Abstract
The objectives were to develop a standardized Ki-67 immunohistochemistry (IHC) method for precise, robust, and reproducible assessment of patients with early breast cancer, and utilize this assay to evaluate patients participating in the monarchE study (NCT03155997). The Ki-67 assay was developed and validated for sensitivity, specificity, repeatability, precision, and robustness using a predefined ≥20% cutoff. Reproducibility studies (intersite and intrasite, interobserver and intraobserver) were conducted at 3 external laboratories using detailed scoring instructions designed for monarchE. Using the assay, patient tumors were classified as displaying high (≥20%) or low (<20%) Ki-67 expression; Kaplan-Meier methods evaluated 2-year invasive disease-free survival rates for these 2 groups among patients treated with endocrine therapy (ET) alone. All analytical validation and reproducibility studies achieved point estimates of >90% for negative, positive, and overall percent agreement. Intersite reproducibility produced point estimate values of 94.7%, 100.0%, and 97.3%. External interobserver reproducibility produced point estimate values of 98.9%, 97.8%, and 98.3%. Among 1954 patients receiving ET alone, 986 (50.5%) had high and 968 (49.5%) had low Ki-67 expression. Patients with high Ki-67 had a clinically meaningful increased risk of developing invasive disease within 2 years compared with those with low Ki-67 [2-y invasive disease-free survival rate: 86.1% (95% confidence interval: 83.1%-88.7%) vs. 92.0% (95% confidence interval: 89.7%-93.9%), respectively]. This standardized Ki-67 methodology resulted in high concordance across multiple laboratories, and its use in the monarchE study prospectively demonstrated the prognostic value of Ki-67 IHC in HR+, HER2− early breast cancer with high-risk clinicopathologic features.
Key Words: assay validation, CDK 4 and 6 inhibitor, early breast cancer, immunohistochemistry, Ki-67
Ki-67 expression has increasingly gained attention as a possible prognostic and predictive marker of responsiveness to chemotherapy or endocrine therapy (ET) among patients with breast carcinoma.1 However, lack of standardized procedures or accepted cutoff definitions have prohibited clinical trial comparisons and limited the application of Ki-67 assessment for clinical use.2 Therefore, Ki-67 immunohistochemistry (IHC) is not routinely performed as part of the diagnostic workup of breast carcinomas in many practice settings, and its use in the management of patients with breast carcinoma is not universally accepted. To minimize variability and facilitate clinical adoption of Ki-67 assessments, an expert group on Ki-67 testing in breast carcinoma has provided guidance on preferred Ki-67 staining and scoring methods.2
Cyclin-dependent kinase 4 and 6 (CDK4 and 6) inhibitors have improved outcomes for patients with hormone receptor-positive (HR+), human epidermal growth factor receptor 2-negative (HER2-) advanced breast cancer when used in combination with ET.3–5 Abemaciclib is a selective CDK4 and 6 inhibitor approved for the treatment of HR+, HER2− advanced breast cancer.3–7 In the phase 3 monarchE trial (NCT03155997), when combined with ET, abemaciclib demonstrated a significant improvement in invasive disease-free survival (IDFS), compared with ET alone, in patients with HR+, HER2− node positive early breast cancer (EBC) at a high risk of recurrence.7 In available outcome analyses, patients with high Ki-67 tumors (≥20%) had a greater risk of recurrence within 2 years than those with low Ki-67 tumors (<20%).6 However, abemaciclib plus ET reduced the risk of developing IDFS events, regardless of Ki-67 index.6
The comparisons presented in this manuscript were conducted to test the hypothesis that a standardized, automated testing system and uniform scoring algorithm to determine Ki-67 expression in formalin-fixed, paraffin-embedded (FFPE) breast carcinoma would enable the global multisite registrational study design of the monarchE phase 3 clinical trial6 to deliver quality biomarker data and allow investigators to draw meaningful conclusions regarding patient outcomes stratified by Ki-67 status (Fig. 1).
FIGURE 1.
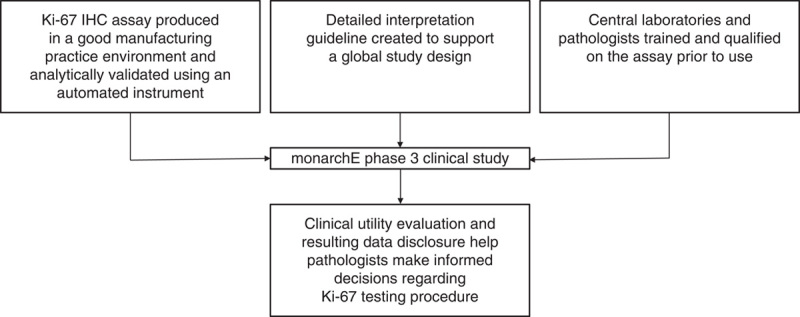
Key objectives of this study were to develop a precise, robust, and reproducible assay paired with clear interpretation guidance to reduce ambiguities in scoring. Training was provided to pathologists from different laboratories, resulting in high concordance between labs. This created a foundation for testing tumor proliferation using Ki-67 IHC in the monarchE phase 3 clinical trial. IHC indicates immunohistochemistry.
