. 2010; 25:372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yan P., Li Q., Wang L., Lu P., Suzuki K., Liu Z., Lei J., Li W., He X., Wang S.et al.. FOXO3-Engineered human ESC-Derived vascular cells promote vascular protection and regeneration. Cell Stem Cell. 2019; 24:447–461. [DOI] [PubMed] [Google Scholar]
- 70. Brugiolo M., Botti V., Liu N., Müller-McNicoll M., Neugebauer K.M.. Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. Nucleic Acids Res. 2017; 45:10452–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ruprecht N., Hungerbuhler M.N., Bohm I.B., Heverhagen J.T.. Improved identification of DNA double strand breaks: gamma-H2AX-epitope visualization by confocal microscopy and 3D reconstructed images. Radiat. Environ. Biophys. 2019; 58:295–302. [DOI] [PubMed] [Google Scholar]
- 72. Ren X., Hu B., Song M., Ding Z., Dang Y., Liu Z., Zhang W., Ji Q., Ren R., Ding J.et al.. Maintenance of nucleolar homeostasis by CBX4 alleviates senescence and osteoarthritis. Cell Rep. 2019; 26:3643–3656. [DOI] [PubMed] [Google Scholar]
- 73. Cheng F., Wang S., Song M., Liu Z., Liu P., Wang L., Wang Y., Zhao Q., Yan K., Chan P.et al.. DJ-1 is dispensable for human stem cell homeostasis. Protein Cell. 2019; 10:846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu Z., Li W., Geng L., Sun L., Wang Q., Yu Y., Yan P., Liang C., Ren J., Song M.et al.. Cross-species metabolomic analysis identifies uridine as a potent regeneration promoting factor. Cell Discov. 2022; 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lei J., Jiang X., Li W., Ren J., Wang D., Ji Z., Wu Z., Cheng F., Cai Y., Yu Z.R.et al.. Exosomes from antler stem cells alleviate mesenchymal stem cell senescence and osteoarthritis. Protein Cell. 2021; 10.1007/s13238-021-00860-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Thomas C.A., Tejwani L., Trujillo C.A., Negraes P.D., Herai R.H., Mesci P., Macia A., Crow Y.J., Muotri A.R.. Modeling of TREX1-Dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell. 2017; 21:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ke Q., Li W., Lai X., Chen H., Huang L., Kang Z., Li K., Ren J., Lin X., Zheng H.et al.. TALEN-based generation of a cynomolgus monkey disease model for human microcephaly. Cell Res. 2016; 26:1048–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang F., Zhang W., Yang Q., Kang Y., Fan Y., Wei J., Liu Z., Dai S., Li H., Li Z.et al.. Generation of a hutchinson-gilford progeria syndrome monkey model by base editing. Protein Cell. 2020; 11:809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang H., Li J., Ren J., Sun S., Ma S., Zhang W., Yu Y., Cai Y., Yan K., Li W.et al.. Single-nucleus transcriptomic landscape of primate hippocampal aging. Protein Cell. 2021; 12:695–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang S., Zheng Y., Li Q., He X., Ren R., Zhang W., Song M., Hu H., Liu F., Sun G.et al.. Deciphering primate retinal aging at single-cell resolution. Protein Cell. 2021; 12:889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhang W., Zhang S., Yan P., Ren J., Song M., Li J., Lei J., Pan H., Wang S., Ma X.et al.. A single-cell transcriptomic landscape of primate arterial aging. Nat. Commun. 2020; 11:2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang W., Wan H., Feng G., Qu J., Wang J., Jing Y., Ren R., Liu Z., Zhang L., Chen Z.et al.. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature. 2018; 560:661–665. [DOI] [PubMed] [Google Scholar]
- 83. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L.et al.. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 84. Vogel M.J., Peric-Hupkes D., van Steensel B.. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007; 2:1467–1478. [DOI] [PubMed] [Google Scholar]
- 85. Hu G., Dong X., Gong S., Song Y., Hutchins A.P., Yao H.. Systematic screening of CTCF binding partners identifies that BHLHE40 regulates CTCF genome-wide distribution and long-range chromatin interactions. Nucleic Acids Res. 2020; 48:9606–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Meth. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Anders S., Pyl P.T., Huber W.. HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England). 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019; 10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 91. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Langmead B., Salzberg S.L.. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and b cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W.. A clustering approach for identification of enriched domains from histone modification chip-Seq data. Bioinformatics (Oxford, England). 2009; 25:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hao Z., Lv D., Ge Y., Shi J., Weijers D., Yu G., Chen J.. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Computer Sci. 2020; 6:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of chip-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Xu H., Gustafson C.L., Sammons P.J., Khan S.K., Parsley N.C., Ramanathan C., Lee H.W., Liu A.C., Partch C.L.. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 c terminus. Nat. Struct. Mol. Biol. 2015; 22:476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kiyohara Y.B., Tagao S., Tamanini F., Morita A., Sugisawa Y., Yasuda M., Yamanaka I., Ueda H.R., van der Horst G.T., Kondo T.et al.. The BMAL1 c terminus regulates the circadian transcription feedback loop. Proc. Natl. Acad. Sci. USA. 2006; 103:10074–10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Langmesser S., Tallone T., Bordon A., Rusconi S., Albrecht U.. Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol. Biol. 2008; 9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Park N., Kim H.D., Cheon S., Row H., Lee J., Han D.H., Cho S., Kim K.. A novel bmal1 mutant mouse reveals essential roles of the C-Terminal domain on circadian rhythms. PLoS One. 2015; 10:e0138661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rowe H.M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P.V., Layard-Liesching H., Verp S., Marquis J.et al.. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010; 463:237–240. [DOI] [PubMed] [Google Scholar]
- 104. Sadaie M., Salama R., Carroll T., Tomimatsu K., Chandra T., Young A.R., Narita M., Perez-Mancera P.A., Bennett D.C., Chong H.et al.. Redistribution of the lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Develop. 2013; 27:1800–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Osanai T., Tanaka M., Izumiyama K., Mikami K., Kitajima M., Tomisawa T., Magota K., Tomita H., Okumura K.. Intracellular protons accelerate aging and switch on aging hallmarks in mice. J. Cell Biochem. 2018; 119:9825–9837. [DOI] [PubMed] [Google Scholar]
- 106. Lu J.Y., Chang L., Li T., Wang T., Yin Y., Zhan G., Han X., Zhang K., Tao Y., Percharde M.et al.. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res. 2021; 31:613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. van Steensel B., Belmont A.S.. Lamina-Associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell. 2017; 169:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Cordaux R., Batzer M.A.. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kazazian H.H. Jr, Moran J.V. Mobile DNA in health and disease. New England J. Med. 2017; 377:361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gorbunova V., Seluanov A., Mita P., McKerrow W., Fenyo D., Boeke J.D., Linker S.B., Gage F.H., Kreiling J.A., Petrashen A.P.et al.. The role of retrotransposable elements in ageing and age-associated diseases. Nature. 2021; 596:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Dou Z., Ghosh K., Vizioli M.G., Zhu J., Sen P., Wangensteen K.J., Simithy J., Lan Y., Lin Y., Zhou Z.et al.. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017; 550:402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jones R.B., Garrison K.E., Wong J.C., Duan E.H., Nixon D.F., Ostrowski M.A.. Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PLoS One. 2008; 3:e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Branchet M.C., Boisnic S., Frances C., Robert A.M.. Skin thickness changes in normal aging skin. Gerontology. 1990; 36:28–35. [DOI] [PubMed] [Google Scholar]
- 114. Jeong D., Qomaladewi N.P., Lee J., Park S.H., Cho J.Y.. The role of autophagy in skin fibroblasts, keratinocytes, melanocytes, and epidermal stem cells. J. Invest. Dermat. 2020; 140:1691–1697. [DOI] [PubMed] [Google Scholar]
- 115. Lexell J. Human aging, muscle mass, and fiber type composition. J. Gerontol. Series A, Biol. Sci. Med. Sci. 1995; 11–16.50. [DOI] [PubMed] [Google Scholar]
- 116. Wilkinson D.J., Piasecki M., Atherton P.J.. The age-related loss of skeletal muscle mass and function: measurement and physiology of muscle fibre atrophy and muscle fibre loss in humans. Ageing Res. Rev. 2018; 47:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Levine D.C., Hong H., Weidemann B.J., Ramsey K.M., Affinati A.H., Schmidt M.S., Cedernaes J., Omura C., Braun R., Lee C.et al.. NAD(+) controls circadian reprogramming through PER2 nuclear translocation to counter aging. Mol. Cell. 2020; 78:835–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Li E., Li X., Huang J., Xu C., Liang Q., Ren K., Bai A., Lu C., Qian R., Sun N.. 2020) BMAL1 regulates mitochondrial fission and mitophagy through mitochondrial protein BNIP3 and is critical in the development of dilated cardiomyopathy. Protein Cell. 11:661–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Hand L.E., Dickson S.H., Freemont A.J., Ray D.W., Gibbs J.E.. The circadian regulator bmal1 in joint mesenchymal cells regulates both joint development and inflammatory arthritis. Arthritis Res. Ther. 2019; 21:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lenain C., de Graaf C.A., Pagie L., Visser N.L., de Haas M., de Vries S.S., Peric-Hupkes D., van Steensel B., Peeper D.S.. Massive reshaping of genome-nuclear lamina interactions during oncogene-induced senescence. Genome Res. 2017; 27:1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Narita M. Cellular senescence and chromatin organisation. Brit. J. Cancer. 