Abstract
Packaging of the eukaryotic genome into chromatin places fundamental physical constraints on transcription. Clarifying how transcription operates within these constraints is essential to understand how eukaryotic gene expression programs are established and maintained. Here we review what is known about the mechanisms of transcription on chromatin templates. Current models indicate that transcription through chromatin is accomplished by the combination of an inherent nucleosome disrupting activity of RNA polymerase and the action of ATP-dependent chromatin remodeling motors. Collaboration between these two types of molecular motors is proposed to occur at all stages of transcription through diverse mechanisms. Further investigation of how these two motors combine their basic activities is essential to clarify the interdependent relationship between genome structure and transcription.
Graphical Abstract
I. Introduction
Elucidating the mechanisms of eukaryotic transcription is a major pursuit with deep implications for understanding the regulation of cellular states, development, and disease. Transcription in eukaryotes occurs in the context of chromatin: the crowded and highly regulated structures that package DNA. The fundamental unit of chromatin is the nucleosome: a highly stable structure, which wraps 145–147 bp of DNA around an octamer of histone proteins[1,2]. Because of the tight DNA wrapping and high stability of nucleosomes, chromatin presents a major barrier to transcription by RNA polymerases (Pols)[3]. Indeed, transcription by bacterial RNA polymerase and eukaryotic RNA Pol II is greatly inhibited on nucleosomal templates in vitro at physiological salt concentrations[4,5]. Furthermore, several factors which directly reshape chromatin structure, including ATP-dependent chromatin remodeling enzymes, also regulate transcription in vitro and in vivo[6,7]. These findings have shaped a view of nucleosomes as generic repressors of transcription, with their regulated disruption enabling transcription.
However, several findings challenge this simple view of the role of nucleosomes and more generally, chromatin structure in transcription. In contrast to Pol II, Pol III and bacteriophage SP6 Pols are only modestly inhibited on nucleosome-containing templates at physiological salt[8,9]. Furthermore, transcription by these polymerases does not necessarily involve nucleosome eviction [8,9]. These observations suggest that transcription need not be incompatible with the presence of nucleosomes. Additionally, at slightly higher salt concentrations, Pol II efficiently transcribes through nucleosomes with only the loss of a single H2A/H2B dimer[4]. However, some sub-nucleosome particles, which physically occlude less DNA, inhibit transcription when compared to a complete nucleosome[10]. Finally, it has been shown that transcriptionally silenced regions can be accessible to digestion by nucleases and further that nucleosome plasticity promotes formation of transcriptionally repressed heterochromatin [11,12]. Together these results shape a more nuanced view of the role of chromatin in transcription as a dynamic platform that has coevolved with RNA polymerase and other nuclear proteins to enable complex, tightly regulated gene expression programs.
Key to understanding how transcription operates on chromatin is a thorough understanding of how chromatin structure is reorganized. The SWI2/SNF2 superfamily of ATP-dependent chromatin remodeling enzymes play an essential role in facilitating DNA-based processes (including transcription) by directly reshaping chromatin at the level of individual nucleosomes[6,7]. Like eukaryotic RNA polymerase, remodelers are large, multi-subunit molecular machines which carry out complex and highly regulated reactions. Compared to RNA polymerase, our understanding of the basic mechanisms of remodelers is less mature. Even less understood is how remodeler activity is coordinated with RNA polymerases and other factors to both faithfully regulate transcription while maintaining genome architecture. However, recent structures of both remodelers and RNA polymerase with nucleosomes, together with prior biochemical data enable models for how these motors might collaborate.
In this review we first briefly summarize our current understanding of the core process of transcription and its regulation by elongation factors. We then discuss how this basic process is influenced by the presence of chromatin. Specifically, we focus on how the molecular motor at the heart of transcription, RNA polymerase, may accomplish its activity despite the physical constraints of the nucleosome. This is both due to an inherent chromatin remodeling capability of RNA polymerase and due to its collaboration with other factors including remodelers. We further compare our understanding of the chromatin remodeling activity of RNA polymerase to that of ATP-dependent chromatin remodelers and highlight open questions. Finally, we review how these ATP-dependent chromatin remodelers and RNA polymerases might collaborate.
II. The nucleosome: beyond a barrier
Although transcription by all three eukaryotic RNA polymerases on chromatin templates is generally inhibited, the nature and strength of this barrier varies. This depends not only on properties of the polymerase and its associated factors but also the structural properties of the nucleosome itself. Since the detailed structure of the canonical nucleosome was solved, a wealth of biophysical studies shed some light on the core physical properties of the nucleosome, which provides insights into how molecular motors disrupt its structure (Figure 1)[13].
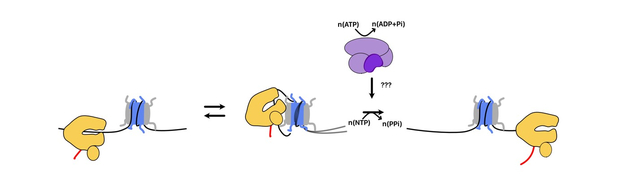
FIGURE 1. Core properties of the nucleosome.
A. Nucleosome core particle with the surface of the histone octamer colored according to the gradient specified below. The unit kT/e refers to the energy associated with placing a unit charge at a specific location. B. Ribbon diagram of one pseudosymmetrical half of the nucleosome. Locations of nucleosomal DNA are specified according to their SuperHelical Location (SHL) with each location being 10 bp from the dyad axis of symmetry. C. Dynamic states of a nucleosome in solution. Nucleosomes exist in equilibrium between many possible states. Transition between states may involve conformational changes in the histone octamer (e.g. during DNA breathing).
The canonical nucleosome is an ~0.2 MDa complex and contains ~147bp of DNA bound to an octamer of histone proteins: 2 copies each of histones H2A, H2B, H3, and H4 (Figure 1B). Each histone possesses both a structured domain, which folds cooperatively with the other histones to form a globular core, and unstructured tails[2]. Histones assemble in a stepwise fashion on DNA with two heterodimers of H3/H4 first depositing to form a core tetramer followed by two heterodimers of H2A/H2B. DNA wraps around the symmetric globular core ~1.7 times with the unstructured tails projecting out of the core[1]. This wrapping is stabilized by electrostatic contacts between basic residues in the histone core and the DNA phosphodiester backbone. With an average pI of ~11, histone proteins have a high net negative charge at physiological pH that facilitates their interaction with DNA [14]. However, a key acidic surface on H2A/H2B, known as the acidic patch, plays an important role as a recognition surface for many chromatin proteins (Figure 1A)[15]. Additionally, wrapping of DNA around the nucleosome introduces a single negative supercoil constrained over the length of the nucleosomal DNA[16]. This is because the > −1 writhe associated with wrapping is compensated by reduced DNA twist within the nucleosome[17]. Topological changes in the DNA are important for the stability of nucleosomes as octamer assembly on positively supercoiled DNA templates is disfavored[18]. Correspondingly, subjecting nucleosomal DNA to torsional stress from positive supercoiling reduces nucleosome stability[19].
The nucleosome is thermodynamically a very stable structure. In vitro nucleosomes remain intact under physiological salt concentrations at low micromolar concentrations even when heated to temperatures as high as 65 °C [20]. At the same time, however, several dynamic transitions can be observed in nucleosomes without the addition of other factors (Figure 1C). DNA near the entry/exit (SHL±7) can transiently peel off the surface off the histone octamer exposing the underlying DNA[21,22]. The histone H2A/H2B dimer can also associate with this unpeeled DNA and undock from the histone core[23]. Electron cryo-microscopy (cryo-EM) structures suggest that such dynamic changes involve intermediates with subtle conformational changes in the histone octamer[24]. Reversible dissociation of the H2A/H2B dimer also occurs and can result in stable sub-nucleosome structures, which wrap less DNA than a full nucleosome[25]. Additionally, it is proposed that the unstructured histone tails may associate with nucleosomal DNA influencing its exposure and dynamics[26,27]. Finally, nucleosomes have also been observed to spontaneously reposition along DNA in a process known as nucleosome sliding[28]. Molecular dynamics simulations suggest that subtle local changes in DNA twist could allow the resetting of histone-DNA contacts around the octamer by ~1bp increments[29]. CryoEM studies combined with histone-histone crosslinking suggest that dynamic rearrangements in histone conformation also facilitate nucleosome sliding[30].
The intrinsic properties of the nucleosome can be modulated by several factors. Sequences that can accommodate the DNA deformations associated with nucleosome formation promote nucleosome stability. Such sequences are often associated with AT dinucleotides at sites where the minor groove of DNA faces the histone octamer and GC dinucleotides at sites where the major groove of DNA faces the histone octamer[31]. This is because AT and GC dinucleotides favor compression of the minor and major grooves respectively, which occurs upon interaction with the histone octamer[32]. In contrast. continuous poly dA:dT tracts seen at some promoters disfavor nucleosome stability[33]. In addition to DNA sequence, post-translational modifications on histones can influence the structure, dynamics, and stability of the nucleosome[34]. Acetylation of histone tails has small effects on DNA unpeeling and histone tail accessibility, while phosphorylation and acetylation of the H3 core may have more substantial effects[34,35]. Several variant histones also exist and can be incorporated into the nucleosome, which alter nucleosome properties. For example, histone H2Az is an H2A variant that is is particularly enriched at the +1 nucleosome relative to the transcription start site (TSS)[36]. H2Az-containing nucleosomes are less stable to mechanical stress, which may facilitate transcription elongation (discussed further below)[37].
III. Transcription through nucleosomes
A. RNA polymerase: a nucleotide dependent chromatin remodeler
In order to understand how RNA polymerase negotiates nucleosome structure, an understanding of the basic structure and mechanism of RNA polymerase is required. Transcription in Eukaryotes is carried out by one of three large (~0.5–1 MDa) multisubunit RNA Polymerases: Pol I, II, and III (Figure 2A)[38,39]. Most protein coding genes are transcribed by RNA Pol II. In contrast Pol I and III mostly transcribe noncoding genes, with rRNA being transcribed by Pol I and 5S rRNA, tRNA and other small RNAs being transcribed by Pol III. All three polymerases contain a core of 10 polymerase-specific subunits that share structural homology to prokaryotic multisubunit RNA polymerases[38]. This structure resembles a “claw” with three pincers called the “clamp”, “lobe”, and “jaw” that surround the DNA (Figure 2A). The clamp and jaw specifically engage the downstream DNA and feed it into the catalytic center [38]. In addition to these core subunits, the three polymerases also possess a flexible heterodimeric “stalk module” that binds to the core clamp module. The stalk module can make interactions with transcriptional regulators and the nascent RNA. Though considered part of the core polymerase, under certain circumstances the stalk module may need to dissociate during transcription by Pol II with consequences on RNA processing[40,41].
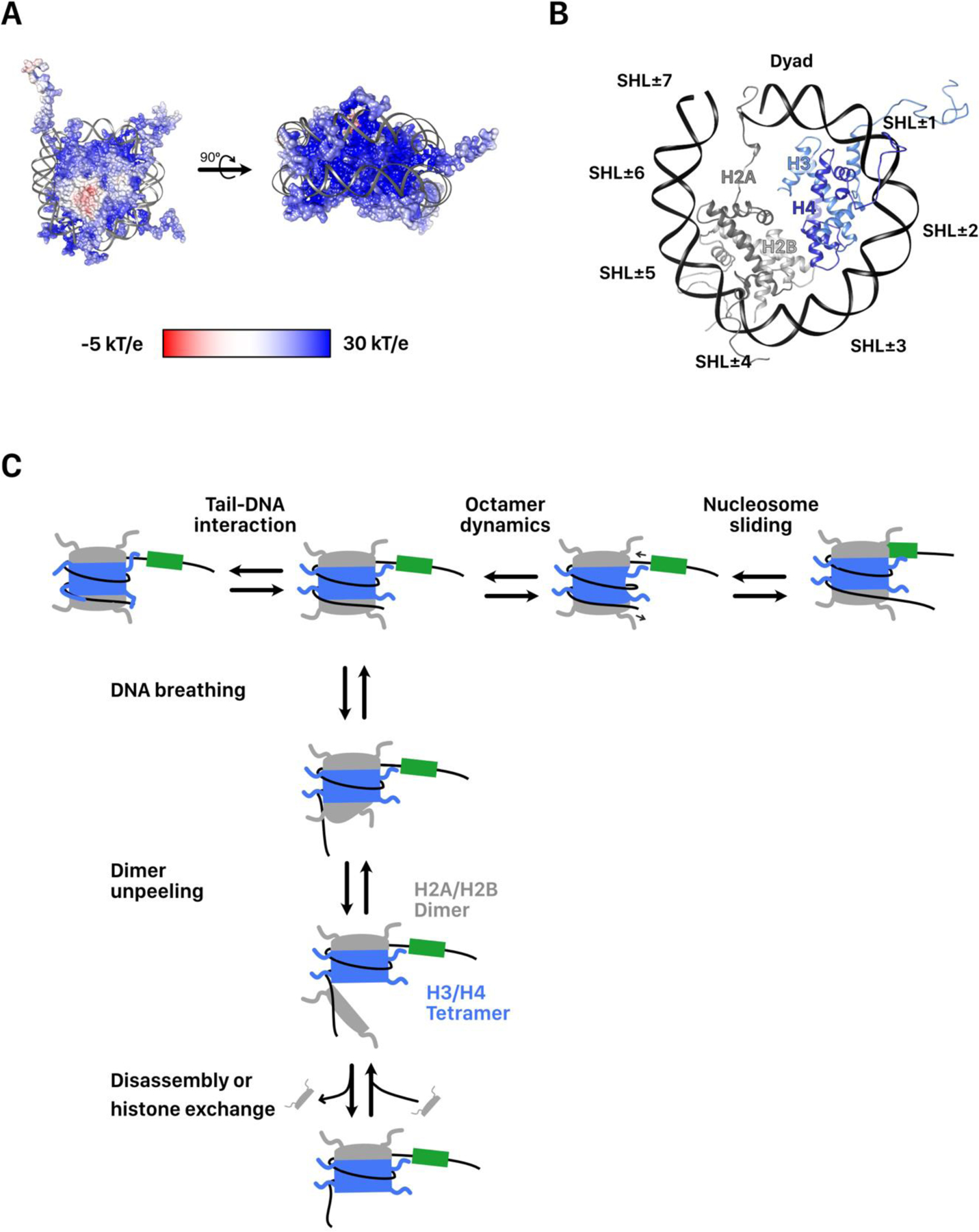
FIGURE 2. Mechanisms of nucleosome remodeling by RNA polymerases.
