Abstract
Detection of low-level DNA mutations can reveal recurrent, hotspot genetic changes of clinical relevance to cancer, prenatal diagnostics, organ transplantation or infectious diseases. However, the high excess of wild-type (WT) alleles, which are concurrently present, often hinders identification of salient genetic changes. Here, we introduce UV-mediated cross-linking minor allele enrichment (UVME), a novel approach that incorporates ultraviolet irradiation (∼365 nm UV) DNA cross-linking either before or during PCR amplification. Oligonucleotide probes matching the WT target sequence and incorporating a UV-sensitive 3-cyanovinylcarbazole nucleoside modification are employed for cross-linking WT DNA. Mismatches formed with mutated alleles reduce DNA binding and UV-mediated cross-linking and favor mutated DNA amplification. UV can be applied before PCR and/or at any stage during PCR to selectively block WT DNA amplification and enable identification of traces of mutated alleles. This enables a single-tube PCR reaction directly from genomic DNA combining optimal pre-amplification of mutated alleles, which then switches to UV-mediated mutation enrichment-based DNA target amplification. UVME cross-linking enables enrichment of mutated KRAS and p53 alleles, which can be screened directly via Sanger sequencing, high-resolution melting, TaqMan genotyping or digital PCR, resulting in the detection of mutation allelic frequencies of 0.001–0.1% depending on the endpoint detection method. UV-mediated mutation enrichment provides new potential for mutation enrichment in diverse clinical samples.
INTRODUCTION
Identification of DNA mutations at low allelic frequencies within an excess of wild-type (WT) alleles is an essential requirement for analysis of clinical samples on several occasions, including cancer, organ transplantation, prenatal diagnostics and infectious diseases (1–6). For example, in liquid biopsy of cancer using circulating free DNA (7–11), low levels of somatic mutations or aberrant methylation serve as biomarkers for early detection (11–13), minimal residual disease detection (14) or tumor response to treatment (15–17), yet the high excess of circulating WT DNA often restricts opportunities for diagnosis and treatment. While next-generation sequencing technologies combined with incorporation of molecular barcodes can provide accurate enumeration of rare mutations on exome or genome-wide levels (18–22), interrogation of samples for specific, hotspot mutations is more practical and cost-effective to perform using PCR genotyping-based approaches directed to DNA targets of interest, such as KRAS (23,24), BRAF (25), EGFR (26,27) and others. Single-step, closed-tube PCR approaches that provide mutation detection directly from human genomic DNA are preferable over multistep methods, to avoid contamination, experimental errors and increased labor. Digital PCR technology, when available, provides a practical and accurate solution for targeted, quantitative detection of low-level alleles (28–31), albeit requires specialized instrumentation and entails elevated operation costs as compared to standard real-time PCR. As an alternative, mutation enrichment methods are often employed to elevate mutation concentrations to levels at which accurate and precise downstream analysis becomes feasible (32). Mutation enrichment methods include enzymatic approaches (33,34), PCR denaturation-based approaches like COLD-PCR (co-amplification at lower denaturation temperature PCR) (35,36) or WT ‘blocker’ approaches that enrich mutated alleles during PCR (37–39), including LNA/PNA-modified oligonucleotides (40). Once included in the reaction, such blockers operate during every PCR cycle. Depending on input DNA quantity and quality, addition of high blocker concentrations can occasionally reduce amplification even for mutated alleles, and this can be problematic when mutation allelic frequencies are low (25). The requirement to adjust blocker concentration leads to sample-dependent optimization to enable adequate WT DNA suppression while still allowing adequate amplification of rare, mutated alleles.
Oligonucleotides incorporating the photoreactive molecule 3-cyanovinylcarbazole nucleoside (CNVK) cross-link to pyrimidines on the opposite DNA strand upon UVA in a rapid reaction with ∼365 nm UVA radiation, which otherwise produces no detectable DNA damage (41). CNVK-modified oligonucleotides have been used to identify downstream miRNA targets (42), or for high-throughput in vitro selection of cDNA display (43). CNVK was also used for distinguishing single-nucleotide polymorphism typing for Japanese rice strains (44).
Here, we introduce UV-mediated cross-linking minor allele enrichment (UVME) to enable efficient detection of low-level mutations in clinical samples. UVME employs UV irradiation for selective photo-cross-linking of WT DNA to prevent its amplification (Figure 1A), thereby enabling preferential enrichment of mutated DNA molecules during subsequent amplification. Oligonucleotide probes matching the sense strand WT target sequence and incorporating a CNVK modification are employed for cross-linking. In its simplest form, UVME is applied directly on sheared genomic DNA by performing DNA denaturation followed by repeated probe hybridization and UV cross-linking, in the absence of enzymes (Figure 1B). This is then followed by a standard PCR reaction and endpoint mutation detection. Alternatively, UV is applied during the PCR annealing phase, at which the CNVK-modified probes cross-link preferentially to the sense strand of WT DNA, as compared to mutant DNA, and inhibit further amplification (Figure 1C). UV-based cross-linking can be applied at any cycle during PCR, thus providing a most flexible approach. Meanwhile, CNVK-modified probes commonly binding a nontarget region of the antisense strand of both WT and mutant DNA are also employed (‘common’ CNVK probes) to inhibit antisense strand amplification for both WT and mutated DNA. The UVME-PCR program is applied in a single tube, directly from human genomic DNA, and includes two stages. First, 10–20 cycles of regular PCR are performed to pre-amplify the target DNA. The UV irradiation is then turned on for 10 s at annealing temperature during each subsequent PCR cycle to induce photo-cross-linking of the CNVK-modified probes to the target DNA. In view of the probe design, the photo-cross-linked duplex of CNVK-modified probes and template DNA suppress the polymerization of both strands of WT DNA but only one strand of mutant DNA during each cycle, thereby resulting in mutation enrichment during PCR. The mutation-enriched PCR product can be used directly for Sanger sequencing or high-resolution melting (HRM) to identify mutations without secondary amplification; alternatively, a PCR-based TaqMan or digital droplet PCR (ddPCR) assay can be performed for endpoint detection (Figure 1C). We validate UVME mutation enrichment in serial dilutions of mutated genomic DNA and clinical samples containing low-level mutations.
Figure 1.
