Abstract
Alternative splicing is frequently involved in the diversification of protein function and can also be modulated for therapeutic purposes. Here we develop a predictive model, called Exon ByPASS (predicting Exon skipping Based on Protein amino acid SequenceS), to assess the criticality of exon inclusion based solely on information contained in the amino acid sequence upstream and downstream of the exon junctions. By focusing on protein sequence, Exon ByPASS predicts exon skipping independent of tissue and species in the absence of any intronic information. We validate model predictions using transcriptomic and proteomic data and show that the model can capture exon skipping in different tissues and species. Additionally, we reveal potential therapeutic opportunities by predicting synthetically skippable exons and neo-junctions arising in cancer cells.
INTRODUCTION
Accurate gene annotations help clarify the relationship between a gene and its potential impact on biology (1–3). With the advent of high-throughput sequencing and sophisticated analytical methods, scientists have discovered enormous complexity resulting from alternative splicing of expressed genes. Splice variants from protein coding transcripts frequently encode distinct protein isoforms, and gene function can vary widely depending on the isoform that is expressed (4,5). In higher eukaryotes, alternative splicing increases the diversity of functions that can be carried out by a limited number of genes, including tissue-specific functions (6,7). Cataloging these isoforms is necessary to fully understand function.
Recently, machine-learning models have been developed to predict all possible splice variants that can arise from the transcriptome. These tools interrogate exon flanking sequences to identify features—sequence elements—that are compatible with exon skipping or other forms of alternative splicing. Tools like AVISPA (8), SpliceAI (9), SplicePort (10), MaxEntScan (11) and ESEFinder (12) look for regulatory elements that direct splicing which allow for alternative isoforms. This approach is limited by our knowledge of the factors that mediate splicing and the complexity of this process. Splicing depends on the position, size and strength of sequence elements as well as the presence and accessibility of splicing factors which are unique to cell types and species (13–15). A model that could identify skippable exons, regardless of RNA context, might be helpful in the discovery of additional targets for oligonucleotide-mediated skipping therapies as well as neo-junctions (neo-antigens derived from skipped exons) that could be targeted in cancer immunotherapies.
To overcome these challenges, we built a model called Exon ByPASS (predicting Exon skipping Based on Protein amino acid SequenceS) that attempts to predict the feasibility of exon skipping based on the resulting protein sequence. We hypothesize that the constraints on protein structure provide sufficient information to predict whether a coding exon is removable and that these constraints should be independent of species and tissue. Based on this hypothesis, we leveraged an abundance of protein sequence information that is available across eukaryotic species to train the model, which aims to measure the probability that a protein will fold and function if the segment encoded by a specific exon is removed. With our model, we identified protein features that constrain exon skipping, and we validated the presence of novel isoforms predicted by Exon ByPASS with transcriptomic and proteomic data. We show that the model captures skipping events independent of tissue, and the model can be applied across species. Importantly, the model finds previously unobserved skipping events that could have significant biological consequences.
MATERIALS AND METHODS
Exon classification
Protein sequence data as well as exon genomic coordinates, exon id and exon rank were pulled and merged from Ensembl’s BioMart using biomaRt v2.44.4 (16). Two hundred two genomes were available from biomaRt at the time of training. First, we ignored all first and last exons within a transcript. Any gene with only one transcript annotation was also set aside. Next, we removed any non-coding transcripts. Exon classification into groups of constitutive and skippable exons was done by an in-house script that uses genomic coordinates to find transcripts that have exons that do or do not occupy the same genomic positions. If an exon was found to occupy the same genomic coordinates in every transcript it was considered constitutive. If an exon was not present in one transcript but was in others, it was considered skippable; however, the flanking exons of the two transcripts had to be sequence-equivalent. This ensured that our classifications were specific to exon skipping and not the result of alternative 5′- or 3′-splice sites. Additionally, this classification avoided calling retained introns as skipped exon. Exons were then classified by frame using coding sequence length. If the coding sequence length was divisible by three then it was considered in-frame. After classification, amino acid sequences were assigned to each exon. If an amino acid codon was split between two exons, the exon with more than one nucleotide was assigned the amino acid. Exon classifications were checked to ensure they were consistent for each gene so that an exon was not constitutive in one isoform and skippable in another isoform.
Model parameters, architecture and training
We used the GPU version of TensorFlow in keras v2.3.0 in R 3.6.1 (https://www.r-project.org/) (https://keras.rstudio.com/). The keras package in R uses reticulate to connect to keras in python. Keras is a neural network API which uses TensorFlow as a backend. We built a sequential keras model with embedding layer that allowed us to stack layers. The input of the model uses one-hot coding representations of amino acids. The model starts with an embedding layer with input_dim = 22, output_dim = 22, and input_length = 100. The next 3 layers are 1D convolutional layers with 1028 filters, window sizes of 4, strides of 1 and with a relu activation function. Then there are 3 additional convolutional layers with the same parameters; however, they only have 512 filters. Next, was a max pooling layer that used a window size of 1 and then a dropout layer which removed 20% of the nodes. Next, there is a bidirectional LSTM layer which takes the input after pooling and dropping out. The LSTM layer output is a 100-unit vector before it is passed to a 2-node dense layer with a sigmoid activation function. The last layer serves as the output layer and is where the probabilities are obtained. To optimize weights, we found that ADAM performed better than the traditional stochastic gradient descent algorithm (17). With ADAM, we used a learning rate of 0.001 with other parameters set to default. Finally, for model training we used a 90:10 training validation split. We used a batch size of 1000 and found that accuracy and mean squared error was optimal after 10 epochs. The model can be found at https://github.com/wavelifescience/ExonByPass and sample datasets can be found at https://zenodo.org/record/5998350#.YgFUiFjML64.
Testing exons in mouse and human
To make predictions on mouse and human exons, which we then used for validation, we pulled amino acid sequences from biomaRt (16). We used GRCh38 and GRCm38 assemblies for testing. We pulled all protein sequences from coding exons except the first and last exons of transcripts. We also pulled genomic coordinates, exon id and transcript rank per exon. The model was loaded into R using the R package keras v2.3.0 (https://keras.rstudio.com/). The sequences were again encoded into 100 × 22 matrix representing 50 amino acids in the exon of interest and 25 amino acids from the up- and downstream exon. The predict_proba function was then used to calculate the skip probability using the model and the exon sequences. The exon probabilities were merged with genomic position, exon id and transcript rank, which allowed us to compare to known annotations and validation data.
Pairwise mutational analysis
Pairwise mutational analysis was performed using an in-house script which loops through all pairwise Ala substitutions in a 100-amino acid sequence. To predict skip probability on the modified sequences, we used predict_proba. Each evaluated exon results in 100 × 100 matrix of predictions. We analyzed 10 000 exons from GRCh38 that had the highest predicted probability in the reference sequence. The resulting 100 × 100 matrices from the 10 000 were averaged. The mean probabilities were then plotted using pheatmap v1.0.12.
