Abstract
The ability of the heterochromatin protein-1 (HP1) to phase separate into droplets suggests new mechanisms of gene organization in the cell nucleus. An accumulating body of work suggests that other nuclear proteins also display phase separation behaviors in vitro. To understand the mechanistic and biological significance of such droplet formation a rigorous biophysical characterization of this behavior is necessary. Herein we describe procedures for imaging HP1 droplets by brightfield microscopy, and two methods to quantify phase separation.
1. INTRODUCTION
A cell densely packed with macromolecules must accomplish a broad range of competing molecular processes. Classic compartmentalization, such as the membrane-enclosed nucleus, provides physical separation between transcription and translation and therefore many points of regulation. Within the nucleus, gene poor and transcriptionally silent heterochromatic regions of the genome are known to be more compact than highly expressed euchromatic regions. Heterochromatin also occupies physically distinct territories than euchromatin. Phase separation, or the emergence of a dilute and concentrated phase of macromolecules in solution, has been proposed to help mediate this distinct packaging (Banani, Lee, Hyman, & Rosen, 2017; Larson & Narlikar, 2018). In particular, heterochromatin protein-1 (HP1) is a major mediator of chromatin compaction and has been shown to form protein-rich droplets in vitro (Larson et al., 2017) (Fig. 1A). Correspondingly, in vivo heterochromatin puncta containing HP1 display dynamic properties that are suggestive of a liquid droplet-like state (Strom et al., 2017).
Fig. 1.
Phase separation of HP1α. (A) Schematic of nPhos-HP1α containing two globular domains (chromodomain, CD and chromoshadow-domain, CSD), two terminal extensions (CTE and NTE), and a disordered hinge region. Phosphorylation of the NTE binds to the positive patch on the hinge of an adjacent dimer, leading to phase-separated droplets. (B) Brightfield microscopy images of HP1α or nPhos-HP1α visualized in a 384-well plate with a 40 × objective.
Molecules of unmodified HP1α are typically miscible up to millimolar concentrations in physiological buffers. Upon phosphorylation of the N-terminal extension (nPhos-HP1α) or the addition of DNA, the solution becomes nonhomogenous and partitions into concentrated and dilute populations (Fig. 1A). The high concentration population will often minimize surface tension by forming round droplets. These manifest in a test tube as a cloudy, turbid solution. The formation of phase-separated droplets is usually positively correlated with the ability of a protein to make multiple contacts with other proteins in three dimensions (Li et al., 2012). This property, also called multivalency, holds true for HP1α, as it typically exists in solution as a dimer but upon N-terminal phosphorylation forms higher order oligomers. In addition to mutlivalency, the presence of disordered regions has also been shown to correlate with droplet formation (Hyman, Weber, & Jülicher, 2014). In HP1α, the N-terminal region and the hinge region are intrinsically disordered. These regions contribute to oligomerization through electrostatic interactions between the N-terminal phosphates of one dimer and a patch of positive residues on the hinge region of another dimer (Larson et al., 2017; Larson & Narlikar, 2018) (Fig. 1A). Increasing the concentration of monovalent salts increases the concentration of HP1α required to form droplets, consistent with charge shielding of these electrostatic interactions. HP1α droplet formation also decreases as a function of increasing temperature (Larson et al., 2017). Therefore, both the ionic strength of the solvent and the temperature are critical aspects to consider and control when examining HP1α phase separation.
In this chapter we describe methods used to visualize phosphorylated-HP1α droplet formation as well as two methods to quantify phase separation. The first measurement method, termed the spin down assay, utilizes the increase in density of the protein-rich droplets, which pellet to the bottom of the tube with centrifugation. The concentration of protein in the supernatant and pellet can then be determined. The second method is an assay that measures the turbidity of the solution. In this method, light scattering caused by the droplets is assayed by measuring absorbance. Together the techniques can allow reproducible quantification of the phase separation properties of a given protein in a given solvent.