UVME-PCR reaction. (A) A UV lamp is attached to a thermal cycler to provide UV irradiation during PCR. The UV lamp can be manually switched on/off any time during PCR cycling to provide UV irradiation to DNA samples. (B) CNVK-modified probes are applied in UVME before PCR. Target-specific CNVK-modified probes are designed to match the WT sense strand while forming a mismatch with mutated sense DNA strands. Common CNVK-modified probes matching both WT and mutated DNA are designed for the antisense DNA strand. When UV is applied, the hybridized CNVK-modified probes cross-link with T or C on the opposite DNA strand at the −1 position. (C) UVME-PCR is applied in two stages. First, 10–20 cycles of regular closed-lid PCR are performed to build up target DNA copies from genomic DNA. Optionally, the pre-amplification can be performed via COLD-PCR to amplify preferentially mutated DNA, which further enhances the mutation enrichment. In the second stage of UVME-PCR, the lid of the PCR machine is opened, and the UV lamp is attached. Following denaturation, CNVK-modified probes and primers bind to their corresponding sequences during the annealing stage. The target-specific CNVK-modified probes hybridize to the sense strand WT DNA preferentially as compared to the sense strand mutated DNA and application of UV at this stage blocks subsequent polymerization. Meanwhile, the common CNVK-modified probes bind equally to both WT and mutant antisense templates and block antisense strand polymerization. Accordingly, UV-mediated cross-linking reduces amplification of both strands of WT DNA but only one strand of mutated DNA, in each PCR cycle, resulting in robust mutation enrichment during PCR.
MATERIALS AND METHODS
Genomic DNA and clinical tissue and circulating DNA samples
Human male genomic DNA (HMC, Promega Corporation) was used as WT DNA. Genomic DNA from KRAS-mutated cell lines containing 100% (PFSK-p53-C275G, A549-KRAS-G12S and SW480-KRAS-G12V) or 50% (H2009-KRAS-G12A and LOVO-KRAS-G13D) mutation was mixed with HMC to create serial dilutions containing 5%, 3%, 1%, 0.3%, 0.1%, 0.03%, 0.01%, 0.003% and 0.001% mutation allelic abundance. Snap-frozen, macro-dissected lung tumor specimens were obtained from the Massachusetts General Hospital Tumor Bank following approval from the Internal Review Board of the Dana-Farber Cancer Institute and kept frozen at −80°C until use. Plasma circulating DNA (cfDNA) from a breast cancer patient was used under approval from the Internal Review Board of the Dana-Farber Cancer Institute. Previously performed exome sequencing on this patient had identified MIER3 mutations at allelic frequency of ∼5% (14). To generate lower level (∼0.5%) mutation allelic frequency, a 10-fold dilution into cfDNA obtained from a healthy volunteer was created. For DNA extraction and ligation-mediated PCR (LM-PCR) protocol, see Supplementary Methods.
Setup for uniform UV irradiation on sections of 64-well plates during open-lid PCR
To enable UV irradiation during PCR cycling, an open-lid PCR approach was used, with a UV light source replacing the PCR machine lid (Supplementary Figure S1). See Supplementary Methods and Supplementary Figures S1 and S2 for the setup used to ensure uniform UV irradiation.
Primers and CNVK probes used for pre-PCR-UVME and UVME-PCR
The CNVK probes were designed against WT sequence with CNVK modification replacing one of the DNA bases and such that a T at position −1 is available in the opposite strand for cross-linking when the probe is hybridized to the targeted DNA. See Supplementary Methods and Table 1 for sequences and PCR conditions.
Table 1.
Primers and CNVK-modified probes for UVME-PCR
| Primer ID | Sequence |
|---|---|
| TP53 F1 | 5′-CCTATCCTGAGTAGTGGTAATCT-3′ |
| TP53 R1 | 5′-TTACCTCGCTTAGTGCTC-3′ |
| KRAS F1 | 5′-CATTATTTTTATTATAAGGCCTGC-3′ |
| KRAS R1 | 5′-CAAAATGATTCTGAATTAGCTGT-3′ |
| MIER3 F1 | 5′-ACCTGATAGCTCCCCTCAAGTTTA-3′ |
| MIER3 R1 | 5′-GACCAATGGTTTCATCAGTGCC-3′ |
| TP53 target-specific CNVK-modified probe | 5′-GGCACAAACACNVKGCAC/phos-3′ |
| TP53 common CNVK-modified probe | 5′-CAGAGGAAGAGAACNVKCTCCG/phos-3′ |
| KRAS target-specific CNVK-modified probe | 5′-CCTACNVKGCCACCAGCTCCA/phos-3′ |
| KRAS common CNVK-modified probe | 5′-TGAAAATGACNVKTGAATATAAACTTGTGG/phos-3′ |
| MIER3 target-specific CNVK-modified probe | 5′-CTCACNVKAGTGTAGGGCCG/phos-3′ |
| MIER3 common CNVK-modified probe | 5′-CGGACNVKCTGCGTGTGC/phos-3′ |
Pre-PCR-UVME
To apply the protocol depicted in Figure 1B, 30 ng fragmented DNA was used in a 10 μl reaction containing 1× AmpliTaqGold™ buffer, 2 mM MgCl2, 0.8 μl GC enhancer, 100 nM CNVK target-specific probes and CNVK common probes. The sample was placed in an Eppendorf Mastercycler™ to go through five cycles of denaturation at 95°C for 2 min, incubation at 52°C for 1 min and 10 s incubation at 52°C with UV irradiation. Treatment for cfDNA toward MIER3 mutation enrichment was done with a similar protocol (see Supplementary Methods). PCR conditions applied to the mutation-enriched samples are also described in Supplementary Methods.