The objectives of this report were to assess the analytical validity and multisite reproducibility of an investigational use only (IUO) Ki-67 IHC assay using rigorous predefined acceptance criteria, and to utilize this standardized methodology to evaluate patients participating in the ET-only arm of monarchE to determine the prognostic value of Ki-67 IHC in HR+, HER2− EBC with high-risk clinicopathologic features.
MATERIALS AND METHODS
Tissue Specimen Preparation
With exception of the monarchE samples, specimens were commercially procured FFPE human breast carcinoma tissue, and consisted of core biopsies and surgical resection tissue, as indicated. Information regarding HR and HER2 status was not available for all commercial specimens. Sections were cut at 4 to 5 µm thickness, placed on positively charged slides, and dried in an oven at 58±2°C for 1 hour. The mounted sections were stored in the dark at 2 to 8°C and stained using the assay within 5 months of sectioning.
IUO Ki-67 IHC Assay
For details on prototype assay design input, see the Supplemental Digital Content (http://links.lww.com/AIMM/A333) which describes Supplemental Methods and Results to provide more detail on assay development. IHC staining procedure was performed on the Dako Omnis (Agilent Technologies, Carpinteria, CA) platform and used a validated automated staining protocol. This modular assay consists of an optimally purified monoclonal mouse antibody (clone MIB-1), produced in a good manufacturing practice environment under strict quality control criteria. Table 1 provides a summary of relevant factors incorporated in the assay development.
TABLE 1.
Summary of Preanalytic, Analytic, and Postanalytic Factors Considered in Development of the Assay
| Setting | Factor | IUO Ki-67 IHC Assay Overview | |
|---|---|---|---|
| Preanalytical | Biopsy type | Core biopsy or surgical resection specimens | |
| Time to fixation | ≤1 h | ||
| Fixation time and fixative | 6-72 h in 10% neutral buffered formalin | ||
| Paraffin embedding | Tissues were infiltrated with melted paraffin, at or below 60°C | ||
| Slide preparation | Tissue section of 4-5 µm were mounted on Dako FLEX IHC Microscope Slides or SuperFrost Plus slides | ||
| Specimens were oven-dried at 58±2°C | |||
| Cut slide storage | Specimens were stained within 5 mo of sectioning when stored in the dark at 2-8°C or within 4 mo of sectioning when stored in the dark at room temperature up to 25°C | ||
| Analytical | Staining instrument | Dako Omnis* | |
| Antigen retrieval | EnVision FLEX Target Retrieval Solution, Low pH (Code GV805). pH of 1x solution 6.1±0.2. Antigen retrieval took place onboard the Dako Omnis instrument at 97°C | ||
| Specific antibody | Purified monoclonal mouse anti-Ki-67, clone MIB-1 (Code GE020) | ||
| Negative Control Reagent | Protein matched, mouse immunoglobulin G isotype control. Negative Control Reagent (Code GE020) | ||
| Detection system | Polymer based—EnVision FLEX Detection System (Code GV800) | ||
| Counterstain | EnVision FLEX Hematoxylin (Code GC808) | ||
| Quality control | Positive and negative control tissues were run for each staining procedure. Control tissues were fixed in the same way as the patient tissue | ||
| Postanalytical (interpretation and scoring) | Tissue specimen criteria | Invasive breast carcinoma | |
| Minimum 200 viable invasive tumor cells present in specimen | |||
| Only well-preserved and well-stained areas of the specimen were used to determine the percentage of positive tumor cells | |||
| Only the invasive cancer component was scored; in situ carcinoma was not scored | |||
| Whole tissue score methodology | The entire tissue section was scored. If hot spots were present in the section, they were an integral part of the final score. For whole tissue evaluation, objectives of 10-40× magnification are appropriate. Only nuclear staining of tumor cells was scored | ||
| The percentage of positively stained tumor cells among the total number of invasive cells across the entire slide was determined. Nuclear staining at all intensities (1-3+) was included. The lower limit of 1+ positivity was evaluated using a high-power (eg, 40×) objective and defined by the following rules: | |||
| Signal must be unequivocally brown | |||
| The staining must correspond to a nucleus | |||
| The staining must cover the whole chromatin distribution within the nucleus | |||
| The staining must correspond to a nonapoptotic cell | |||
| Cutoff | Positive | Negative | |
| ≥20% of invasive tumor cells stained | ˂20% of invasive tumor cells stained | ||
Fully automated staining, from deparaffinization to counterstaining.
