CONSPECTUS:
Over the past decade, we have contributed to the chemistry of microbial natural products and synthetic ligands, related to riboflavin and uracils, that modulate immune cells called mucosal associated invariant T cells (MAIT cells). These highly abundant T lymphocytes were only discovered in 2003 and have become recognized for their importance in mammalian immunology. Unlike other T cells, MAIT cells are not activated by peptide or lipid antigens. In collaboration with immunology and structural biology research groups, we discovered that they are instead activated by unstable nitrogen-containing heterocycles synthesized by bacteria. The most potent naturally occurring activating compound (antigen) is 5-(2-oxopropylideneamino)-D-ribitylaminouracil (5-OP-RU). This compound is an imine (Schiff base) formed through condensation between an intermediate in the biosynthesis of riboflavin (vitamin B2) and a metabolic byproduct of mammalian and microbial glycolysis. Although it is very unstable in water due to intramolecular ring closure or hydrolysis, we were able to develop a non-enzymatic synthesis that yields a pure kinetically stable compound in a nonaqueous solvent. This compound has revolutionized the study of MAIT cell immunology due to its potent activation (EC50 = 2 pM) of MAIT cells and its development into immunological reagents for detecting and characterizing MAIT cells in tissues. MAIT cells are now linked to key physiological processes and disease, including antibacterial defense, tissue repair, regulation of graft-vs-host disease, gastritis, inflammatory bowel diseases, and cancer. 5-OP-RU activates MAIT cells and, like a vaccine, has been shown to protect mice from bacterial infections and cancers. Mechanistic studies on the binding of 5-OP-RU to its dual protein targets, the major histocompatibility complex class I related protein (MR1) and the MAIT cell receptor (MAIT TCR), have involved synthetic chemistry, 2D 1H NMR spectroscopy, mass spectrometry, computer modeling and molecular dynamics simulations, biochemical, cellular, and immunological assays, and protein structural biology. These combined studies have revealed structural influences for 5-OP-RU in solution on protein binding and antigen presentation and potency; informed the development of potent (EC50 = 2 nM) and water stable analogues; led to fluorescent analogues for detecting and tracking binding proteins in and on cells; and enabled discovery of drugs and drug-like molecules that bind MR1 and modulate MAIT cell function. MAIT cells offer new opportunities for chemical synthesis to enhance the stability, potency, selectivity, and bioavailability of small molecule ligands for MR1 or MAIT TCR proteins, and to contribute to the understanding of T cell immunity and the development of prospective new immunomodulating medicines.
Graphical Abstract
1. INTRODUCTION
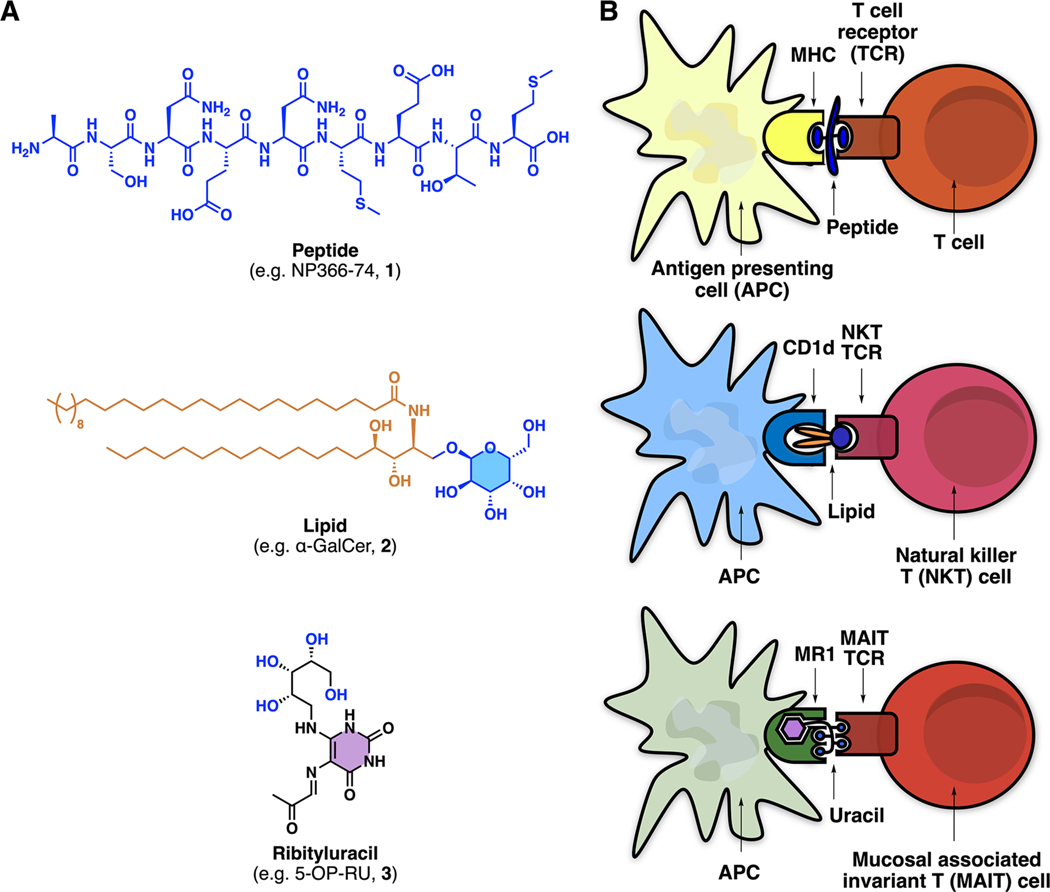
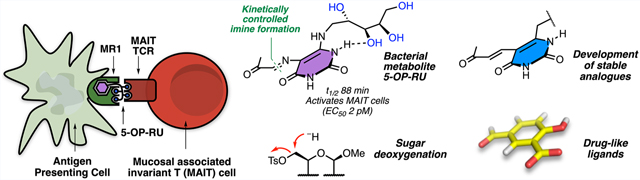
T cells (T lymphocytes) are key cellular mediators of adaptive immunity and are activated by peptide5 or lipid6 antigens (e.g., 1 or 2; Figure 1A). A new type of highly abundant unconventional T cell discovered in 2003 is the mucosal associated invariant T cell (MAIT cell), which is not activated by peptides or lipids7 and is becoming progressively more recognized for its importance in mammalian immunology and disease.8 The riboflavin biosynthesis pathway in bacteria was implicated in 2012 as the source of naturally occurring ligands for mammalian MAIT cells,7 but the structures of the most potent naturally occurring antigens were not reported until 2014 as water-unstable heterocyclic uracil derivatives, such as 5-(2-oxopropylideneamino)-D-ribitylaminouracil (5-OP-RU, 3; Figure 1A).1
Figure 1.
Three compound classes that activate T lymphocytes. (A) Examples of peptide, lipid, and ribityluracil antigens that activate T cells. (B) A peptide, lipid, or ribityluracil antigen first binds to an antigen-presenting protein (MHC, CD1, or MR1, respectively) expressed on an antigen-presenting cell (APC) and then interacts with the T cell receptor (TCR) expressed on a T cell, NKT cell or MAIT cell, respectively. Adapted with permission from ref 9. Copyright 2012 Springer Nature.
The mechanism by which they activate T cells differs from most ligand−receptor binary complexes (e.g., ligand complexes of G-protein coupled receptors). First, the antigen (peptide, lipid, or uracil) binds to an antigen-presenting protein (MHC, CD1, or MR1, respectively; Figure 1B) expressed on an antigen-presenting cell (APC; Figure 1B), which is usually a dendritic cell, macrophage, or B cell. This antigen−protein complex engages a second protein (T cell receptor) expressed on a T cell, NKT cell, or MAIT cell for peptide, lipid, or heterocyclic antigens, respectively. This complex mechanism enables sophisticated control over the degree of antigen presentation to the TCR, with both antigen and antigen-presenting protein cooperatively interacting with the TCR to trigger T cell activation. Despite mechanistic similarities, the three antigen classes interact differently with both antigen-presenting proteins and TCRs (Figure 1B). Peptides sandwich between TCR and MHC proteins using their side chains and main chains to interact with both proteins. Lipids such as α-GalCer embed their lipid chains in CD1d and use their cyclic sugar moiety to interact with NKT TCR. By contrast, ribityluracils such as 5-OP-RU use their uracil moiety to interact with MR1 and their ribityl group to interact with both MR1 and MAIT TCR, as discussed ahead.
Discovery of this third type of antigen, exemplified by the nitrogen-containing heterocycle 5-OP-RU, has helped catalyze a recent explosion of MAIT cell biology (MAIT publications/ year: 41/2005, 170/2012, 6200/2020). The availability of synthetic 5-OP-RU has enabled potent activation of MAIT cells, and production of antigen−MR1 tetramers for the detection and immunological characterization of mammalian MAIT cells in physiology and disease. In turn, this has led to MAIT cells being implicated in antimicrobial defense (bacterial,10–12 viral13), tissue repair,14,15 cancer,16 and inflammatory diseases.17 Here, we describe the identification of ligands that bind MR1 protein and activate or inhibit MAIT cells.
2. DISCOVERY OF ANTIGENS FOR MAIT CELLS
Nonstimulatory MR1 Ligands Derived from Vitamin B9
The discovery of bacterial ligands that bind MR1 and modulate MAIT cells was the result of an interdisciplinary collaboration between the McCluskey (immunology, University of Melbourne), Rossjohn (structural biology, Monash University), and Fairlie (chemistry, University of Queensland) research groups.18 It required exchange of materials, protocols, information, and sharing of expertise between laboratories in two cities and three universities to enable efficient identification and characterization of the highly reactive antigens.
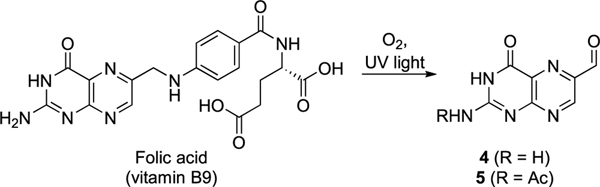
The first breakthrough was the discovery that 6-formylpterin (6-FP, 4), a photodegradation product of folic acid (vitamin B9) in protein buffer, could refold MR1 (Scheme 1).7,9 A crystal structure of 4 bound to MR1 and MAIT TCR showed that its aldehyde formed a reversible covalent bond (Schiff base) with the Lys43 amine side chain in MR1 (PDB: 4GUP). Although 6-FP could bind, refold, and upregulate MR1 expressed in APCs, it could not stimulate MAIT TCR activation. Instead, it weakly inhibited activation of MAIT TCR by bacterial supernatants. Acetyl-6-formylpterin (Ac-6-FP, 5) also upregulated MR1, inhibiting MAIT cell activation more potently (IC50, 0.1 μM). Although nonstimulatory, 4 and 5 provided the first clues to the structural requirements for MR1 binding and refolding.