Disorder prediction, physiochemical properties and InterPro annotations
For protein disorder predictions, we used DISpro. DISpro uses an energy estimation function to calculate interactions between residues; residues more favorable to folding generally have more contact. We used default parameters when running DISpro. Predictions were again returned per amino acid, and the average was taken across each exon for each 25 amino acid segments. Amino acid hydrophobicity was calculated based on the Rose scale (18), and amino acid flexibility was calculated based on the Karplus scale (19). Hydrophobicity and flexibility were averaged across 100 amino acid input sequence using a 5 amino acid window. The 25th, 26th, 75th and 76th amino acids are excluded due to amino acid bias from the acceptor and donor nucleotides. Net charge is defined as the charge of amino acids at pH 7. Asp, Arg, Lys and Glu were considered to be charged and were averaged over the input sequence using a 5 amino acid window. To find InterPro annotation over exon 7 in APAF1, we used biomaRt to pull positions (20). We used ggribbon to visualize annotations.
Identifying skipped exon junction counts in mouse
Published RNA-seq experiments were retrieved using SRA toolkit (21) (https://www.ncbi.nlm.nih.gov/books/NBK56551/). This dataset contains 12 different mouse tissues sampled every 6 h for 2 days, for a total of 96 RNA-seq samples. Fastq files were adaptor trimmed with bbduk v38.73 and quality checked with FastQC v0.11.5 (https://sourceforge.net/projects/bbmap/) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Hisat2 v2.1.0 was used to align reads to GRCm38 (22). Aligned bam files were sorted and indexed using samtools v1.9 (23). Using Rsubread v2.4.0, we counted reads that would span junctions of all coding regions within the GRCm38 genome (24). The genomic coordinates for junction reads, which was an output of Rsubread, was used find specific junction reads that skip single exons. These junction reads were then compared with known skipped exons in annotations and predicted skipped exons. The total skipped exon junction counts per exon was taken from all 96 samples. Sashimi plots were taken from a single sample from the specified tissues. We used the python package ggsashimi (25) to make sashimi plots. Annotations for these plots were extracted from GRCm38 annotations.
Comparing to ExonSkipDB
To find evidence to support predicted skipped exons from human RNA-seq data, we used ExonSkipDB (https://ccsm.uth.edu/ExonSkipDB/). GRCh37 coordinates were converted to GRCh38 using biomaRt. The exon skipped exons where then filtered by frame. Finally, the skipped exons were compared with annotations and predictions using genomic coordinates.
Mouse to human orthologs
We used biomaRt to find orthologs and annotations of orthologs. We use only orthologs with 80% sequence similarity at the nucleotide level. Gene order scores >75 and genome alignment score >75 were also considered as ortholog pairs. First, we obtained orthologous genes using gene ids from biomart and looked at differences in the annotation of the genes between human and mouse. To ensure that only orthologous exons were compared, we examined transcripts in which the total number of exons were the same, and we only called exons as skipped if their rank was equivalent in mouse and human. We also ensured that skipped exons had >70% sequence similarity at the protein level in mouse and human. Finally, we categorized skipped exons that were annotated in human but not in mouse and were predicted to be skippable by our model. We used sankeyMATIC to plot the breakdown on the orthologs of predicted and annotated in-frame skipped mouse exons (http://sankeymatic.com/).
Annotation track plots
Track plots were generated in R using Gviz v 1.30.3 using mouse GRCm38 and human GRCh38 (26). Transcripts were filtered by coding sequences. BiomaRt was used to pull transcript coordinates and annotations. For more detail see exon_annotation_plot.R in github.
AUC
Receiver operating characteristic curve (ROC) analysis was performed in R using pROC v 1.16.2 (27). For the mouse RNA-seq AUC, we used all in-frame skipped exons with at least one junction count as a true case in the response vector. We compared RNA-seq junction counts to annotations in mm10 and predictions by our model. For annotations, the predictor vector was encoded into binary: for each exon, 0 represented annotated constitutive exons, and 1 represented annotated skipped exons. For our model, the predictor vector was just the probability that an exon could be skipped. For ExonSkipDB AUC, we used all in-frame skipped exons identified by ExonSkipDB and all hg38 annotated in-frame skipped exons as true cases in the response vector. We compared this to our model predictions as well as known skipped exon annotations. For annotations, the predictor vector was encoded into binary as described above. For our model, the predictor vector was the probability that an exon could be skipped. The resulting ROC objects were then plotted using ggroc.
Proteomics analysis with Philosopher v3.2.9 and MSFragger v2.3
We used a standard ftp to download mzML per iTRAQ multiplexed samples from the 2012 Breast Cancer Study using the CPTAC data portal (28,29). We used MSFragger based search and Philosopher to demultiplex and to match peptide spectra derived from skipped exon junctions (30,31). To generate the theoretical skipped exon peptide database, we merged the protein sequences of the up- and downstream exons while excluding the exon of interest for every annotated exon. We did this for every exon except the first and last exons of a transcript and any exons from non-coding transcripts. Additionally, decoy sequences were added to estimate and account for false-positive rates. We used Philosopher pipeline and standard parameters to perform a closed database search. Parameters can be found in Supplementary Table S4. We used the combined peptide output to search for junction peptides. After the search, we filtered for any peptides that spanned the junction between the upstream or downstream protein sequence. At least one amino acid had to match the end of the upstream exon or the beginning of the downstream exon to ensure that the peptide spanned the exon junction. To quantify peptides per treatment we used peptides per multiplexed sample.
MARIA HLA class II predictions
For prediction by classification as HLA class II presenting antigens, we used MARIA (https://maria.stanford.edu/) (32). The identified skipped exon peptide sequences from the CPTAC Breast Cancer Study were used as the sequence input. We also assumed all genotypes to be HLA-DRB1*01:01 alleles. We used TCGA’s BRCA studies for reference gene expression. After MARIA, classification confidence scores >95% were considered as presenting peptides.
Exon ByPASS validation by exon skipping oligonucleotides
PubMed was searched for publications involving exon skipping or splice switching. Obvious off-topic publications and reviews were removed. The gene and exon were then identified from the publication title and abstract. Publications were additionally filtered by any skipping strategies that involved targeting cryptic splice sites. This left 51 unique exons that have been experientially validated with exon skipping oligonucleotides.
RESULTS
Exon ByPASS
Deep neural networks are a good choice when trying to infer complex interactions in sequence data (8,9,33–36). Accordingly, we have built a model, called Exon ByPASS, using deep neural networks based on a CNN-LSTM (convolution neural network—long-short-term memory) (Figure 1A) (37,38). Exon ByPASS predicts the likelihood that an exon of interest can be skipped or is constitutive based on the resulting protein sequence. Previous methods to predict exon criticality have been based on nucleotide sequence (8–12); however, Exon ByPASS considers the possibility that exon skipping constraints can be imposed at the protein level.
Figure 1.
Exon ByPASS predicts exon skipping from protein sequence. (A) Overview of Exon ByPASS, illustrating the 100 × 22 amino acid matrix, with underlying exons encoded into matrix before being passed through a series of convolutional neural network layers. The last layer in the CNN is a fully connected layer which gets passed to an LSTM layer. Finally, the probability that the exon is skipped is relayed to two output neurons. (B) Data from 202 annotated genomes in the Ensembl database were used for training. Exons were labeled as constitutive and skippable for training. Exons encoding only one protein isoform or determined to be constitutive and in-frame were removed from training. Data meeting input criteria were divided, with 90% used for training and 10% used for validation; CNN, convolution neural network; LSTM, long-short-term memory.