A. Cartoon representations of the three eukaryotic RNA polymerases. The lobe, clamp, and stalk domains that are shared between all three polymerases are labeled on Pol II. The TFIIS like domain of the lobe and the LB module shared by Pol I and III are labeled on Pol I. Finally, the unique CTD of Pol II is also shown. B. Core mechanism of elongation shared by RNA polymerases. The green triangle is used as a reference point for polymerase translocation. Polymerase can either translocate 1 nt forward or pause and backtrack. DNA unwinding is associated with torsional stress ahead of and behind the transcription bubble. Stress behind the bubble can be absorbed by formation of a stable RNA-DNA hybrid called an R-Loop. Backtracked polymerase can be resolved by cleavage of the nascent transcript by TFIIS or the TFIIS-like domains of Pol I and III. Forward elongation is biased by irreversible incorporation of a complementary nucleotide. C. Mechanism of polymerase passage through the nucleosome. The green box is used as a reference location on the DNA. As polymerase translocates into the nucleosome core, it unpeels DNA at the entry side until it reaches the dyad. Once at the dyad, further passage requires DNA unpeeling at the exit side and loosening of histone DNA contacts downstream of the dyad. Further passage is associated with either octamer transfer upstream of its current site or dissociation of an H2A/H2B dimer from the exit side. Elongation factors assist entrance into the nucleosome and may bias the outcome of Pol II passage. D. Polymerase passage through a hexasome by Pol II is associated with complete disassembly of the hexasome.
While many structural aspects are shared across all three eukaryotic RNA polymerases there are some key differences. The largest subunit of Pol II contains a repetitive extension at its C-terminus called the CTD, which is phosphorylated or dephosphorylated in specific patterns at different stages of transcription to help recruit transcriptional regulators[42]. Pol I and III utilize specific lobe-binding (LB) modules, which resemble a Pol II initiation factor TFIIF[38]. The LB module binds to the core subunits A12.2 in Pol I and C11 in Pol III. These core subunits contain domains that resemble the elongation factor TFIIS that is typically used by Pol II and stimulate elongation using a similar mechanism[43]. Pol III also uniquely contains a heterotrimeric module that assists in transcription initiation[44].
To enable efficient elongation, RNA polymerase must function as a processive molecular motor, efficiently translocating along DNA. Several single-molecule studies have helped uncover the mechanical properties of RNA polymerases. Pol II can transcribe against forces of up to ~6.5 pN while Pol I can withstand somewhat higher forces (~9.5 pN)[45]. Translocation is thought to function through a brownian ratchet mechanism, where the polymerase can freely diffuse one-dimensionally along the RNA•DNA hybrid forward or backward relative to the 3’ end of the nascent RNA (Figure 2B)[46,47]. The irreversible incorporation of a complementary ribonucleotide provides the energy to bias forward translocation. Despite such a mechanism for directionality, pausing and backtracking of the polymerase is frequent and often represents a rate-limiting barrier to transcription[46]. Pause locations are influenced by properties of the DNA sequence being transcribed, the secondary structure of the nascent RNA, and by barriers such as nucleosomes[46,48]. While backtracking and pausing are often reversible, in some instances pausing can be irreversible and require the action of other factors to be overcome. As a result, elongation factors have evolved that physically associate with polymerase and prevent or resolve backtracked pauses. The elongation factor TFIIS helps resolve irreversibly backtracked/paused polymerase by catalyzing the endonucleolytic cleavage of the extruded 3’-end of the nascent RNA (Figure 2B). This resets the 3’ end of the nascent RNA at the pause location and allows further rounds of elongation.
The unwinding of DNA by RNA polymerases generates torsional stress and is a major source of DNA supercoiling throughout the genome[49]. Transcription generates negative torsional stress upstream, which is expected to stabilize nucleosomes, and positive stress downstream of the polymerase, which is expected to destabilize nucleosomes (Figure 2B). If unresolved, torsional stress can hinder both the continued elongation and the stability of the chromatin template[50]. As a result, relief of DNA supercoiling by topoisomerases plays an important role in continued transcription and maintenance of chromatin structure[50,51]. Additionally, stable RNA:DNA hybrids formed between the nascent RNA and the DNA upstream of the polymerase, known as R-loops, may also play a role in absorbing negative torsional stress (Figure 2B)[49].
Eukaryotic RNA polymerases have been long appreciated as remodelers of nucleosome structure[52]. Early experiments of transcription on short nucleosome-containing templates suggested two mechanisms of remodeling linked to the type of polymerase transcribing. Transcription by Pol III is only modestly inhibited at physiological salt concentration and transfers intact histone octamers largely in cis upstream of its original position[9]. Like Pol III, nucleosomes also present only a modest barrier to transcription to Pol I under physiological conditions, although nucleosome transfer activity has not yet been reported[53]. In contrast, Pol II is almost completely inhibited at physiological salt concentrations[4]. At slightly higher salt concentrations, which loosens histone DNA-contacts, Pol II is able to more efficiently transcribe through the nucleosome with the dissociation of a single H2A/H2B dimer and without net change in nucleosome position[4,54]. This matches well with in vivo observations that H2A/H2B dimers are more readily exchanged at active genes than H3/H4[55]. Interestingly, E. coli RNA polymerase transcribes through nucleosomes with very similar properties as Pol II suggesting that nucleosome organization may have evolved to capitalize on an essential and ancient property of multisubunit RNA polymerases [5].
More recent experiments suggest that the mechanisms of Pol II and III may fundamentally be more similar than different. On longer DNA templates, Pol II transfers nucleosomes in cis upstream of its original location[56,57]. Apparent nucleosome transfer intermediates with Pol II have been visualized using atomic force microscopy and CryoEM[57,58]. Also, with TFIIF and TFIIS, Pol II more efficiently transcribes through nucleosomes raising the possibility that the TFIIF-like LB module or the TFIIS-like domains in the lobes of Pol I and III are responsible for their higher efficiency[59]. Indeed, deletion of the LB modules or the TFIIS-like domain from Pol I reduces its ability to transcribe through a nucleosome[60]. In light of these observations, it might be more useful to think of the Pol III and Pol II mechanisms as two potential pathways of nucleosome disruption that might be taken by eukaryotic polymerases (Figure 2C).
Of the three polymerases, the mechanism of transcription through chromatin is most understood for Pol II. Analyses of the pause sites within the nucleosome provide some clues for how this polymerase remodels nucleosomes[10,61]. At low salt concentrations, Pol II is predominantly paused at the entry side of the nucleosome, suggesting that the tight wrapping of the nucleosome blocks transcription. At higher salt concentrations, Pol II pause sites progressively invade further into the nucleosome up until just before the dyad. Pol II paused prior to the nucleosome dyad exposes entry DNA to cleavage by restriction enzymes[10]. Together, these results suggest that entry of Pol II into the nucleosome is associated with unpeeling of nucleosomal DNA at the entry side. Optical trapping experiments combined with kinetic modeling have supported the notion that RNA polymerase takes advantage of spontaneous DNA breathing in order to enter and ratchet through the nucleosome[35,56]. Strangely, however, hexasomes lacking a single H2A/H2B dimer at the entry side, and thus unwrapped, are transcribed less-efficiently than full nucleosomes[10]. This suggests that the entry side dimer may be important in establishing intermediates required for passage through the nucleosome. In contrast, hexasomes lacking a dimer on the nucleosome exit side are transcribed more efficiently than nucleosomes and are disassembled by Pol II passage (Figure 2D)[10]. Hexasomes, produced by Pol II passage are more efficiently transcribed than nucleosomes, suggesting that Pol II preferentially displaces the exit side dimer[62]. This is consistent with genome-wide observations that hexasomes within the gene body primarily lack an H2A/H2B dimer on the promoter-distal side of the nucleosome[63].
Raising salt concentration or adding elongation factors shifts the location of Pol II pauses further into the nucleosome up until SHL −2/−1 [61]. Beyond this location, few nucleosome-dependent pause sites are observed. This suggests that entering the nucleosome represents a greater barrier than exiting the nucleosomes. Interestingly, Pol II pausing in vivo is enriched right before the nucleosome dyad, further suggesting that transiting specifically through the dyad may be the dominant barrier to elongation[61]. Consistent with this idea, mutating a histone-DNA contact near the dyad both reduces pausing near the dyad and improves transcription efficiency[35,64]. What structural changes are necessary to allow Pol II to pass through the dyad? Since removal of the exit side dimer facilitates passage of Pol II, transiting beyond the dyad may also involve unpeeling of DNA from the exit side. Consistent with this possibility, increasing the length of DNA exiting the nucleosome, which promotes DNA breathing, allows Pol II to progress deeper into the nucleosome[65]. Pol II progression may allosterically loosen histone-DNA contacts ahead of the polymerase. Indeed, Pol II arrested near SHL −2 shows increased restriction site accessibility downstream of the polymerase near SHL+1[10]. Additionally, DNA sequences near SHL+2 appear to influence the ability of polymerase to transit. Interestingly, sequences that promote pol II passage do not necessarily destabilize nucleosomes. These observations suggest that Pol II passage may require the formation of a specific sequence-dependent intermediate to pass beyond the dyad.
Recent high resolution CryoEM snapshots of Pol II transcribing on nucleosomal templates provide new clarity to earlier biochemical observations[58,66–68]. As Pol II initially approaches the nucleosome, DNA at the entry site unpeels from the octamer consistent with biochemical and biophysical observations[58,66]. In this initial encounter the lobe and clamp regions of the core make additional contacts with nucleosomal DNA at the dyad [66]. Further progression peels more entry side DNA from the surface of the octamer. Once Pol II reaches SHL-1, the lobe region of Pol II makes contact with the H2A/H2B dimer, providing an explanation for the retention of the entry side dimer. Major pause sites internal to the nucleosome are immediately adjacent to histone-DNA contacts, suggesting that breaking these contacts is required for further progression. The precise relative motion of Pol II with respect to histones and/or DNA between nucleosomal pause sites is unclear and is an important area of study[69–71]. Some reconstructions of the SHL −1 pause also revealed the presence of an additional DNA molecule replacing the region of DNA unpeeled from the octamer, which may represent a histone transfer intermediate. Consistent with this, close inspection of the raw Cryo-EM micrographs show evidence for nucleosomes invading this complex[58].
B. Regulation of RNA polymerase-mediated nucleosome remodeling
Several features of nucleosome structure play an important role in controlling elongation through the nucleosome. As already mentioned, DNA sequence can influence RNA polymerase progression through the nucleosome[61]. Histone tails repress the ability of Pol II to transit through the nucleosome largely through their effect on restricting DNA breathing[35,72]. Several post translational modifications of the histone proteins play positive roles in Pol II passage. Acetylation of histone tails improves Pol II passage by modestly increasing DNA breathing[35]. Histone modifications may also directly affect core histone-DNA interactions. Poly ADP-ribosylation of histones, which is associated with elongation, appears to globally loosen histone-DNA contacts[73]. Importantly, not all transcription-associated histone modifications play a direct role in the progression of Pol II through the nucleosome, as has been suggested for H3K4 trimethylation[74]. Instead, these may regulate transcription by impacting higher order chromatin organization or the recruitment of additional regulators. The incorporation of certain histone variants can also facilitate passage through the nucleosome (as discussed later).
As mentioned before, elongation factors promote the progression of polymerases through the nucleosome. Pol II-nucleosome structures suggest that transcription intermediates can accommodate association of several elongation factors including NELF, Paf1, and TFIIS[66]. Transcription of nucleosome-containing templates with Pol II and elongation factors both shifts pause sites deeper into the nucleosome and improves overall transit efficiency. CryoEM structures of Pol II-nucleosome intermediates with the elongation factors Spt4/5 and Elf1 has also suggested that elongation factors may directly assist in the stabilization of Pol II intermediates through contacts with the histones. Spt4/5 and Elf1 cooperatively promote Pol II passage both by affecting the orientation of the Pol II clamp on the nucleosome and by direct contacts with the histone proteins[67,75]. Direct histone contacts may be a widespread mechanism of elongation factors on the nucleosome. Indeed, contacts between Paf1 and the H2A/H2B acidic patch have been detected and the elongation factor Spt6 has been shown to have histone chaperone activity[76,77]. Interestingly, despite the higher fraction of complexes stalled at SHL-1, no octamer transfer intermediates were detected in their CryoEM reconstructions[67]. It is possible that Spt4/5 and Elf1 cooperate to bias elongation through a pathway that does not involve octamer transfer (Figure 2C).
In addition to specific elongation factors, general histone chaperones also play a key role in transcription, and particularly the H2A/H2B chaperone FACT (FAcilitates Chromatin Transcription). FACT improves elongation in vitro and its depletion in vivo produces major elongation defects by all three polymerases[53,78]. Including FACT alone with Pol II transcription reactions promotes passage through the nucleosome and the formation of hexasomes[78,79]. However, dissociation of the dimer is not required for FACT function as neither covalent crosslinking of the histone octamer, nor destabilization of the histone dimer-tetramer interface inhibits FACT-dependent stimulation[79]. This has led to a model that FACT assists with Pol II passage mainly by facilitating disruption of nucleosomal DNA. Consistent with this, cryoEM structures combined with Hydrogen-Deuterium exchange experiments suggest that FACT binds to the DNA-binding surface of H2A-H2B, promoting DNA unpeeling[80]. A recent cryoEM structure of FACT bound to a nucleosome with Pol II arrested at SHL-4 shows FACT bound at SHL+1 instead of at the dyad [75]. As a result, it is possible that FACT interactions with the nucleosome may be highly plastic and adapt in response to Pol II passage. Interestingly, a kinetic analysis of FACT-assisted elongation has suggested that FACT reduces barriers throughout the nucleosome, but particularly between SHL −5 and SHL+2[79]. FACT has also been implicated in the reassembly of nucleosome structure in the wake of Pol II passage[81]. This reassembly function may be linked to the action of other factors (such as remodelers with nucleosome assembly activity) or elongation-associated histone modifications (such as H2B ubiquitination)[82,83].On a sub-nucleosome, FACT adopts multiple conformations linked to the presence or absence of a second H2A-H2B dimer [80]. This may facilitate the displacement or retention of the dimer during Pol II progression. With Pol II, FACT makes contact with the entry-side dimer, which may further stabilize its retention during Pol II passage[75]
Local changes in DNA topology mediated by RNA polymerase play an important role in influencing transcription through the nucleosome. DNA nicks within the nucleosome, which relax torsional stress, can both promote and inhibit transcription through the nucleosome depending on their location[84]. With prokaryotic polymerase, nicks between the entry side and SHL-3 inhibit passage, while nicks between SHL-3 and SHL+3 facilitate passage. Nicks near the entry side of the nucleosome likewise inhibit Pol II nucleosome passage[84]. This supports a role for the buildup of local torsional stress in regulating Pol II passage through the nucleosome. Higher resolution Pol II-nucleosome structures are needed to visualize whether this stress results in changes in DNA topology. Also, while it is clear that the buildup of torsional stress is critical for elongation, it is unclear how further accumulation of stress at different locations influences progression through the nucleosome. Further reconstitution studies of polymerase progression should be done on topologically constrained nucleosomal templates to better understand how DNA torsion and topology alters transcription on chromatin.