2. PURIFICATION OF nPhos-HP1α
The nPhos-HP1α protein is prepared by coexpressing the kinase CKII and HP1α in bacteria. HP1α contains phosphorylated serine sites in both the N-terminus and hinge domains. To restrict phosphorylation to the N-terminus, we use a HP1α plasmid that has the serine residues in the hinge region mutated to alanines. Further, the construct contains a N-terminal 6 ×-his tag followed by a TEV cleavage domain that allows for purification via a cobalt column and subsequent cleavage by TEV protease to remove the His tag. Following cleavage, nPhos-HP1α is run over two columns: an anion exchange column (MonoQ) followed by a sizing column (Superdex 75). The anion exchange step removes the TEV protease and separates HP1α species that are not fully phosphorylated and the sizing column separates leftover contaminants. The resulting nPhos-HP1α protein is ~99% phosphorylated as shown by mass spectrometry (Larson et al., 2017).
Notes
nPhos-HP1α phase separation is mediated by electrostatic interactions, and therefore is extremely sensitive to the salt concentration. Throughout the purification we use buffer salt concentrations ranging from 150 to 800 mM KCl, as described below. We do not lower the salt concentration below 150 mM KCl. Salt concentration should be closely monitored as phase separation during concentration steps over a 10K spin concentrator results in protein loss due to permanent sticking of the protein to the concentrator membrane.
Solutions, Reagents, and Equipment
CKII, in a pRSF-Duet vector (kanamycin resistance)
HP1α, in a pBH4 vector (carbenicillin resistance)
E. coli. Rosetta (DE3) competent cells (chloramphenicol resistance)
- 2 × LB broth
- 20 g/L tryptone
- 10 g/L yeast extract
- 10 g/L NaCl
Kanamycin
Carbenicillin
Chloramphenicol
C3 Emulsiflex (Avestin)
Cobalt-NTA affinity resin (Clontech)
FPLC
Mono-Q 4.6/100 PE anion exchange column (GE)
Superdex-75 16/60 size-exclusion column
Amicon Ultracel-10K spin concentrator
Lysis Buffer
20 mM HEPES pH 7.5
300 mM NaCl
10% Glycerol
7.5 mM imidazole
Elution Buffer
20 mM HEPES pH 7.5
150 mM KCl
400 mM imidazole
Dialysis Buffer
20 mM HEPES pH 7.5
150 mM KCl
3 mM DTT
MonoQ High Salt Buffer
20 mM HEPES pH 7.5
1 M KCl
1 mM DTT
MonoQ Low Salt Buffer
20 mM HEPES pH 7.5
1 mM DTT
Size Exclusion Chromatography (SEC) Buffer
20 mM HEPES pH 7.5
200 mM KCl
1 mM DTT
10% Glycerol
Procedure
Growing Cells
Perform a single transformation into Rosetta competent cells with the CKII and HP1α plasmids as per the vendor protocol. As Rosetta cells contain a plasmid with chloramphenicol resistance, cells will be selected on LB plates with 25 μg/mL chloramphenicol, 25 μg/mL kanamycin, and 50 μg/mL carbenicillin. After plating cells, let sit for 12h at 37°C.
The next day make a starter culture by picking a single colony and placing it into 50 mL LB with 25 μg/mL chloramphenicol, 25 μg/mL kanamycin, and 50 μg/mL carbenicillin. Let grow while shaking overnight for 6–12h.
Insert the starter culture media into 1L 2 × LB supplemented with 25 μg/mL chloramphenicol, 25 μg/mL kanamycin, and 50 μg/mL carbenicillin. Let grow while shaking at 37°C until an OD of 0.4 is reached.
Place cells at 18°C and let grow while shaking until an OD of 0.8 is reached.
Induce protein expression in cells by adding isopropy-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.3 mM. Let grow while shaking at 18°C for 14h.
Collect cells by spinning at 4000 × g for 30min. Remove the media and transfer the cells to a 50-mL conical tube. Cells can be lysed immediately or frozen in liquid nitrogen and stored at −80°C for future use.