UVME-PCR
The amplicon sequences, in silico Blast and primer location are listed in Supplementary Table S1. For experiments with KRAS mutations, 90 ng of serial dilution of mutated genomic DNA from A549, SW480, H2009 or LOVO into WT DNA was used. DNA was added in a final of 25 μl PCR reaction containing 1.2 μM CNVK target-specific probes and CNVK common probes, 0.8× LC Green (BioFire Diagnostics), 1× AmpliTaqGold buffer, 0.8 mM dNTP, 0.2 mM forward and reverse primers, 2 mM MgCl2, 2 μl GC enhancer, 0.06 μl Phusion™ polymerase (Thermo Fisher Scientific) and 0.125 μl AmpiTaq™ polymerase (Thermo Fisher Scientific). Fifty microliters of mineral oil (Sigma-Aldrich) was overlaid on the PCR master mix to prevent evaporation during open-lid PCR. The tubes were vortexed and spun down before placing in the PCR well plates. The PCR reaction was performed with an initial activation for 2 min at 95°C followed by 10 cycles of closed-lid PCR (95°C for 30 s, 57°C for 30 s and 72°C for 10 s); then, without interrupting the program, the Eppendorf machine lid was opened, the UV source and cover were attached, and the cycling continued with another 50 cycles of open-lid PCR with 10 s UV irradiation applied after the initial 10 s at 57°C during each cycle. The UVME-PCR products were placed on ice or stored at −20°C before dilution for downstream assays.
For a direct 1:1 comparison of enrichment obtained via pre-PCR-UVME and UVME-PCR, as well as for sequential reactions using both pre-PCR-UVME and UVME-PCR approaches applied on the same samples, see Supplementary Methods.
To combine UVME-PCR reactions with temperature-tolerant fast COLD-PCR (UVME-TT-fast-COLD-PCR) for KRAS mutations, the closed-lid pre-amplification stage of the reaction was increased to a total of 20 cycles and conducted as follows: an initial denaturation for 1 min at 95°C followed by 9 cycles of closed-lid PCR (81.2°C for 30 s, 57°C for 30 s and 72°C for 10 s), 11 cycles of closed-lid PCR (82.2°C for 30 s, 57°C for 30 s and 72°C for 10 s) and 50 cycles of open-lid PCR with UV irradiation as described earlier.
For experiments with p53 mutations, a total of 10 ng, 30 ng, 90 ng and 1 μg of DNA at 1%, 0.3–0.1%, 0.03–0.01% and 0.003–0.001% mutation dilutions, respectively, was employed. Similar protocols as with KRAS were used, with minor differences; Phusion™ polymerase was not included in the reaction and 2.4 μM CNVK-modified probes were used in the UVME-PCR reaction. The PCR reaction was conducted with initial activation of AmpliTaq polymerase at 95°C for 10 min followed by 20 cycles of closed-lid PCR (95°C for 30 s, 60°C for 30 s and 72°C for 10 s), 50 cycles of open-lid PCR (95°C for 30 s, 50°C for 30 s and 72°C for 10 s) with 10 s UV irradiation applied after the initial 10 s at 50°C during each cycle, and 72°C for 7 min.
For experiments in circulating DNA targeting MIER3, the same PCR reaction as for KRAS was set up with 30 ng LM-PCR obtained from cfDNA using a modified thermal protocol as follows: an initial enzyme activation for 2 min at 95°C, 8 cycles of closed-lid PCR (95°C for 30 s, 56.8°C for 30 s and 72°C for 10 s) and then 50 cycles of open-lid PCR with 10 s UV irradiation applied after the initial 10 s at 56.8°C during each cycle.
Direct Sanger sequencing, TaqMan real-time genotyping, ddPCR and HRM
Following pre-PCR-UVME or UVME-PCR, standard downstream assays were used to quantify the resulting mutation fraction and to verify the enrichment obtained (see Supplementary Methods, Supplementary Tables S2–S4 and Supplementary Figure S3).
Statistical analysis
PRISM 9 (GraphPad) software was applied for statistical analysis. Statistical significance was determined via the unpaired Student’s t-test. Samples marked with an asterisk represent those with values significantly different (P < 0.05) from the values of corresponding WT controls run in parallel. Otherwise, samples are marked as ‘ns’ (P > 0.05, nonsignificant). At least two independent experiment repeats were performed for each group.
RESULTS
Mutation enrichment via pre-PCR-UVME and UVME-PCR
To provide proof of principle for pre-PCR-UVME and UVME-PCR mutation enrichment, genomic DNA samples with KRAS exon 2 mutations were investigated. A single CNVK-modified probe was employed for enrichment of four common KRAS mutations, G12S, G12V, G12A and G13D, that represent the most frequently encountered KRAS mutations in cancer samples (23,45).
For pre-PCR-UVME mutation enrichment, HMC and KRAS-mutant DNA from cell line A549-G12S, SW480-G12V, H2009-G12A or LOVO-G13D were fragmented and mixed to generate KRAS mutation samples with mutations of ∼0.1% and 1% abundance. Pre-PCR-UVME was performed along with no-UV controls that underwent the exact same protocol by omitting the UV irradiation. Samples were then PCR amplified and the mutation abundance was examined via ddPCR. When the UV was applied, enrichment was observed for all four KRAS mutations converting the original ∼0.1% and ∼1% abundance to ∼3–7% and ∼20–40%, respectively (Supplementary Figure S4).
Next, in a single-tube approach, the UVME-PCR reaction was applied on genomic DNA with 1% KRAS mutations using the four KRAS-mutant cell lines. The reaction products were then used for nested PCR employing allele-specific FAM/HEX-labeled TaqMan probes for mutation identification (23). Using TaqMan probes, the PCR threshold difference (delta-Ct, FAM–HEX, with and without UV irradiation) can be used to quantify the relative amounts of WT and mutated DNA and the enrichment in the presence versus absence of UV irradiation. In the presence of UV irradiation, there is an evident shift of the TaqMan growth curves for a 1% mutation abundance relative to WT DNA (Supplementary Figure S5). To investigate the enrichment when using a single CNVK probe directed to the sense strand, versus two distinct CNVK probes directed to sense and antisense strands (Figure 1C), the experiments were repeated employing either a single or two probes per reaction. The data indicate that including only the target-specific CNVK-modified probe directed to the sense strand enriches all four KRAS mutations, from 1% to ∼10–20%; however, using the sense strand probe plus a CNVK-modified probe directed to the antisense strand that binds equally to WT and mutated DNA (‘common’ CNVK probe) increases the mutation abundance even more (from 1% to 15–40%, Supplementary Figure S6) indicating a higher mutation enrichment when both probes are used. Accordingly, a combination of target-specific sense and common antisense probes was used in this investigation.