Several other properties of the polymerase, and the nascent RNA may play a key role in regulating transcription through the nucleosome. Notably phosphorylation of the CTD does not appear to directly impact chromatin transcription[85]. However, association of factors with the CTD, which can be linked to its phosphorylation state, is likely to allosterically influence core polymerase properties (as has been proposed to occur during termination of transcription)[86,87]. Adjacent polymerases may also influence nucleosome remodeling properties. Transcription with tandem polymerases improves overall transcription efficiency through the nucleosome[62,88]. This is in part due to the remodeling activity of the leading polymerase but also due to prevention of backtracking of the leading polymerase by the trailing polymerase. Furthermore, multiple polymerases may cooperate through the accumulation of torsional stress, which can both destabilize nucleosome structure and additionally directly improve polymerase elongation [89,90]. Finally, nascent RNA can both positively and negatively impact transcription through the nucleosome through at least two types of mechanisms. Nascent RNA secondary structure can influence polymerase pausing and backtracking [35,48] and R-loops may contribute directly to remodeling of the nucleosome as they are expected to generally destabilize nucleosome structure[91,92].
IV. ATP-dependent chromatin remodelers
A. Diverse activities mediated by diverse motors
Although possessing considerable remodeling activity on its own, RNA polymerase is fundamentally limited in its remodeling capabilities. On the mechanistic side, this may reflect fundamental mechanical limitations imposed by the polymerase machinery. However, on the biological side, by being dependent on other factors RNA polymerases open themselves up for many regulatory possibilities. ATP-dependent chromatin remodelers are among the many factors that regulate RNA polymerase action on chromatin. Remodelers range in size from small single subunit motors to large multi-subunit complexes (Figure 3A)[6]. They are able to catalyze a wide range of activities including most spontaneous transitions seen with the nucleosome alone (Figure 1C). However, remodelers do not necessarily act as “heat”, simply lowering the activation barrier to thermally inaccessible states[7]. This is because the irreversibility of ATP-hydrolysis allows structural changes produced by remodelers to be directional. For example, the SWR complex catalyzes the exchange of canonical H2A/H2B dimers with H2Az/H2B dimers but does not effectively catalyze the opposite reaction (Figure 3B)[93]. Remodelers are also capable of creating stable non-canonical nucleosome structures not observed spontaneously. SWI/SNF-family remodelers catalyze the formation of stable structures called remosomes which contain an intact histone octamer but wrap ~30bp more DNA than canonical nucleosomes and with overall weaker histone-DNA contacts (Figure 3C)[94,95]. Some remodelers can also slide nucleosomes in order to displace DNA-bound proteins or other nucleosomes (Figure 3D/E)[96,97]. The wider range of transformations produced by remodelers when compared to RNA polymerase is likely linked to their fundamental activity of DNA translocation, which, as we discuss below, is a highly flexible means of disrupting nucleosome structure.
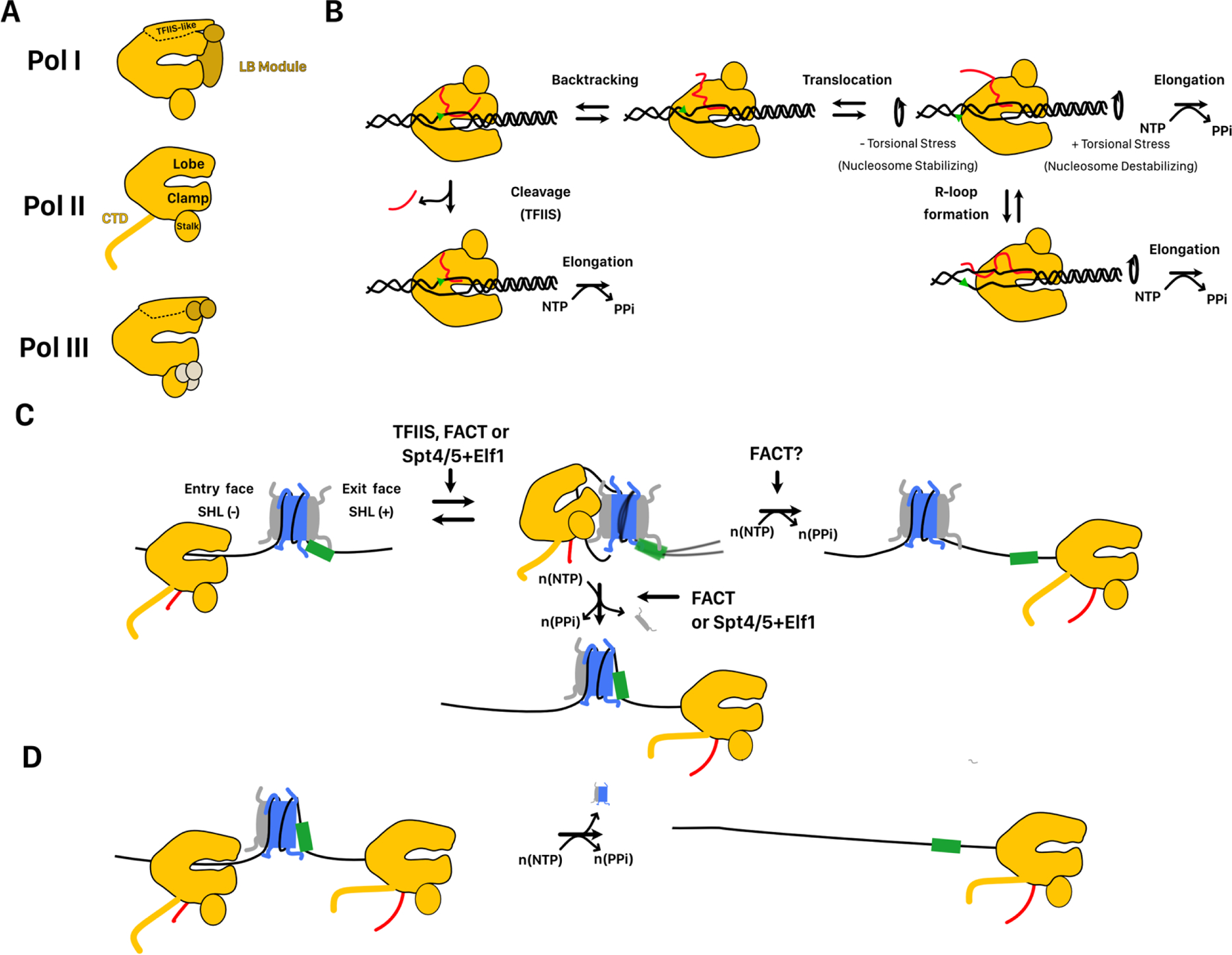
FIGURE 3. Mechanisms of nucleosome remodeling by ATP-dependent chromatin remodelers.
A. Cartoon representations of selected remodelers from S. cerevisiae. In dark purple is the ATP hydrolysing subunit. Remodelers range in size from the single subunit remodeler CHD1 to the mega dalton sized 17-subunit RSC complex. B. Directional histone exchange catalyzed by the SWR complex. H2A-H2B is exchanged for H2Az-H2B but not the reverse reaction. C. Remosome formation catalyzed by the RSC complex. The structure of remosomes is unclear but involves ~30bp of additional DNA wrapped around the core and looser histone-DNA contacts throughout the nucleosome. D. Nucleosome disassembly catalyzed by the RSC complex. RSC sliding of a nucleosome into an adjacent nucleosome causes its disassembly. This may involve an overlapping dinucleosome intermediate recently described. The green box is used as a reference location on the DNA. E. Transcription factor disruption by the RSC complex. RSC sliding of a nucleosome through a DNA-bound transcription factor causes its displacement. F. DNA translocation by SWI2/SNF2 family chromatin remodelers binding at SHL 2. The green triangle is used as a reference location on DNA. Binding at SHL2 stabilizes twist defects at that location, allowing breaking of histone DNA contacts. Binding, hydrolysis, and/or release of a nucleotide allows the resetting of histone-DNA contacts. Further rounds of translocation with additional contacts is expected to enable more complex activities, such as histone exchange. G. Regulation of remodeler activity by autoinhibitory domains. After binding the nucleosomes, remodelers are held inactive by contacts with autoinhibitory domains on either the ATP hydrolyzing subunit or accessory subunits. Contacts with substrate cues relieve this autoinhibition and allow remodeling activity. The active state may be associated with increased conformational dynamics in the histone octamer. The green box is used as a reference point on the DNA.
B. Structures and mechanisms of ATP-dependent Chromatin Remodeling Enzymes
Common to all remodelers is the presence of a single subunit containing an ATP-hydrolyzing domain with homology to the yeast protein SNF2 from the SWI/SNF complex. Remodelers are further divided into families based on differences within this subunit[98]. These differences appear to determine the range of reactions catalyzed by the ATPase complexes. For example, complexes containing ISWI-family ATPases appear to only catalyze nucleosome sliding and nucleosome assembly, while SWI/SNF-family ATPase complexes catalyze a diverse range of reactions[7]. The ATPase domain of remodelers has two RecA-like lobes, which contact the phosphodiester backbone of DNA. ATP binding and hydrolysis occurs at the interface between the two lobes. Changes in the relative conformations of the RecA-like lobes driven by the ATPase cycle is proposed to power DNA translocation.
Biochemical and structural studies are highlighting both the commonalities and differences amongst remodelers in how they engage and transform nucleosomes. The ATPase domain of most remodelers associates with nucleosomal DNA at SHL±2 (Figure 3F), with the only exception so far being the INO80 complex, which associates at SHL±6[99,100]. Interestingly, structures of a truncated SNF2 ATPase subunit from the SWI/SNF complex suggest that this protein can associate with the nucleosome at both SHL±2 and SHL±6 suggesting that additional interactions outside of the ATPase domain may determine where the ATPase binds to the nucleosome[101]. From either of these locations, DNA translocation along the phosphodiester backbone appears to power the breaking of histone-DNA contacts required for nucleosome remodeling. This is supported by the fact that single-stranded DNA gaps at these locations, which prevent translocation along DNA also prevent remodeling[102–105]. Binding of the SWI2/SNF2 ATPase to the phosphodiester backbone of nucleosomal DNA at SHL±2 appears to amplify local deformations in the conformation of the DNA double helix caused by nucleosome formation (Figure 3F)[106]. Such effects may exert sufficient torsional stress to locally destabilize histone-DNA contacts. Indeed, on naked DNA several remodelers have been shown to generate negative superhelical torsion[107]. However, it is important to note that remodeling of nucleosomes assembled on non-specifically nicked DNA, which globally relaxes superhelical torsion, is not necessarily inhibited [108,109]. This indicates that large-scale generation of superhelical torsion throughout the nucleosome is not necessarily a core requirement for remodeling. Beyond the scale of monucleosomes, it has been shown that human and yeast SWI/SNF complexes can promote supercoiling changes in closed circular nucleosomal arrays [110–112]. Such changes can in principle be coupled with the torsional changes induced by RNA polymerases.
Despite the core similarities between remodelers, how translocation by the ATPase domains of remodelers is harnessed to create specific remodeling outcomes is likely to differ substantially and many of the details remain unclear. Nucleosome sliding has been proposed to occur by the propagation of helical twist defects around the nucleosome in a manner similar to what has been proposed for uncatalyzed nucleosome sliding (Figure 3F)[29,113]. The resetting of histone-DNA contacts around the nucleosome may only require single translocation events. Indeed, fundamental ~1–2bp increments of translocation can be observed during nucleosome sliding catalyzed by multiple remodelers[114,115]. Larger-scale disruptions of nucleosome structure such as nucleosome disassembly or histone exchange, which by necessity involve more extensive breaking of histone-DNA contacts, are likely to require more units of translocation and additional remodeler-nucleosome contacts to stabilize intermediates. For example, histone exchange by the SWR complex relies on contacts between its noncatalytic subunit Swc6 and flanking DNA to enable the loosening of contacts between the exiting H2A-H2B dimer and nucleosomal DNA[116].
Remodelers also rely on dynamics in specific regions of the histone octamer core. The requirement for dynamics has been inferred from the combination of structural studies using techniques such as NMR and cryo-EM and from functional studies where restraining histone dynamics using site-specific disulfide crosslinking inhibits remodeling activity[117–119]. However, the histone dynamics required for remodeling appear to vary considerably between different remodelers. For example, H3-H4 crosslinking inhibits remodeling by the ACF complex but does not affect remodeling by the INO80 complex[117]. The use of octamer dynamics by remodelers may be a major distinguishing feature of their remodeling mechanisms when compared to RNA polymerases. So far, restraining histone octamer dynamics by crosslinking only appears to modestly, if at all, affect polymerase passage through the nucleosome[53,79].
Outside of the core SNF2-related catalytic subunit, remodelers are frequently associated with several accessory proteins, which differ substantially between families (Figure 3A). These proteins often play integral roles in promoting or regulating the core activity of remodelers[120]. In the case of the SWI/SNF family BAF complex, several alternative complexes exist with different compositions each with different roles in promoting tissue specific gene expression[121]. Some subunits may also have additional catalytic activities including histone modifying or ATPase activity. For example, in addition to a remodeling ATPase, the NURD complex contains a histone deacetylase subunit and members of the INO80 family of remodelers contain the hetero-hexameric AAA+ ATPase Rvb1/2[122,123]. While the histone deacetylase subunit of NURD is thought to act in concert with the remodeling ATPase to de-acetylate nucleosomes, the Rvb1/Rvb2 ATPase in the INO80 complexes is thought to be involved in complex assembly[124]. It is also possible that accessory subunits dynamically associate with the complex during a remodeling reaction. Some evidence for this possibility is suggested by ChIP-exo data, which has captured subcomplexes of INO80-family remodelers on chromatin[125].
C. Principles of regulation
The process of remodeling can be divided into three stages in a manner analogous to transcriptional regulation. Like RNA polymerase, to initiate their activity remodelers must first be recruited to their relevant genomic locations at the proper time. After finding its target nucleosome, remodelers must commit to their remodeling activity. In the case of some activities, such as nucleosome sliding, this not only involves the decision to activate remodeling but also a commitment to a particular direction for the activity. Finally, after disrupting nucleosome structure, remodelers must have a means to sense the formation of an appropriate product and terminate further activity.