Cell Lysis
Resuspend the cell pellet with 30 mL of lysis buffer supplemented with the protease inhibitors 1 mM phenylmethanesulfonyl fluoride, 1 μg/mL pepstatin A, 2 μg/mL aprotinin, and 3 μg/mL leupeptin. Vortex and mix with a serological pipette until cells are completely suspended in solution with no residual chunks. A dounce homogenizer is used if vortexing does not result in a homogenous sample.
Lyse the cells by passing through a C3 Emulsiflex.
Spin down the lysate at 25,000 × g for 30min. Remove supernatant (clarified lysate) into a fresh tube.
Cobalt Column
Take 2 mL cobalt bead slurry per liter of culture being purified and transfer into a fresh 50mL conical. Add water up to top of conical and spin at 1000 × g for 3 min. Remove water and repeat one more time.
Remove excess water and place 20mL lysis buffer into tube. Spin at 1000 × g for 3 min and remove excess buffer.
Add clarified lysate into beads and let incubate at 4°C for 45 min while rotating.
Place bead-lysate mixture into a gravity column. Let lysate flow through, but do not let beads run dry.
Wash beads with 50 × the bead volume of lysis buffer. Do not add any protease inhibitors to this lysis buffer as this will interfere with the subsequent TEV cleavage step.
Collect protein by adding 10mL of elution buffer to the column. Let slowly flow into a 50-mL conical tube.
Add TEV protease into sample at a final concentration of 3mg/mL.
Dialyze sample into 2L of dialysis buffer overnight at 4°C.
MonoQ Column
Equilibrate the monoQ column with 15% high salt solution (therefore 150 mM KCl; made by mixing 15% High salt MonoQ buffer with 85% low salt MonoQ buffer). Inject sample onto column.
Perform salt gradient from 15% high salt (150 mM KCl) to 80% high salt (800 mM KCl; made by mixing 80% High salt MonoQ buffer with 20% low salt MonoQ buffer) over 15-column volumes. Fully phosphorylated nPhos-HP1α should come off the column at 380 mM KCl. There is often multiple peaks prior to the fully phosphorylated species that represent HP1α that is not fully phosphorylated and therefore has a lower net negative charge. We do not collect these species.
Collect the nPhos-HP1α fractions and concentrate in a Amicon 10K spin concentrator until the sample is under 500 μL.
S75 Sizing Column
Equilibrate the column with SEC buffer.
Inject the nPhos-HP1α sample onto the column. The protein normally comes off at 7 mL of a 25-mL column.
Collect the fractions containing nPhos-HP1α and concentrate down to a concentration greater than 500 μM.
Flash-freeze 50 μL aliquots in liquid nitrogen then store at −80°C until further use.
3. PREPARING nPhos-HP1α SAMPLE FOR EXPERIMENTS
Prior to performing the experiments described below, we dialyze nPhos-HP1α overnight into a salt concentration where phase separation does occur (75mM KCl). After dialysis, recovery of protein off the membrane is facilitated by letting the solution sit at room temperature for a few hours. Phase separation is promoted at lower temperatures, and droplets tend to be stickier than miscible protein. Although nPhos-HP1α will phase separate at both 4°C and 22°C, the protein droplets better release from the membrane at room temperature. Concentration measurements can then be taken by an absorbance measurement at 280 nm (A280) on a NanoDrop Spectrophotometer as described below in detail.
Notes
At room temperature (22°C) droplet formation of nPhos-HP1α is visible at 200 μM in a buffer containing 75 mM KCl, 20 mM HEPES, pH 7.5, and 1 mM DTT.
If using dialysis tubing with width larger than 10 mm, it is advisable to dialyze a larger volume of protein sample to avoid losing sample. Note that concentration should be maintained above 400 μM.