Scheme 1. MR1 Ligands 6-Formylpterin (4) and Acetyl-6-formylpterin (5) That Weakly Inhibit MAIT Cell Activation.
Stimulatory Ligands Derived from Vitamin B2 (Riboflavin)
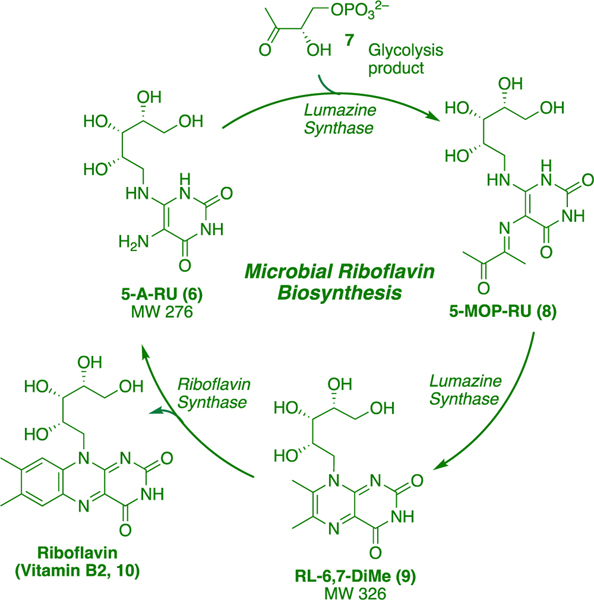
A second breakthrough came through genetic studies on Gram-positive and Gram-negative bacteria, which showed that only bacteria possessing enzymes for riboflavin biosynthesis could produce ligands that bound MR1 and stimulated MAIT TCR.1 MR1 could be refolded by the supernatant of Salmonella typhimurium, a riboflavin-producing bacterium. The molecular formulas of ligands that bound and refolded MR1 were established through EI-MS and isotopic mass distribution as C12H18N4O7 (m/z 329.1099, negative mode) and C11H16N4O7 (m/z 315.0944, negative mode). We examined the riboflavin biosynthetic pathway (Scheme 2) for compounds matching these formulas. The key biosynthetic intermediate 5-A-RU (6) condenses with glycolysis product 3,4-dihydroxy-2-butanone-4-phosphate (7) to give 5-(1-methyl-2-oxopropylideneamino)-6-D-ribitylaminouracil (5-MOP-RU, 8), a process catalyzed by lumazine synthase. This intermediate readily undergoes dehydrative ring closure (also catalyzed by lumazine synthase) to form bicyclic 6,7-dimethyl-8-D-ribityllumazine (RL-6,7-DiMe, 9). Riboflavin synthase then catalyzes the disproportionation of two molecules of 9 to give riboflavin (10) and 5-A-RU (6), which re-enters the catalytic cycle. However, none of these compounds matched the molecular formula of the bacterial isolate.
Scheme 2. Biosynthesis of Riboflavin (10).
Chemical Synthesis of Riboflavin Biosynthetic Intermediates and Metabolites
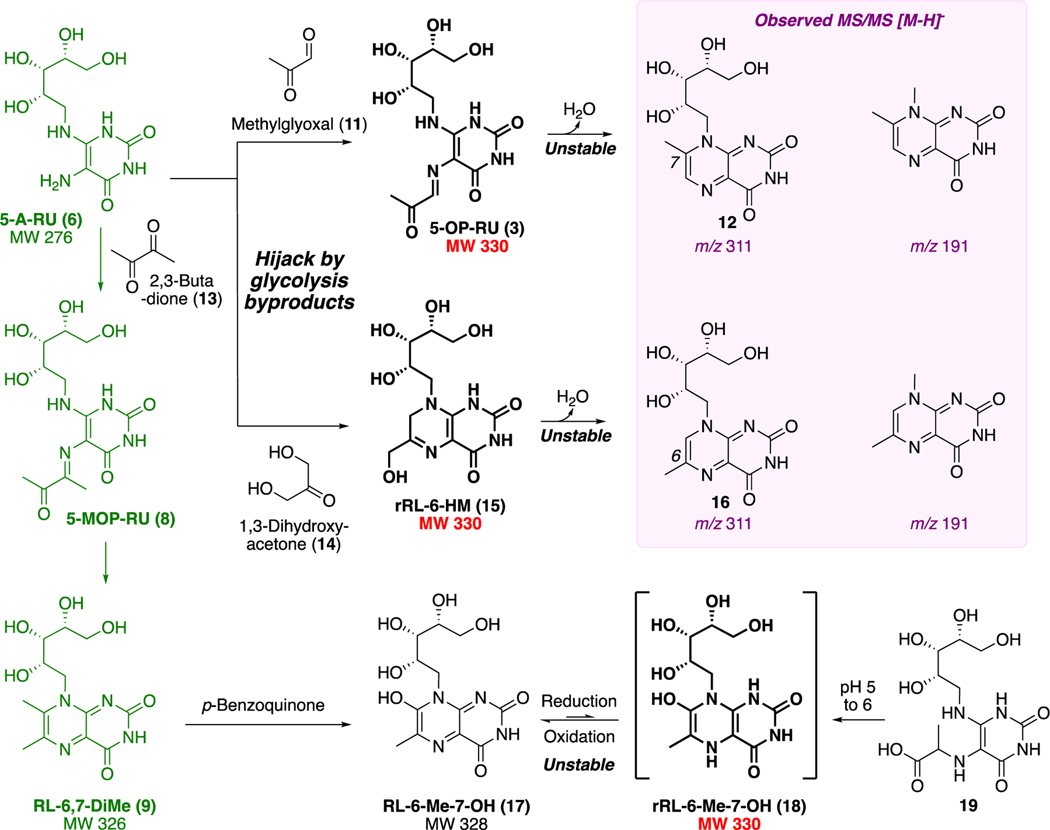
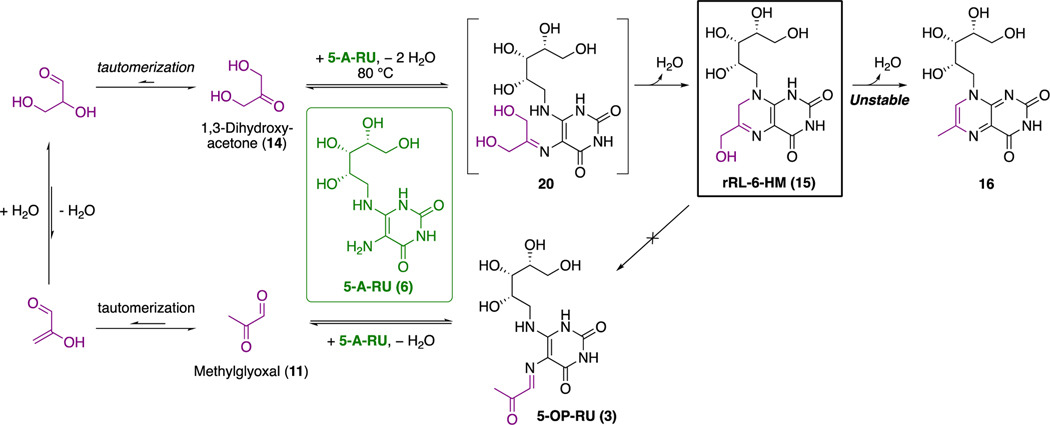
To explore other compounds in the pathway as putative antigens, we took advantage of the knowledge that key intermediate 5-A-RU (6) was also known to non-enzymatically produce RL-6,7-DiMe (9; MW 326),19 which can undergo further benzoquinone-mediated oxidation to metabolite 17 (RL-6-Me-7-OH; MW 328; Scheme 3). A two-electron reduction of 17 could theoretically give 18 (rRL-6-Me-7-OH; MW 330). Alternatively, cyclocondensation of 6 with glycolysis byproduct20 1,3-dihydroxyacetone (14) could yield 15 (rRL-6-HM; MW 330). Therefore, we undertook chemical syntheses of 9, 15, 17, and 18 in search of the potent MAIT-activating antigen. Reaction of pH-adjusted aqueous 5-A-RU (6) with 1,3-dihydroxyacetone (14) gave 15 in situ, which was prone to aromatizing oxidation or dehydration to form 16. Switching the dicarbonyl electrophile to 2,3-butanedione (13) gave 9 (RL-6,7-DiMe), which was oxidized to 17 (RL-6-Me-7-OH). However, we could not isolate 18 (rRL-6-Me-7-OH), as it was very susceptible to oxidative rearomatization to RL-6-Me-7-OH (17) and only transiently detectable during reduction of 17 or ring closure of acid 19 (Scheme 3). All three synthetic compounds (9, 15, and 17) activated MAIT TCR, measured by upregulation of surface marker CD69, interferon-γ, and tumor necrosis factor, but samples of rRL-6-HM (15) were the most potent by orders of magnitude.
Scheme 3. Putative Antigens Derived from Riboflavin Biosynthetic Intermediatesa.
aUnstable 3, 15, and 18 have molecular formulas matching the stimulatory ligand in the bacterial supernatant; 3 and 15 have near identical MS/MS fragments (violet). Data from ref 7. and ref 1.
Unexpected Capture of 5-OP-RU by MR1
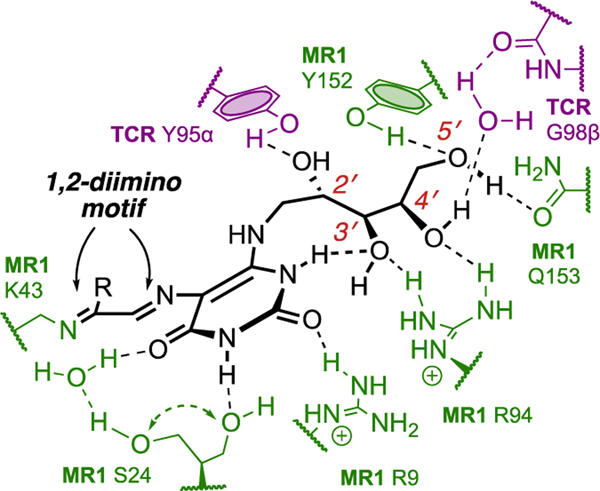
Compound 15 was generated over 5−20 min from pH-adjusted aqueous 5-A-RU (6) and 1,3-dihydroxyacetone (14), liberated from its commercially available dimer in situ by heating to 80 °C. Product 15 was unstable and not isolated completely pure, but it possessed the same MS/MS fragmentation pattern and properties (ability to refold MR1 and activate MAIT TCR) as the antigen in the bacterial supernatant. It was therefore used for cocrystallization with MR1 and MAIT TCR proteins. Instead of 15, the structure of the ternary complex (Figure 2) showed that MR1 and MAIT TCR had preferentially captured a trace amount of the highly unusual ring-opened imine isomer 5-OP-RU (3), one of the possible condensation products of 6 and 11 (Scheme 3). Like 4 and 5,7 5-OP-RU (3) was covalently bonded to the side chain amine of Lys43 in MR1, forming an imine (Figure 2). Although ligand-protein interactions forming imines (Schiff bases) are known in biology (pyridoxal phosphate, fructose 1,6-bisphosphate, retinal),21 the covalent linkage between the uracil and MR1 (Figure 2) involves two consecutive Schiff bases, creating a 1,2-diimino motif (-NH═CR1−CR2═N-) unprecedented in biological chemistry.