How remodelers achieve genomic specificity is unclear but is likely to involve the cooperative recognition of several chromatin features in addition to recruitment to specific genomic sequences by sequence specific DNA binding factors. In some cases, this may be achieved directly through subunits of the remodeling complex. Many remodelers contain histone reader domains in either the ATPase or accessory subunits which can recruit the remodeler to nucleosomes with a specific post translational modification state. For example, the SWR complex’s accessory protein Bdf1 contains a bromodomain which recruits it to acetylated nucleosomes[126,127]. However, targeting of remodelers to specific loci may often depend on its interaction with other factors that provide specificity. Remodelers can directly associate with sequence-specific transcription factors or other reader-domain proteins (like HP1) to execute their function [128,129]. Recruitment can not only play an important role in selecting the appropriate nucleosome to disrupt, but also the directionality of activity. For example, transcription factor binding to the ISW2 complex helps set the direction of nucleosome sliding by this complex[130].
After associating with their target nucleosomes, remodelers must dynamically integrate information about their substrate nucleosome and chromatin context to decide whether to proceed with remodeling. To accomplish this, a common theme across remodelers is regulation through auto-inhibition. In the absence of their substrate, the ATPase subunit is held in an inactive conformation often through interactions with specific autoinhibitory domains (Figure 3G)[131–133]. Binding to the appropriate nucleosome substrate can disrupt this inactive conformation. However, in some cases, even after binding the nucleosome, autoinhibitory domains may still prevent either ATP hydrolysis and/or the coupling of hydrolysis to remodeling[134,135]. Overcoming such auto-inhibition may require a post-binding conformational change that enables contacts between the complex and specific substrate cues (Figure 3G). Common stimulatory substrate cues include accessible extranucleosomal DNA, the N-terminal tail of histone H4, and the H2A-H2B acidic patch. Sensing of these cues can be accomplished by direct interaction of the ATPase subunit with each cue, as appears to be the case for ISWI-family remodelers[136]. Alternatively, accessory subunits may also bind to these motifs and allosterically activate the ATPase domain. For example, the non-catalytic SMARCB1 subunit of BAF complexes contacts the acidic patch in order to activate remodeling[137]. Modification of substrate cues provides a simple but potent means to control remodeler activity both positively and negatively. For example, post translational modification of the acidic patch reduces remodeling by ISWI-family remodelers[138].
Another interesting means of regulation is the association of multiple remodeling complexes to the same nucleosome. Because the nucleosome is two-fold pseudosymmetric, two complexes can in principle engage the same superhelical location on opposite faces of the nucleosome. For example, processive nucleosome sliding by the ACF complex is promoted by the cooperative association of two active complexes to a single nucleosome[139]. It has been proposed that a coordinated conformational switch prevents a futile tug of war between the two motors on the nucleosome[140]. Coordinating this switch may be achieved in part through asymmetric allosteric conformational changes in the octamer core[141]. This behavior may not be unique to ACF as the INO80 complex has also been reported to cooperatively associate with and remodel single nucleosomes[142]. It remains unclear what would result if two different remodeling complexes were to attempt to remodel the same nucleosome. Interestingly, the locations of several different remodeling families frequently overlap in ChIP-seq datasets, though further study will be needed to assess actual co-occupancy[143].
The changes that specify termination of remodeling activity are very poorly understood. Loss of substrate cues directly associated with the remodeling reaction catalyzed might provide a means to directly communicate termination of remodeling. For example, H2Az-only containing nucleosomes do not stimulate ATP hydrolysis by SWR, likely because they lack a substrate cue unique to the L2 loop of canonical H2A which is recognized by the Swc5 subunit[144]. Alternatively, substrate cues may emerge after remodeling which inhibit further remodeling activity. Some ISWI-family remodelers have been demonstrated to bind adjacent nucleosomes in addition to its substrate nucleosome, which may control its activity[145,146]. Competition between remodelers and other proteins for binding substrate cues may also provide an important means for terminating remodeling. For example, the H2A/H2B acidic patch, which is a common substrate cue for remodelers, is both bound by many non-remodeler proteins and by nucleosomes in trans via the H4 N terminal tail[15].
V. Interface of remodelers and RNA polymerases at different stages of transcription
A. Regulation of promoter chromatin architecture
Chromatin remodelers play an integral role in creation of promoter chromatin architecture by all three polymerases. This involves first the creation of a nucleosome depleted region (NDR) where sequence-specific and general transcription factors can assemble followed by the precise positioning of nucleosomes upstream and downstream of the transcription start site. Creation of an NDR appears to be an obligate feature of transcription initiation, since binding of TBP to DNA, which is required for all three polymerases to initiate, is greatly inhibited in the presence of nucleosomes[147,148]. However, chromatin architecture at the promoter plays additional key roles in coordinating events during initiation. Precise positioning of both the +1 and −1 nucleosome has been implicated in the cooperative assembly of the pre-initiation complex for Pol II and III-dependent genes [149–152]. This positioning of the +1 nucleosome may also be particularly important for transcription start site choice at some Pol II-dependent genes[152].
Our best understanding of how nucleosome-positioning is achieved at promoters comes from several elegant studies of S. cerevisiae Pol II promoter architecture (Figure 4A). Using a library of genomic DNA, purified histones, remodelers, and/or transcription factors, nucleosome positioning at Pol II promoters has been faithfully reconstituted[153]. Careful analysis of positioning experiments with different combinations of remodelers has delineated at least two general pathways through which remodelers cooperate to create promoter architecture (Figure 4A). One pathway relies on the binding of sequence-specific transcription factors to create a barrier against which the +1 and −1 ucleosomes are positioned by the ISW2 complex. The second pathway relies on solely the ability of the INO80 complex to sense local DNA topology associated with promoter-proximal sequences to position the +1 and −1 nucleosomes[153,154]. Changes in DNA topology associated with RNA polymerase loading may cooperate with remodelers in further refining promoter chromatin architecture. Interestingly, neither histone modifications nor histone tails appear to substantially influence +1 positioning by INO80 as native, recombinant, and tailless histones are roughly equally sufficient for +1 positioning[154]. Nucleosomes downstream of the +1 nucleosome are then arranged in an evenly spaced nucleosome array, which is accomplished by remodelers with spacing activity such as ISW1b [155].
FIGURE 4. Chromatin remodeling near the transcription start site.
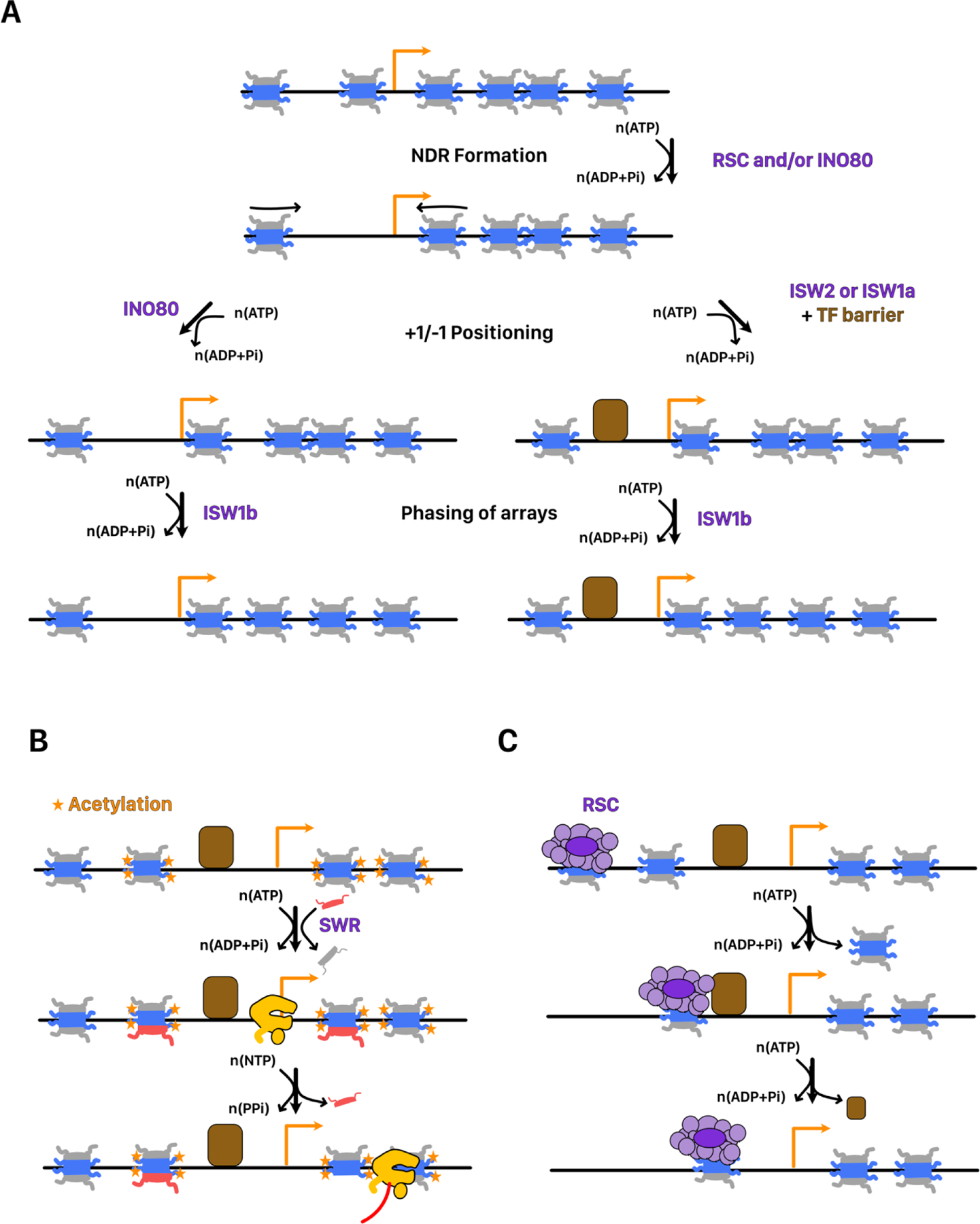
A. Two pathways for promoter chromatin architecture in S. cerevisiae. The NDR is first formed by the action of the RSC and/or INO80 complex recognizing sequence features near the promoter. The +1/−1 nucleosome is then positioned either by the INO80 complex using local DNA topology to position the nucleosome or by the ISW2 or ISW1a complexes positioning nucleosomes relative to a bound transcription factor. After establishment of the +1 nucleosome, ISW1b establishes a phased nucleosome array relative to the +1 nucleosome. B. Establishment of H2Az nucleosomes at the +1/−1 nucleosomes. SWR recognizes histone acetylation and possible promoter DNA sequences to exchange H2A with H2Az dimers. H2Az has a marked preference for incorporation on the promoter distal side of the nucleosome. Transcription by Pol II may disassemble H2Az containing nucleosomes during passage. C. RSC action at “fragile” nucleosome containing NDRs. RSC-bound “fragile” nucleosomes may be translocated to maintain a clear NDR or displace bound transcription factors.
The +1 nucleosome is also associated with an enrichment in the histone variant H2Az, particularly for poised or inducible genes[36,156]. This is thought to reduce the initial barrier to Pol II immediately after promoter escape. In cerevisiae, the SWR complex is recruited to the +1 nucleosome in a manner that depends heavily on its Swc2 subunit binding promoter-proximal DNA and to a lesser extent on Bdf1 binding histone acetylation (Figure 4B)[125,127,157]. It is currently unclear whether these features are sufficient to directly recruit SWR to its substrate nucleosomes, or whether SWR depends on other factors for its recruitment. The opposite exchange reaction has been proposed to be catalyzed by INO80, but this activity remains debated[105,158–160]. Recent live-cell imaging experiments suggest instead that Pol II transcription directly displaces H2Az[160]. Interestingly, H2Az is enriched on the promoter distal side of the +1 nucleosome, which may facilitate its displacement by polymerase[161].
After initiation, remodelers may also play a direct role in recycling transcription factors bound at the promoters. SWI/SNF-family remodelers have been shown to displace nucleosomes and transcription factors from DNA by sliding nucleosomes through them[162,163]. Based on the specific methods used, it has been proposed that in S. cerevisiae wide NDRs may be associated with with non-histone proteins or with a transient RSC-bound MNase-sensitive “fragile” nucleosome that can be used to displace nucleosomes and recycle transcription factors (Figure 4C) [164] [165]. Consistent with a role in transcription factor displacement, rapid depletion of a subunit of the RSC complex increases the dwell time of the Ace1p transcription factor on chromatin[166].
Remodelers also play a key role in preventing transcription initiation by occluding NDRs. When recruited by specific transcription factors, the remodeler ISW2 in cerevisiae occludes NDRs by repositioning promoter nucleosomes [167,168]. In mammals, Pol I transcription is repressed by positioning of promoter nucleosomes over the NDR via the NoRC complex[169]. Additionally, remodelers play a role in preventing cryptic transcription. Deletion of several remodelers involved in even nucleosome spacing downstream of the TSS leads to the widespread upregulation of cryptic antisense transcripts[170–172]. Widespread nucleosome spacing over ORFs may be a general mechanism to repress cryptic promoters, which if allowed to proceed into active gene bodies can lead to transcription conflicts and inhibition of elongation.
B. Elongation
Because of the intrinsic nucleosome remodeling activity of the transcriptional machinery, it is possible that elongating RNA polymerase alone may be sufficient to transcribe through the gene body. Elongating polymerase is likely to be most self-sufficient at highly transcribed loci, where continued Pol II passage and accumulated supercoiling destabilizes nucleosome structure enough for elongation to proceed unimpeded [19,50,62,88,90]. However, at lowly expressed genes, or during a pioneering round of transcription, Pol II passage is likely to require assistance in moving through nucleosomes. Indeed, even in the presence of several elongation factors, reconstituted transcription reactions on nucleosomal templates under physiological conditions proceed at only a fraction of the 2kb/min average elongation rate observed in vivo [173–175]. As a result, remodelers likely play a key role in both promoting the progression of polymerases through the gene body and maintaining chromatin structure after Pol II passage (Figure 5). Remodelers can, in principle, facilitate elongation by disassembling nucleosomes ahead of the polymerase completely or to hexasomes (Figure 5A). In vitro, inclusion of RSC and the H2A/H2B chaperone NAP1 in transcription reactions promotes the formation of a hexasome which improves transcription efficiency by Pol II[176]. Incorporation of histone variants that present a lower barrier to polymerase by remodelers, like H2Az or the mammalian-specific variant H2A.B, has also been suggested to improve elongation (Figure 4B)[177]. Remodelers can also reposition nucleosomes off of sequences that present a high energetic barrier to nucleosomal passage (Figure 5B). Repositioning of nucleosomes off of a high barrier sequence by ISW2 in vitro has been demonstrated to improve elongation efficiency[178]. Finally, remodelers may create stable conformational rearrangements in the nucleosome that facilitate Pol II passage (Figure 5C). The remosome produced by RSC could represent one such rearrangement[94,95]. Interestingly, RSC has recently been shown to not only be associated with promoter nucleosomes, but also with nucleosomes within the bodies of highly transcribed genes[179].