UVME-PCR application on serial dilutions of KRAS mutation-containing DNA, followed by diverse downstream assays
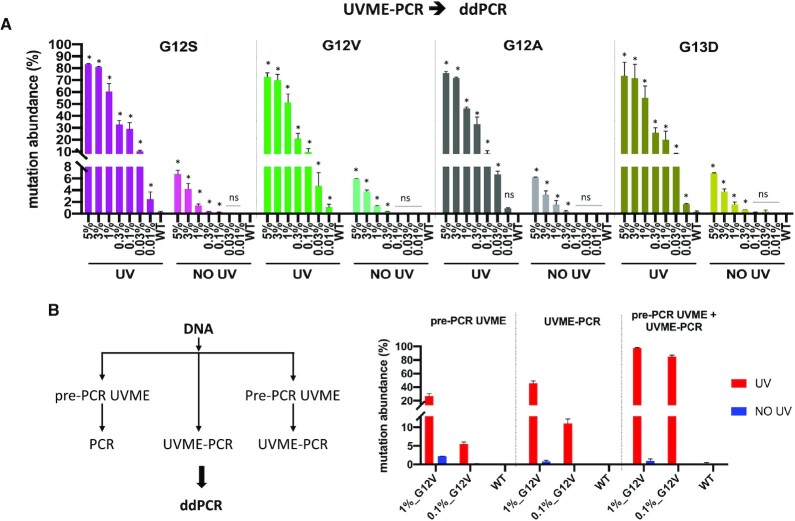
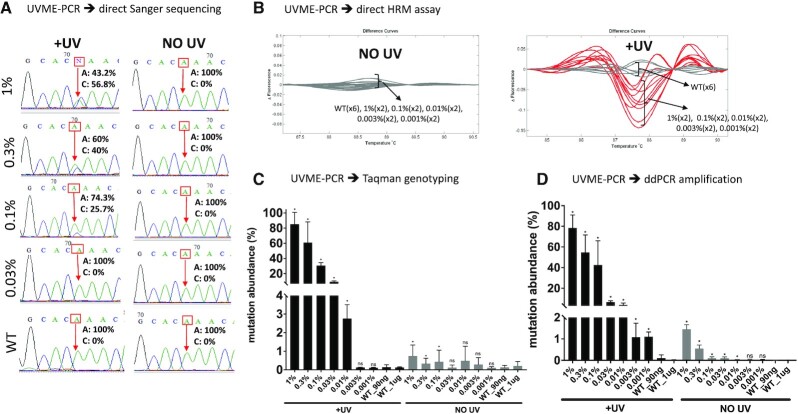
UVME-PCR was next applied on serial dilutions of KRAS mutation-containing DNA to identify limits of detection (LODs), depending on the downstream assay used to identify the mutation. UVME reactions with 90 ng input genomic DNA were used for 5–0.01% allelic frequencies. Sanger sequencing and HRM were then performed directly on UVME-PCR products without secondary amplification to identify mutations. The data indicate detection down to 0.1–0.3% MAF (minor allele frequency) via Sanger sequencing (Supplementary Figure S7) and 0.1% MAF via HRM (Supplementary Figure S8) for the four KRAS mutations in the presence of UV, while no mutations were detected in the absence of UV for MAF ≤ 5% or WT DNA. UVME-PCR products were also examined via TaqMan genotyping assays. The data indicate an LOD of 0.03% (G12S, G12V and G12A) and 0.1% (G13D) after UVME-PCR, while in the absence of UV the LOD is 1% (G12S, G12S and G12V), 3% (G12A) and 5% (G13D) (Supplementary Figure S9). To precisely evaluate the mutation abundance, ddPCR was also employed on UVME-PCR products. Data indicate an LOD down to MAF ∼ 0.01–0.03% and up to 250-fold enrichment for four KRAS mutations (Figure 2A). In the absence of UV, the ddPCR LOD is ∼0.1%, essentially dictated by the number of droplets per run in the standard Bio-Rad ddPCR format. Additionally, when plotted for the lower level mutations (<1%), the starting mutation abundance indicates a correlation (R2 ∼ 0.95) to the final mutation abundance for all four KRAS mutations studied (Supplementary Figure S10). Therefore, following UVME, the ddPCR result can be used for approximate quantification of the input mutation fraction. In summary, the KRAS mutation dilution experiments indicate that UVME-PCR mutation enrichment applied as shown in Figure 1 leads to KRAS mutation enrichments of ∼50–250-fold and an LOD of 0.01–0.3% depending on the downstream assay used.
Figure 2.
Application of UVME-PCR to enrich KRAS exon 2 mutations (G12S, G12V, G12A and G13D) and combination of pre-PCR-UVME and UVME-PCR. (A) UVME-PCR was tested with serial dilutions of genomic DNA from four KRAS-mutated cell lines (A549-G12S, SW480-G12V, H2009-G12A and LOVO-G13D) into WT DNA. A single pair of KRAS target-specific CNVK-modified probes and KRAS common CNVK-modified probes was used for all four mutations. UVME-PCR products screened via ddPCR quantify mutations down to 0.01–0.03% MAF for all four KRAS mutations. (B) Pre-PCR-UVME, UVME and combination of pre-PCR-UVME and UVME-PCR were performed on 30 ng sheared gDNA containing 0.1% or 1% KRAS G12V mutation. The final product screened via ddPCR showed that UVME-PCR has somewhat better enrichment efficiency than pre-PCR-UVME, while performing sequential pre-PCR-UVME and UVME-PCR could further boost the enrichment.
To directly compare the enrichment efficiency of pre-PCR-UVME and UVME-PCR and the effect of combining both approaches, 30 ng sheared gDNA containing 0.1% or 1% G12V was subjected to pre-PCR-UVME only, UVME-PCR only or pre-PCR-UVME followed by UVME-PCR. The final KRAS mutation abundance was quantified via ddPCR. The pre-PCR-UVME approach provided enrichment of 25- and 60-fold at 1% and 0.1% respectively, while the UVME-PCR approach showed enrichment of 45- and 100-fold exceeding the performance of pre-PCR-UVME (Figure 2B). Furthermore, combination of sequential pre-PCR-UVME and UVME-PCR results in very high enrichment, reaching a final mutation frequency of 85% and 98% from an original 0.1% and 1% original mutation frequency, respectively (Figure 2B). Accordingly, while each approach on its own provides enrichment that enables traditional approaches (Sanger sequencing and HRM) to detect mutations on the order of 0.1–1%, combining both approaches provides even higher enrichment, essentially delivering mostly mutated alleles.