Figure 2.
Hydrogen bonds between 5-OP-RU (black) and protein residues of MAIT TCR (violet) and MR1 (green) as revealed by the crystal structure (PDB: 4NQC). Data from ref 1.
In addition to the imine being prone to hydrolysis, 5-OP-RU (3) was poised for a 6-exo-trig aromatizing cyclocondensationbetween the carbonyl and nucleophilic vinylogous amide nitrogen to give 12. LCMS indicated that an isomer of 15 was indeed present as a low-level impurity in the sample of 15 used for crystallization (only later confirmed as 5-OP-RU after its successful synthesis, vide infra) and that trace amounts of methylglyoxal (11) could also be formed in situ during synthesis of 15 (Schemes 3 and 4).
Scheme 4. Proposed Origin of 5-OP-RU (3) During the Synthesis of 15a.
aTautomerization22 and in situ dehydration of 1,3-dihydroxyacetone (14) to trace methylglyoxal (11) at 80 °C could produce 5-OP-RU (3) during the synthesis of rRL-6-HM (15).
Synthesis of 5-OP-RU
We sought to synthesize pure 5-OP-RU (3) from 5-A-RU (6) and glycolysis byproduct23 methylglyoxal (11; Scheme 3). Several challenges needed to be overcome. First, reaction of diamine 6 with dicarbonyl 11 was known to form bicyclic lumazine 12,24 with 3 postulated to be a fleeting intermediate that simultaneously forms and degrades. Second, the condensation could form multiple regioisomers. We attempted to use LCMS and NMR spectroscopy to detect the compound in situ during its transient formation from 5-A-RU (6) and methylglyoxal (11) in water (5-A-RU is insoluble in many solvents). However, we detected only transient formation of trace amounts of 5-OP-RU, and the accumulation of lumazine 12 and its 6-methyl regioisomer (16). Varying the temperature, concentration, and pH all changed the reaction rate and regioisomeric distribution, but did not enable isolation of 5-OP-RU from water.
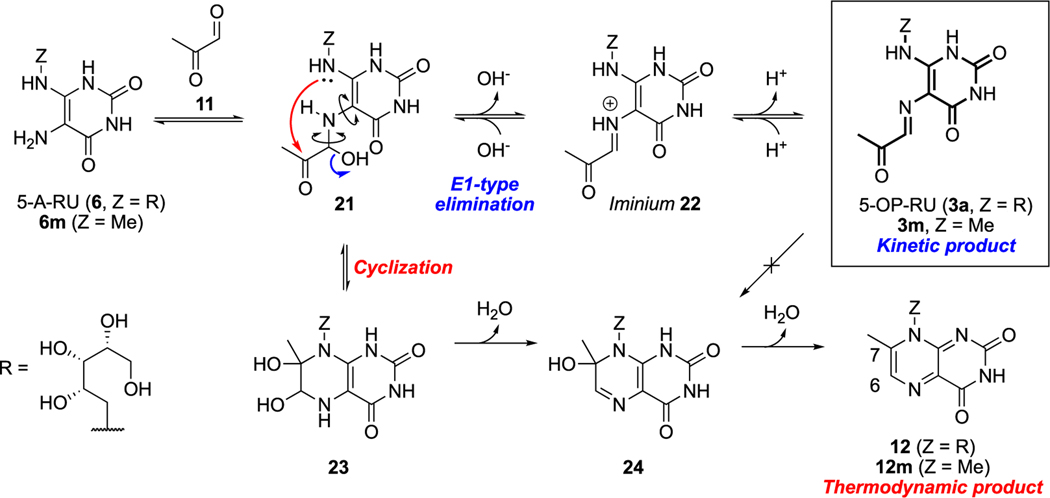
Reexamination of the mechanism of 5-OP-RU (3) formation and degradation led to the next breakthrough (Scheme 5). We noted that although 5-OP-RU cyclized readily, the E-configuration of the imine held the carbonyl and nucleophilic nitrogen apart. Thus, for 5-OP-RU to cyclize, the imine must first rehydrate to hemiaminal 21. This enables rotation of both C−N bonds highlighted in 21 (Scheme 5), allowing intramolecular condensation. Therefore, 5-OP-RU was not a reactive intermediate but rather a kinetically stable product that was thermodynamically unstable but potentially isolatable under carefully controlled conditions.
Scheme 5. Mechanism of Formation (Blue) and Degradation (Red) of 5-OP-RU (3a) and Model Compound 3ma.
aData from ref 2.
To investigate this possibility, we focused on controlling the fate of hemiaminal 21, the intermediate from which 5-OP-RU (kinetic product) or the lumazine (thermodynamic product via 23 and 24) would competitively form. We hypothesized that, as imine formation in polar solvents typically proceeds via the iminium intermediate (22), aprotic polar solvents might stabilize the cation while avoiding solvent attack on iminium 22, thus diverting the reaction toward 5-OP-RU.
Thus, we condensed methylglyoxal with model compound 6m in solvents of increasing dielectric constants, namely, nitromethane-d3, DMF-d7, and DMSO-d6 (Scheme 5). While 6m was sparingly soluble in nitromethane, and DMF gave a mixture of imine 3m and lumazine 12m, DMSO exclusively yielded 3m, providing evidence for our mechanistic proposal. DMSO was then similarly employed to deliver 5-OP-RU (3a) as a single product, unambiguously characterized by 2D NMR spectroscopy.
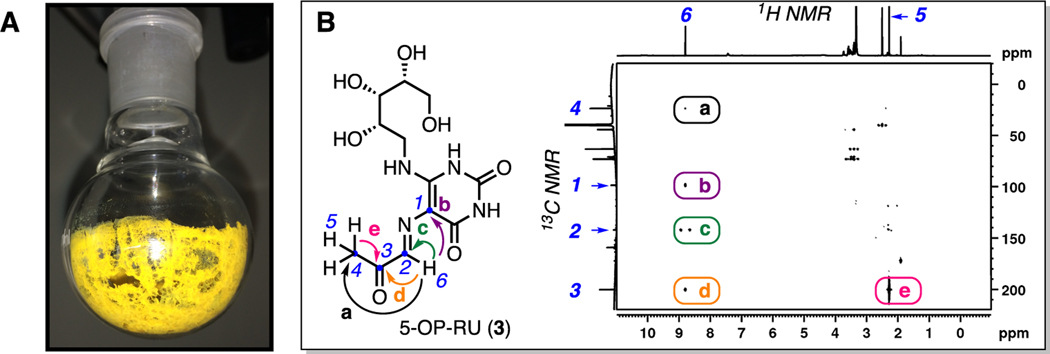
Although 5-OP-RU is reasonably pure and characterizable in situ in DMSO-d6, we purified 5-OP-RU further by rapid reversed phase HPLC using an acetonitrile−water gradient to definitively prove its structure and acquire it in pure solid form (Figure 3A). Speed, neutralization of product fractions, and lyophilization under acid-free conditions were critical to success. The solution structure of 5-OP-RU was established in 2013 by HRMS and 2D-NMR spectroscopy, with the 1H−13C HMBC NMR spectrum establishing the structure of the distinctive iminocarbonyl group and its connectivity to the uracil ring (Figure 3B). The material matched the crystal structure of proteinbound 5-OP-RU obtained from the sample of 15, but pure 5-OP-RU was remarkably more potent in activating MAIT cells (EC50, 2 pM),2 thus securing the identity of the bacterial MAIT cell antigen. This extremely potent antigen is indefinitely stable in DMSO at −20 °C, with a half-life of 88 min under aqueous conditions at 37 °C, pH 5.4.2 The m/z 315 species from the bacterial supernatant was attributed to its aldehyde homologue 5-OE-RU, which was similarly synthesized from 5-A-RU and glyoxal (another glycolysis byproduct23) and characterized spectroscopically and crystallographically (PDB 4NQE),1 but it was much less stable2 and more difficult to purify.
Figure 3.
(A) Kinetically stable 5-OP-RU after HPLC purification, and (B) characterization of 5-OP-RU (3) by 1H−13C HMBC NMR spectroscopy in DMSO-d6. Key correlations (colored arrows and round boxes, labeled a−e) established the connectivity of atoms (numbered 1−6) in the iminocarbonyl group. Data from ref 2.
Impact of 5-OP-RU
Synthetic 5-OP-RU (3) has made an indelible impact on MAIT cell immunology. The MR1−(5-OP-RU) binary complex is recognized by MAIT TCR (and slight variants), which is uniquely expressed on MAIT cells. Synthetic 5-OP-RU has been used to form fluorophoric immunological tetramers,25,26 consisting of four MR1−(5-OP-RU) complexes tethered together. These reagents selectively bind MAIT TCRs and are regarded as the gold standard27 for identifying and staining MAIT cells, since even antibodies raised against MAIT TCR exhibit nonspecific binding to other TCRs. These tetramers have been distributed worldwide8 through licensing of our patents to the U.S. National Institutes of Health (NIH) Tetramer Facility.28
While early studies relied on bacteria or supernatant mixtures as the antigen source, availability of pure 5-OP-RU (3) allowed MAIT cells to be activated with much greater efficiency and precision. This enabled collaborators to discover the requirement of cofactors or co-stimulants for the expansion of MAIT cell numbers (e.g., toll-like receptor 2 agonists),29 lung accumulation (CXCL16),30 augmented effector function (α-ketoglutarate),31 and vaccination (IL-23).11 It was used to interrogate the intracellular pathway32,33 of antigen capture and presentation by MR1, and to study an MR1 mutation in a person without MAIT cells.34 Other collaborators used 5-OP-RU to study MAIT cell activation in gastritis,17 graft-vs-host disease,35 and antibiotic resistsance.36 Similarly, the ability for 5-OP-RU to selectively activate MAIT cells has been used to detect MAIT cell (or MAIT-like) populations in animals in the absence of species-specific MR1 tetramers.37–39 Landmark studies found that 5-OP-RU controls MAIT cell development in the thymus,40–42 imprints MAIT cells in early life, and promotes tissue repair.14
Medicinal applications are also being explored. MAIT cells are attractive vaccine targets,43 and 5-OP-RU (3) can confer antibacterial protection against Legionella longbeachae and Francisella tularensis infections in mice.10–12 5-OP-RU used as a treatment during chronic Mycobacterium tuberculosis infection in mice increased MAIT cell expansion and reduced bacterial loads.44 5-OP-RU also conferred protection in mouse models of B16F10 (melanoma) and E0771 (breast) metastasized lung cancer,45 suggesting the potential for 5-OP-RU in cancer immunotherapy. Collectively, these findings highlight the importance of this natural product as a tool for MAIT cell immunology and immunotherapeutics.