FIGURE 5. Chromatin remodeling during elongation.
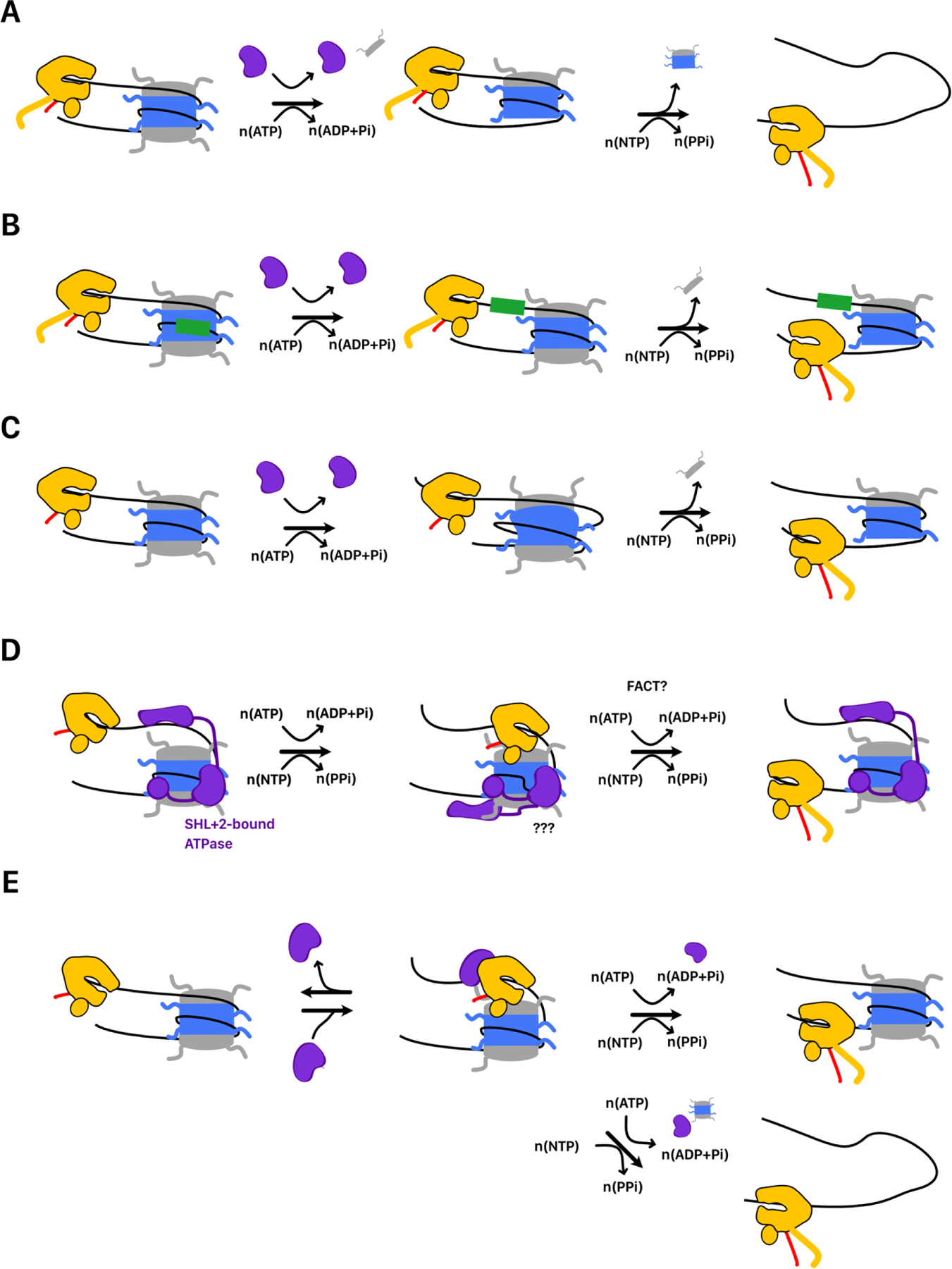
Remodelers can promote Pol II passage by A. disassembling nuclesomes, B. translocating nucleosomes off of high-barrier sequences, or C. creating stable conformational rearrangements in nucleosome structure (e.g. remosomes). D. Speculative model for promotion of elongation. Binding of extranucleosomal DNA by CHD1’s DNA binding domain stabilizes an unpeeled state that facilitates Pol II entry into the nucleosome. DNA translocation by the ATPase domain at SHL+2 may loosen histone DNA contacts needed to allow Pol II passage through the dyad. Contacts with FACT or other elongation factors (e.g. Paf1) may facilitate Pol II passage without CHD1 displacement. E. ATP-dependent elongation activity of CSB (Rad26). CSB binds Nucleosome-associated Pol II. Translocation of the DNA upstream of Pol II by CSB prevents backtracking and promotes passage through the nucleosome. This results in either nucleosome disassembly or full nucleosome survival.
Alternatively, remodelers may facilitate transcription elongation by directly acting on the elongating Pol II-nucleosome complex (Figure 5D). CHD1 is a single-subunit chromatin remodeler that is required for efficient elongation[75,180–182]. CHD1 possess a DNA-Binding Domain (DBD) which engages extranucleosomal DNA and sets the directionality of nucleosome sliding as well as an ATPase domain that engages at SHL+2[183]. While nucleosome sliding could account for its role in elongation, we propose that CHD1 may also more directly assist in the disruption of nucleosome structure as Pol II transits the nucleosome. Biochemical data has suggested that CHD1 engages extranucleosomal DNA beyond SHL+7 to direct the sliding reaction[183]. However, Cryo-EM and biochemical data have revealed that CHD1 can also engage extranucleosomal DNA near SHL −7 with its ATPase domain still bound at SHL+2[184–186]. In this conformation, the DBD unpeels DNA at SHL-7 from the octamer surface[185–187]. This stable unpeeling may facilitate the entry of Pol II into the nucleosome. Pol II passage through the dyad requires disruption of DNA near SHL+2 which could be enabled by CHD1’s ATPase domain. As Pol II progresses through the nucleosome, it may displace the DBD from SHL-7, allowing interactions at SHL+7. Consistent with this, a structure with CHD1 bound to a Pol-II transcribed nucleosome arrested at SHL-4.5 suggest the DBD undocks from SHL-7 as Pol II enters the nucleosome [75]. Unlike many remodelers, CHD1 does not depend on the H2A/H2B acidic patch to activate remodeling, which could allow it to cooperate with Paf1 or FACT in facilitating Pol II passage[188]. Interestingly CHD1 can form a complex with both FACT and Paf1 and deletion of CHD1 suppresses FACT phenotypes[180,181,189]. How long CHD1 can remain associated with the nucleosome during Pol II passage is not fully understood. Recent cryo-EM structures of Pol II-nucleosome complexes suggest that binding of CHD1 and FACT to a transcribed nucleosome is mutually exclusive[75]. However, additional conformational rearrangements in the remodeler not detected in the captured states and/or contacts with elongation factors could allow CHD1 to remain associated with the nucleosome during Pol II passage.
Finally, remodelers may also facilitate elongation by treating elongating polymerase as a substrate. The protein CSB (Cockayne Syndrome B or Rad26 in S. cerevisiae) possesses nucleosome sliding and disassembly activity but can also directly associate with RNA polymerase[190,191]. Recently it was shown that CSB stimulates transcription in an ATP-dependent manner that is separate from nucleosome sliding[192]. CSB accomplishes this by associating with both DNA at the transcription bubble and the DNA immediately upstream of the polymerase and preventing backtracking. CSB activity promotes progression of Pol II through the nucleosome with either nucleosome survival or complete disassembly (Figure 5E). It is unclear whether this is a mechanism unique to CSB or whether other remodelers could also treat RNA polymerase as a substrate.
C. Transcriptional Termination
Remodelers may also play important roles in transcription termination by Pols I and II[193–195]. One general theme is the creation of accessible sites for termination factors to assemble at the 3’ end of ORFs. Near Pol II termination sites at coding genes is a nucleosome depleted region surrounded by an evenly spaced array of nucleosomes [150,196] In the S. cerevisiae genome, 3’ NDRs are found only weakly and are sensitive to the degree of MNase digestion. Additionally, S. cerevisiae 3’ NDRs correlate with closely spaced tandem genes and are not generally found at convergent genes[165]. This suggests that in S. cerevisiae the 3’NDRs may result simply from an extended 5’ NDRs of neighboring genes. However, in D. melanogaster S2 cells and human T cells, which both have substantially less densely packed genomes than S cerevisiae, the 3’ NDR is quite pronounced [197,198]. In humans the 3’ NDR also appears to correlate with polyadenylation site usage, which may allow the cleavage and polyadenylation factor (CPF) to access and process the nascent RNA during termination [197]. Formation of the 3’ NDR may be caused in part by DNA supercoiling due to elongating polymerase[199]. The 3’ NDR also possesses higher A/T content, which intrinsically disfavors nucleosomes[165,197]. However, in S. cerevisiae the NDR relies on the action of RSC, ISW1, and CHD1 and depletion of these remodelers can also cause termination defects[195]. How remodelers are recruited to these sites is unclear but direct interactions between remodelers and CPF, as has been reported for SWI/SNF complexes, could be responsible[200].
Following termination sites, slowing elongating polymerase plays a key role in the process of termination as some mechanisms of Pol I and Pol II disassembly rely on kinetic competition between factors traveling along the nascent RNA transcript and further elongation[87]. For Pol I and Pol II coding transcription, termination can be achieved by the 5’−3’ exonuclease Rat1 which degrades the elongating transcript and, upon reaching the polymerase, displaces it from DNA. In contrast, Pol II noncoding transcripts are released by the RNA helicase Sen1, which translocates along the nascent transcript until it reaches and dissociates the elongating polymerase. Nucleosomes positioned by remodelers near the 3’ NDR may function as a roadblock to slow elongating polymerase to facilitate termination. Mutating histone-DNA contacts at the entry/exit sites of nucleosomes in S. cerevisiae results in widespread transcriptional termination defects and changes in pol II occupancy consistent with an increased elongation rate[201].
In addition to these regulated termination pathways, there also exists pathways for clearing polymerase when it is terminally stalled by various roadblocks within the gene body. This is particularly relevant when the polymerase encounters DNA lesions that cannot be traversed. In this case, the stalled polymerase is polyubiquitinated, displaced, and degraded by the proteasome[202]. INO80 has been shown to associate with the AAA+ ATPase Cdc48 and assist in the displacement of stalled Pol II in a manner that depends on INO80’s ATPase activity and ubiquitination of Pol II[203]. However, it is unclear whether INO80 assists in displacement through its nucleosome remodeling activity or through an undiscovered mechanism.
VI. Outlook
Our understanding of the mechanisms of chromatin remodeling by both RNA polymerases and specialized ATP-dependent remodelers has advanced considerably in recent years. While new insights will continue to emerge from studying both of these motors’ actions on chromatin in isolation, a major gap is how much the mechanism of remodeling by these two classes of motors is altered when both are associated with the same nucleosome. How and under what conditions do remodelers and polymerase directly synergize to disrupt chromatin structure? When their activities conflict, whose activity prevails and what determines who results as the “winner?” The continuing application of single molecule methodologies could be very useful as a means to assess what happens when these motors meet on a single nucleosome. Understanding the role of R-loops in regulating chromatin structure and remodelers is also a rich area of investigation. Interestingly, it was recently suggested that INO80 associates with and promotes the resolution of R-loops in cancer cells[204].
Chromatin-based regulation of transcription also occurs on larger scales beyond that of the single nucleosome scale. Although chromatin was initially proposed to form a regular higher-order structure, multiple lines of evidence have converged on a view that chromatin structure is far more heterogeneous than initially anticipated[205]. In particular, the finding that nucleosome arrays can form liquid-like condensates underscores the complex and dynamic nature of higher-levels of chromatin organization[206]. While we are beginning to appreciate the myriad ways such condensates can influence genome compartmentalization, the environments within condensates may also dramatically influence both the properties and mechanisms of nuclear motors [207]. Further clarifying the range of packaging states adopted by chromatin and clarifying which of their physico-chemical properties are compatible with transcription and chromatin remodeling are essential areas of investigation. Interestingly, the RNA polymerase CTD has been demonstrated to form liquid-like condensates, which may regulate Pol II’s localization or activity [208]. Integrating insights from these disparate approaches holds immense promise toward our comprehensive understanding of the relationship between genome structure and transcription.
Research Highlights.
Eukaryotic RNA polymerases are chromatin remodelers that can disassemble or transfer nucleosomes
SWI2/SNF2 family ATP-dependent chromatin remodelers have diverse activities and mechanisms
Remodelers and RNA polymerase collaborate at all stages of transcription to promote gene expression and maintain chromatin architecture
Further study of the mechanisms of how these two motors collaborate is required
Acknowledgements
We thank members of the Narlikar lab for helpful discussions while preparing this manuscript. This work was supported by a grant from the NIH to GJN (R35GM127020) and an NSF predoctoral and UCSF discovery fellowship to NG.