UVME-TT-fast-COLD-PCR to boost enrichment for the Tm-decreasing KRAS mutations G12S, G12V and G13D
The mutation enrichment by UVME-PCR relies on selective UV cross-linking at the annealing step, while the enrichment by COLD-PCR is based on selective melting during denaturation (35,46–50). Thus, we explored combination of these two enrichment methods to further boost the mutation enrichment achieved during a single PCR reaction from genomic DNA. As a proof of principle, 90 μg input HMC DNA containing 0.01% of the Tm-decreasing mutations G12S (G > A), G12V (G > T) and G13D (G > A) was employed in the UVME-TT-fast-COLD-PCR. Instead of a regular denaturation temperature of 95°C, critical denaturation temperatures of 81.2°C (9 cycles, step 1) and 82.2°C (11 cycles, step 2) were used to selectively amplify the mutated DNA copies during the pre-amplification stage of UVME-PCR. The products were then screened via TaqMan genotyping (Supplementary Figure S11A) and ddPCR (Supplementary Figure S11B). The data indicate an increase in mutation enrichment via UVME-TT-fast-COLD-PCR, as compared to UVME-PCR alone for both TaqMan and ddPCR approaches. On the other hand, WT samples run in parallel demonstrated a somewhat higher background mutation rate when UVME-TT-fast-COLD-PCR was applied. Accordingly, while the mutation enrichment increased, the overall LOD showed no clear improvement following serial addition of COLD-PCR.
UVME-PCR application to a p53 hotspot mutation (p53-C275G)
To apply UVME-PCR to a second DNA target, other than KRAS, the p53 mutation C275G that is frequently encountered in lung cancer (51,52) was investigated. Genomic DNA from PFSK cell line containing a homozygous p53-C275G mutation was serially diluted into WT DNA, from 1% to 0.001%, using a total of 1 μg DNA per PCR reaction. This corresponds to ∼330 000 genomic copies, which is expected to contain ∼3–4 mutant molecules in the reaction at the 0.001% level. Varying amounts of input DNA up to 1 μg per reaction were used as input in UVME-PCR reactions. The reaction products were then directly screened via Sanger sequencing, HRM, TaqMan genotyping and ddPCR. Sanger sequencing indicated a limit of 0.1% MAF in the presence of UV irradiation, while no mutation could be detected in the absence of UV for the dilutions tested (Figure 3A). HRM analysis indicated presence of mutations down to 0.001% in the presence of UV, while no difference was evident in samples without UV (Figure 3B). TaqMan genotyping indicated detection down to 0.01% indicating a ∼300-fold mutation enrichment (Figure 3C) and ddPCR indicated detection down to MAF ∼ 0.001%, corresponding to 1000-fold mutation enrichment (Figure 3D). In summary, using high-input genomic DNA (1 μg) per reaction, UVME-PCR enrichment plus HRM/ddPCR for the hotspot p53 mutation C275G enables detection down to 1 in 105 alleles.
Figure 3.
Application of UVME-PCR to a p53 mutation, C275G. UVME-PCR was tested with serial dilutions of genomic DNA containing a hotspot tp53 mutation (PFSK cell line, C275G) into WT DNA. Serial dilutions of PFSK into HMC DNA down to 0.001% MAF were formed, with the lowest dilutions using an input of 1 μg DNA. (A) UVME-PCR followed by Sanger sequencing reveals that mutations in dilutions down to 0.1% were detectable following UV irradiation, while no mutations are evident without UV application. (B) When UVME-PCR was followed by HRM, mutations down to 0.001% were discriminated from WT samples following UV irradiation. (C) UVME-PCR followed by TaqMan genotyping shows that application of UV increases mutation abundance from 0.01–1% to 3–80%, respectively. (D) UVME followed by ddPCR detects mutations down to 0.001% and increases mutation abundance from 0.001–1% to 1–80%, respectively.
Application of UVME-PCR in clinical tumor samples and plasma circulating DNA
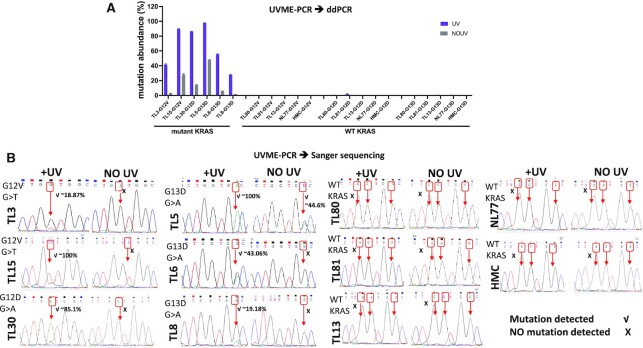
For a proof-of-principle application of UVME-PCR in clinical samples, DNA from 10 lung tumors was used. Six of these samples (TL3, TL30, TL5, TL6, TL15 and TL8) had been previously shown to harbor low-level KRAS mutations using a sensitive PNA-PCR-based approach (47,48), while four samples (TL80, TL13, TL81 and NL77) along with HMC DNA were WT. These samples were re-examined from genomic DNA using regular ddPCR and mutations at ∼1–45% mutation abundance were ascertained for the six samples previously found to be positive (Figure 4A). When UVME-PCR was applied prior to ddPCR, the mutation abundance detected via ddPCR was increased to ∼30–90% (Figure 4A). No mutations for the remaining four samples or HMC DNA were detected. Samples were then also examined via Sanger sequencing as the endpoint detection method. The mutations were detected in the six positive samples via UVME–Sanger sequencing (Figure 4B). No mutations could be detected via Sanger sequencing in the absence of UV cross-linking (no UV) in five out of six positive samples. The data indicate that UVME mutation enrichment enabled Sanger sequencing-based detection of mutations in low-purity clinical samples and magnified ddPCR mutation signals as compared to regular ddPCR.
Figure 4.