Input comprises amino acid sequences upstream and downstream of an exon, allowing us to consider determinants within and adjacent to the exon of interest. After exploring several input lengths, we found that including at least 25 amino acids derived from the upstream exon and 25 derived from the downstream exon provided the most consistent results. Additionally, we used the first 25 amino acids and the last 25 amino acids from the exon of interest. Including the sequences from the flanking exon, the input length totals to 100 amino acids. We padded Exon ByPASS inputs with <100 amino acid inputs, adjusting to achieve a total input of 100 amino acids per exon.
Amino acids sequences are encoded into a 100 × 22 format, where there are 100 amino acid positions with 22 possible identities at each position, including stop codons and ambiguous amino acids (Figure 1A). The data pass through several convolutional layers, a fully connected layer and an LSTM layer. The model output is binary, classifying each exon as skippable or constitutive. We trained the model with the Ensembl (release 100) BioMart database of amino acid sequences containing 202 vertebrate genomes (Figure 1B) (16). Input exons were divided by existing annotations into constitutive (all isoforms contain the exon), skippable (one or more isoforms exclude the exon while retaining adjacent exons) or one annotation (only one isoform has been annotated). Constitutive exons were divided into in-frame and out-of-frame categories. Exons that are constitutive and in-frame were excluded from training and validation to avoid training on skippable exons that have not yet been identified, and these exons are what we considered our test set. Genes with only one transcript annotation were similarly excluded to avoid training on understudied or new genomes that could bias training results. In total, we identified ∼18 million constitutive exons and ∼1 million skippable exons that were divided into a training set and validation set at a ratio of 90% to 10%, respectively. After 10 epochs of training, Exon ByPASS reached 99% accuracy on training data and 93% accuracy on validation data. Exon ByPASS performed similarly to alternative splicing models such SpliceAI (accuracy = 95%) (9); however, it is difficult to directly compare Exon ByPASS to other models because Exon ByPASS considers only exon skipping and evaluates exons as a whole while other models consider all aspects of alternative splicing and evaluate gains or losses in acceptor sites, donor sites and other splicing motifs.
Predicting skippable exons in mouse
To validate existing skipped exons and find novel skippable exons using protein sequence, we applied our model to mouse coding sequences (Genome Reference Consortium mouse build 38, mm10; https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/). We used the model to calculate probabilities for all mouse exons using criteria provided for the model. We classified exons with probabilities >0.5 as skippable and ≤0.5 as constitutive. A majority were predicted to be constitutive (Figure 2A,B). Compared with existing mm10 annotations, we predicted more exons than expected to be skippable (Figure 2A,B; 29,370 annotated as skippable; 89,262 predicted as skippable).
Figure 2.
Validation of predicted skipped exons in mouse using RNA-seq data. (A) Histogram showing probability that an exon is skippable based on protein sequence derived from mm10 coding regions. Constitutive exons have a skip probability ≤0.5 and skippable exons have a skip probability >0.5. (B) Pie chart showing exons that are annotated as constitutive (navy blue) or skippable (light blue) in mm10. Of the 410,069 analyzed exons, 7.2% are annotated as skippable and 92.8% are annotated as constitutive. (C) Pie chart showing exons that are predicted as constitutive (navy blue) or skippable (light blue). Of the 410,069 analyzed exons, 21.8% are predicted as skippable and 78.2% are predicted as constitutive. Pie chart showing the reading frame for exons from the predicted constitutive cohort. 75% are out-of-frame exons (navy blue), and 25% are in-frame exons (light blue). Pie chart showing the reading from for exons predicted to be skippable. >99% are in-frame exons (light blue), and <1% are out-of-frame exons (navy blue). (D) Scheme of data used to validate predicted skipped exons. Mouse RNA-seq data has 12 tissue types with 8 timepoints per tissue (21). These data were used to identify skipped exon junctions reads which are evidence for skipped exons. (E) Distribution of skipped exon-junction counts for predicted constitutive (navy blue) and predicted skippable (light blue) exons. The x-axis is the log of the sum of the skipped exon-junction counts in each sample for all identified skipped exon junctions. (F) Scatter plot showing all identified skipped exon junctions. Each point is a unique skipped exon junction. The y-axis represents the log of the sum of the skipped exon-junction counts in each sample for all identified skipped exon junctions. The x-axis depicts probability of exon skipping based on Exon ByPASS. Exons that are out-of-frame (navy blue) and in-frame (light blue) are represented.
Next, we confirmed that predicted skippable exons retained the reading frame of the transcript, and that reading frame is not a singular feature driving selection of skippable exons (Figure 2C). Differences between Exon ByPASS predictions and annotations arose for test exons that were excluded during training (Figure 1B). Because these exons were omitted from the training data, we predict Exon ByPASS will more accurately classify these exons than existing annotations. To validate predicted skippable exons, we assessed a diverse dataset comprising transcriptomes of 12 mouse tissues sampled at 8 timepoints over 2 days (21). Skipped exon-junction counts were used to gauge performance of Exon ByPASS (Figure 2D).
First, we compared the distribution of skipped exon-junction counts from predicted skippable and constitutive exons (Figure 2E). Most skipped exon-junction counts are predicted as skippable: 85% of these were correctly predicted as skippable. For the 15% that were misclassified, we found that most counts derive from out-of-frame exons (Figure 2F), which could be from transcripts destined for non-sense mediated decay or that are in midst of RNA processing, and additional exon skipping will eventually create an in-frame transcript. Overall, we found substantial agreement between predicted skipped exons and skipped exons validated with junction counts. Next, we looked for instances where the mm10 annotation missed exon skipping events detectable in the RNA-seq data that was predicted by Exon ByPASS. In total, we found 3557 skipped exons in the RNA-seq data that were not annotated but that were predicted by Exon ByPASS (Supplementary Table S1). Including these exons increases the total counts mapping to in-frame skipped exons by >4%. We found that Exon ByPASS predictions outperformed classification of exon skipping events in the RNA-seq dataset mm10 annotations (model AUC = 0.86 (light blue), mm10 AUC = 0.68 (navy blue), DeLong’s test: P < 2.2e-16) (Supplementary Figure S1).