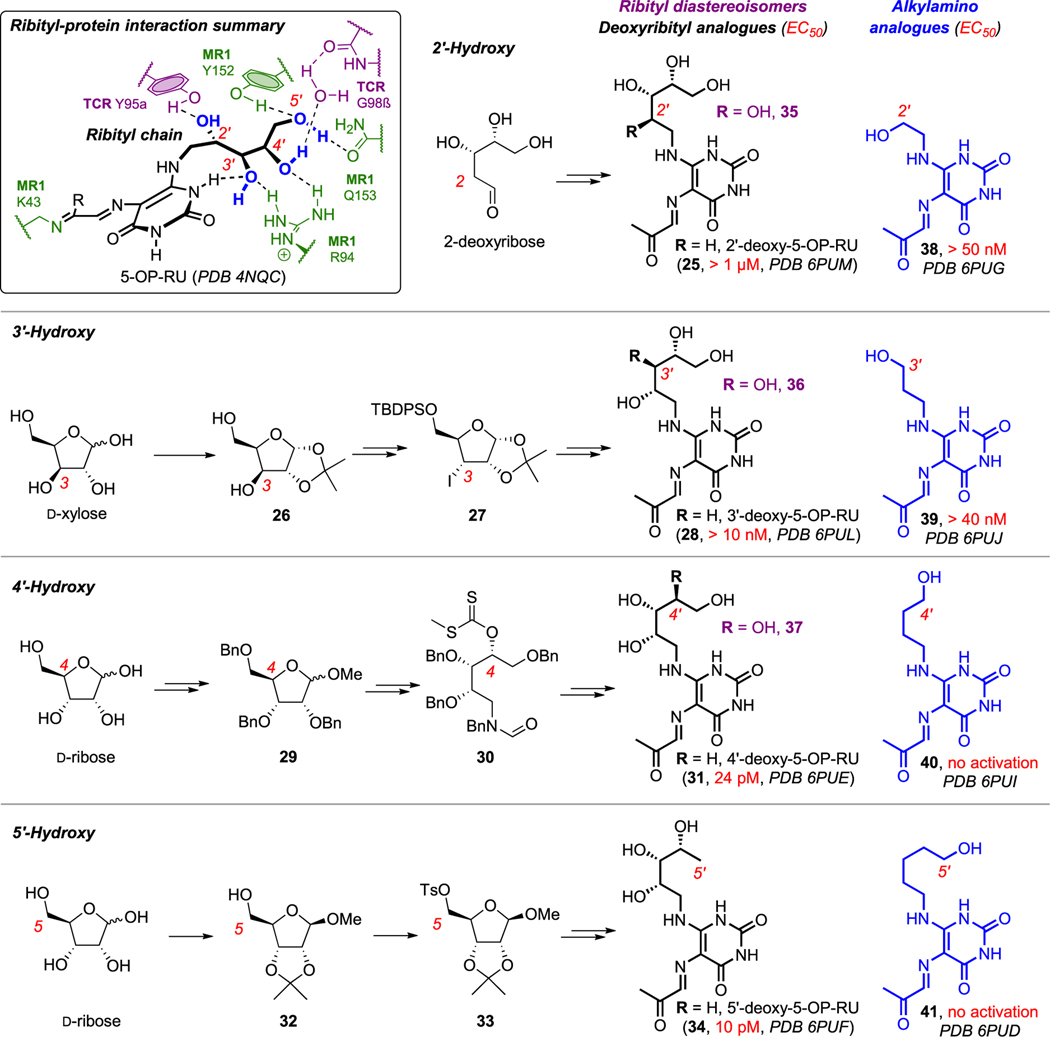
3. ROLE OF THE RIBITYL CHAIN
In view of the remarkable immunogenicity of 5-OP-RU (3), and the known roles of sugars in mediating cell adhesion and recognition, we sought to explore the immunological importance of the 5-OP-RU ribityl chain, which resides at the interface between MR1 and MAIT TCR. DFT calculations and molecular dynamics (MD) simulations suggested different influences from each hydroxyl group, so we synthesized all four deoxyribityl analogues of 5-OP-RU by merging the corresponding deoxyribitylamines into the synthesis of 5-OP-RU (black, Scheme 6).3
Scheme 6. Summary of Interactions between the Ribityl Chain of 5-OP-RU with MR1 and MAIT TCR (Inset) 1 and Synthetic Analogues of 5-OP-RU for Investigating the Roles of Ribityl Hydroxyl Groups in MAIT Cell Activation via Cellular Assays and Protein−Ligand Crystal Structuresa.
aPDB codes: 4NQC, 6PUM, 6PUG, 6PUL, 6PUJ, 6PUE, 6PUI, 6PUF, and 6PUD. Data from ref 3 and ref 46.
The 2-deoxy analogue of 5-OP-RU (25) was accessed from commercial 2-deoxyribose. The 3-deoxy analogue (28) was synthesized from D-xylose (the C3-OH epimer of D-ribose); this epimer’s stereochemistry enabled facile regioselective protection (26) to allow C3-OH radical deoxygenation via iodide 27. Meanwhile, 4- and 5-deoxy analogues (31 and 34) were synthesized from D-ribose via orthogonal protection (29 and 32) and then radical deoxygenation of 30 or hydride displacement of 33, respectively. We similarly synthesized four monohydroxylalkyl analogues (blue, 38−41) from commercial aminoalcohols.
Next, we examined the analogues for MR1 binding and MAIT TCR activation through cellular assays and protein−ligand crystal structures (PDB codes in Scheme 6).46 Loss of the 2′-OH led to considerable reduction of MAIT TCR activation potency. Although it formed the only direct hydrogen bond with MAIT TCR (Scheme 6 inset), its removal surprisingly did not ablate activity. This could be partially due to 3′-OH compensating through a new interaction with MAIT TCR Tyr95α, as observed in MD simulations in water and supported by the solid-state crystal structure. Loss of 3′-OH also significantly reduced potency. Although it does not interact with TCR (Scheme 6 inset),1 MD simulations suggest it is important for restricting the ribityl conformation and projecting other hydroxyls for interaction with MR1 and MAIT TCR. It does so through an intramolecular H-bond between 3′-OH and a uracil NH, conceptually similar to conformation-stabilizing intramolecular H-bonds in cyclic peptides.47 Conversely, although 4′- and 5′-OH interact with MR1 R94, Q153, Y152, and TCR G98β (through water; Scheme 6 inset),1 loss of either 4′- or 5′-OH had little effect on MAIT TCR activation. Using protein crystal structures of 31 and 34, MD simulations revealed that these hydroxyl groups could attenuate activation potency by hindering protein−protein interactions. Monohydroxylalkyl analogues were significantly less potent than deoxy analogues, but hydroxyethyl (38) and hydroxypropyl (39) analogues were the most potent within that series, consistent with 2′-OH and 3′-OH interacting with MAIT TCR Tyr95α.
Others have determined activities for other diastereoisomeric48,49 sugar and alkyl50 analogues (Scheme 6, purple and blue, respectively). Their reported biological data show trends similar to our findings, but should be interpreted cautiously. For example, one study systematically inverted stereochemistry at each ribityl hydroxyl group and similarly found that 2′-OH and 3′-OH epimers (35 and 36) but not 4′-OH (37) lost potency.49 However, they reported EC50 100 nM for 5-OP-RU (3), compared to our EC50 2 pM.2 Another study on sugar analogues found that the 2′-OH epimer (35) was less active than 5-OPRU.48 However, all of those compounds exhibited comparable activities, including 5-OP-RU (3), its 2-deoxy analogue (25), and 2′-OH, 3′-OH, and 4′-OH epimers (35−37), likely because they were all tested at a single high concentration (10 μM). Another study found that hydroxyethyl and hydroxypropyl analogues 38 and 39 were the most potent hydroxyalkyl analogues, but 38 was reported to be equipotent with 5-OP-RU (EC50 ~ 1 μM).50
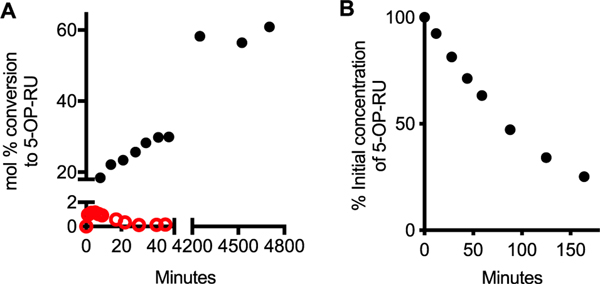
We attribute these discrepancies to the very low conversion to 5-OP-RU (or analogues) in water relative to DMSO (Figure 4, Scheme 5).2 Although we reported early MAIT TCR activation data for 5-OP-RU derived from its synthesis in water (EC50 ~10 μM),1 researchers need to know that aqueous preparations result in extremely low yields and rapid degradation of 5-OP-RU and analogues (Figure 4).2 We recommend the use of DMSO for substantially higher yields (Figure 4A), purity, and quantitation by NMR spectroscopy,51 facilitating comparison with true antigen potency.2 Water exposure should be minimized to limit 5-OP-RU degradation, although small amounts of water in DMSO can be tolerated. We found that NMR quantified51 5-OP-RU, generated in situ from 5-A-RU52 and methylglyoxal (1.1 eq, 40% aqueous solution) in dry DMSO-d6 as described,2 can be used directly in assays without further purification, with no reduction of activity compared to lyophilized material from HPLC purification, provided that the final DMSO concentration in aqueous media does not affect cell viability. Importantly, once 5-OP-RU binds MR1, it is sequestered, stabilized, and no longer susceptible to degradation.
Figure 4.