Clinical tissue samples were applied in UVME-PCR. UVME-PCR was applied to DNA extracted from 10 lung tumor tissue samples previously shown to harbor medium to low KRAS G12D, G12V or G13D mutations and four samples with WT KRAS. Human male genomic DNA was also screened as a control. (A) ddPCR was first applied to document the mutation abundance in the absence of UVME enrichment and mutations in the 1–40% abundance were demonstrated. ddPCR applied following UVME-PCR resulted in an increase in mutation abundance. No mutations were detected for the four WT samples and HMC DNA. (B) UVME-PCR followed by Sanger sequencing depicts mutations for all six samples harboring KRAS mutations; mutations could not be detected in five out of six positive samples in the absence of UV irradiation, using Sanger sequencing.
Application of pre-PCR-UVME or UVME-PCR on cfDNA was investigated following the workflow depicted in Supplementary Figure S12A. cfDNA from a metastatic breast cancer patient was obtained and a mutation on MIER3 p.H548D, which was previously detected via whole-exome sequencing, was selected as a target. After UVME treatment, the MIER3 mutation was enriched ∼30-fold, from an initial allelic frequency of ∼0.5% to ∼14% in the presence of UV (Supplementary Figure S12B). Pre-PCR-UVME and UVME-PCR were also performed on a LM-PCR product obtained from the same cfDNA sample. Similar MIER3 fold enrichment was obtained in the presence of UV irradiation (Supplementary Figure S12C and D). No MIER3 mutation was evident in similarly treated healthy donor cfDNA samples. In summary, UVME can be applied for a range of clinical samples, including tissue and circulating DNA. Application on FFPE samples or damaged DNA is also considered potentially possible, depending on sample degradation and formalin fixation conditions (53) or damage by DNA damaging agents (54,55).
DISCUSSION
We demonstrate that introducing photochemistry either prior to or during PCR enables a novel ‘WT DNA blocker approach’ that can be applied for enrichment of mutations directly from genomic DNA in a closed-tube format. When applied during PCR, the ability to apply UV at any point during cycling provides the flexibility to apply target pre-amplification without UV using regular PCR in the first few cycles. This pre-amplification boosts mutated DNA copies and then switches to WT DNA-specific UV cross-linking to amplify selectively mutation-containing DNA alleles. Unlike mutation enrichment approaches that require a separate pre-amplification step prior to enrichment (56,57), the ability to combine pre-amplification plus enrichment steps in a single tube directly from genomic DNA has practical advantages such as convenience, speed and reduced contamination risk. As with all approaches that employ pre-amplification, there is a risk of introducing PCR errors that can then be enriched during the UV cross-linking stage and result in false positives. An indication for increased PCR noise was evident when COLD-PCR was applied for 20 cycles of pre-amplification (Supplementary Figure S11).
Meanwhile, as a simpler alternative to UV-based cross-linking during PCR and to obviate PCR errors altogether, UV-based selective WT DNA cross-linking was also applied directly on genomic DNA prior to PCR, in the absence of enzymes. This strategy has several advantages: (i) It enables existing amplification and detection strategies without change following the initial UV cross-linking phase. This potentially includes isothermal amplification of selectively cross-linked DNA instead of PCR, for point-of-care applications. (ii) It bypasses the need for customized instrumentation since UV cross-linking can in principle be applied via a simpler setup instead of PCR machine. (iii) It increases the potential for multiplexity: since UVME entails a simple hybridization step without PCR, one can multiplex CNVK-modified probes against multiple targets for selective cross-linking. Moreover, sequential application of pre-PCR-UVME plus UVME-PCR results in even higher enrichment efficiency, enabling a 0.1% mutation dilution to become 85%, representing an ∼850-fold mutation enrichment. In summary, the ability to apply WT DNA selective cross-linking either before or during PCR or in combination provides unique advantages for UVME and the ability to apply mutation enrichment in conjunction with many existing technologies in a variety of settings.
While pre-PCR-UVME is straightforward to perform, the application of UV cross-linking during PCR is tedious in the absence of a PCR machine that incorporates a UV lamp. To prove the principle, here we performed open-lid PCR and applied UV to PCR tubes using an external UV lamp. While this approach worked well as a proof of principle, it is tedious since the UV is operated manually during PCR. Further, open-lid PCR requires higher number of cycles relative to closed, heated-lid PCR to achieve a given amplification. Accordingly, incorporation of the UV source within a heated-lid PCR cycler and automated control are anticipated instrumentation developments to enable widespread UVME application. Implementation of UVME-PCR in a real-time PCR mode would provide additional advantages, such as automatically switching the UV on/off when the reaction reaches certain PCR thresholds using real-time PCR based either on DNA-binding dyes or on oligonucleotide probes (58).
DATA AVAILABILITY
All sequences of probes and primers used in the present investigation are described in the supplementary file accompanying the manuscript. The data supporting the results of this study are available in the paper and its supplementary information.
Supplementary Material
Contributor Information
Ka Wai Leong, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women’s Hospital, Harvard Medical School, 450 Brookline Avenue, Boston, MA 02115, USA.
Fangyan Yu, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women’s Hospital, Harvard Medical School, 450 Brookline Avenue, Boston, MA 02115, USA.