Exon ByPASS predicts skipping independent of tissue
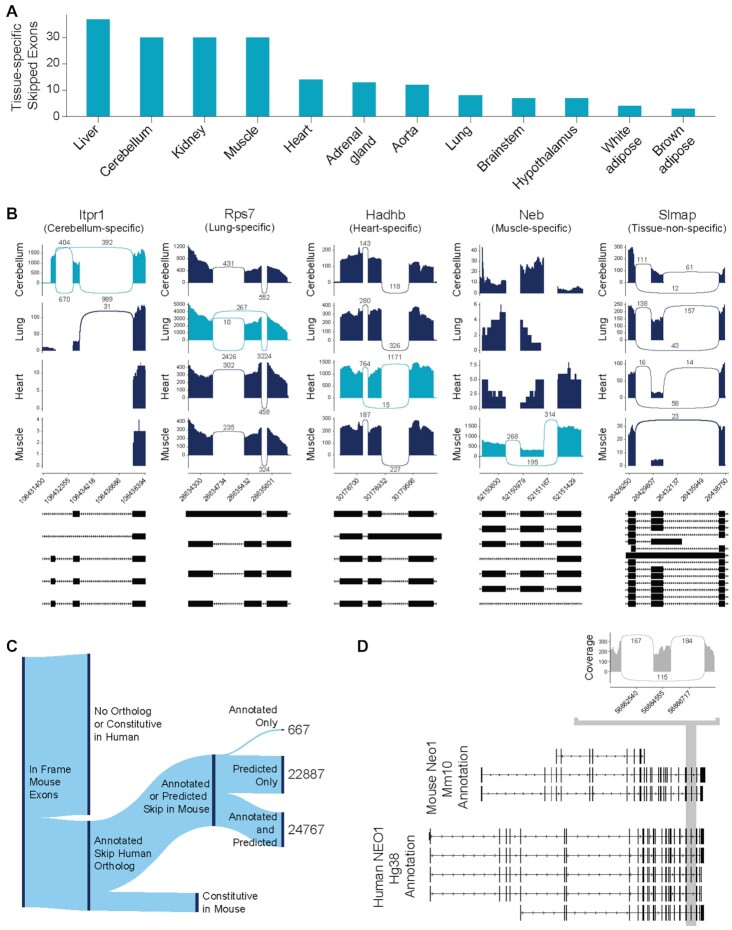
Alternative splicing is largely regulated by tissue-specific factors (6,7,39–41). To examine Exon ByPASS’s ability to predict tissue-independent exon skipping events, we surveyed the mouse RNA-seq dataset (21), considering data from the different tissues separately. As expected, we found exon skipping events across all tissue types (Supplementary Figure S2A,B). For each tissue, we identified at least 20,000 unique skipped exons and, depending on the tissue, 3–6% of these exons had mean counts per million reads (CPM) >1. We also found 195 skipped exons that were predicted to be skipped by Exon ByPASS but were not annotated in mm10 and only skipped in one unique tissue type (Figure 3A). We identified a majority of unannotated exons in liver, cerebellum, kidney and muscle, which is consistent with complex alternative splicing that has been previously associated with these tissues, particularly the cerebellum (39–41). Existing annotations may have also been limited by biased sampling from data used in NCBI pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_euk/process/).
Figure 3.
Exon ByPASS predicts exon skipping independent of tissue and can reconcile human and mouse databases. (A) The number of tissue-specific unannotated but predicted skipped exons with junction counts in RNA-seq data detected in each tissue. (B) Sashimi plots for representative unannotated but predicted skipped exons showing cerebellum-specific splicing for Itpr1 (inositol 1,4,5-trisphosphate receptor type 1) exon 12, lung-specific splicing for Rps7 (ribosomal protein S7) exon 3, heart-specific splicing for Hadhb (hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta) exon 12, muscle-specific splicing for Neb exon 15, and non-tissue-specific splicing for Slmap exon 16. For each panel, expression in representative samples of cerebellum, lung, heart and muscle tissue are depicted. Annotations are based on mm10. (C) Sankey plot showing the breakdown for in-frame mouse exons. Two-thirds of the in-frame mouse exons do not have a human ortholog. The rest have a human ortholog and are annotated as skippable in hg38. Of the annotated skippable exons in humans, 23,554 are also annotated as skippable in mouse. This leaves 24,767 exons that are not annotated as skippable in mouse but that are annotated as skippable in human. (D) Sashimi plot of junction reads for mouse Neo1 exon 26, with 115 reads that skip exon 26 in mouse. Mm10 annotation for Neo1 is shown, with exon 23 highlighted. Hg38 annotation for NEO1 is shown, with region orthologous to mouse Neo1 exon 26 highlighted.
For each tissue, we found at least 1,400 skipped exon junctions that were unannotated but predicted as skipped exon junctions by Exon ByPASS (Figure 3B and Supplementary Figure S2C). In cerebellum, we discovered 2,591 unannotated but predicted skipped exons; for brainstem, we found 2,437 (Supplementary Figure S2D). These observations are consistent with known high levels of alternative splicing in neuronal tissue (39–41). Many of these newly identified skipped exons are not associated with minor isoforms, as 3–6% of them have a mean CPM > 1 (Supplementary Figure S2C,D). In addition to having the most unannotated but predicted skipped exons, cerebellum also had the most highly expressed unannotated but predicted skipped exons, with 141 having mean CPM > 1. The most prevalent skipped exon of those that were unannotated was exon 15 from Neb (nebulin), which was detected in muscle (Figure 3B). Junctions reads skipping exon 15 of Neb have a mean >100 CPM across all muscle tissue samples. Additionally, we identified junction counts for 341 additional unannotated but predicted skipped exons in all 12 tissues.
Model predictions reconcile human and mouse databases
Although we found RNA-seq reads that validate many unannotated skippable exons predicted by Exon ByPASS, confirming all possible skipped exons would require sampling transcriptomes under all conditions that can impact splicing (3,42). To better assess the scope of skippable exons that remain unannotated, we compared annotations in mm10 to better characterized annotations in hg38 and then to Exon ByPASS predictions (Figure 3C). Of 169,895 in-frame mouse exons, we found 24,767 exons which are predicted and not annotated to be skipped despite being annotated as orthologs that are skipped in hg38. More than half of the mouse exons that are orthologs of annotated human skipped exons have been missed by mm10 but were predicted by our model. One example of this is illustrated in Figure 3D. In cerebellum, exon 26 of Neo1 (neogenin 1) is skipped in 115 junction counts. In the mm10 annotation, exon 26 is not skipped; however, in the hg38 annotation, the orthologous exon for NEO1 is predicted to be skipped. In addition to Neo1, we confirmed that 540 exons, which were predicted skippable by Exon ByPASS and have been annotated as skipped in hg38 but have not been comparably annotated in mm10, and are confirmed as skipped in RNA-seq data.
Predicting human skippable exons
We next applied our predictions to human protein sequences from hg38 (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/). Again, we used Exon ByPASS to calculate skip probabilities for all exons according to model criteria (Figure 4A). As expected, we found most exons were constitutive (Figure 4B). However, we found a higher proportion of exons that were annotated and predicted to be skippable in human than mouse. Exon ByPASS predicted 73,041 more exons to be skippable than were annotated as skippable in hg38 (Figure 4C). High numbers of both predicted skipped exons (99%) and predicted constitutive exons (25%) retain the reading frame, indicating that the reading frame cannot be the sole feature driving predictions (Figure 4C).
Figure 4.