(A) Formation of 5-OP-RU in DMSO at 22 °C (black) vs PBS at 37 °C (red). (B) Degradation of 5-OP-RU (pH 7.4 PBS, 37 °C). Data from ref 2.
The literature qualitatively corroborates our findings, emphasizes the importance of MAIT TCR Tyr95α engagement for TCR activation, and demonstrates the exceptional power of MD simulations for rationalizing structure−activity relationships in these dynamic systems. Although protein crystal structures containing deoxyribityl analogues were all very similar, MD simulations in a water environment uniquely correlated MR1-TCR SPR affinity and MAIT TCR activation potencies with ligand and MR1 residue flexibility.3,46 Together, these newly revealed determinants of potency and use of computer modeling and MD simulations3,46 to account for flexibility in solution provide important new clues for analogue design.
4. STABLE ANALOGUES OF 5-OP-RU
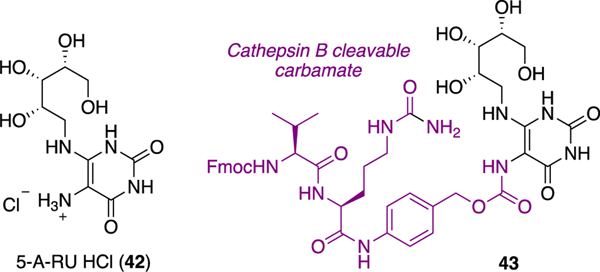
Instability of 5-OP-RU (3) and analogues in water2 is a significant limitation for immunologists, pharmacologists, and medicinal chemists in probing MAIT cell biology and potential applications of activators as vaccines and immunostimulants, or inhibitors as anti-inflammatory agents.43 Other groups have addressed the instability of precursor 5-A-RU (6, prone to aerial oxidation) by isolating its hydrochloride salt (42)27 or forming a prodrug53 (43) (Figure 5), both of which can very weakly activate the MAIT TCR after reacting with cellular metabolites to form trace amounts of 5-OP-RU and 5-OE-RU in situ. However, we focused instead on stabilizing the antigen 5-OP-RU (Scheme 7).
Figure 5.
Stabilizing 5-A-RU via its HCl salt (42) or carbamate prodrug (43).
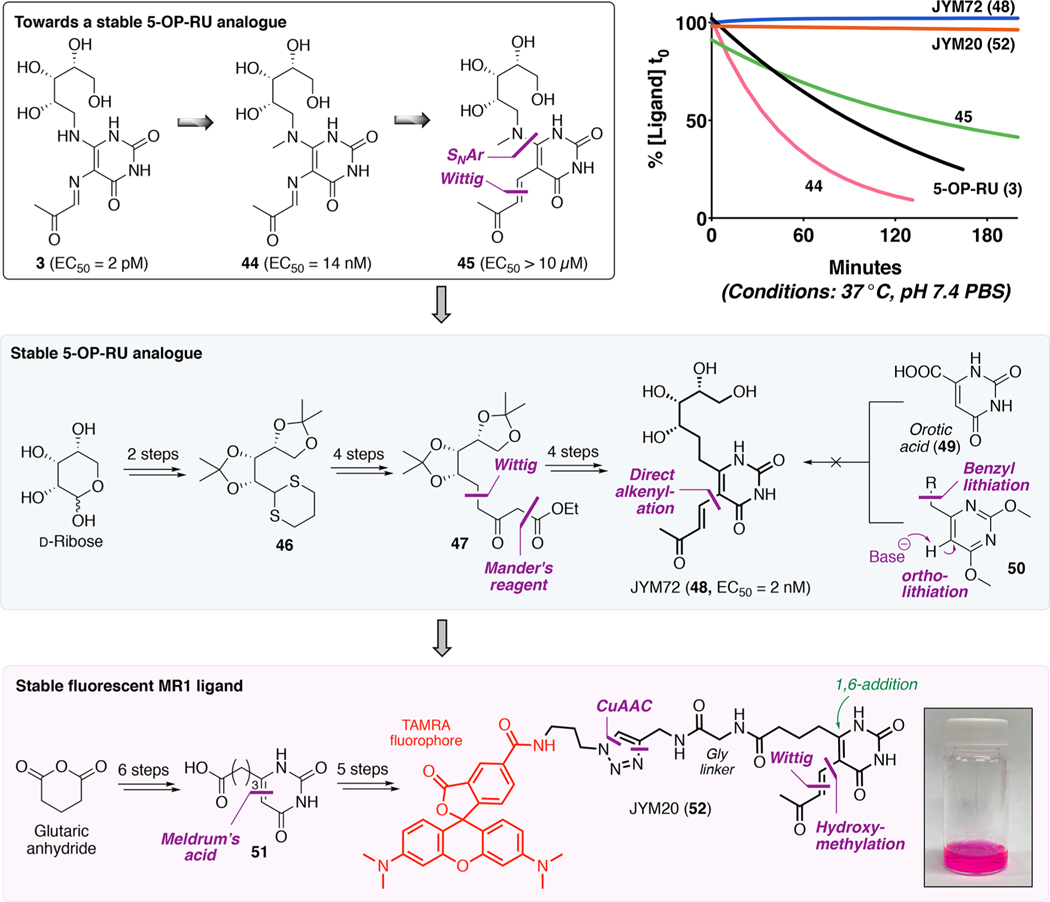
Scheme 7. Development of Stable 5-OP-RU Analogue JYM72 (48) and Stable Fluorescent MR1 Ligand JYM20 (52)a.
The 5-OP-RU protein crystal structure1 indicated that the two exocyclic nitrogens, implicated in its degradation (Scheme 7), did not directly contact the proteins (although the vinylogous amide formed a water-bridged hydrogen bond with MR1 Tyr62), which gave logical starting points for modifications. Initially, we methylated the vinylogous amide to block cyclization (44 and 45; Scheme 7). Surprisingly, this reduced both stability and antigen potency, which we attributed to sterically induced torsional twisting of the ribityl and carbonyl groups, as supported by calculations.2
To solve this problem, we designed JYM72 (48) to replace both nitrogen atoms with carbon. Although routes via orotic acid (49) or ortho-lithiation (50) failed, the analogue could be synthesized in 10 steps from D-ribose. The target enone side chain posed a challenge, but a pragmatic approach with excess base and 4-(trimethylsilyl)but-3-yn-2-one was used to construct the all-carbon backbone in moderate yields. This analogue was stable in water and functionally comparable to 5-OP-RU (3), including its ability to bind MR1, activate MAIT TCR, expand MAIT cells in mouse lung, and induce cytokines ex vivo.2 However, it was 1000-fold less potent than 5-OP-RU in vitro and in vivo,2 possibly because the sp2-nitrogen to sp3-carbon replacement increased ribityl chain flexibility, affecting its projection in the MR1-MAIT TCR complex (PDB: 6PUK).46 Nonetheless, JYM72 (48) was a water-stable and potent antigen (EC50 2 nM) that illustrates the potential for rational development of stable antigens.
MHC and CD1 antigen-presenting proteins typically present intracellularly derived antigens, but MR1 presents extracellular bacterial antigens. This may be important for restricting MAIT cell activation to when APCs are also present, thereby enabling co-stimulatory responses. However, it was unclear whether extracellular antigens bound to MR1 on the cell surface or after entering antigen-presenting cells and encountering intracellular MR1. To investigate this, we developed a chemically stable fluorescent analogue of 48, namely, JYM20 (52), enabling the cellular location of MR1 binding to be visualized by microscopy.32 JYM20 lacked the MAIT TCR-binding ribityl chain of JYM72, greatly simplifying its synthesis (via 51) and permitting stepwise installation of the enone side chain. This side chain doubly activates the uracil ring for 1,6-addition by amines, so we introduced it after the glycyl linker, which incorporates two polar amides to increase water solubility.
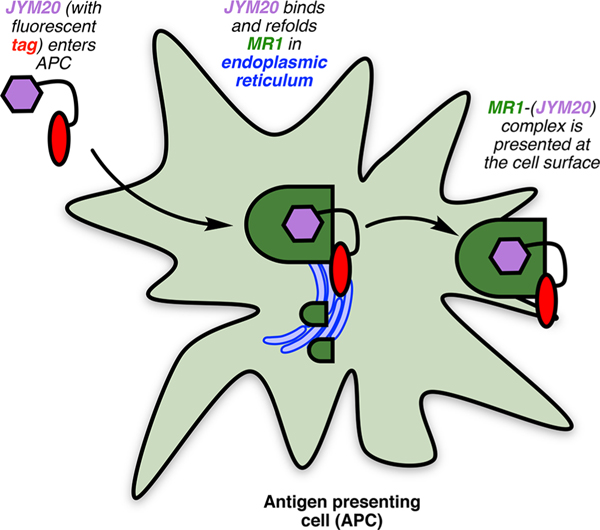
Despite the simplifications, JYM20 (52) behaved like 5-OP-RU (3) in cell uptake kinetics, MR1 binding, and appearance at the cell surface.32 Without the ribityl hydroxyls, it did not activate MAIT TCR but did inhibit 5-OP-RU-mediated activation, consistent with competitive binding to MR1.32 Microscopy and biochemical experiments indicated that JYM20 (52) first enters cells to bind MR1 in the endoplasmic reticulum (Figure 6),32 refolding MR1 to enable translocation of the MR1−ligand complex to the APC surface for MAIT TCR engagement.32 This illustrates the distinctiveness of the MAIT cell activation process, although the mechanism of cell entry and which specific APCs present antigens to MAIT cells in different tissues are not yet established. Nevertheless, we envisage that JYM20 (52) would find general utility for characterizing other aspects of MR1 biology, and as an archetype of other tagged MR1-binding ligands incorporating bespoke payloads.
Figure 6.
Cellular pathway of MAIT cell ligand presentation.
In summary, more stable potent activators of MAIT cells are desired with a longer duration of action, ease of handling, storage, and distribution. In view of the potent immunostimulating properties of 5-OP-RU (3), but lack of aqueous stability, an equipotent stable analogue could represent a key advance, but the challenge is to confer stability without sacrificing potency. Our success in developing stable ligands equipotent with proteins that bind to other cell surface receptors54 gives us confidence this goal can be reached.