IHC indicates immunohistochemistry; IUO, investigational use only.
Heat-induced epitope retrieval was performed using diluted EnVision FLEX Target Retrieval Solution, Low pH (50x; Code GV805). Deparaffinization, rehydration, and target retrieval were performed on-board. After incubation with the primary monoclonal mouse anti-human Ki-67 antibody, clone MIB-1, or the Negative Control Reagent (NCR; mouse immunoglobulin G isotype control), specimens were incubated with ready-to-use visualization reagent consisting of a secondary antibody and horseradish peroxidase coupled to a dextran polymer backbone. The enzymatic conversion of the subsequently added 3,3′-diaminobenzidine tetrahydrochloride chromogen resulted in a visible reaction product at the antigen site. Specimens were then counterstained with hematoxylin and coverslipped.
Scoring Interpretation
Assay results were interpreted using a light microscope. A minimum of 200 viable invasive tumor cells was required for scoring. All viable invasive tumor cells were evaluated and included in the Ki-67 scoring assessment. Carcinoma in situ was not scored. Only nuclear staining was considered for positive staining evaluation in tumor cell nuclei. A tumor cell was considered positive when the signal was unequivocally brown and covered the whole chromatin distribution within the nucleus. For Ki-67 protein expression determination, intensity grades of 1+ (weak) to 3+ (strong) were reported. Nonspecific staining was recorded using a 0 to 3+ scale in 0.25 increments. Cytoplasmic and/or membrane staining, if present, was excluded from scoring. The Ki-67 score was determined as:
Breast carcinoma specimens stained with the NCR were required to have ˂1+ intensity nonspecific background staining, for the same specimens stained with the Ki-67 antibody to be considered valid. Tumors were classified as positive Ki-67 (≥20%) or negative Ki-67 (<20%). An overview of the scoring methodology is provided in Table 1.
Internal observers and external laboratory pathologists were trained and tested on the scoring algorithm and guidelines prior to use. In the initial project phase, hot spots were analyzed in an exploratory manner. A hot spot was defined as the area corresponding to the field of vision in a 20× objective with the highest percentage of positive tumor nuclei in the section. For details on assay sensitivity, specificity, robustness, tumor heterogeneity, and precision studies, see the Supplemental Digital Content, which describes Supplemental Methods (http://links.lww.com/AIMM/A333).
External Reproducibility Study
Intersite and intrasite reproducibility studies were conducted at 3 external Clinical Laboratory Improvement Amendments (CLIA)-certified laboratories (referred to as “sites”). One trained and certified technician from each laboratory performed 5 automated IHC staining runs using the assay over 5 nonconsecutive days. Each run contained replicate sections from the same set of breast carcinoma specimens (n=30), with 1 slide stained each with the NCR and the Ki-67 primary antibody. Efforts were made to balance the proportion of positive (≥20%) and negative (<20%) specimens and to include ~20% to 25% considered to be in the near cutoff range (10% to 30%). Each set of blinded and randomized replicate sections was evaluated by a single trained and certified pathologist at each of 3 external sites with a minimum 14-day washout period between each evaluation.
Interobserver and intraobserver reproducibility across pathologists from different laboratories were assessed through blinded and randomized slide evaluation at 3 external CLIA-certified laboratories. Samples were prestained with the assay and sent to 3 external sites for evaluation. Efforts were made to balance the proportion of positive/negative specimens and include ~20% to 25% of specimens considered to be in the near cutoff range. One certified pathologist at each site performed 3 independent Ki-67 staining evaluations on the same set of breast carcinoma specimens (n=60), representing a dynamic Ki-67 expression range. A minimum 14-day washout period between each read was implemented. The pathologists participating in the external interobserver/intraobserver study differed from those scoring the intersite/intrasite reproducibility study.