Validation of predicted skipped exons in humans using ExonSkipDB. (A) Histogram showing probability that an exon is skippable based on protein sequence derived from hg38 coding regions. Constitutive exons have a skip probability ≤0.5 and skippable exons have a skip probability >0.5. (B) Pie chart showing exons that are annotated as constitutive (navy blue) or skippable (light blue) in hg38. Of the 622,195 analyzed exons, 12.9% are annotated as skippable and 87.1% are annotated as constitutive. (C) Pie chart showing exons that are predicted as constitutive (navy blue) or skippable (light blue). Of the 622,195 analyzed exons, 24.6% are predicted as skippable and 75.4% are predicted as constitutive. Pie chart showing the reading frame for exons from the predicted constitutive cohort. 76.4% are out-of-frame exons (navy blue), and 23.6% are in-frame exons (light blue). Pie chart showing the reading frame for exons predicted to be skippable. >99% are in-frame exons (light blue), and <1% are out-of-frame exons (navy blue). (D) Scheme of data used to validate predicted skipped exons. ExonSkipDB (43) used GTEx and TCGA to identify skipped exons. Additional validated skipped exons were found from VastDB (44). (E) Upset plots showing the overlap of skipped exons as predicted by Exon ByPASS, annotated in hg38, present in GTEx, present in TCGA or present in VastDB. The overlap of predicted but not annotated exons identified by GTEx and TCGA or VastDB is shown (light blue). (F) Violin plot showing skip probability with respect to TCGA (light blue) and GTEx (navy blue). The dashed line demarcates a skip probability of 0.5. (G) Receiver operator curve (ROC) showing Exon ByPass's classification of all hg38 exons. Exon ByPASS predictions compared with skipped exons in TCGA and hg38 annotation (light blue) and compared with skipped exons in GTEx and hg38 annotation (navy blue) are shown.
To validate predicted skippable exons, we evaluated ExonSkipDB (43) and VastDB (44), a database of skipped exons (Figure 4D). Exons in ExonSkipDB derive from RNA-seq data from two major initiatives: Genotype-Tissue Expression project (GTEx) and The Cancer Genome Atlas program (TCGA) (6,45). VastDB uses extensive RNA-seq data and provides additional exons that can be used as a gold standard for comparison. After filtering for in-frame skipped exons, we looked for overlap between predicted skipped exons and exons found to be skipped in GTEx,TCGA or VastDB. As expected, predicted skippable exons overlap substantially with hg38 annotations (Figure 4E, 28,434 exons). Surprisingly, the next most substantial overlap was between predicted skipped exons and exons found to be skipped in VastDB (19,450 exons). Additionally, a large overlap was observed between predicted skipped exons and exons found in GTEx and TCGA (6,979), None of these >25,000 exons are identified in the current hg38 annotation as skippable.
Next, we considered the skip probabilities for all in-frame skipped exons from GTEx and TCGA (Figure 4F). Our model correctly identified more than two-thirds of these exons, with a mean skip probability >0.5. We also observed that Exon ByPASS had >0.5 mean skip probability for all in-frame skipped exons identified by VastDB (Supplementary Figure S3). Next, we compared exon skip probabilities from Exon ByPASS to hg38 annotations and found that the model increased area under the receiver operating characteristic curve (AUC) compared with hg38 annotation (DeLong’s test: GTEx’s P value < 2.2e-16, TCGA’s P value < 2.2e-16; Figure 4G).
Finally, we examined the proportion of Exon ByPASS predictions with respect to PSI (percent spliced in) of exons found in VastDB. By using PSI, which is provided for all in-frame skipped exons by VastDB, we can assess Exon ByPASS accuracy compared to exon-skipping frequency (Supplementary Figure S4). As expected, in both mouse and human, the more frequently an exon is skipped, the more likely Exon ByPASS will call it skippable.
Sequence position and structure contribute to skip probability
To examine the potential driving factors of exon skipping, we looked at the properties of input amino acid sequences from the top 10,000 predicted skippable exons and top 10,000 predicted constitutive exons in hg38. To compare predicted constitutive and skippable exons, we first compared the frequency of each amino acid (Supplementary Figure S5). Predicted skippable exons were found to have a much higher proportion of glycine (Gly) and proline (Pro). Both amino acids have distinct structural features. Gly, without a side chain, is highly flexible, so it is generally not found in helices (46). Pro, whose sidechain in connected to its backbone, is rigid and lacks the hydrogen-bonding potential necessary for helices (47). Because we observed differences in structurally unique amino acids, we compared predicted disorder of constitutive and skippable exons. We used DISpro to calculate a disorder score for 25 amino acids segments across input sequences of predicted skippable exons predicted constitutive exons (48). We found that across each 25 amino acid segment, skippable exons encoded significantly more disordered regions than constitutive exons (Welch’s t-test P-value < 2.2e-16, Figure 5A). To further refine the distinction observed by disorder score, we looked at the physiochemical properties across the input amino acid sequence, including average hydrophobicity, flexibility and net charge across the input sequence. Net charge showed no difference between constitutive and skippable exons. Hydrophobicity is consistently higher at all input positions in constitutive exons and flexibility is consistently higher at all input positions in skippable exons (Supplementary Figure S6). However, there are regions within the input sequences where the difference between constitutive and skippable decreases or increases.
Figure 5.
Amino acids physiochemical properties and amino acid substitutions impact skip probability. (A) Disorder score for the input from the last 25 amino acids of the upstream exon, the first 25 amino acids of the exon of interest, the last 25 amino acids of the exon on interest, and the first 25 amino acids of the downstream exon in the top 10,000 predicted skippable exons and top 10,000 predicted constitutive exons in human genome (hg38). (B) Hydrophobicity, flexibility, and net charge ([Arg + Lys]-[Asp + Glu]) were averaged over a 5 amino acid window across the input sequences for the top 10,000 predicted skippable exons and top 10 000 predicted constitutive exons in human genome (hg38). Plotted error represents standard error. (C) Heatmap of change in probability across amino acid sequence for the averaged 100 × 100 matrix. Pairwise substitution of Ala into predictable skippable human genome (hg38) exons. 10,000 hg38 skippable exons and the change in skip probability when pairwise Ala substitutions are introduced.
To evaluate specific positional features driving exon skipping predictions, we perturbed protein sequences at specific positions. To gauge amino acid sequence associations, we used a pairwise mutational scheme and measured the impact on skip probability. Similar approaches have been used to find sequences that are important features in other neural network models (10). For this evaluation, we selected the 10,000 highest scoring skippable exons from the human genome (Genome Reference Consortium human build 38, hg38: www.ncbi.nlm.nih.gov/assembly/GCF_000001405.26/). For each of the 10,000 exons, we used the 100-position input (as described above) for predictions, but input data were modified iteratively with two alanine (Ala) substitutions within the input sequence. We then quantified the change in skip probability between pairwise substituted sequences and the wild-type sequence for each exon. Because we examined the most highly skippable exons, Ala substitutions rendered them more constitutive. This pairwise substitution resulted in a 100 × 100 matrix for each exon. These 100 × 100 matrices created from pairwise substitution of the 10,000 skippable exons were averaged to create a single matrix that highlights the regions of input that most significantly impact the likelihood an exon can be skipped (Figure 5). The largest perturbations come from pairwise Ala substitutions encoded by the upstream and downstream exons. Large changes in probability were also observed when pairwise substitutions were made in sequences encoded by the upstream exon alone, or in combination with substitutions in the first half of the exon of interest. Surprisingly, minimal changes in probability were observed when pairwise substitutions were made in sequences encoded by the exon of interest. Similarly, minimal changes were observed when single Ala substitutions were made in the input.