Footnotes
Declarations of interest: none
References
- [1].Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution., Nature. 389 (1997) 251–260. 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- [2].McGinty RK, Tan S, Nucleosome Structure and Function, Chem Rev. 115 (2014) 2255–2273. 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Izban MG, Luse DS, Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing., Gene Dev 5 (1991) 683–696. 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- [4].Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM, Nucleosome Remodeling Induced by RNA Polymerase II Loss of the H2A/H2B Dimer during Transcription, Mol Cell. 9 (2002) 541–552. 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- [5].Walter W, Kireeva ML, Studitsky VM, Kashlev M, Bacterial Polymerase and Yeast Polymerase II Use Similar Mechanisms for Transcription through Nucleosomes, J Biol Chem. 278 (2003) 36148–36156. 10.1074/jbc.m305647200. [DOI] [PubMed] [Google Scholar]
- [6].Clapier CR, Cairns BR, The Biology of Chromatin Remodeling Complexes, Annu Rev Biochem. 78 (2009) 273–304. 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- [7].Zhou CY, Johnson SL, Gamarra NI, Narlikar GJ, Mechanisms of ATP-Dependent Chromatin Remodeling Motors, Annual Review of Biophysics. 45 (2016) 153–181. 10.1146/annurev-biophys-051013-022819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Studitsky VM, Clark DJ, Felsenfeld G, Overcoming a nucleosomal barrier to transcription, Cell. 83 (1995) 19–27. 10.1016/0092-8674(95)90230-9. [DOI] [PubMed] [Google Scholar]
- [9].Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G, Mechanism of Transcription Through the Nucleosome by Eukaryotic RNA Polymerase, Science. 278 (1997) 1960–1963. 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- [10].Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM, Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II, Nat Struct Mol Biol. 16 (2009) 1272–1278. 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chereji RV, Eriksson PR, Ocampo J, Prajapati HK, Clark DJ, Accessibility of promoter DNA is not the primary determinant of chromatin-mediated gene regulation, Genome Res. 29 (2019) 1985–1995. 10.1101/gr.249326.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ, HP1 reshapes nucleosome core to promote phase separation of heterochromatin, Nature. 575 (2019) 390–394. 10.1038/s41586-019-1669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhou K, Gaullier G, Luger K, Nucleosome structure and dynamics are coming of age, Nat Struct Mol Biol. 26 (2018) 3–13. 10.1038/s41594-018-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chung SY, Hill WE, Doty P, Characterization of the histone core complex, Proc National Acad Sci. 75 (1978) 1680–1684. 10.1073/pnas.75.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].McGinty RK, Tan S, Recognition of the nucleosome by chromatin factors and enzymes., Current Opinion in Structural Biology. 37 (2016) 54–61. 10.1016/j.sbi.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Richmond TJ, Davey CA, The structure of DNA in the nucleosome core., Nature. 423 (2003) 145–150. 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- [17].Segura J, Joshi RS, Díaz-Ingelmo O, Valdés A, Dyson S, Martínez-García B, Roca J, Intracellular nucleosomes constrain a DNA linking number difference of −1.26 that reconciles the Lk paradox, Nat Commun. 9 (2018) 3989. 10.1038/s41467-018-06547-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Clark DJ, Felsenfeld G, Formation of nucleosomes on positively supercoiled DNA., Embo J. 10 (1991) 387–395. 10.1002/j.1460-2075.1991.tb07960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sheinin MY, Li M, Soltani M, Luger K, Wang MD, Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss, Nat Commun. 4 (2013) 2579. 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Taguchi H, Horikoshi N, Arimura Y, Kurumizaka H, A method for evaluating nucleosome stability with a protein-binding fluorescent dye, Methods. 70 (2014) 119–126. 10.1016/j.ymeth.2014.08.019. [DOI] [PubMed] [Google Scholar]
- [21].Polach KJ, Widom J, Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation., Journal of Molecular Biology. 254 (1995) 130–149. 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- [22].Li G, Levitus M, Bustamante C, Widom J, Rapid spontaneous accessibility of nucleosomal DNA., Nature Structural & Molecular Biology. 12 (2005) 46–53. 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- [23].Böhm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Tóth K, Luger K, Langowski J, Nucleosome accessibility governed by the dimer/tetramer interface., Nucleic Acids Research. 39 (2011) 3093–3102. 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bilokapic S, Strauss M, Halic M, Histone octamer rearranges to adapt to DNA unwrapping., Nature Structural & Molecular Biology. 25 (2018) 101–108. 10.1038/s41594-017-0005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Arimura Y, Tachiwana H, Oda T, Sato M, Kurumizaka H, Structural analysis of the hexasome, lacking one histone H2A/H2B dimer from the conventional nucleosome., Biochemistry. 51 (2012) 3302–3309. 10.1021/bi300129b. [DOI] [PubMed] [Google Scholar]
- [26].Polach KJ, Lowary PT, Widom J, Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes., Journal of Molecular Biology. 298 (2000) 211–223. 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- [27].Li Z, Kono H, Distinct Roles of Histone H3 and H2A Tails in Nucleosome Stability, Sci Rep-Uk. 6 (2016) 31437. 10.1038/srep31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Flaus A, Owen-Hughes T, Dynamic properties of nucleosomes during thermal and ATP-driven mobilization., Molecular and Cellular Biology. 23 (2003) 7767–7779. 10.1128/mcb.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brandani GB, Niina T, Tan C, Takada S, DNA sliding in nucleosomes via twist defect propagation revealed by molecular simulations, Nucleic Acids Res. 46 (2018) gky158–. 10.1093/nar/gky158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bilokapic S, Strauss M, Halic M, Structural rearrangements of the histone octamer translocate DNA., Nature Communications. 9 (2018) 1330. 10.1038/s41467-018-03677-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lowary PT, Widom J, New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning., Journal of Molecular Biology. 276 (1998) 19–42. 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- [32].Wu B, Mohideen K, Vasudevan D, Davey CA, Structural Insight into the Sequence Dependence of Nucleosome Positioning, Structure. 18 (2010) 528–536. 10.1016/j.str.2010.01.015. [DOI] [PubMed] [Google Scholar]
- [33].Segal E, Widom J, Poly(dA:dT) tracts: major determinants of nucleosome organization, Current Opinion in Structural Biology. 19 (2009) 65–71. 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bowman GD, Poirier MG, Post-Translational Modifications of Histones That Influence Nucleosome Dynamics, Chem Rev. 115 (2015) 2274–2295. 10.1021/cr500350x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bintu L, Ishibashi T, Dangkulwanich M, Wu Y-Y, Lubkowska L, Kashlev M, Bustamante C, Nucleosomal Elements that Control the Topography of the Barrier to Transcription, Cell. 151 (2012) 738–749. 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD, Histone Variant H2A.Z Marks the 5′ Ends of Both Active and Inactive Genes in Euchromatin, Cell. 123 (2005) 233–248. 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rudnizky S, Bavly A, Malik O, Pnueli L, Melamed P, Kaplan A, H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes, Nat Commun. 7 (2016) 12958. 10.1038/ncomms12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cramer P, Armache K-J, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn C-D, Lehmann E, Leike K, Sydow JF, Vannini A, Structure of Eukaryotic RNA Polymerases, Annu Rev Biophys. 37 (2008) 337–352. 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- [39].Roeder RG, 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms, Nat Struct Mol Biol. 26 (2019) 783–791. 10.1038/s41594-019-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mosley AL, Hunter GO, Sardiu ME, Smolle M, Workman JL, Florens L, Washburn MP, Quantitative Proteomics Demonstrates That the RNA Polymerase II Subunits Rpb4 and Rpb7 Dissociate during Transcriptional Elongation, Mol Cell Proteomics. 12 (2013) 1530–1538. 10.1074/mcp.m112.024034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Duek L, Barkai O, Elran R, Adawi I, Choder M, Dissociation of Rpb4 from RNA polymerase II is important for yeast functionality, Plos One. 13 (2018) e0206161. 10.1371/journal.pone.0206161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zaborowska J, Egloff S, Murphy S, The pol II CTD: new twists in the tail, Nat Struct Mol Biol. 23 (2016) 771–777. 10.1038/nsmb.3285. [DOI] [PubMed] [Google Scholar]
- [43].Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P, Evolution of Two Modes of Intrinsic RNA Polymerase Transcript Cleavage, J Biol Chem. 286 (2011) 18701–18707. 10.1074/jbc.m111.222273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vorländer MK, Khatter H, Wetzel R, Hagen WJH, Müller CW, Molecular mechanism of promoter opening by RNA polymerase III, Nature. 553 (2018) 295–300. 10.1038/nature25440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lisica A, Engel C, Jahnel M, Roldán É, Galburt EA, Cramer P, Grill SW, Mechanisms of backtrack recovery by RNA polymerases I and II, Proc National Acad Sci. 113 (2016) 2946–2951. 10.1073/pnas.1517011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nudler E, RNA Polymerase Backtracking in Gene Regulation and Genome Instability, Cell. 149 (2012) 1438–1445. 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dangkulwanich M, Ishibashi T, Liu S, Kireeva ML, Lubkowska L, Kashlev M, Bustamante CJ, Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism, Elife. 2 (2013) e00971. 10.7554/elife.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M, Nature of the Nucleosomal Barrier to RNA Polymerase II, Mol Cell. 18 (2005) 97–108. 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- [49].Corless S, Gilbert N, Effects of DNA supercoiling on chromatin architecture, Biophysical Rev. 8 (2016) 245–258. 10.1007/s12551-016-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Teves SS, Henikoff S, Transcription-generated torsional stress destabilizes nucleosomes, Nat Struct Mol Biol. 21 (2014) 88–94. 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N, Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures, Nat Struct Mol Biol. 20 (2013) 387–395. 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Studitsky VM, Walter W, Kireeva M, Kashlev M, Felsenfeld G, Chromatin remodeling by RNA polymerases, Trends Biochem Sci. 29 (2004) 127–135. 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- [53].Birch JL, Tan BC, Panov KI, Panova TB, Andersen JS, Owen- Hughes TA, Russell J, Lee S, Zomerdijk JCBM, FACT facilitates chromatin transcription by RNA polymerases I and III, Embo J. 28 (2009) 854–865. 10.1038/emboj.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kulaeva OI, Studitsky VM, Mechanism of histone survival during transcription by RNA polymerase II, Transcription. 1 (2010) 85–88. 10.4161/trns.1.2.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Thiriet C, Hayes JJ, Replication-independent core histone dynamics at transcriptionally active loci in vivo, Gene Dev. 19 (2005) 677–682. 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C, Nucleosomal Fluctuations Govern the Transcription Dynamics of RNA Polymerase II, Science. 325 (2009) 626–628. 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C, The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes, Nat Struct Mol Biol. 18 (2011) 1394–1399. 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kujirai T, Ehara H, Fujino Y, Shirouzu M, Sekine S, Kurumizaka H, Structural basis of the nucleosome transition during RNA polymerase II passage, Science. 362 (2018) 595–598. 10.1126/science.aau9904. [DOI] [PubMed] [Google Scholar]
- [59].Izban MG, Luse DS, Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates., J Biological Chem 267 (1992) 13647–55. file:///Users/ngamarra/Desktop/J.%20Biol.%20Chem.-1992-Izban-13647-55.pdf. [PubMed] [Google Scholar]
- [60].Merkl PE, Pilsl M, Fremter T, Schwank K, Engel C, Längst G, Milkereit P, Griesenbeck J, Tschochner H, RNA polymerase I (Pol I) passage through nucleosomes depends on Pol I subunits binding its lobe structure, J Biol Chem. 295 (2020) 4782–4795. 10.1074/jbc.ra119.011827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bondarenko VA, Steele LM, Újvári A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM, Nucleosomes Can Form a Polar Barrier to Transcript Elongation by RNA Polymerase II, Mol Cell. 24 (2006) 469–479. 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- [62].Kulaeva OI, Hsieh F-K, Studitsky VM, RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones, Proc National Acad Sci. 107 (2010) 11325–11330. 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ramachandran S, Ahmad K, Henikoff S, Transcription and Remodeling Produce Asymmetrically Unwrapped Nucleosomal Intermediates, Mol Cell. 68 (2017) 1038–1053.e4. 10.1016/j.molcel.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hsieh F, Fisher M, Újvári A, Studitsky VM, Luse DS, Histone Sin mutations promote nucleosome traversal and histone displacement by RNA polymerase II, Embo Rep. 11 (2010) 705–710. 10.1038/embor.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Crickard JB, Lee J, Lee T-H, Reese JC, The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome, Nucleic Acids Res. 45 (2017) gkx220–. 10.1093/nar/gkx220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Farnung L, Vos SM, Cramer P, Structure of transcribing RNA polymerase II-nucleosome complex, Nat Commun. 9 (2018) 5432. 10.1038/s41467-018-07870-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine S, Structural insight into nucleosome transcription by RNA polymerase II with elongation factors, Science. 363 (2019) eaav8912. 10.1126/science.aav8912. [DOI] [PubMed] [Google Scholar]
- [68].Kujirai T, Kurumizaka H, Transcription through the nucleosome, Curr Opin Struc Biol. 61 (2020) 42–49. 10.1016/j.sbi.2019.10.007. [DOI] [PubMed] [Google Scholar]