5. DRUG-LIKE LIGANDS
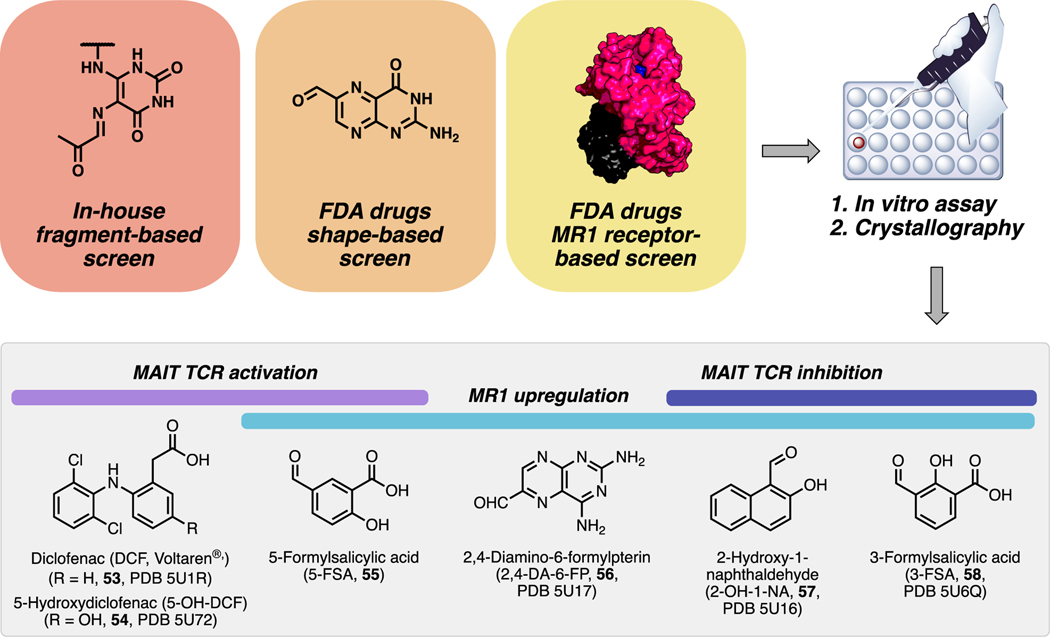
MR1 has multiple tyrosines that form a hydrophobic cleft in which uracils and pterins bind, so we probed whether drugs and drug-like molecules could also bind MR1. We used in silico virtual screening to examine molecular fragments similar to 5-OP-RU from a library of 6000 commercial compounds (Figure 7).4 We also used a library of FDA drugs to perform a shapebased screen on Ac-6-FP (5) and a receptor-based screen on the 5-OP-RU binding site in MR1. Of 81 compounds assayed for binding MR1 and modulating MAIT TCR, some hit compounds were studied using protein crystallography (PDB codes in Figure 7).
Figure 7.
Drugs and drug-like compounds that bind MR1 and activate or inhibit MAIT TCR or upregulate MR1 expression in antigen-presenting cells, and PDB codes for ligand−protein crystal structures (PDB 5U1R, 5U72, 5U17, 5U16, and 5U6Q). Data from ref 4.
A diverse range of putative MR1-binding ligands, including salicylates, diclofenac, and flavonoids weakly modulated MAIT cells (Figure 7).4 New ligands that upregulated MR1 expression included 55−58. Compounds 57 and 58 also inhibited 5-OP-RU-mediated activation of the MAIT TCR. MR1 upregulating ligand 55 modestly activated MAIT TCR, while noncovalent anti-inflammatory drug diclofenac 53 and major metabolite 54 activated the TCR more potently without triggering MR1 upregulation. Interestingly, these ligands activated distinct MAIT TCR variants derived from different MAIT cell subsets, suggesting the potential for future subset-specific chemical modulation.
This study was the first to show that drugs and drug-like molecules could activate MR1 and the MAIT TCR. As expected from unoptimized hits, most of these ligands lacked potency. For example, Ac-6-FP (5; Scheme 1) was a more potent upregulator of MR1 and inhibitor of MAIT TCR, while 55 was ~107 times less potent than 5-OP-RU. Nonetheless, ligands such as 55 (activator; EC50 ~ 20 μM) and 58 (inhibitor; IC50 ~ 10 μM) could be drug-like leads for chemical elaboration. Despite their low potencies, some drug concentrations might be clinically relevant and help explain side effects. For example, MR1 upregulator 56 is a known photodegradation product of chemotherapeutic drugs aminopterin and methotrexate. The extent of formation of 56 in vivo is unknown, but a chemotherapeutic dose of methotrexate can result in higher drug concentrations in plasma than required for MR1 upregulation. Also, oral 53 can produce drug concentrations that would be sufficient for activating the MAIT TCR in vitro, which may reconcile some drug complications such as hepatotoxicity. These preliminary studies provide a glimpse into the future for MR1 and MAIT cell modulation with drug-like compounds.
6. CONCLUSION AND PERSPECTIVE
The seminal discovery of potent antigens for MAIT cells has heralded a new class of T lymphocyte modulators and, along with MR1-antigen tetramers developed therefrom, is enabling the activation and detection of MAIT cells, respectively. These heterocyclic pyrimidine antigens raise potential opportunities for developing new drugs, vaccines, and immunological tools for interrogating a new area of T cell immunity.
New efficacious and functionally distinct ligands are still needed to characterize MR1 and MAIT cell biology and assess the therapeutic potential of modulating them. However, ligand potency is extremely sensitive to subtle chemical modifications, making development of superior compounds challenging. Structure−activity modeling has suggested that very small changes to bond lengths and dihedral angles can dramatically affect potency.2,46 Furthermore, protein−ligand crystal structures are often extremely similar for ligands with disparate activities, so computationally expensive and complex MD simulations might be necessary for deeper insights into dynamic protein−antigen interactions. This sensitivity is expected, given that T cell activation must be highly regulated to prevent hyperreactivity, and reflects complex processes controlling cell uptake, intracellular ligand−protein trafficking, protein refolding, and cell surface expression.32,33 The densely decorated 5OP-RU presents synthetic challenges with selective functionalization often requiring circuitous routes. Removing or masking reactive groups can confer stability, but electrophiles that covalently bond to MR1 Lys43 have been required so far for potent antigenicity and are not easily incorporated.
Despite the challenges, organic synthesis will surely deliver new ligands that can significantly shape the future of MAIT cell immunology. It is not yet clear which diseases require activators or inhibitors of MAIT cells, but both will be useful investigative tools. Compounds 5, 25, and 58 lack potency, but have been patented for treating skin conditions.55 MAIT cell activation has also been associated with COVID-19 disease severity.56 Activation seems unlikely to be via MR1, although SARS-CoV-2-infected macrophages induce MAIT cell cytotoxicity in an MR1-dependent manner.57 One strategy toward inhibitors is to first develop potent stable 5-OP-RU analogues. Not only might these be potentially useful vaccine adjuvants43 and cancer immunotherapeutics,45 but they are potentially also tight-binding ligands that offer clues to more drug-like scaffolds, tagging58 of 5-OP-RU for monitoring cellular uptake, and structure−activity relationships that lead to stable and effective modulators of MR1 and MAIT TCR proteins.
Lastly, a better understanding of MR1-binding requirements might inform the discovery of other naturally occurring antigens of MAIT cells or other MR1-restricted T cells.59,60 For example, MR1-mediated activation of MC7.G.5 T cells are able to kill most cancer cell lines without affecting healthy cells,61 indicating promise for cancer immunotherapy62 if specific antigens can be identified. In conclusion, MAIT cells offer new opportunities for chemical synthesis to contribute to the understanding of T cell immunity and the development of new immunomodulating medicines.
ACKNOWLEDGMENTS
We thank NHMRC (Grants SPRF1027369, SPRF1117017, APP1125493), ARC (Centre of Excellence in Advanced Molecular Imaging Grant CE140100011), U.S. National Institutes of Health (RO1 Grant AI148407–01A1), University of Queensland (Grant UQECR1834385 to J.Y.W.M.), and Australian Government Research Training Program (scholarship 2015−2020) for support. We also thank our immunology and pharmacology collaborators (many of whom could not be mentioned in the limited reference list) for support, especially Prof. James McCluskey and Prof. Jamie Rossjohn, whose groups were our primary collaborators during the chemical studies outlined.
Biographies
Jeffrey Mak completed his Ph.D. in the total synthesis of caged diterpenoid natural products with Prof. Craig Williams in 2012 before joining the Fairlie group at the Institute for Molecular Bioscience. His research interests are focused on the application of organic synthesis and chemical principles in the solution of diverse biological problems, including in immunity, drug development, and the synthesis of bioactive natural products. He has lectured undergraduate synthetic chemistry at UQ since 2017 and was promoted to Research Fellow in 2020.
Ligong Liu pursued undergraduate studies at Nankai University, China, postgraduate research at Beijing Medical University, China and Central Queensland University, Australia, and postdoctoral research at Central Queensland University and the Australian National University in Canberra (1997−2001). He spent 7 years in the Australian biotechnology industry (Alchemia, Progen Pharmaceuticals) specializing in carbohydrate and oligosaccharide-based therapeutics. From 2008, he was Senior Research Officer at the Institute for Molecular Bioscience, University of Queensland, Australia. Research interests are in chemical synthesis, medicinal chemistry, drug design and development, combinatorial chemistry, and modulators of enzymes/growth factors/GPCRs associated with inflammatory disorders and cancers.
David Fairlie is Senior Principal Research Fellow of the National Health and Medical Research Council. His undergraduate, postgraduate, and postdoctoral studies were at Adelaide, Australian National, New South Wales, and Stanford and Toronto Universities. At University of Queensland, he led the Chemistry Group in the Centre for Drug Design and Development and is Professor and Head of the Division of Chemistry and Structural Biology at the Institute for Molecular Bioscience. His research in medicinal/organic chemistry, drug discovery, protein/peptide mimics and biochemistry/pharmacology of GPCRs, enzymes, and protein−protein interactions has permitted mechanistic studies of chemical, immunological, and biological reactions and of disease development and drug action, particularly in inflammation, infection, immunity, and cancer.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.accounts.1c00359
Notes
The authors declare the following competing interest(s): J.Y.W.M., L.L., and D.P.F. are inventors on patents describing MAIT and MR1 ligands and MR1-ligand tetramers.
Contributor Information
Jeffrey Y. W. Mak, Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072, Australia; Australian Research Council Centre of Excellence in Advanced Molecular Imaging, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072, Australia.
Ligong Liu, Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072, Australia; Australian Research Council Centre of Excellence in Advanced Molecular Imaging, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072, Australia.
David P. Fairlie, Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072, Australia; Australian Research Council Centre of Excellence in Advanced Molecular Imaging, Institute for Molecular Bioscience and Centre for Inflammation and Disease Research, Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072, Australia.
REFERENCES
- (1). Corbett AJ; Eckle SBG; Birkinshaw RW; Liu L; Patel O; Mahony J; Chen ZJ; Reantragoon R; Meehan B; Cao HW; Williamson NA; Strugnell RA; Van Sinderen D; Mak JYW; Fairlie DP; Kjer-Nielsen L; Rossjohn J; McCluskey J. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 2014, 509, 361–365. • A landmark discovery of the naturally occurring antigens for mucosal associated invariant T cells (MAIT). The uracil derivatives (e.g., 5-OP-RU) were produced during bacterial biosynthesis of riboflavin, and differ from peptide and glycolipid antigens known to activate other T cells.