Exon ByPASS identifies exons amenable to exon-skipping oligonucleotides
Since Exon ByPASS relies on protein data to make predictions, the model may predict compatible protein sequences after an exon is skipped regardless of whether it occurs naturally. Thus, there is a possibility that Exon ByPASS can predict exons that can be skipped through synthetic means, which would be a useful tool in the discovery of exon-skipping targets for oligonucleotide therapeutics. These therapeutic oligonucleotides can be used to skip over exons containing disease-causing mutations in order to restore protein expression or normal protein function (49). To examine this application of Exon ByPASS, we conducted an extensive literature search for oligonucleotides that mediate exon skipping. We found 51 exons that were validated experimentally as skippable using oligonucleotides. Of these 51 exons, Exon ByPASS correctly predicted 41 of them as skippable, which was 10 more than the number of exons found in VastDB and 33 more than are annotated as skippable (Supplementary Table S2). High concordance between experimentally confirmed skippable exons and Exon ByPASS makes Exon ByPASS a viable tool for the prediction of therapeutic exon skipping targets.
Proteomic validation of predicted exons
Finally, we extended our validation to consider proteomic data from CPTAC (Clinical Proteomics Tumor Analysis Consortium) (28), focusing on the Cancer Proteome Study of Breast Tissue. The study consisted of 105 proteome samples from breast cancer tissue as well as a 3 proteome samples from normal breast tissue (50). To compare predictions and annotations, we first created a theoretical peptide database containing all possible peptides derived from skipped junctions that could arise from the human proteome consistent with input criteria for Exon ByPASS (Figure 6A). Using Philosopher and MsFragger, we compared this theoretical database to proteomic data from above (30,31). In total, we found 81 unique peptides that are indicative of skipped exons (Figure 6B). Of these, 54 peptides matched predicted skipped exons, whereas only 40 matched annotated skipped exons. Of the 14 peptides that are predicted but not annotated, the mean skip probability was 0.937, indicating that the skipped exons are highly predictable.
Figure 6.
Peptides from breast cancer samples overlap with predicted skipped exons. (A) Scheme of theoretical peptide database created to compare to proteomics data from CPTAC study of breast tissue. The database contains all possible peptide junctions that could arise from skipped exons. (B) Bar plot showing unique peptides derived from skipped exon junctions. All skipped exon junctions (black) were not filtered by annotations or model predictions. Predicted (light blue) skipped exon junctions are skipped exon junctions where exon skip probability is >0.5. Annotated (navy blue) skipped exon junctions are skipped exon junctions that are annotated in hg38. (C) Annotation of APAF1 in hg38 with amino acid sequences for exon 6, exon 7 and exon 8 shown. (D) Dot plot showing the abundance of the predicted APAF1 peptide in healthy (light blue) and cancer tissue (navy blue) per multiplexed sample. (E) Image of cryo-EM structure of APAF1 (55). The position of exon 7 (light blue) and ATP (multicolored spheres) are indicated.
The potential impact that the discovery of unannotated skipped exons can have on cancer treatment merits attention. Neoantigens—those produced specifically by cancer cells—may be a source for novel targets for immunotherapies (51,52). Cancer can impact splicing, so skipped exons are a likely source for neoantigens (53). Neo-junctions (neoantigens derived from skipped exons) have been shown to occur at a higher frequency than neoantigens derived from single nucleotide variant (SNV) somatic mutations and are more likely to be shared by patients (29). To this end, we explored the CPTAC Study of Breast Tissue to identify neoantigens produced by unannotated skippable exons. To identify potential neo-junctions, we compared annotated and predicted peptide junctions and used MARIA (MHC Analyzer with Recurrent Integrated Architecture) to find HLA class II presenting peptides (32). We found 9 peptide junctions in the CPTAC study that have not been annotated as skipped but were predicted (i) as skippable by Exon ByPASS and (ii) as HLA class II presented peptides by MARIA (Supplementary Table S3).
For example, we found the peptide ‘VQPDGVTLEYNPYSWNLVAQSNFEALQDFFR.’ This peptide maps to an unannotated skipped CTSA (cathepsin A) transcript derived from the end of exon 4 and the start of exon 6. The presence of this peptide suggests CTSA exon 5 is skipped in breast cancer cells. Exon ByPASS predicts CTSA exon 5 as highly skippable (probability > 0.9), and MARIA predicts this peptide as likely for HLA class II presenting (normalized score = 99.299). This peptide is likely missed by databases derived from hg38 and would not be considered as a potential neoantigen for the development of immunotherapies.
Skipping of APAF1 exon 7 is prevalent in breast cancer samples
In addition to expanding the neo-junction search space, identifying proteins derived from unannotated skipped exons could reveal novel protein structure and function. We once again interrogated the CPTAC Study of Breast Tissue, looking for peptides derived from skipped exons in genes that could be drivers of cancer progression. Comparing peptides found in cancer samples to those found in healthy samples, we identified a peptide (SVTDSVMGSPLVVSLIGALLR) that was enriched in cancer samples. This peptide aligns to the APAF1 (apoptotic peptidase activating factor 1) protein and corresponds to the end of exon 6 and the start of exon 8 (Figure 6C), suggesting the presence of an alternatively spliced APAF1 transcript missing exon 7. This variant of APAF1 is not annotated in hg38, but it was predicted by Exon ByPASS (exon 7 skip probability = 0.953). Proteomics data from multiple cancer samples support the presence of this peptide (Figure 6D), indicating that APAF1 likely undergoes alternative splicing.
APAF1 is activated by p53 and supports p53-mediated apoptosis by promoting the activation of caspase-9 (54–56). Although this isoform of APAF1 has not been studied, the missing exon 7 corresponds to a region within the NB-ARC domain of the protein, which contains an ATP-binding pocket (55) (Supplementary Figure S7 and Figure 6E). This pocket hydrolyses ATP, inducing a conformational change that promotes the formation of the active apoptosome (55). The peptide encoded by exon 7 sits near bound ATP (Figure 6E) and may impact ATP binding and hydrolysis. An APAF1 isoform that lacks this peptide could have compromised apoptosome activity, which would compromise its ability to activate caspase-9 and apoptosis, potentially enabling cells expressing this isoform to escape programmed cell death.
DISCUSSION
We built Exon ByPASS to test the hypothesis that constraints on protein structure hold enough information to predict whether a coding exon can be removed. After training, Exon ByPASS reached 99% accuracy and 93% accuracy on validation data from the Ensembl database (release 100), indicating that protein sequence is sufficient to predict exon criticality. To the best of our knowledge, Exon ByPASS is the first alternative splicing model that relies solely on protein features.
In addition to predicting known skippable exons, Exon ByPASS predicted novel exon skipping events, even in well-annotated mouse and human genomes (mm10 and hg38, respectively). Using an extensive mouse RNA-seq dataset (21), we assessed the predictions of Exon ByPASS in the context of mouse annotations (mm10). This evaluation identified 3,557 unannotated exons that were predicted to be skippable by Exon ByPASS and that are supported by the RNA-seq dataset. Using a similar approach, we evaluated Exon ByPASS using extensive human RNA-seq datasets (43,44) and found many unannotated exons that are predicted to be skippable by Exon ByPASS and that are supported by the data sets. In a parallel analysis of proteomic data, we identified peptides that likely derive from exon-skipping events predicted by ByPASS that are not annotated. This analysis revealed novel peptides enriched in cancer samples that could serve as neoantigens for immunotherapy, as well as a novel peptide from APAF1 that could result from an unannotated alternative splicing event in cancer samples and help to explain how these cells could escape programmed cell death. Finally, we used exon-skipping oligonucleotide literature to show that Exon ByPASS can predict synthetically skippable exons as well.