- [69].Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR, Active RNA Polymerases: Mobile or Immobile Molecular Machines?, Plos Biol. 8 (2010) e1000419. 10.1371/journal.pbio.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lee K, Hsiung CC-S, Huang P, Raj A, Blobel GA, Dynamic enhancer–gene body contacts during transcription elongation, Gene Dev. 29 (2015) 1992–1997. 10.1101/gad.255265.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cook PR, Marenduzzo D, Transcription-driven genome organization: a model for chromosome structure and the regulation of gene expression tested through simulations, Nucleic Acids Res. 46 (2018) gky763. 10.1093/nar/gky763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Újvári A, Hsieh F-K, Luse SW, Studitsky VM, Luse DS, Histone N-terminal Tails Interfere with Nucleosome Traversal by RNA Polymerase II, J Biol Chem. 283 (2008) 32236–32243. 10.1074/jbc.m806636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Martinez-Zamudio R, Ha HC, Histone ADP-Ribosylation Facilitates Gene Transcription by Directly Remodeling Nucleosomes, Mol Cell Biol. 32 (2012) 2490–2502. 10.1128/mcb.06667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D, Histone H2B Monoubiquitination Functions Cooperatively with FACT to Regulate Elongation by RNA Polymerase II, Cell. 125 (2006) 703–717. 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- [75].Farnung L, Ochmann M, Engehom M, Cramer P, Structural basis of nucleosome transcription mediated by Chd1 and FACT, BioRxiv. (2020). 10.1101/2020.11.30.403857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Bortvin A, Winston F, Evidence That Spt6p Controls Chromatin Structure by a Direct Interaction with Histones, Science. 272 (1996) 1473–1476. 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- [77].Cucinotta CE, Hildreth AE, McShane BM, Shirra MK, Arndt KM, The nucleosome acidic patch directly interacts with subunits of the Paf1 and FACT complexes and controls chromatin architecture in vivo, Nucleic Acids Research. 47 (2019) 8410–8423. 10.1093/nar/gkz549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D, FACT facilitates transcription-dependent nucleosome alteration., Science (New York, N.Y.). 301 (2003) 1090–1093. 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- [79].Hsieh F-K, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM, Histone chaperone FACT action during transcription through chromatin by RNA polymerase II, Proc National Acad Sci. 110 (2013) 7654–7659. 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Liu Y, Zhou K, Zhang N, Wei H, Tan YZ, Zhang Z, Carragher B, Potter CS, D’Arcy S, Luger K, FACT caught in the act of manipulating the nucleosome, Nature. 577 (2020) 426–431. 10.1038/s41586-019-1820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Jamai A, Puglisi A, Strubin M, Histone Chaperone Spt16 Promotes Redeposition of the Original H3-H4 Histones Evicted by Elongating RNA Polymerase, Mol Cell. 35 (2009) 377–383. 10.1016/j.molcel.2009.07.001. [DOI] [PubMed] [Google Scholar]
- [82].Lee J-S, Garrett AS, Yen K, Takahashi Y-H, Hu D, Jackson J, Seidel C, Pugh BF, Shilatifard A, Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1, Gene Dev. 26 (2012) 914–919. 10.1101/gad.186841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jeronimo C, Angel A, Poitras C, Collin P, Mellor J, Robert F, FACT is recruited to the +1 nucleosome of transcribed genes and spreads in a Chd1-dependent manner, Biorxiv. (2020) 2020.08.20.259960. 10.1101/2020.08.20.259960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Pestov NA, Gerasimova NS, Kulaeva OI, Studitsky VM, Structure of transcribed chromatin is a sensor of DNA damage, Sci Adv. 1 (2015) e1500021. 10.1126/sciadv.1500021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Liu YV, Clark DJ, Tchernajenko V, Dahmus ME, Studitsky VM, Role of C- terminal domain phosphorylation in RNA polymerase II transcription through the nucleosome, Biopolymers. 68 (2003) 528–538. 10.1002/bip.10302. [DOI] [PubMed] [Google Scholar]
- [86].Phatnani HP, Greenleaf AL, Phosphorylation and functions of the RNA polymerase II CTD, Gene Dev. 20 (2006) 2922–2936. 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- [87].Porrua O, Boudvillain M, Libri D, Transcription Termination: Variations on Common Themes, Trends Genet. 32 (2016) 508–522. 10.1016/j.tig.2016.05.007. [DOI] [PubMed] [Google Scholar]
- [88].Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD, Synergistic action of RNA polymerases in overcoming the nucleosomal barrier, Nat Struct Mol Biol. 17 (2010) 745–752. 10.1038/nsmb.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sheinin MY, Li M, Soltani M, Luger K, Wang MD, Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss, Nat Commun. 4 (2013) 2579. 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Kim S, Beltran B, Irnov I, Jacobs-Wagner C, Long-Distance Cooperative and Antagonistic RNA Polymerase Dynamics via DNA Supercoiling, Cell. 179 (2019) 106–119.e16. 10.1016/j.cell.2019.08.033. [DOI] [PubMed] [Google Scholar]
- [91].Dunn K, Griffith JD, The presence of RNA in a double helix inhibits its interaction with histone protein, Nucleic Acids Res. 8 (1980) 555–566. 10.1093/nar/8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Fazzio TG, Regulation of chromatin structure and cell fate by R-loops, Biochem Soc Symp. 7 (2016) 00–00. 10.1080/21541264.2016.1198298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Luk E, Ranjan A, FitzGerald PC, Mizuguchi G, Huang Y, Wei D, Wu C, Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome., Cell. 143 (2010) 725–736. 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Shukla MS, Syed SH, Montel F, Faivre-Moskalenko C, Bednar J, Travers A, Angelov D, Dimitrov S, Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer, Proc National Acad Sci. 107 (2010) 1936–1941. 10.1073/pnas.0904497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Shukla MS, Syed SH, Boopathi R, Simon EB, Nahata S, Ramos L, Dalkara D, Moskalenko C, Travers A, Angelov D, Dimitrov S, Hamiche A, Bednar J, Generation of Remosomes by the SWI/SNF Chromatin Remodeler Family, Scientific Reports. 9 (2019) 14212. 10.1038/s41598-019-50572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Li M, Hada A, Sen P, Olufemi L, Hall MA, Smith BY, Forth S, McKnight JN, Patel A, Bowman GD, Bartholomew B, Wang MD, Dynamic regulation of transcription factors by nucleosome remodeling., ELife. 4 (2015). 10.7554/elife.06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B, SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes., Molecular Cell. 38 (2010) 590–602. 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Flaus A, Martin DMA, Barton GJ, Owen-Hughes T, Identification of multiple distinct Snf2 subfamilies with conserved structural motifs., Nucleic Acids Research. 34 (2006) 2887–2905. 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhou CY, Johnson SL, Gamarra NI, Narlikar GJ, Mechanisms of ATP-Dependent Chromatin Remodeling Motors, Annual Review of Biophysics. 45 (2016) 153–181. 10.1146/annurev-biophys-051013-022819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, Lee T-H, Bartholomew B, INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers., Nature Communications. 8 (2017) 15616. 10.1038/ncomms15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Liu X, Li M, Xia X, Li X, Chen Z, Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure, Nature. 544 (2017) 440–445. 10.1038/nature22036. [DOI] [PubMed] [Google Scholar]
- [102].Saha A, Wittmeyer J, Cairns BR, Chromatin remodeling by RSC involves ATP-dependent DNA translocation., Genes & Development. 16 (2002) 2120–2134. 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zofall M, Persinger J, Kassabov SR, Bartholomew B, Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome., Nature Structural & Molecular Biology. 13 (2006) 339–346. 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- [104].Ranjan A, Wang F, Mizuguchi G, Wei D, Huang Y, Wu C, H2A histone-fold and DNA elements in nucleosome activate SWR1-mediated H2A.Z replacement in budding yeast., ELife. 4 (2015) e06845. 10.7554/elife.06845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Brahma S, Udugama MI, Kim J, Hada A, Bhardwaj SK, Hailu SG, Lee T-H, Bartholomew B, INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers., Nature Communications. 8 (2017) 15616. 10.1038/ncomms15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Li M, Xia X, Tian Y, Jia Q, Liu X, Lu Y, Li M, Li X, Chen Z, Mechanism of DNA translocation underlying chromatin remodelling by Snf2, Nature. 567 (2019) 409–413. 10.1038/s41586-019-1029-2. [DOI] [PubMed] [Google Scholar]
- [107].Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T, Generation of superhelical torsion by ATP-dependent chromatin remodeling activities., Cell. 103 (2000) 1133–1142. [DOI] [PubMed] [Google Scholar]
- [108].Längst G, Becker PB, ISWI Induces Nucleosome Sliding on Nicked DNA, Molecular Cell. 8 (2001) 1085–1092. 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- [109].Aoyagi S, Hayes JJ, hSWI/SNF-catalyzed nucleosome sliding does not occur solely via a twist-diffusion mechanism., Molecular and Cellular Biology. 22 (2002) 7484–7490. 10.1128/mcb.22.21.7484-7490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Guyon JR, Narlikar GJ, Sullivan EK, Kingston RE, Stability of a human SWI-SNF remodeled nucleosomal array., Molecular and Cellular Biology. 21 (2001) 1132–1144. 10.1128/mcb.21.4.1132-1144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Jaskelioff M, Gavin IM, Peterson CL, Logie C, SWI-SNF-Mediated Nucleosome Remodeling: Role of Histone Octamer Mobility in the Persistence of the Remodeled State, Mol Cell Biol. 20 (2000) 3058–3068. 10.1128/mcb.20.9.3058-3068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR, Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex, Nature. 370 (1994) 477–481. 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- [113].Winger J, Nodelman IM, Levendosky RF, Bowman GD, A twist defect mechanism for ATP-dependent translocation of nucleosomal DNA., ELife. 7 (2018) 391. 10.7554/elife.34100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Deindl S, Hwang WL, Hota SK, Blosser TR, Prasad P, Bartholomew B, Zhuang X, ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps., Cell. 152 (2013) 442–452. 10.1016/j.cell.2012.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Harada BT, Hwang WL, Deindl S, Chatterjee N, Bartholomew B, Zhuang X, Stepwise nucleosome translocation by RSC remodeling complexes., ELife. 5 (2016) 3653. 10.7554/elife.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Willhoft O, Ghoneim M, Lin C-L, Chua EYD, Wilkinson M, Chaban Y, Ayala R, McCormack EA, Ocloo L, Rueda DS, Wigley DB, Structure and dynamics of the yeast SWR1-nucleosome complex, Science (New York, N.Y.). 362 (2018) eaat7716. 10.1126/science.aat7716. [DOI] [PubMed] [Google Scholar]
- [117].Sinha KK, Gross JD, Narlikar GJ, Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler., Science (New York, N.Y.). 355 (2017) eaaa3761. 10.1126/science.aaa3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Armache JP, Gamarra N, Johnson SL, Leonard JD, Wu S, Narlikar GJ, Cheng Y, Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome, ELife. 8 (2019) e46057. 10.7554/eLife.46057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Gamarra N, Narlikar G, Histone dynamics within the nucleosome octamer play a critical role in SNF2h-mediated nucleosome sliding, BioRxiv. (2020) 2020.03.14.992073. 10.1101/2020.03.14.992073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Oppikofer M, Bai T, Gan Y, Haley B, Liu P, Sandoval W, Ciferri C, Cochran AG, Expansion of the ISWI chromatin remodeler family with new active complexes., EMBO Reports. 18 (2017) 1697–1706. 10.15252/embr.201744011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE, Zullow HJ, McKenzie ZM, Kubiak RL, St R. Pierre, A.M. Valencia, S.J. Poynter, S.H. Cassel, J.A. Ranish, C. Kadoch, Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes, Cell. 175 (2018) 1272–1288.e20. 10.1016/j.cell.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Jónsson ZO, Dhar SK, Narlikar GJ, Auty R, Wagle N, Pellman D, Pratt RE, Kingston R, Dutta A, Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes., The Journal of Biological Chemistry. 276 (2001) 16279–16288. 10.1074/jbc.m011523200. [DOI] [PubMed] [Google Scholar]
- [123].Allen HF, Wade PA, Kutateladze TG, The NuRD architecture, Cell Mol Life Sci. 70 (2013) 3513–3524. 10.1007/s00018-012-1256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Zhou CY, Stoddard CI, Johnston JB, Trnka MJ, Echeverria I, Palovcak E, Sali A, Burlingame AL, Cheng Y, Narlikar GJ, Regulation of Rvb1/Rvb2 by a Domain within the INO80 Chromatin Remodeling Complex Implicates the Yeast Rvbs as Protein Assembly Chaperones., Cell Reports. 19 (2017) 2033–2044. 10.1016/j.celrep.2017.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Yen K, Vinayachandran V, Pugh BF, SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes., Cell. 154 (2013) 1246–1256. 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Krogan NJ, Keogh M-C, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF, A Snf2 Family ATPase Complex Required for Recruitment of the Histone H2A Variant Htz1, Mol Cell. 12 (2003) 1565–1576. 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- [127].Durant M, Pugh BF, NuA4-Directed Chromatin Transactions throughout the Saccharomyces cerevisiae Genome▿ †, Mol Cell Biol. 27 (2007) 5327–5335. 10.1128/mcb.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Xiao H, Sandaltzopoulos R, Wang H-M, Hamiche A, Ranallo R, Lee K-M, Fu D, Wu C, Dual Functions of Largest NURF Subunit NURF301 in Nucleosome Sliding and Transcription Factor Interactions, Mol Cell. 8 (2001) 531–543. 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- [129].Eskeland R, Eberharter A, Imhof A, HP1 Binding to Chromatin Methylated at H3K9 Is Enhanced by Auxiliary Factors, Mol Cell Biol. 27 (2006) 453–465. 10.1128/mcb.01576-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Donovan DA, Crandall JG, Truong VN, Vaaler AL, Bailey TB, Dinwiddie D, McKnight LE, McKnight JN, Basis of Specificity for a Conserved and Promiscuous Chromatin Remodeling Protein, Biorxiv. (2020) 2020.05.25.115584. 10.1101/2020.05.25.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Hauk G, McKnight JN, Nodelman IM, Bowman GD, The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor., Molecular Cell. 39 (2010) 711–723. 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Xia X, Liu X, Li T, Fang X, Chen Z, Structure of chromatin remodeler Swi2/Snf2 in the resting state., Nature Structural & Molecular Biology. 23 (2016) 722–729. 10.1038/nsmb.3259. [DOI] [PubMed] [Google Scholar]
- [133].Yan L, Wang L, Tian Y, Xia X, Chen Z, Structure and regulation of the chromatin remodeller ISWI, Nature. 540 (2016) 466–469. 10.1038/nature20590. [DOI] [PubMed] [Google Scholar]
- [134].Clapier CR, Cairns BR, Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes, Nature. 492 (2012) 280–284. 10.1038/nature11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Hwang WL, Deindl S, Harada BT, Zhuang X, Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA., Nature. 512 (2014) 213–217. 10.1038/nature13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Dao HT, Dul BE, Dann GP, Liszczak GP, Muir TW, A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing, Nat Chem Biol. 16 (2019) 134–142. 10.1038/s41589-019-0413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Mashtalir N, Suzuki H, Farrell DP, Sankar A, Luo J, Filipovski M, D’Avino AR, St R. Pierre, A.M. Valencia, T. Onikubo, R.G. Roeder, Y. Han, Y. He, J.A. Ranish, F. DiMaio, T. Walz, C. Kadoch, A Structural Model of the Endogenous Human BAF Complex Informs Disease Mechanisms, Cell. (2020). 10.1016/j.cell.2020.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Dann GP, Liszczak GP, Bagert JD, Müller MM, Nguyen UTT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R, Pollock SB, Diehl KL, Allis CD, Muir TW, ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference, Nature. 548 (2017) 607–611. 10.1038/nature23671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ, The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes., Nature. 462 (2009) 1016–1021. 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Leonard JD, Narlikar GJ, A Nucleotide-Driven Switch Regulates Flanking DNA Length Sensing by a Dimeric Chromatin Remodeler, Molecular Cell. (2015). 10.1016/j.molcel.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Gamarra N, Johnson SL, Trnka MJ, Burlingame AL, Narlikar GJ, The nucleosomal acidic patch relieves auto-inhibition by the ISWI remodeler SNF2h, ELife. 7 (2018) e35322. 10.7554/elife.35322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Willhoft O, McCormack EA, Aramayo RJ, Bythell-Douglas R, Ocloo L, Zhang X, Wigley DB, Crosstalk within a functional INO80 complex dimer regulates nucleosome sliding, Elife. 6 (2017) e25782. 10.7554/elife.25782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Morris SA, Baek S, Sung M-H, John S, Wiench M, Johnson TA, Schiltz RL, Hager GL, Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions., Nature Structural & Molecular Biology. 21 (2014) 73–81. 10.1038/nsmb.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Huang Y, Sun L, Pierrakeas L, Dai L, Pan L, Luk E, Zhou Z, Role of a DEF/Y motif in histone H2A-H2B recognition and nucleosome editing, Proceedings of the National Academy of Sciences. 117 (2020) 3543–3550. 10.1073/pnas.1914313117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ, Structure and mechanism of the chromatin remodelling factor ISW1a., Nature. 472 (2011) 448–453. 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- [146].Bhardwaj SK, Hailu SG, Olufemi L, Brahma S, Kundu S, Hota SK, Persinger J, Bartholomew B, Dinucleosome specificity and allosteric switch of the ISW1a ATP-dependent chromatin remodeler in transcription regulation, Nat Commun. 11 (2020) 5913. 10.1038/s41467-020-19700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Imbalzano AN, Kwon H, Green MR, Kingston RE, Facilitated binding of TATA-binding protein to nucleosomal DNA., Nature. 370 (1994) 481–485. 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- [148].Kramm K, Engel C, Grohmann D, Transcription initiation factor TBP: old friend new questions, Biochem Soc T. 47 (2019) 411–423. 10.1042/bst20180623. [DOI] [PubMed] [Google Scholar]