- (2). Mak JYW; Xu W; Reid RC; Corbett AJ; Meehan BS; Wang H; Chen Z; Rossjohn J; McCluskey J; Liu L; Fairlie DP Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat. Commun. 2017, 8, 14599. • Full paper describing a kinetically controlled synthesis of the thermodynamically unstable MAIT cell antigen 5-OP-RU, its chemical stability and antigen potency, and development of a thermodynamically stable antigen.
- (3). Ler GJM; Xu W; Mak JYW; Liu L; Bernhardt PV; Fairlie DP Computer modelling and synthesis of deoxy and monohydroxy analogues of a ribitylaminouracil bacterial metabolite that potently activates human T cells. Chem. - Eur. J. 2019, 25, 15594–15608. • A systematic investigation of the roles of ribityl hydroxyl groups in 5-OP-RU on MR1 binding and MAIT cell activation, using computer modeling, solution structures, molecular dynamics simulations and synthesis, enabling biological characterization through assays and protein−ligand crystal structures.
- (4). Keller AN; Eckle SBG; Xu W; Liu L; Hughes VA; Mak JYW; Meehan BS; Pediongco T; Birkinshaw RW; Chen Z; Wang H; D’Souza C; Kjer-Nielsen L; Gherardin NA; Godfrey DI; Kostenko L; Corbett AJ; Purcell AW; Fairlie DP; McCluskey J; Rossjohn J. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat. Immunol. 2017, 18, 402–411. • First demonstration that MAIT cells and antigen-presenting protein MR1 could be modulated by ligands unrelated to vitamin B2 (riboflavin) or B9 (folic acid). This structural plasticity highlights the potential for drugs and drug-like compounds to modulate MR1 and MAIT cells.
- (5).Wieczorek M; Abualrous ET; Sticht J; Álvaro-Benito M; Stolzenberg S; Noé F; Freund C. Major histocompatibility complex (MHC) class I and MHC class II proteins: Conformational plasticity in antigen presentation. Front. Immunol. 2017, 8, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yoo HJ; Kim NY; Kim JH Current understanding of the roles of CD1a-restricted T cells in the immune system. Mol. Cells 2021, 44, 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kjer-Nielsen L; Patel O; Corbett AJ; Le Nours J; Meehan B; Liu LG; Bhati M; Chen ZJ; Kostenko L; Reantragoon R; Williamson NA; Purcell AW; Dudek NL; McConville MJ; O’Hair RAJ; Khairallah GN; Godfrey DI; Fairlie DP; Rossjohn J; McCluskey J. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012, 491, 717–723. [DOI] [PubMed] [Google Scholar]
- (8).Provine NM; Klenerman P. MAIT cells in health and disease. Annu. Rev. Immunol. 2020, 38, 203–228. [DOI] [PubMed] [Google Scholar]
- (9).Chua WJ; Hansen TH Vitamins prime immunity. Nature 2012, 491, 680–681. [DOI] [PubMed] [Google Scholar]
- (10).Wang H; D’Souza C; Lim XY; Kostenko L; Pediongco TJ; Eckle SBG; Meehan BS; Shi M; Wang N; Li S; et al. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 2018, 9, 3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wang H; Kjer-Nielsen L; Shi M; D’Souza C; Pediongco T; Cao H; Kostenko L; Lim X; Eckle S; Meehan B; et al. IL-23 costimulates antigen-specific MAIT cell activation and enables vaccination. Sci. Immunol. 2019, 4, No. eaaw0402. [DOI] [PubMed] [Google Scholar]
- (12).Zhao Z; Wang H; Shi M; Zhu T; Pediongco T; Lim XY; Meehan BS; Nelson AG; Fairlie DP; Mak JYW; et al. Francisella tularensis induces Th1 like MAIT cells conferring protection against systemic and local infection. Nat. Commun. 2021, 12, 4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).van Wilgenburg B; Loh L; Chen Z; Pediongco TJ; Wang H; Shi M; Zhao Z; Koutsakos M; Nüssing S; Sant S; et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat. Commun. 2018, 9, 4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Constantinides MG; Link VM; Tamoutounour S; Wong AC; Perez-Chaparro PJ; Han S-J; Chen YE; Li K; Farhat S; Weckel A; Krishnamurthy SR; Vujkovic-Cvijin I; Linehan JL; Bouladoux N; Merrill ED; Roy S; Cua DJ; Adams EJ; Bhandoola A; Scharschmidt TC; Aubé J; Fischbach MA; Belkaid Y. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 2019, 366, No. eaax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hinks TSC; Marchi E; Jabeen M; Olshansky M; Kurioka A; Pediongco TJ; Meehan BS; Kostenko L; Turner SJ; Corbett AJ; Chen Z; Klenerman P; McCluskey J. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 2019, 28, 3249–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yan J; Allen S; McDonald E; Das I; Mak JYW; Liu L; Fairlie DP; Meehan BS; Chen Z; Corbett AJ; Varelias A; Smyth MJ; Teng MWL MAIT cells promote tumor initiation, growth, and metastases via tumor MR1. Cancer Discovery 2020, 10, 124–141. [DOI] [PubMed] [Google Scholar]
- (17).D’Souza C; Pediongco T; Wang H; Scheerlinck J-PY; Kostenko L; Esterbauer R; Stent AW; Eckle SBG; Meehan BS; Strugnell RA; et al. Mucosal-associated invariant T cells augment immunopathology and gastritis in chronic Helicobacter pylori infection. J. Immunol. 2018, 200, 1901–1916. [DOI] [PubMed] [Google Scholar]
- (18).Kjer-Nielsen L; Corbett AJ; Chen Z; Liu L; Mak JY; Godfrey DI; Rossjohn J; Fairlie DP; McCluskey J; Eckle SB An overview on the identification of MAIT cell antigens. Immunol. Cell Biol. 2018, 96, 573–587. [DOI] [PubMed] [Google Scholar]
- (19).Kis K; Kugelbrey K; Bacher A. Biosynthesis of riboflavin. The reaction catalyzed by 6,7-dimethyl-8-ribityllumazine synthase can proceed without enzymatic catalysis under physiological conditions. J. Org. Chem. 2001, 66, 2555–2559. [DOI] [PubMed] [Google Scholar]
- (20).Lunt SY; Vander Heiden MG Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [DOI] [PubMed] [Google Scholar]
- (21).Ritter E; Przybylski P; Brzezinski B; Bartl F. Schiff bases in biological systems. Curr. Org. Chem. 2009, 13, 241–249. [Google Scholar]
- (22).Chen S-H; Hiramatsu H. Tautomer structures in ketose−aldose transformation of 1,3-dihydroxyacetone studied by infrared electroabsorption spectroscopy. J. Phys. Chem. B 2019, 123, 10663–10671. [DOI] [PubMed] [Google Scholar]
- (23).Wang Y; Ho CT Flavour chemistry of methylglyoxal and glyoxal. Chem. Soc. Rev. 2012, 41, 4140–4149. [DOI] [PubMed] [Google Scholar]
- (24).Maley GF; Plaut GWE Isolation, synthesis, and metabolic properties of 6,7-dimethyl-8-ribityllumazine. J. Biol. Chem. 1959, 234, 641–647. [PubMed] [Google Scholar]
- (25).Rahimpour A; Koay HF; Enders A; Clanchy R; Eckle SBG; Meehan B; Chen Z; Whittle B; Liu L; Fairlie DP; Goodnow CC; McCluskey J; Rossjohn J; Uldrich AP; Pellicci DG; Godfrey DI Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J. Exp. Med. 2015, 212, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Reantragoon R; Corbett AJ; Sakala IG; Gherardin NA; Furness JB; Chen ZJ; Eckle SBG; Uldrich AP; Birkinshaw RW; Patel O; Kostenko L; Meehan B; Kedzierska K; Liu LG; Fairlie DP; Hansen TH; Godfrey DI; Rossjohn J; McCluskey J; Kjer-Nielsen L. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 2013, 210, 2305–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Li K; Vorkas CK; Chaudhry A; Bell DL; Willis RA; Rudensky A; Altman JD; Glickman MS; Aubé J. Synthesis, stabilization, and characterization of the MR1 ligand precursor 5amino-6-D-ribitylaminouracil (5-A-RU). PLoS One 2018, 13, No. e0191837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Corbett AJ; McCluskey J; Kjer-Nielsen L; Chen Z; Rossjohn J; Patel O; Birkinshaw RW; Eckle SBG; Fairlie DP; Liu L; Mak JYW Immunological reagents comprising MR1 ligands and uses therefor. Patent WO 2015149130 A1, 2015.