Together, these data suggest that protein sequences can be used to predict skippable exons and that this approach is applicable across species. The interpretation of ‘omics data sets notably suffers from incomplete annotations, as these approaches rely on comparisons to a reference data set. As the number of transcriptomic and proteomic studies increases, the quality of our annotated references becomes increasingly important. Exon ByPASS has the potential to improve annotations of reference genomes across species. Furthermore, Exon ByPASS can be used in the search for therapeutics. A literature search in PubMed suggests that only 51 exons have been confirmed as skippable with oligonucleotides. Exon ByPASS can be used to search for additional exons that may be amenable to therapeutic skipping approaches. Additionally, Exon ByPASS can be used to search for neo-antigens from skipped exons in cancer cells. Regardless of genetic changes that disrupt splicing, Exon ByPASS will reveal compatible protein sequences when exons are skipped, providing a reliable set of protein sequences that can produce neo-junctions, which can potentially be exploited for immunotherapies.
Our analysis with Exon ByPASS revealed several characteristics of amino acid sequences in proximity to the splice junctions that are significantly associated with exon criticality. Notably, sequences that are compatible with alternative splicing tend to have a lower average hydrophobicity and, relatedly, a higher average flexibility. It has been reported that low average hydrophobicity is a characteristic of intrinsically disordered regions and that these regions are often associated with splice junctions, our findings are consistent with this observation (57–60). Our pairwise amino acid substitution analysis identified a reduced likelihood of alternative splicing following perturbation of amino acid pairs upstream and downstream of the skipped exon (Figure 5). In the skipped isoform, these pairs would be separated by approximately 25–30 amino acids and may reflect the important role of loops in globular protein structures (61,62). Interestingly, these features differ from those suggested by RNA-based models, which tend to emphasize features immediately adjacent to the junctions. Although we identified several features that correlate with exon criticality, our understanding remains incomplete. We know what positions and which amino acids influence exon skipping; however, we do not know how these amino acids interact with the rest of the protein to accommodate exon skipping. More investigations at the protein level should provide additional insights into exon criticality. One avenue for investigation could be to compare structures of proteins derived from splice variants. By comparing enough protein structures, additional governing features would likely emerge. Unfortunately, very few proteins in the Protein Data Bank have structures for more than one isoform (60). Additional structures could help provide insight into the protein properties required for exon skipping.
Others have developed machine-learning models to predict alternative splicing (8–12). Some of these models work well, achieving both high sensitivity and specificity. They can also predict other alternative splicing outcomes. For example, MaxEntScan and SpliceAI can find both 5′ and 3′ splice sites for an exon based on RNA sequence (9,11). MaxEntScan uses a probabilistic model to evaluate splice site potential given a short RNA sequence. SpliceAI uses a neural network model to predict splice junctions given a long RNA sequence. Both models predict exon skipping, intron retention, as well as alternative 5′- or 3′-splice sites based on input RNA sequence. Because of the limitations imposed by the training set, Exon ByPASS is not suitable for predicting intron retention or finding alternative 5′- or 3′-splice sites. These limitations were necessary to ensure correct classification in the training set; however, alternate strategies could be used to extend protein sequence modeling to these other forms of alternative splicing.
In conclusion, Exon ByPASS can reliably predict exon criticality from protein sequence. Exon ByPASS can be applied to all protein-coding genes, across tissues and, based on work reported here, may have applications in therapeutic discovery.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Amy Donner and Eric Smith for editorial and graphical support, respectively. Additionally, the authors would like to thank Heta Desai for guidance on proteomics data analysis.
Contributor Information
Jigar Desai, Wave Life Sciences, Cambridge, MA 02138, USA.
Christopher Francis, Wave Life Sciences, Cambridge, MA 02138, USA.
Kenneth Longo, Wave Life Sciences, Cambridge, MA 02138, USA.
Andrew Hoss, Wave Life Sciences, Cambridge, MA 02138, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wave Life Sciences. Funding for open access charge: Wave Life Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Frazer K.A. Decoding the human genome. Genome Res. 2012; 22:1599–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chanock S. Toward mapping the biology of the genome. Genome Res. 2012; 22:1612–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S.et al.. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park E., Pan Z., Zhang Z., Lin L., Xing Y.. The expanding landscape of alternative splicing variation in human populations. Am. J. Human Genet. 2018; 102:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The GTEx Consortium The GTEx consortium atlas of genetic regulatory effects across human tissues. Science. 2020; 369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B.. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008; 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barash Y., Vaquero-Garcia J., González-Vallinas J., Xiong H., Gao W., Lee L.J., Frey B.J.. AVISPA: a web tool for the prediction and analysis of alternative splicing. Genome Biol. 2013; 14:R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B.et al.. Predicting splicing from primary sequence with deep learning. Cell. 2019; 176:535–548. [DOI] [PubMed] [Google Scholar]
- 10. Dogan R.I., Getoor L., Wilbur W.J., Mount S.M.. SplicePort–An interactive splice-site analysis tool. Nucleic Acids Res. 2007; 35:W285–W291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 12. Cartegni L. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003; 31:3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen M., Manley J.L.. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009; 10:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wahl M.C., Will C.L., Lührmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 15. Pleiss J.A., Whitworth G.B., Bergkessel M., Guthrie C.. Transcript specificity in yeast Pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007; 5:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smedley D., Haider S., Ballester B., Holland R., London D., Thorisson G., Kasprzyk A.. BioMart – biological queries made easy. BMC Genomics. 2009; 10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kingma D.P., Ba J.. Adam: a method for stochastic optimization. 2017; arXiv doi:30 Jan 2017, preprint: not peer reviewedhttps://arxiv.org/abs/1412.6980.