Assessment of monarchE Samples
The primary study endpoint in monarchE was IDFS in the intention-to-treat population, using the Standardized Definitions for Efficacy End Points in Adjuvant Breast Cancer Trials (STEEP) criteria.8 A key secondary endpoint was to evaluate IDFS among all patients from the intention-to-treat population who had a Ki-67 percent positive ≥20%. Exploratory analyses investigated IDFS among patients who received ET alone, by Ki-67 high (≥20%) versus low (<20%). Samples submitted for Ki-67 testing from monarchE were assessed using assay scoring guidelines according to the clinical study protocol.7
In the monarchE clinical trial, patients were enrolled and assigned to 1 of 2 cohorts based on clinicopathologic features: cohort 1 enrolled patients with either ≥4 positive axillary lymph nodes (ALN), or 1 to 3 positive ALN and at least one of the following features: tumor size ≥5 cm or histologic grade 3. Cohort 2 began enrollment 1 year after cohort 1 and enrolled patients with 1 to 3 ALN, tumor size <5 cm, grade <3, and a centrally determined Ki-67 index ≥20%. Ki-67 was also determined centrally in all cohort 1 patients who had a suitable breast tumor tissue sample, but a Ki-67 index was not an enrollment requirement. For all patients, Ki-67 testing was performed on untreated breast tumor tissue. Patients included in analyses in this manuscript were all from cohort 1.
Statistical Analysis
Comparisons between IHC status (positive/negative) of each test condition and the consensus (most frequently occurring diagnostic observation within a specimen) were made for each specimen. Agreement parameters were calculated using comparisons to consensus pooled across samples. Negative percent agreement (NPA), positive percent agreement (PPA), and overall percent agreement (OA) were calculated for each validation study (interday, interinstrument, interlot, repeatability, robustness, and reproducibility), with corresponding 2-sided 95% bootstrap confidence intervals (CIs). Lower-bound CIs (LBCI) were reported for NPA, PPA, and OA measurements. If a given parameter (NPA, PPA, and/or OA) resulted in zero discordant comparisons, CIs were computed with the Wilson score method. Locally estimated scatterplot smoothing (LOESS) was applied to external interobserver reproducibility data to evaluate trends in specimen scoring across the dynamic Ki-67 expression range. LOESS curves were used to compare scoring trends across multiple observers and across multiple reads within each observer. Kaplan-Meier methods evaluated 2-year IDFS rates (95% CIs) in the ET-alone arm of monarchE, among those with high versus low Ki-67 expression.
RESULTS
Definition of Positive Ki-67 Staining
To achieve a high degree of scoring reproducibility, clear and comprehensive scoring guidelines were developed. Since any staining intensity ≥1+ was considered positive, distinguishing between staining intensity 0 and 1+ was critical. Therefore, emphasis was placed on defining and training recognition of the lower staining threshold. When assessing if tumor cells were negative versus weakly positive, a cell exhibiting a “gray”, rather than an unequivocally brown nucleus, was deemed as insufficiently meeting the lower threshold and excluded from the numerator when determining the Ki-67 score (Fig. 2).
FIGURE 2.
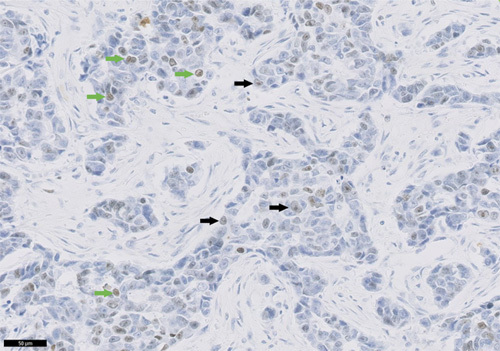
Expression levels in breast carcinoma tissue bank specimens stained with the investigational use only Ki-67 assay. Breast carcinoma specimen stained with Ki-67 primary antibody exhibiting both negative and weak positive staining. Negative cells showing gray staining defined as being below the threshold for positivity are indicated with black arrows. Cells with weak positive 1+ staining are indicated with green arrows (20× objective; scale bar is 50 μm).
Assay Performance
Assay validation showed a high degree of reproducibility, including high interobserver reproducibility (OA point estimate, 97.2%; 95% LBCI, 95.4%). Intrablock and intracase heterogeneity studies also demonstrated high OA with point estimates (95% LBCI) of 96.5% (92.4%) and 96.0% (90.0%), respectively. For results describing the prototype comparison with a previously utilized laboratory developed test,9 and sensitivity, specificity, robustness, tumor heterogeneity, and precision studies, see the Supplemental Digital Content (http://links.lww.com/AIMM/A333) which provides more detailed results describing assay development.
Investigational Assay External Reproducibility
Intersite and intrasite assay reproducibility was performed by testing staining and scoring reproducibility across and within 3 qualified external sites on 15 replicates of unstained slide sets (n=30). The analyses were performed on 450 observations (comparisons to consensus), and the consensus determined as the majority call for the sample across all 15 observations (intersite) and 5 observations for the sample-site (intrasite) combination. Intersite reproducibility achieved NPA, PPA, and OA point estimates (95% LBCI) of 94.7% (88.4%), 100.0% (98.3%), and 97.3% (94.2%), respectively (Fig. 3). Intrasite reproducibility was evaluated by testing the staining and scoring reproducibility within a site across 5 testing runs. The consensus for intrasite reproducibility analysis was determined as the majority call for the sample across all 5 observations within a given site. Intrasite reproducibility achieved NPA, PPA, and OA point estimates (95% LBCI) of 100.0% (98.2%), 98.8% (96.9%), and 99.3% (98.2%), respectively (Fig. 3). As all intersite/intrasite reproducibility study parameters met predefined acceptance criteria, this study demonstrated the assay was reproducible across multiple sites, and within the same site, over multiple days and runs.