- (29).Chen Z; Wang H; D’Souza C; Sun S; Kostenko L; Eckle SBG; Meehan BS; Jackson DC; Strugnell RA; Cao H; Wang N; Fairlie DP; Liu L; Godfrey DI; Rossjohn J; McCluskey J; Corbett AJ Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. 2017, 10, 58–68. [DOI] [PubMed] [Google Scholar]
- (30).Yu H; Yang A; Liu L; Mak JYW; Fairlie DP; Cowley S. CXCL16 Stimulates antigen-induced MAIT cell accumulation but trafficking during lung infection is CXCR6-independent. Front. Immunol. 2020, 11, 1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Howson LJ; Li J; von Borstel A; Barugahare A; Mak JYW; Fairlie DP; McCluskey J; Turner SJ; Davey MS; Rossjohn J. Mucosal-associated invariant T cell effector function is an intrinsic cell property that can be augmented by the metabolic cofactor αketoglutarate. J. Immunol. 2021, 206, 1425–1435. [DOI] [PubMed] [Google Scholar]
- (32).McWilliam HEG; Mak JYW; Awad W; Zorkau M; CruzGomez S; Lim HJ; Yan Y; Wormald S; Dagley LF; Eckle SBG; et al. Endoplasmic reticulum chaperones stabilize ligand-receptive MR1 molecules for efficient presentation of metabolite antigens. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 24974–24985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).McWilliam HEG; Eckle SBG; Theodossis A; Liu L; Chen Z; Wubben JM; Fairlie DP; Strugnell RA; Mintern JD; McCluskey J; Rossjohn J; Villadangos JA The intracellular pathway for the presentation of vitamin B-related antigens by the antigenpresenting molecule MR1. Nat. Immunol. 2016, 17, 531–537. [DOI] [PubMed] [Google Scholar]
- (34).Howson LJ; Awad W; von Borstel A; Lim HJ; McWilliam HEG; Sandoval-Romero ML; Majumdar S; Hamzeh AR; Andrews TD; McDermott DH; Murphy PM; Le Nours J; Mak JYW; Liu L; Fairlie DP; McCluskey J; Villadangos JA; Cook MC; Turner SJ; Davey MS; Ojaimi S; Rossjohn J. Absence of mucosal-associated invariant T cells in a person with a homozygous point mutation in MR1. Sci. Immunol. 2020, 5, No. eabc9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Varelias A; Bunting MD; Ormerod KL; Koyama M; Olver SD; Straube J; Kuns RD; Robb RJ; Henden AS; Cooper L; Lachner N; Gartlan KH; Lantz O; Kjer-Nielsen L; Mak JY; Fairlie DP; Clouston AD; McCluskey J; Rossjohn J; Lane SW; Hugenholtz P; Hill GR Recipient mucosal-associated invariant T cells control GVHD within the colon. J. Clin. Invest. 2018, 128, 1919–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Boulouis C; Sia WR; Gulam MY; Teo JQM; Png YT; Phan TK; Mak JYW; Fairlie DP; Poon IKH; Koh TH; Bergman P; Lim CM; Wang L-F; Kwa ALH; Sandberg JK; Leeansyah E. Human MAIT cell cytolytic effector proteins synergize to overcome carbapenem resistance in Escherichia coli. PLoS Biol. 2020, 18, No. e3000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Edmans MD; Connelley TK; Jayaraman S; Vrettou C; Vordermeier M; Mak JYW; Liu L; Fairlie DP; Maze EA; Chrun T; Klenerman P; Eckle SBG; Tchilian E; Benedictus L. Identification and phenotype of MAIT cells in cattle and their response to bacterial infections. Front. Immunol. 2021, 12, 627173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Juno JA; Wragg KM; Amarasena T; Meehan BS; Mak JYW; Liu L; Fairlie DP; McCluskey J; Eckle SBG; Kent SJ MAIT cells upregulate alpha4beta7 in response to acute simian immunodeficiency virus/simian HIV infection but are resistant to peripheral depletion in pigtail macaques. J. Immunol. 2019, 202, 2105–2120. [DOI] [PubMed] [Google Scholar]
- (39).Leeansyah E; Hey YY; Sia WR; Ng JHJ; Gulam MY; Boulouis C; Zhu F; Ahn M; Mak JYW; Fairlie DP; Kwa ALH; Sandberg JK; Wang L-F MR1-restricted T cells with MAIT-like characteristics are functionally conserved in the pteropid bat Pteropus alecto. iScience 2020, 23, 101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Legoux F; Bellet D; Daviaud C; El Morr Y; Darbois A; Niort K; Procopio E; Salou M; Gilet J; Ryffel B; Balvay A; Foussier A; Sarkis M; El Marjou A; Schmidt F; Rabot S; Lantz O. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 2019, 366, 494–499. [DOI] [PubMed] [Google Scholar]
- (41).Koay H-F; Su S; Amann-Zalcenstein D; Daley SR; Comerford I; Miosge L; Whyte CE; Konstantinov IE; d’Udekem Y; Baldwin T; et al. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci. Immunol. 2019, 4, No. eaay6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Koay HF; Gherardin NA; Enders A; Loh L; Mackay LK; Almeida CF; Russ BE; Nold-Petry CA; Nold MF; Bedoui S; Chen Z; Corbett AJ; Eckle SB; Meehan B; d’Udekem Y; Konstantinov IE; Lappas M; Liu L; Goodnow CC; Fairlie DP; Rossjohn J; Chong MM; Kedzierska K; Berzins SP; Belz GT; McCluskey J; Uldrich AP; Godfrey DI; Pellicci DG A threestage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol. 2016, 17, 1300–1311. [DOI] [PubMed] [Google Scholar]
- (43).Downey AM; Kaplonek P; Seeberger PH MAIT cells as attractive vaccine targets. FEBS Lett. 2019, 593, 1627–1640. [DOI] [PubMed] [Google Scholar]
- (44).Sakai S; Kauffman KD; Oh S; Nelson CE; Barry CE; Barber DL MAIT cell-directed therapy of Mycobacterium tuberculosis infection. Mucosal Immunol. 2021, 14, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Petley E; Koay H-F; Henderson M; Sek K; Todd K; Keam S; Lai J; House I; Zethoven M; Chen A; et al. MAIT cells regulate NK cell mediated tumor immunity. Nat. Commun. 2021, 12, 4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Awad W; Ler G; Xu W; Keller A; Mak J; Lim X; Liu L; Eckle S; Le Nours J; McCluskey J; et al. The molecular basis underpinning the potency and specificity of MAIT cell antigens. Nat. Immunol. 2020, 21, 400–411. [DOI] [PubMed] [Google Scholar]
- (47).Nielsen DS; Hoang HN; Lohman R-J; Hill TA; Lucke AJ; Craik DJ; Edmonds DJ; Griffith DA; Rotter CJ; Ruggeri RB; Price DA; Liras S; Fairlie DP Improving on nature: making a cyclic heptapeptide orally bioavailable. Angew. Chem., Int. Ed. 2014, 53, 12059–12063. [DOI] [PubMed] [Google Scholar]
- (48).Braganza CD; Shibata K; Fujiwara A; Motozono C; Sonoda K-H; Yamasaki S; Stocker BL; Timmer MSM The effect of MR1 ligand glyco-analogues on mucosal-associated invariant T (MAIT) cell activation. Org. Biomol. Chem. 2019, 17, 8992–9000. [DOI] [PubMed] [Google Scholar]
- (49).Matsuoka T; Motozono C; Hattori A; Kakeya H; Yamasaki S; Oishi S; Ohno H; Inuki S. The effects of 5-OP-RU stereochemistry on its stability and MAIT-MR1 axis. ChemBioChem. 2021, 22, 672–678. [DOI] [PubMed] [Google Scholar]
- (50).Braganza CD; Motozono C; Sonoda K-H; Yamasaki S; Shibata K; Timmer MSM; Stocker BL Agonistic or antagonistic mucosal-associated invariant T (MAIT) cell activity is determined by the 6-alkylamino substituent on uracil MR1 ligands. Chem. Commun. 2020, 56, 5291–5294. [DOI] [PubMed] [Google Scholar]
- (51).Wider G; Dreier L. Measuring protein concentrations by NMR spectroscopy. J. Am. Chem. Soc. 2006, 128, 2571–2576. [DOI] [PubMed] [Google Scholar]
- (52).5-A-RU is highly prone to aerial oxidation, and its synthesis and handling should be conducted under an inert atmosphere.
- (53).Lange J; Anderson RJ; Marshall AJ; Chan STS; Bilbrough TS; Gasser O; Gonzalez-Lopez C; Salio M; Cerundolo V; Hermans IF; Painter GF The chemical synthesis, stability, and activity of MAIT cell prodrug agonists that access MR1 in recycling endosomes. ACS Chem. Biol. 2020, 15, 437–445. [DOI] [PubMed] [Google Scholar]
- (54).Reid RC; Yau M-K; Singh R; Hamidon JK; Reed AN; Chu P; Suen JY; Stoermer MJ; Blakeney JS; Lim J; Faber JM; Fairlie DP Downsizing a human inflammatory protein to a small molecule with equal potency and functionality. Nat. Commun. 2013, 4, 2802. [DOI] [PubMed] [Google Scholar]
- (55).Gasser O; Parata K; Naidoo K. Methods of treating or preventing skin conditions. Patent WO 2020080957 A1, 2019.
- (56).Parrot T; Gorin J-B; Ponzetta A; Maleki KT; Kammann T; Emgård J; Perez-Potti A; Sekine T; Rivera-Ballesteros O; Gredmark-Russ S; Rooyackers O; Folkesson E; Eriksson LI; Norrby-Teglund A; Ljunggren H-G; Björkström NK; Aleman S; Buggert M; Klingström J; Strålin K; Sandberg JK MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 2020, 5, No. eabe1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Flament H; Rouland M; Beaudoin L; Toubal A; Bertrand L; Lebourgeois S; Rousseau C; Soulard P; Gouda Z; Cagninacci L; Monteiro AC; Hurtado-Nedelec M; Luce S; Bailly K; Andrieu M; Saintpierre B; Letourneur F; Jouan Y; Si-Tahar M; Baranek T; Paget C; Boitard C; Vallet-Pichard A; Gautier J-F; Ajzenberg N; Terrier B; Pene F; Ghosn J; Lescure X; Yazdanpanah Y; Visseaux B; Descamps D; Timsit J-F; Monteiro RC; Lehuen A. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol. 2021, 22, 322–335. [DOI] [PubMed] [Google Scholar]
- (58).Yvorra T; Steinmetz A; Retailleau P; Lantz O; Schmidt F. Synthesis, biological evaluation and molecular modelling of new potent clickable analogues of 5-OP-RU for their use as chemical probes for the study of MAIT cell biology. Eur. J. Med. Chem. 2021, 211, 113066. [DOI] [PubMed] [Google Scholar]
- (59).Koay H-F; Gherardin NA; Xu C; Seneviratna R; Zhao Z; Chen Z; Fairlie DP; McCluskey J; Pellicci DG; Uldrich AP; Godfrey DI Diverse MR1-restricted T cells in mice and humans. Nat. Commun. 2019, 10, 2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Le Nours J; Gherardin NA; Ramarathinam SH; Awad W; Wiede F; Gully BS; Khandokar Y; Praveena T; Wubben JM; Sandow JJ; Webb AI; von Borstel A; Rice MT; Redmond SJ; Seneviratna R; Sandoval-Romero ML; Li S; Souter MNT; Eckle SBG; Corbett AJ; Reid HH; Liu L; Fairlie DP; Giles EM; Westall GP; Tothill RW; Davey MS; Berry R; Tiganis T; McCluskey J; Pellicci DG; Purcell AW; Uldrich AP; Godfrey DI; Rossjohn J. A class of γδ T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science 2019, 366, 1522–1527. [DOI] [PubMed] [Google Scholar]
- (61).Crowther MD; Dolton G; Legut M; Caillaud ME; Lloyd A; Attaf M; Galloway SAE; Rius C; Farrell CP; Szomolay B; Ager A; Parker AL; Fuller A; Donia M; McCluskey J; Rossjohn J; Svane IM; Phillips JD; Sewell AK Genome-wide CRISPR− Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat. Immunol. 2020, 21, 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Vacchini A; Chancellor A; Spagnuolo J; Mori L; De Libero G. MR1-restricted T cells are unprecedented cancer fighters. Front. Immunol. 2020, 11, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]