G Mike Makrigiorgos, Department of Radiation Oncology, Dana-Farber Cancer Institute and Brigham and Women’s Hospital, Harvard Medical School, 450 Brookline Avenue, Boston, MA 02115, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 CA221874, in part]; Dana-Farber Cancer Institute (in part). Funding for open access charge: Dana-Farber Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kobayashi S., Boggon T.J., Dayaram T., Janne P.A., Kocher O., Meyerson M., Johnson B.E., Eck M.J., Tenen D.G., Halmos B.. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005; 352:786–792. [DOI] [PubMed] [Google Scholar]
- 2. Hoffmann C., Minkah N., Leipzig J., Wang G., Arens M.Q., Tebas P., Bushman F.D.. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007; 35:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lo Y.M., Corbetta N., Chamberlain P.F., Rai V., Sargent I.L., Redman C.W., Wainscoat J.S.. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997; 350:485–487. [DOI] [PubMed] [Google Scholar]
- 4. Snyder T.M., Khush K.K., Valantine H.A., Quake S.R.. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl Acad. Sci. U.S.A. 2011; 108:6229–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong S.M., Traverso G., Johnson C., Geng L., Favis R., Boynton K., Hibi K., Goodman S.N., D’Allessio M., Paty P.et al.. Detecting colorectal cancer in stool with the use of multiple genetic targets. J. Natl. Cancer Inst. 2001; 93:858–865. [DOI] [PubMed] [Google Scholar]
- 6. Galbiati S., Brisci A., Lalatta F., Seia M., Makrigiorgos G.M., Ferrari M., Cremonesi L.. Full COLD-PCR protocol for noninvasive prenatal diagnosis of genetic diseases. Clin. Chem. 2011; 57:136–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M., Thornton K., Agrawal N., Sokoll L., Szabo S.A.et al.. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008; 14:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thierry A.R., Mouliere F., El Messaoudi S., Mollevi C., Lopez-Crapez E., Rolet F., Gillet B., Gongora C., Dechelotte P., Robert B.et al.. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014; 20:430–435. [DOI] [PubMed] [Google Scholar]
- 9. Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C., Modlin L.A., Liu C.L., Neal J.W., Wakelee H.A., Merritt R.E.et al.. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014; 20:548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M.et al.. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014; 6:224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diehl F., Li M., Dressman D., He Y., Shen D., Szabo S., Diaz L.A., Goodman S.N., David K.A., Juhl H.et al.. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl Acad. Sci. U.S.A. 2005; 102:16368–16373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu M.C., Oxnard G.R., Klein E.A., Swanton C., Seiden M.V., Liu M.C., Oxnard G.R., Klein E.A., Smith D., Richards D.et al.. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020; 31:745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen J.D., Li L., Wang Y., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A.et al.. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018; 359:926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Parsons H.A., Rhoades J., Reed S.C., Gydush G., Ram P., Exman P., Xiong K., Lo C.C., Li T., Fleharty M.et al.. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin. Cancer Res. 2020; 26:2556–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kimura T., Holland W.S., Kawaguchi T., Williamson S.K., Chansky K., Crowley J.J., Doroshow J.H., Lenz H.J., Gandara D.R., Gumerlock P.H.. Mutant DNA in plasma of lung cancer patients: potential for monitoring response to therapy. Ann. N. Y. Acad. Sci. 2004; 1022:55–60. [DOI] [PubMed] [Google Scholar]
- 16. Misale S., Yaeger R., Hobor S., Scala E., Janakiraman M., Liska D., Valtorta E., Schiavo R., Buscarino M., Siravegna G.et al.. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012; 486:532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chabon J.J., Hamilton E.G., Kurtz D.M., Esfahani M.S., Moding E.J., Stehr H., Schroers-Martin J., Nabet B.Y., Chen B., Chaudhuri A.A.et al.. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020; 580:245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Newman A.M., Lovejoy A.F., Klass D.M., Kurtz D.M., Chabon J.J., Scherer F., Stehr H., Liu C.L., Bratman S.V., Say C.et al.. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016; 34:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wan J.C.M., Heider K., Gale D., Murphy S., Fisher E., Mouliere F., Ruiz-Valdepenas A., Santonja A., Morris J., Chandrananda D.et al.. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 2020; 12:eaaz8084. [DOI] [PubMed] [Google Scholar]
- 20. McDonald B.R., Contente-Cuomo T., Sammut S.-J., Odenheimer-Bergman A., Ernst B., Perdigones N., Chin S.-F., Farooq M., Mejia R., Cronin P.A.et al.. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 2019; 11:eaax7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B.. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl Acad. Sci. U.S.A. 2011; 108:9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A.. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. U.S.A. 2012; 109:14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amicarelli G., Shehi E., Makrigiorgos G.M., Adlerstein D.. FLAG assay as a novel method for real-time signal generation during PCR: application to detection and genotyping of KRAS codon 12 mutations. Nucleic Acids Res. 2007; 35:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shi C., Eshleman S.H., Jones D., Fukushima N., Hua L., Parker A.R., Yeo C.J., Hruban R.H., Goggins M.G., Eshleman J.R.. LigAmp for sensitive detection of single-nucleotide differences. Nat. Methods. 2004; 1:141–147. [DOI] [PubMed] [Google Scholar]
- 25. How-Kit A., Lebbe C., Bousard A., Daunay A., Mazaleyrat N., Daviaud C., Mourah S., Tost J.. Ultrasensitive detection and identification of BRAF V600 mutations in fresh frozen, FFPE, and plasma samples of melanoma patients by E-ice-COLD-PCR. Anal. Bioanal. Chem. 2014; 406:5513–5520. [DOI] [PubMed] [Google Scholar]
- 26. Kuang Y., Rogers A., Yeap B.Y., Wang L., Makrigiorgos M., Vetrand K., Thiede S., Distel R.J., Janne P.A.. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin. Cancer Res. 2009; 15:2630–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guha M., Castellanos-Rizaldos E., Makrigiorgos G.M.. DISSECT method using PNA-LNA clamp improves detection of T790m mutation. PLoS One. 2013; 8:e67782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fitarelli-Kiehl M., Yu F., Ashtaputre R., Leong K.W., Ladas I., Supplee J., Paweletz C., Mitra D., Schoenfeld J.D., Parangi S.et al.. Denaturation-enhanced droplet digital PCR for liquid biopsies. Clin. Chem. 2018; 64:1762–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vogelstein B., Kinzler K.W.. Digital PCR. Proc. Natl Acad. Sci. U.S.A. 1999; 96:9236–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taly V., Huggett J.. Digital PCR, a technique for the future. Biomol. Detect. Quantif. 2016; 10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. dMIQE Group, Huggett J.F. The digital MIQE guidelines update: minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 2020; 66:1012–1029. [DOI] [PubMed] [Google Scholar]