FIGURE 3.
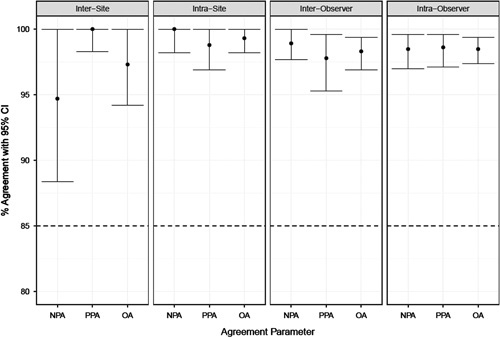
Summary of percent agreement for assay external reproducibility studies performed at 3 external sites. Left 2 graphs demonstrate intersite and intrasite reproducibility, which measures assay staining and scoring interpretation. Right 2 graphs demonstrate inter- and intraobserver reproducibility, which measures scoring interpretation only. Horizontal dashed lines indicate acceptance criteria for the external reproducibility studies. CI indicates confidence interval; OA, overall percent agreement; NPA, negative percent agreement; PPA positive percent agreement.
Interobserver and intraobserver precision was evaluated externally by testing scoring reproducibility between and within each of the 3 certified pathologists from different laboratories, who each performed 3 independent Ki-67 evaluations on 60 prestained specimens; analyses were performed on 540 observations, and the consensus was the majority call for the sample across all 9 observations (interobserver), or majority call across 3 observations for the sample-observer combination (intraobserver). Interobserver reproducibility achieved NPA, PPA, and OA point estimates (95% LBCI) of 98.9% (97.7%), 97.8% (95.3%), and 98.3% (96.9%), respectively (Fig. 3). Intraobserver reproducibility achieved NPA, PPA, and OA point estimates (95% LBCI) of 98.5% (97.0%), 98.6% (97.1%), and 98.5% (97.4%), respectively (Fig. 3). Observers demonstrated similar trends in scoring across the dynamic range. Score variability increased as Ki-67 expression range increased, but there was not marked interobserver variability (Fig. 4). These results demonstrate the ≥20% cutoff was reproducible within and between pathologists from different laboratories using breast carcinoma specimens stained and scored with the assay.
FIGURE 4.
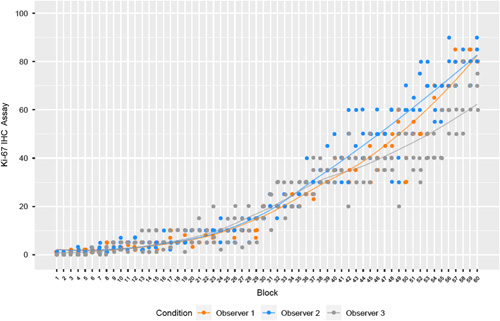
LOESS plot of external reproducibility interobserver Ki-67 continuous scores grouped by observer. LOESS lines demonstrate average trends over interobserver data using locally weighted regression. IHC indicates immunohistochemistry; LOESS, locally estimated scatterplot smoothing.
Investigational Assay Use in the monarchE Phase 3 Clinical Study
In the monarchE study 1,954 patients from cohort 1 received standard of care ET without the addition of abemaciclib. Of these patients, 986 (50.5%) had high Ki-67 expression (≥20%) and 968 (49.5%) had low Ki-67 expression (<20%) as defined by the assay. Kaplan-Meier curves of the 2-year IDFS rate for high versus low Ki-67 populations are shown in Figure 5. Patients with high Ki-67 had a clinically meaningful increased risk of developing invasive disease within 2 years compared with those with low Ki-67 [2-y IDFS rate: 86.1% (95% CI: 83.1-88.7%) vs. 92.0% (95% CI: 89.7-93.9%), respectively]. Within the short follow-up duration available for monarchE, data suggest having high Ki-67 expression (≥20%), in addition to existing high-risk clinical pathologic features, increased the likelihood of developing recurrent disease, compared with patients with low Ki-67 expression (<20%).
FIGURE 5.