- 18. Rose G., Geselowitz A., Lesser G., Lee R., Zehfus M.. Hydrophobicity of amino acid residues in globular proteins. Science. 1985; 229:834–838. [DOI] [PubMed] [Google Scholar]
- 19. Karplus P.A., Schulz G.E.. Prediction of chain flexibility in proteins: a tool for the selection of peptide antigens. Naturwissenschaften. 1985; 72:212–213. [Google Scholar]
- 20. Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.-Y., El-Gebali S., Fraser M.I.et al.. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019; 47:D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang R., Lahens N.F., Ballance H.I., Hughes M.E., Hogenesch J.B.. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. USA. 2014; 111:16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. [DOI] [PMC free article] [PubMed]
- 24. Liao Y., Smyth G.K., Shi W.. The r package rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019; 47:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garrido-Martín D., Palumbo E., Guigó R., Breschi A.. ggsashimi: sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput. Biol. 2018; 14:e1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Helaers R., Bareke E., De Meulder B., Pierre M., Depiereux S., Habra N., Depiereux E.. gViz, a novel tool for the visualization of co-expression networks. BMC Res Notes. 2011; 4:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M.. pROC: an open-source package for r and S+ to analyze and compare ROC curves. BMC Bioinf. 2011; 12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ellis M.J., Gillette M., Carr S.A., Paulovich A.G., Smith R.D., Rodland K.K., Townsend R.R., Kinsinger C., Mesri M., Rodriguez H.et al.. Connecting genomic alterations to cancer biology with proteomics: the NCI clinical proteomic tumor analysis consortium. Cancer Discov. 2013; 3:1108–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kahles A., Lehmann K.-V., Toussaint N.C., Hüser M., Stark S.G., Sachsenberg T., Stegle O., Kohlbacher O., Sander C., Rätsch G.et al.. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell. 2018; 34:211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kong A.T., Leprevost F.V., Avtonomov D.M., Mellacheruvu D., Nesvizhskii A.I.. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods. 2017; 14:513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma K., Vitek O., Nesvizhskii A.I.. A statistical model-building perspective to identification of MS/MS spectra with peptideprophet. BMC Bioinf. 2012; 13:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen B., Khodadoust M.S., Olsson N., Wagar L.E., Fast E., Liu C.L., Muftuoglu Y., Sworder B.J., Diehn M., Levy R.et al.. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 2019; 37:1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alipanahi B., Delong A., Weirauch M.T., Frey B.J.. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015; 33:831–838. [DOI] [PubMed] [Google Scholar]
- 34. Lv Z., Ding H., Wang L., Zou Q.. A convolutional neural network using dinucleotide One-hot encoder for identifying DNA N6-Methyladenine sites in the rice genome. Neurocomputing. 2021; 422:214–221. [Google Scholar]
- 35. Kim H.K., Lee S., Kim Y., Park J., Min S., Choi J.W., Huang T.P., Yoon S., Liu D.R., Kim H.H.. High-throughput analysis of the activities of xCas9, SpCas9-NG and spcas9 at matched and mismatched target sequences in human cells. Nat. Biomed. Eng. 2020; 4:111–124. [DOI] [PubMed] [Google Scholar]
- 36. Rives A., Meier J., Sercu T., Goyal S., Lin Z., Guo D., Ott M., Zitnick C.L., Ma J., Fergus R.. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences synthetic biology. PNAS. 2021; 118:e2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krizhevsky A., Sutskever I., Hinton G.E.. ImageNet classification with deep convolutional neural networks. Commun. ACM. 2017; 60:84–90. [Google Scholar]
- 38. Hochreiter S., Schmidhuber J.. Long short-term memory. Neural Comput. 1997; 9:1735–1780. [DOI] [PubMed] [Google Scholar]
- 39. Noh S.-J., Lee K., Paik H., Hur C.-G.. TISA: Tissue-specific alternative splicing in human and mouse genes. DNA Res. 2006; 13:229–243. [DOI] [PubMed] [Google Scholar]
- 40. Xu Q. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002; 30:3754–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yeo G., Holste D., Kreiman G., Burge C.B.. Variation in alternative splicing across human tissues. Genome Biol. 2004; 5:R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Team T.M.P., Temple G., Gerhard D.S., Rasooly R., Feingold E.A., Good P.J., Robinson C., Mandich A., Derge J.G., Lewis J.et al.. The completion of the mammalian gene collection (MGC). Genome Res. 2009; 19:2324–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim P., Yang M., Yiya K., Zhao W., Zhou X.. ExonSkipDB: functional annotation of exon skipping event in human. Nucleic Acids Res. 2019; 48:D907–D896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tapial J., Ha K.C.H., Sterne-Weiler T., Gohr A., Braunschweig U., Hermoso-Pulido A., Quesnel-Vallières M., Permanyer J., Sodaei R., Marquez Y.et al.. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017; 27:1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. The Cancer Genome Atlas Research Network Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R.M., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M.. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013; 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Högel P., Götz A., Kuhne F., Ebert M., Stelzer W., Rand K.D., Scharnagl C., Langosch D. Glycine perturbs local and global conformational flexibility of a transmembrane helix. Biochemistry. 2018; 57:1326–1337. [DOI] [PubMed] [Google Scholar]
- 47. Morgan A.A., Rubenstein E.. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One. 2013; 8:e53785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng J., Sweredoski M.J., Baldi P.. Accurate prediction of protein disordered regions by mining protein structure data. Data Min Knowl Disc. 2005; 11:213–222. [Google Scholar]
- 49. Li D., Mastaglia F.L., Fletcher S., Wilton S.D.. Precision medicine through antisense oligonucleotide-mediated exon skipping. Trends Pharmacol. Sci. 2018; 39:982–994. [DOI] [PubMed] [Google Scholar]
- 50. The cancer genome atlas network comprehensive molecular portraits of human breast tumours. Nature. 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schumacher T.N., Schreiber R.D.. Neoantigens in cancer immunotherapy. Science. 2015; 348:69–74. [DOI] [PubMed] [Google Scholar]
- 52. Jiang T., Shi T., Zhang H., Hu J., Song Y., Wei J., Ren S., Zhou C.. Tumor neoantigens: from basic research to clinical applications. J. Hematol. Oncol. 2019; 12:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Slansky J.E., Spellman P.T.. Alternative splicing in tumors — a path to immunogenicity?. N. Engl. J. Med. 2019; 380:877–880. [DOI] [PubMed] [Google Scholar]
- 54. Qin H., Srinivasula S.M., Wu G., Fernandes-Alnemri T., Alnemri E.S., Shi Y.. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999; 399:549–557. [DOI] [PubMed] [Google Scholar]
- 55. Zhou M., Li Y., Hu Q., Bai X., Huang W., Yan C., Scheres S.H.W., Shi Y.. Atomic structure of the apoptosome: mechanism of cytochrome c - and dATP-mediated activation of Apaf-1. Genes Dev. 2015; 29:2349–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saleh A., Srinivasula S.M., Acharya S., Fishel R., Alnemri E.S.. Cytochrome c and dATP-mediated oligomerization of apaf-1 is a prerequisite for procaspase-9 activation. J. Biol. Chem. 1999; 274:17941–17945. [DOI] [PubMed] [Google Scholar]
- 57. Ellis J.D., Barrios-Rodiles M., Çolak R., Irimia M., Kim T., Calarco J.A., Wang X., Pan Q., O’Hanlon D., Kim P.M.et al.. Tissue-Specific alternative splicing remodels protein-protein interaction networks. Mol. Cell. 2012; 46:884–892. [DOI] [PubMed] [Google Scholar]
- 58. Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Çolak R.et al.. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012; 338:1587–1593. [DOI] [PubMed] [Google Scholar]
- 59. Romero P.R., Zaidi S., Fang Y.Y., Uversky V.N., Radivojac P., Oldfield C.J., Cortese M.S., Sickmeier M., LeGall T., Obradovic Z.et al.. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. 2006; 103:8390–8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang P., Yan B., Guo J.-t., Hicks C., Xu Y.. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc. Natl. Acad. Sci. 2005; 102:18920–18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Berezovsky I.N., Trifonov E.N.. Loop fold nature of globular proteins. Protein Eng. 2001; 14:403–407. [DOI] [PubMed] [Google Scholar]
- 62. Berezovsky I.N., Guarnera E., Zheng Z.. Basic units of protein structure, folding, and function. Prog. Biophys. Mol. Biol. 2017; 128:85–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.