- 32. Milbury C.A., Li J., Makrigiorgos G.M.. PCR-based methods for the enrichment of minority alleles and mutations. Clin. Chem. 2009; 55:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song C., Liu Y., Fontana R., Makrigiorgos A., Mamon H., Kulke M.H., Makrigiorgos G.M.. Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic Acids Res. 2016; 44:e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuery C.J., Impey H.L., Roberts N.J., Applegate T.L., Ward R.L., Hawkins N.J., Sheehan C.A., O’Grady R., Todd A.V.. Detection of rare mutant alleles by restriction endonuclease-mediated selective-PCR: assay design and optimization. Clin. Chem. 2000; 46:620–624. [PubMed] [Google Scholar]
- 35. Li J., Wang L., Mamon H., Kulke M.H., Berbeco R., Makrigiorgos G.M.. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 2008; 14:579–584. [DOI] [PubMed] [Google Scholar]
- 36. Li J., Wang L., Janne P.A., Makrigiorgos G.M.. Coamplification at lower denaturation temperature-PCR increases mutation-detection selectivity of TaqMan-based real-time PCR. Clin. Chem. 2009; 55:748–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu L.R., Chen S.X., Wu Y., Patel A.A., Zhang D.Y.. Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification. Nat. Biomed. Eng. 2017; 1:714–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Milbury C.A., Li J., Makrigiorgos G.M.. Ice-COLD-PCR enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations. Nucleic Acids Res. 2011; 39:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Murphy D.M., Bejar R., Stevenson K., Neuberg D., Shi Y., Cubrich C., Richardson K., Eastlake P., Garcia-Manero G., Kantarjian H.et al.. NRAS mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia. 2013; 27:2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun X., Hung K., Wu L., Sidransky D., Guo B.. Detection of tumor mutations in the presence of excess amounts of normal DNA. Nat. Biotechnol. 2002; 20:186–189. [DOI] [PubMed] [Google Scholar]
- 41. Yoshimura Y., Fujimoto K.. Ultrafast reversible photo-cross-linking reaction: toward in situ DNA manipulation. Org. Lett. 2008; 10:3227–3230. [DOI] [PubMed] [Google Scholar]
- 42. Yoshimura Y., Ohtake T., Okada H., Fujimoto K.. A new approach for reversible RNA photocrosslinking reaction: application to sequence-specific RNA selection. ChemBioChem. 2009; 10:1473–1476. [DOI] [PubMed] [Google Scholar]
- 43. Mochizuki Y., Suzuki T., Fujimoto K., Nemoto N.. A versatile puromycin-linker using cnvK for high-throughput in vitro selection by cDNA display. J. Biotechnol. 2015; 212:174–180. [DOI] [PubMed] [Google Scholar]
- 44. Fujimoto K., Yamada A., Yoshimura Y., Tsukaguchi T., Sakamoto T.. Details of the ultrafast DNA photo-cross-linking reaction of 3-cyanovinylcarbazole nucleoside: cis–trans isomeric effect and the application for SNP-based genotyping. J. Am. Chem. Soc. 2013; 135:16161–16167. [DOI] [PubMed] [Google Scholar]
- 45. How Kit A., Mazaleyrat N., Daunay A., Nielsen H.M., Terris B., Tost J.. Sensitive detection of KRAS mutations using enhanced-ice-COLD-PCR mutation enrichment and direct sequence identification. Hum. Mutat. 2013; 34:1568–1580. [DOI] [PubMed] [Google Scholar]
- 46. Castellanos-Rizaldos E., Liu P., Milbury C.A., Guha M., Brisci A., Cremonesi L., Ferrari M., Mamon H., Makrigiorgos G.M.. Temperature-tolerant COLD-PCR reduces temperature stringency and enables robust mutation enrichment. Clin. Chem. 2012; 58:1130–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Milbury C.A., Li J., Makrigiorgos G.M.. COLD-PCR-enhanced high-resolution melting enables rapid and selective identification of low-level unknown mutations. Clin. Chem. 2009; 55:2130–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Milbury C.A., Li J., Liu P., Makrigiorgos G.M.. COLD-PCR: improving the sensitivity of molecular diagnostics assays. Expert Rev. Mol. Diagn. 2011; 11:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pinzani P., Santucci C., Mancini I., Simi L., Salvianti F., Pratesi N., Massi D., De Giorgi V., Pazzagli M., Orlando C.. BRAFV600E detection in melanoma is highly improved by COLD-PCR. Clin. Chim. Acta. 2011; 412:901–905. [DOI] [PubMed] [Google Scholar]
- 50. Pritchard C.C., Akagi L., Reddy P.L., Joseph L., Tait J.F.. COLD-PCR enhanced melting curve analysis improves diagnostic accuracy for KRAS mutations in colorectal carcinoma. BMC Clin. Pathol. 2010; 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li J., Milbury C.A., Li C., Makrigiorgos G.M.. Two-round coamplification at lower denaturation temperature-PCR (COLD-PCR)-based Sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma. Hum. Mutat. 2009; 30:1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Olivier M., Eeles R., Hollstein M., Khan M.A., Harris C.C., Hainaut P.. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 2002; 19:607–614. [DOI] [PubMed] [Google Scholar]
- 53. Wang F., Wang L., Briggs C., Sicinska E., Gaston S.M., Mamon H., Kulke M.H., Zamponi R., Loda M., Maher E.et al.. DNA degradation test predicts success in whole-genome amplification from diverse clinical samples. J. Mol. Diagn. 2007; 9:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Makrigiorgos G.M., Berman R.M., Baranowska-Kortylewicz J., Bump E., Humm J.L., Adelstein S.J., Kassis A.I.. DNA damage produced in V79 cells by DNA-incorporated iodine-123: a comparison with iodine-125. Radiat. Res. 1992; 129:309–314. [PubMed] [Google Scholar]
- 55. Makrigiorgos G., Adelstein S.J., Kassis A.I.. Auger electron emitters: insights gained from in vitro experiments. Radiat. Environ. Biophys. 1990; 29:75–91. [DOI] [PubMed] [Google Scholar]
- 56. Liu Q., Guo X., Xun G., Li Z., Chong Y., Yang L., Wang H., Zhang F., Luo S., Deng Z.et al.. Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations. Nucleic Acids Res. 2021; 49:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Song J., Hegge J.W., Mauk M.G., Chen J., Till J.E., Bhagwat N., Azink L.T., Peng J., Sen M., Mays J.et al.. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics. Nucleic Acids Res. 2019; 48:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tyagi S., Kramer F.R.. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996; 14:303–308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences of probes and primers used in the present investigation are described in the supplementary file accompanying the manuscript. The data supporting the results of this study are available in the paper and its supplementary information.