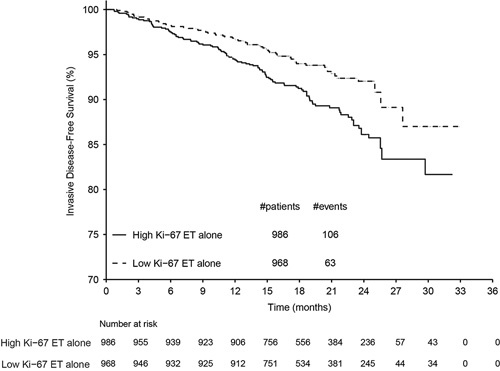
Kaplan-Meier plot demonstrating the prognostic value of the assay in the monarchE phase 3 study. Among patients with high clinicopathologic risk factors, patients whose tumors displayed high Ki-67 (≥20%) had an even greater risk of recurrence than those with low Ki-67 tumors (<20%); 2-year IDFS rate: 86.1% (95% confidence interval: 83.1-88.7) versus 92.0% (95% confidence interval: 89.7-93.9), respectively. ET indicates endocrine therapy.
DISCUSSION
Ki-67 IHC is a biomarker widely used for assessments of tumor proliferation. However, its broad clinical adoption for breast carcinoma management has been limited by lack of standardization and established cutoffs.2 Despite some oncologists considering Ki-67 IHC in breast carcinoma treatment decisions, the American Society of Clinical Oncology has not endorsed the use of Ki-67 to determine whether a patient should receive chemotherapy or to guide adjuvant ET selection.10 The International Ki-67 in Breast Cancer Working Group (IKWG) has acknowledged MIB-1 as the most widely validated Ki-67 antibody.2 Therefore, this assay was developed utilizing the MIB-1 clone, which was produced under stringent quality control conditions.
This assay was designed to run on the Dako Omnis platform. An initial feasibility study by Komforti et al,11 performed on 2 Agilent automated staining platforms demonstrated no inherent bias between instruments. However, analytical validation studies have only been performed using the IUO Ki-67 IHC assay on the Dako Omnis. It should be noted that recent results from the United Kingdom National External Quality Assessment Scheme (UK NEQAS) highlight the importance of matching primary antibody clone choice and other methodological factors to ensure quality outcomes.12
Internal observers and external laboratory pathologists were extensively trained and certified on using the assay scoring algorithm and testing guidelines before conducting precision studies. International recommendations for Ki-67 assessment in breast cancer have proposed counting a minimum of 500 malignant invasive cells and including data from hot spots in the overall score.13 Another study, using a 15% Ki-67 cutoff, concluded counting 500 to 1000 cells was necessary to achieve an acceptable error rate.14 To maximize available patient specimens while maintaining a high degree of scoring reproducibility, this assay scoring algorithm was developed without a minimum counting requirement but stipulated at least 200 viable invasive tumor cells must be present in the specimen for assessment. The scoring methodology proved to be robust and consistent, even when using nonserial sections. Although conventional wisdom accepts “intratumoral heterogeneity,” including spatial and temporal heterogeneity, can be responsible for variability in Ki-67 scoring,14–16 the data from this study suggest the impact is not universal. The results reported herein demonstrate spatial heterogeneity within a given breast carcinoma of a degree that interferes with accurate assessment of “positive” (Ki-67 ≥20%) or “negative” (Ki-67 <20%) tumors is infrequent. In this study, intrablock and intracase heterogeneity studies demonstrated high OA, as reported above.
The IKWG recommendations have evolved since the IUO assay was developed in 2017. The updated consensus recommendations suggest Ki-67 of ≤5% or ≥30% can be used to estimate prognosis in ER+, HER2−, T1-2, N0-1 breast cancer.2 In the monarchE clinical study, this assay used a prospectively defined cutoff of ≥20% to distinguish high versus low Ki-67 tumors6,7; results demonstrated that within a 2-year follow-up, among patients with high clinicopathologic risk factors, those with high Ki-67 tumors (vs. low) had an even greater risk of recurrence. This outcome does not conflict with the updated IKWG recommendation2; rather, it confirms the prognostic value of Ki-67 and establishes the ≥20% cutoff as a clinically relevant decision point for patients with EBC and high risk clinicopathologic factors.
While these results demonstrated high scoring reproducibility across laboratories, there may be opportunities to further improve scoring performance. Inherent limitations exist with manual interpretation approaches. Scoring of the entire slide was chosen over hot spot scoring, since additional variability, potentially due to different scorers identifying different areas of the tumor as hot spots, was observed when using the hot spot method. Establishment of validated digital image analysis approaches may mitigate these potential limitations in the future.