2007; 96:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Tomimatsu K., Bihary D., Olan I., Parry A.J., Schoenfelder S., Chan A.S.L., Slater G.S.C., Ito Y., Rugg-Gunn P.J., Kirschner K.et al.. Locus-specific induction of gene expression from heterochromatin loci during cellular senescence. Nat. Aging. 2021; 2:31–45. [DOI] [PubMed] [Google Scholar]
- 123. Olan I., Parry A.J., Schoenfelder S., Narita M., Ito Y., Chan A.S.L., Slater G.S.C., Bihary D., Bando M., Shirahige K.et al.. Transcription-dependent cohesin repositioning rewires chromatin loops in cellular senescence. Nat. Commun. 2020; 11:6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Shumaker D.K., Dechat T., Kohlmaier A., Adam S.A., Bozovsky M.R., Erdos M.R., Eriksson M., Goldman A.E., Khuon S., Collins F.S.et al.. Mutant nuclear lamin a leads to progressive alterations of epigenetic control in premature aging. 2006; 103:8703–8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Narita M., Nũnez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W.. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003; 113:703–716. [DOI] [PubMed] [Google Scholar]
- 126. Chiang M., Michieletto D., Brackley C.A., Rattanavirotkul N., Mohammed H., Marenduzzo D., Chandra T.. Polymer modeling predicts chromosome reorganization in senescence. Cell Rep. 2019; 28:3212–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Dou Z., Xu C., Donahue G., Shimi T., Pan J.A., Zhu J., Ivanov A., Capell B.C., Drake A.M., Shah P.P.et al.. Autophagy mediates degradation of nuclear lamina. Nature. 2015; 527:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chandra T., Kirschner K., Thuret J.-Y., Pope BenjaminD., Ryba T., Newman S., Ahmed K., Samarajiwa ShamithA., Salama R., Carroll Tet al.. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol. Cell. 2012; 47:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Van Meter M., Kashyap M., Rezazadeh S., Geneva A.J., Morello T.D., Seluanov A., Gorbunova V.. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014; 5:5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Judd J., Sanderson H., Feschotte C.. Evolution of mouse circadian enhancers from transposable elements. Genome Biol. 2021; 22:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Koike N., Yoo S.H., Huang H.C., Kumar V., Lee C., Kim T.K., Takahashi J.S.. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012; 338:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Park J., Zhu Q., Mirek E., Na L., Raduwan H., Anthony T.G., Belden W.J.. BMAL1 associates with chromosome ends to control rhythms in TERRA and telomeric heterochromatin. PLoS One. 2019; 14:e0223803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Simon M., Van Meter M., Ablaeva J., Ke Z., Gonzalez R.S., Taguchi T., De Cecco M., Leonova K.I., Kogan V., Helfand S.L.et al.. LINE1 derepression in aged wild-type and SIRT6-Deficient mice drives inflammation. Cell Metab. 2019; 29:871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., Tang B., Dong L., Ding N., Zhang Q.et al.. GSA: genome sequence Archive. Genom. Proteom. Bioinform. 2017; 15:14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Aging Atlas Consortium Aging atlas: a multi-omics database for aging biology. Nucleic Acids Res. 2020; 49:D825–D830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data obtained in this study have been deposited in the Genome Sequence Archive (134) in the National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation) of the Chinese Academy of Sciences, under accession number HRA001460 and CRA005216 that are publicly accessible at http://bigd.big.ac.cn/gsa-human or http://bigd.big.ac.cn/gsa. Data of BMAL1 interacting candidates that identified by LC-MS/MS data have been uploaded in the ProteomeXchange under the dataset identifier PXD030570. The differentially expressed genes identified in tissues derived from BMAL1–/– cynomolgus monkey were uploaded to the Aging Atlas (AA, https://ngdc.cncb.ac.cn/aging/index) database (135).