- [149].Zhao X, Pendergrast PS, Hernandez N, A Positioned Nucleosome on the Human U6 Promoter Allows Recruitment of SNAPc by the Oct-1 POU Domain, Mol Cell. 7 (2001) 539–549. 10.1016/s1097-2765(01)00201-5. [DOI] [PubMed] [Google Scholar]
- [150].Jiang C, Pugh BF, Nucleosome positioning and gene regulation: advances through genomics, Nat Rev Genet. 10 (2009) 161–172. 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Kumar Y, Bhargava P, A unique nucleosome arrangement, maintained actively by chromatin remodelers facilitates transcription of yeast tRNA genes, Bmc Genomics. 14 (2013) 402. 10.1186/1471-2164-14-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Lai WKM, Pugh BF, Genome-wide uniformity of human ‘open’ pre-initiation complexes, Genome Res. 27 (2017) 15–26. 10.1101/gr.210955.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P, Genomic Nucleosome Organization Reconstituted with Pure Proteins, Cell. 167 (2016) 709–721.e12. 10.1016/j.cell.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Oberbeckmann E, Krietenstein N, Niebauer V, Wang Y, Schall K, Moldt M, Straub T, Rohs R, Hopfner K-P, Korber P, Eustermann S, Genome information processing by the INO80 chromatin remodeler positions nucleosomes, Biorxiv. (2020) 2020.11.03.366690. 10.1101/2020.11.03.366690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P, Genomic Nucleosome Organization Reconstituted with Pure Proteins, Cell. 167 (2016) 709–721.e12. 10.1016/j.cell.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L, Variant Histone H2A.Z Is Globally Localized to the Promoters of Inactive Yeast Genes and Regulates Nucleosome Positioning, Plos Biol. 3 (2005) e384. 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Ranjan A, Mizuguchi G, FitzGerald PC, Wei D, Wang F, Huang Y, Luk E, Woodcock CL, Wu C, Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement., Cell. 154 (2013) 1232–1245. 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL, Global Regulation of H2A.Z Localization by the INO80 Chromatin-Remodeling Enzyme Is Essential for Genome Integrity, Cell. 144 (2011) 200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Wang F, Ranjan A, Wei D, Wu C, Comment on “A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme,” Science. 353 (2016) 358–358. 10.1126/science.aad5921. [DOI] [PubMed] [Google Scholar]
- [160].Ranjan A, Nguyen VQ, Liu S, Wisniewski J, Kim JM, Tang X, Mizuguchi G, Elalaoui E, Nickels TJ, Jou V, English BP, Zheng Q, Luk E, Lavis LD, Lionnet T, Wu C, Live-cell single particle imaging reveals the role of RNA polymerase II in histone H2A.Z eviction, Elife. 9 (2020) e55667. 10.7554/elife.55667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Rhee HS, Bataille AR, Zhang L, Pugh BF, Subnucleosomal Structures and Nucleosome Asymmetry across a Genome, Cell. 159 (2014) 1377–1388. 10.1016/j.cell.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B, SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes., Molecular Cell. 38 (2010) 590–602. 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Li M, Hada A, Sen P, Olufemi L, Hall MA, Smith BY, Forth S, McKnight JN, Patel A, Bowman GD, Bartholomew B, Wang MD, Dynamic regulation of transcription factors by nucleosome remodeling., ELife. 4 (2015). 10.7554/elife.06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Brahma S, Henikoff S, RSC-Associated Subnucleosomes Define MNase-Sensitive Promoters in Yeast, Mol Cell. 73 (2019) 238–249.e3. 10.1016/j.molcel.2018.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Chereji RV, Ocampo J, Clark DJ, MNase-Sensitive Complexes in Yeast: Nucleosomes and Non-histone Barriers, Mol Cell. 65 (2017) 565–577.e3. 10.1016/j.molcel.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Mehta GD, Ball DA, Eriksson PR, Chereji RV, Clark DJ, McNally JG, Karpova TS, Single-Molecule Analysis Reveals Linked Cycles of RSC Chromatin Remodeling and Ace1p Transcription Factor Binding in Yeast, Mol Cell. 72 (2018) 875–887.e9. 10.1016/j.molcel.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T, The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p., Cell. 103 (2000) 423–433. 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- [168].Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T, Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression., Molecular and Cellular Biology. 21 (2001) 6450–6460. 10.1128/mcb.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Li J, Längst G, Grummt I, NoRC- dependent nucleosome positioning silences rRNA genes, The EMBO Journal. 25 (2006) 5735–5741. 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Whitehouse I, Rando OJ, Delrow J, Tsukiyama T, Chromatin remodelling at promoters suppresses antisense transcription., Nature. 450 (2007) 1031–1035. 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- [171].Pointner J, Persson J, Prasad P, Norman-Axelsson U, Strålfors A, Khorosjutina O, Krietenstein N, Svensson JP, Ekwall K, Korber P, CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe., The EMBO Journal. 31 (2012) 4388–4403. 10.1038/emboj.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Hennig BP, Bendrin K, Zhou Y, Fischer T, Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription., EMBO Reports. 13 (2012) 997–1003. 10.1038/embor.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Mason PB, Struhl K, Distinction and Relationship between Elongation Rate and Processivity of RNA Polymerase II In Vivo, Mol Cell. 17 (2005) 831–840. 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- [174].Hsieh F-K, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM, Histone chaperone FACT action during transcription through chromatin by RNA polymerase II, Proc National Acad Sci. 110 (2013) 7654–7659. 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Farnung L, Ochmann M, Engehom M, Cramer P, Structural basis of nucleosome transcription mediated by Chd1 and FACT, BioRxiv. (2020). 10.1101/2020.11.30.403857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Kuryan BG, Kim J, Tran NNH, Lombardo SR, Venkatesh S, Workman JL, Carey M, Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro, Proc National Acad Sci. 109 (2012) 1931–1936. 10.1073/pnas.1109994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Soboleva TA, Nekrasov M, Ryan DP, Tremethick DJ, Histone variants at the transcription start-site, Trends Genet. 30 (2014) 199–209. 10.1016/j.tig.2014.03.002. [DOI] [PubMed] [Google Scholar]
- [178].Gaykalova DA, Nagarajavel V, Bondarenko VA, Bartholomew B, Clark DJ, Studitsky VM, A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation, Nucleic Acids Res. 39 (2011) 3520–3528. 10.1093/nar/gkq1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Biernat E, Kinney J, Dunlap K, Rizza C, Govind CK, The RSC complex remodels nucleosomes in transcribed coding sequences and promotes transcription in Saccharomyces cerevisiae, BioRxiv. (2020) 2020.03.11.987974. 10.1101/2020.03.11.987974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF, RNA Polymerase II Elongation Factors of Saccharomyces cerevisiae: a Targeted Proteomics Approach, Mol Cell Biol. 22 (2002) 6979–6992. 10.1128/mcb.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM, Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes, Embo J. 22 (2003) 1846–1856. 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Skene PJ, Hernandez AE, Groudine M, Henikoff S, The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1, Elife. 3 (2014) e02042. 10.7554/elife.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].McKnight JN, Jenkins KR, Nodelman IM, Escobar T, Bowman GD, Extranucleosomal DNA binding directs nucleosome sliding by Chd1., Molecular and Cellular Biology. 31 (2011) 4746–4759. 10.1128/mcb.05735-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Nodelman IM, Bleichert F, Patel A, Ren R, Horvath KC, Berger JM, Bowman GD, Interdomain Communication of the Chd1 Chromatin Remodeler across the DNA Gyres of the Nucleosome, Molecular Cell. 0 (2017) 711–723. 10.1016/j.molcel.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Farnung L, Vos SM, Wigge C, Cramer P, Nucleosome-Chd1 structure and implications for chromatin remodelling., Nature. 550 (2017) 539–542. 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Sundaramoorthy R, Hughes AL, El-Mkami H, Norman DG, Ferreira H, Owen-Hughes T, Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome., ELife. 7 (2018) 977. 10.7554/elife.35720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Tokuda JM, Ren R, Levendosky RF, Tay RJ, Yan M, Pollack L, Bowman GD, The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome, Nucleic Acids Res. 46 (2018) gky206–. 10.1093/nar/gky206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Levendosky RF, Sabantsev A, Deindl S, Bowman GD, The Chd1 chromatin remodeler shifts hexasomes unidirectionally., ELife. 5 (2016) 3302. 10.7554/elife.21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Biswas D, Dutta-Biswas R, Stillman DJ, Chd1 and yFACT Act in Opposition in Regulating Transcription▿ †, Mol Cell Biol. 27 (2007) 6279–6287. 10.1128/mcb.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Wang W, Xu J, Limbo O, Fei J, Kassavetis GA, Chong J, Kadonaga JT, Russell P, Li B, Wang D, Molecular basis of chromatin remodeling by Rhp26, a yeast CSB ortholog, Proc National Acad Sci. 116 (2019) 201818163. 10.1073/pnas.1818163116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Xu J, Lahiri I, Wang W, Wier A, Cianfrocco MA, Chong J, Hare AA, Dervan PB, DiMaio F, Leschziner AE, Wang D, Structural basis for the initiation of eukaryotic transcription-coupled DNA repair, Nature. 551 (2017) 653–657. 10.1038/nature24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Xu J, Wang W, Xu L, Chen J-Y, Chong J, Oh J, Leschziner AE, Fu X-D, Wang D, Cockayne syndrome B protein acts as an ATP-dependent processivity factor that helps RNA polymerase II overcome nucleosome barriers, Proc National Acad Sci. 117 (2020) 25486–25493. 10.1073/pnas.2013379117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Alén C, Kent NA, Jones HS, O’Sullivan J, Aranda A, Proudfoot NJ, A Role for Chromatin Remodeling in Transcriptional Termination by RNA Polymerase II, Mol Cell. 10 (2002) 1441–1452. 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- [194].Németh A, Perez-Fernandez J, Merkl P, Hamperl S, Gerber J, Griesenbeck J, Tschochner H, RNA polymerase I termination: Where is the end?, Biochimica Et Biophysica Acta Bba - Gene Regul Mech. 1829 (2013) 306–317. 10.1016/j.bbagrm.2012.10.007. [DOI] [PubMed] [Google Scholar]
- [195].Ocampo J, Chereji RV, Eriksson PR, Clark DJ, Contrasting roles of the RSC and ISW1/CHD1 chromatin remodelers in RNA polymerase II elongation and termination, Genome Res. 29 (2019) 407–417. 10.1101/gr.242032.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Spies N, Nielsen CB, Padgett RA, Burge CB, Biased Chromatin Signatures around Polyadenylation Sites and Exons, Mol Cell. 36 (2009) 245–254. 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Spies N, Nielsen CB, Padgett RA, Burge CB, Biased Chromatin Signatures around Polyadenylation Sites and Exons, Mol Cell. 36 (2009) 245–254. 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Chereji RV, Kan T-W, Grudniewska MK, Romashchenko AV, Berezikov E, Zhimulev IF, Guryev V, Morozov AV, Moshkin YM, Genome-wide profiling of nucleosome sensitivity and chromatin accessibility in Drosophila melanogaster, Nucleic Acids Res. 44 (2016) 1036–1051. 10.1093/nar/gkv978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Durand-Dubief M, Svensson JP, Persson J, Ekwall K, Topoisomerases, chromatin and transcription termination, Biochem Soc Symp. 2 (2011) 66–70. 10.4161/trns.2.2.14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Yu S, Jordán-Pla A, Gañez-Zapater A, Jain S, Rolicka A, Östlund Farrants A-K, Visa N, SWI/SNF interacts with cleavage and polyadenylation factors and facilitates pre-mRNA 3′ end processing, Nucleic Acids Res. 46 (2018) gky438–. 10.1093/nar/gky438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Hildreth AE, Ellison MA, Francette AM, Seraly JM, Lotka LM, Arndt KM, The nucleosome DNA entry-exit site is important for transcription termination and prevention of pervasive transcription, Elife. 9 (2020) e57757. 10.7554/elife.57757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Somesh BP, Reid J, Liu W-F, Søgaard TMM, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Multiple Mechanisms Confining RNA Polymerase II Ubiquitylation to Polymerases Undergoing Transcriptional Arrest, Cell. 121 (2005) 913–923. 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- [203].Lafon A, Taranum S, Pietrocola F, Dingli F, Loew D, Brahma S, Bartholomew B, Papamichos-Chronakis M, INO80 Chromatin Remodeler Facilitates Release of RNA Polymerase II from Chromatin for Ubiquitin-Mediated Proteasomal Degradation, Mol Cell. 60 (2015) 784–796. 10.1016/j.molcel.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Prendergast L, McClurg UL, Hristova R, Berlinguer-Palmini R, Greener S, Veitch K, Hernandez I, Pasero P, Rico D, Higgins JMG, Gospodinov A, Papamichos-Chronakis M, Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability, Nat Commun. 11 (2020) 4534. 10.1038/s41467-020-18306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Maeshima K, Ide S, Babokhov M, Dynamic chromatin organization without the 30-nm fiber, Curr Opin Cell Biol. 58 (2019) 95–104. 10.1016/j.ceb.2019.02.003. [DOI] [PubMed] [Google Scholar]
- [206].Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK, Organization of Chromatin by Intrinsic and Regulated Phase Separation., Cell. 179 (2019) 470–484.e21. 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Zhang Y, Narlikar GJ, Kutateladze TG, Enzymatic Reactions inside Biological Condensates, J Mol Biol. (2020). 10.1016/j.jmb.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, Darzacq X, Zweckstetter M, RNA polymerase II clustering through carboxy-terminal domain phase separation, Nat Struct Mol Biol. 25 (2018) 833–840. 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]