CONCLUSION
In summary, the development and analytical validation of an investigational assay demonstrated reproducibility across different laboratories, and was sensitive, specific, precise, and robust. Use of the assay to assess patients in the monarchE phase 3 clinical study identified a group of patients, among those with high-risk clinicopathologic factors, who displayed high Ki-67 expression and experienced a clinically meaningful increased risk of developing distant recurrence when treated with ET alone. In this setting, assessment of Ki-67 may provide additional information to assist individual physicians with understanding risk and evaluating patient prognosis. These results provide significant contributions toward standardizing Ki-67 IHC assessments and will assist pathologists in making informed decisions regarding local Ki-67 testing procedures. Since completion of these studies, the IUO assay received premarket approval by the United States Food and Drug Administration as a companion diagnostic assay.17 Instructions for use of the commercial version Ki-67 IHC MIB-1 pharmDx (Dako Omnis), the assay Interpretation Manual,18 as well as detailed information about the assay and opportunities to train on the scoring algorithm, are available at Agilent Technologies Inc. (https://www.agilent.com/en/product/pharmdx/ki-67-ihc-mib-1-pharmdx-dako-omnis/ki-67-ihc-mib-1-pharmdx-dako-omnis-1963684).
Supplementary Material
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.appliedimmunohist.com.
ACKNOWLEDGMENTS
The authors would like to thank Anna Strickland, Susan Swift, Kelianne Heinz, Tricia Marquez, and Luella Roma for their contributions to the validation studies; and to acknowledge the Agilent Histology team, led by Jillian Russell, and the tissue procurement team led by Lenka van Alphen. The authors gratefully recognize the contributions of Dr Lauren Jacobson who gave input from a pathologist’s perspective. The authors would also like to thank Ivonne Villalobos (University of Southern California) for administrative coordination and data management, as well as Angela Santiago (University of Southern California) for technical support. Funding of this study was provided by Eli Lilly and Company (Indianapolis, IN). Michael Press was supported by Breast Cancer Research Foundation, Tower Cancer Research Foundation, and a gift from Dr Richard Balch. Some of the tissue samples were supplied by BioIVT (Hicksville, NY). Tissue samples for preanalytical variable and analytical validation studies were provided by the Cooperative Human Tissue Network which is funded by the National Cancer Institute. Other investigators may have received specimens from the same subjects. Eli Lilly and Company contracted with Syneos Health for writing and editorial assistance from Andrea Metti, PhD, MPH and Dana Schamberger, MA. The authors thank Dr Bryce Portier for critical review of this manuscript.
Footnotes
M.D.P. and G.B.N.: equal contribution, and co-first authorship.
C.G. and A.M.G. co-last authorship.
This work was supported by Eli Lilly and Company.
M.D.P. is an employee and company stockholder at Agilent Technologies; she also reports her spouse, Gregory Cherryholmes, is an employee and company stockholder for Agilent Technologies. G.B.N. is an employee and company stockholder at Agilent Technologies; she also reports her spouse, Jon St. Onge, is an employee and company stockholder at Agilent Technologies. Y.G. is former employee of Agilent technologies, and company stockholder. A.T.W. owns stock in Agilent Technologies. G.G. has nothing to disclose. S.T.-F. is an employee and company stockholder at Agilent Technologies. M.C. has nothing to disclose. M.M. is a former employee and minor company stockholder at Eli Lilly and Company, and a current employee at ImmunoGen; all work for this manuscript was completed while he was employed at Eli Lilly and Company. M.F.P. reports receiving honoraria from Science Branding Communications; serving on a Scientific Advisory Board and receiving consulting fees from Biocartis, Eli Lilly and Company, Merck, AstraZeneca, and Novartis; serving as a consultant with consulting fees from Zymeworks Inc; serving as a consultant and having private equity from TORL Biotherapeutics LLC; serving on a Medical Advisory Board and receiving consulting fees from Cepheid; and serving on a Pathology Advisory Board and receiving consulting fees from Eli Lilly and Company, USA. C.G. is an employee and company stockholder at Agilent Technologies. A.M.G. is an employee and company stockholder at Eli Lilly and Company; he also reports his spouse is an employee of Eli Lilly and Company. D.H. declares no conflict of interest.
Contributor Information
Monika D. Polewski, Email: monika.polewski@agilent.com.
Gitte B. Nielsen, Email: gitte.nielsen@agilent.com.
Ying Gu, Email: russellgu76@gmail.com.
Aaron T. Weaver, Email: atweaver89@gmail.com.
Gavin Gegg, Email: gcgegg91@gmail.com.
Siena Tabuena-Frolli, Email: siena.tabuena-frolli@agilent.com.
Mariana Cajaiba, Email: maricajaiba@gmail.com.
Debra Hanks, Email: drdeb@earthlink.net.
Michael Method, Email: m.method@icloud.com.
Michael F. Press, Email: michael.press@med.usc.edu.
Claudia Gottstein, Email: claudia.gottstein@agilent.com.
Aaron M. Gruver, Email: gruver_aaron_m@lilly.com.
REFERENCES
- 1.Smith I, Robertson J, Kilburn L, et al. Long-term outcome and prognostic value of Ki-67 after periopertative endocrine therapy in postmenopausal women with hormone-sensitive early breast cancer (POETIC): an open-label, multicentre, parallel-group, randomised, phase 3 trial. Lancet Oncol. 2020;21:1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nielsen TO, Leung SC, Rimm DL, et al. Assessment of Ki67 in breast cancer: updated recommendations from the international Ki67 in breast cancer working group. J Natl Cancer Inst. 2021;113:808–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickler MN, Tolaney SM, Rugo HS, et al. MONARCH 1, a phase II study of abemaciclib, a CDK4 and CDK6 inhibitor, as a single agent, in patients with refractory HR(+)/HER2(−) metastatic breast cancer. Clin Cancer Res. 2017;23:5218–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston S, Martin M, Di Leo A, et al. MONARCH 3 final PFS: a randomized study of abemaciclib as initial therapy for advanced breast cancer. NPJ Breast Cancer. 2019;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sledge GW, Jr, Toi M, Neven P, et al. MONARCH 2: abemaciclib in combination with fulvestrant in women with HR+/HER2− advanced breast cancer who had progressed while receiving endocrine therapy. J Clin Oncol. 2017;35:2875–2884. [DOI] [PubMed] [Google Scholar]
- 6.Harbeck N, Rastogi P, Martin M, et al. Adjuvant abemaciclib combined with endocrine therapy for high-risk early breast cancer: updated efficacy and Ki-67 analysis from the monarchE study. Ann Oncol. 2021;32:1571–1581. [DOI] [PubMed] [Google Scholar]
- 7.Johnston S, Harbeck N, Hegg R, et al. Abemaciclib combined with endocrine therapy for the adjuvant treatment of HR+, HER2−, node-positive, high-risk, Early breast cancer (monarchE). J Clin Oncol. 2020;38:3987–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudis CA, Barlow WE, Costantino JP, et al. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25:2127–2132. [DOI] [PubMed] [Google Scholar]
- 9.Hurvitz SA, Martin M, Press MF, et al. Potent cell-cycle inhibition and upregulation of immune response with abemaciclib and anastrozole in neoMONARCH, phase II neoadjuvant study in HR(+)/HER2(−) breast cancer. Clin Cancer Res. 2020;26:566–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris LN, Ismaila N, McShane LM, et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice Guideline Summary. J Oncol Pract. 2016;12:384–389. [DOI] [PubMed] [Google Scholar]
- 11.Komforti M, Downs-Kelly E, Badve S. Analytic comparison study of IUO Ki-67 clinical trial assay on Dako Omnis and Dako AutoStainer Link48 (ASL48). Presented at: College of American Pathologists (CAP) Meeting; Septmeber 25–28, 2021; Chicago, IL.
- 12.Parry S, Dowsett M, Dodson A. UK NEQAS ICC & ISH Ki-67 data reveal differences in performance of primary antibody clones. Appl Immunohistochem Mol Morphol. 2021;29:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowsett M, Nielsen TO, A’Hern R, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103:1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denkert C, Budczies J, von Minckwitz G, et al. Strategies for developing Ki67 as a useful biomarker in breast cancer. Breast. 2015;24(suppl 2):S67–S72. [DOI] [PubMed] [Google Scholar]
- 15.Nassar A, Radhakrishnan A, Cabrero IA, et al. Intratumoral heterogeneity of immunohistochemical marker expression in breast carcinoma: a tissue microarray-based study. Appl Immunohistochem Mol Morphol. 2010;18:433–441. [DOI] [PubMed] [Google Scholar]
- 16.Tramm T, Kyndi M, Sorensen FB, et al. Influence of intra-tumoral heterogeneity on the evaluation of BCL2, E-cadherin, EGFR, EMMPRIN, and Ki-67 expression in tissue microarrays from breast cancer. Acta Oncol. 2018;57:102–106. [DOI] [PubMed] [Google Scholar]
- 17.Ki-67 IHC PharmDx Premarket Approval (PMA). US Food and Drug Administration. Available at: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P210026. Accessed December 1, 2021.
- 18.Agilent Technologies, Inc. Ki-67 IHC MIB-1 pharmDx (Dako Omnis) Interpretation manual—breast carcinoma. 2021. Available at: https://www.agilent.com/en/product/pharmdx/ki-67-ihc-mib-1-pharmdx-dako-omnis/ki-67-ihc-mib-1-pharmdx-dako-omnis-1963684. Accessed December 1, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.appliedimmunohist.com.