Solutions, Reagents, and Equipment
NanoDrop Spectrophotometer
Dialysis Tubing, 10kDa molecular weight cutoff, 10 mm width
Dialysis clips
Dialysis Buffer Components
20 mM HEPES pH 7.5
75 mM KCl
1 mM DTT
Procedure
- Dialyze phosphorylated nPhos-HP1α overnight at 4°C, preferably 50 μL with a concentration ~400 μM or higher.
- Boil dialysis tubing in water.
- Squeeze out excess water and immerse in buffer. Squeeze out buffer and repeat a few times.
- Clip one end of tubing with dialysis clip. Pipette 50 μL nPhos-HP1α into tubing. Clip other end.
- Place into 1L of dialysis buffer with stir bar at bottom. Let incubate at 4°C overnight while stirring.
Let dialyzed protein and buffer sit at room temperature for ~3h. Take out of dialysis tubing and pipette against bag to resuspend any protein sticking to the sides. Transfer to a fresh tube. Protein solution should look turbid, i.e. cloudy.
Dilute and take 280 nm absorbance measurement. To measure the concentration of solution it is important to dilute the turbid solution to a concentration where visible droplet formation does not occur. For this we recommend the following protocol. First, mix the solution with a pipette tip, then take 5 μL of the mixed solution and dilute into a volume of dialysis buffer that results in a clear solution. We typically dilute 1:10, but additional dilution might be required depending on the concentration of your stock solution. Use Beer’s law to calculate concentration of solution (extinction coefficient of HP1α is 29,500 M−1 cm−1).
4. IMAGING DROPLETS
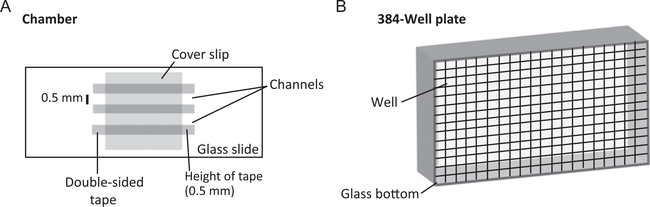
It is difficult to determine why a solution of purified protein has become cloudy. Common purification issues, such as protein precipitation that occurs when proteins unfold and begin to nonspecifically stick to one another, can also cause turbidity. While these precipitations, unlike droplet formation, are often irreversible, it is important to visualize the solution using a brightfield microscope before making any conclusions. To visualize droplets we either use 384-well plates or chambers crafted out of glass slides, double-sided tape and glass coverslips (Fig. 2). To prevent nonspecific sticking of protein to the glass we first coat the surface as described below.
Fig. 2.
Tools for droplet visualization. Schematic of chamber setup (A), and 384-well plates with imaging quality glass bottom (B).
4.1. Coating Glass Surfaces With PEG Silane
One common issue with visualizing proteins is the wetting of protein onto glass coverslips due to the dipole moments of silicon dioxide attracting the charged protein. Many procedures are utilized to block the glass surface, such as washing with a relatively neutral protein like BSA or casein before the addition of the turbid protein solution. We utilize a surface reactive PEG derivative, PEG silane that reacts with the silica and blocks the surface from wetting of nPhos-HP1α.
Two different imaging setups are used. The first is a 384-well plate with an imaging quality glass bottom and the second is a set of chambers created from glass slides, cover slips, and double-sided tape.
Notes
PEG silane is reactive with moisture and light sensitive. Resuspend fresh powder before each use and keep materials in the dark during wash steps.
4.1.1. Well Plate Protocol
Notes
The thickness of the glass bottom needed depends on the type of microscopy being used. A thickness of 0.15 mm allows visualization with both brightfield and confocal microscopy. However, thicker glass is often more cost-effective and can be easily used with a brightfield microscope. Avoid glass that has surface treatments such as collagen for cell culture.
Solutions and Reagents
384-Well plate with imaging quality bottom
2% Hellmanex detergent
1 M NaOH
Distilled water (dH20)
95% Ethanol
PEG silane powder
Procedure
Pipette 20 μL of 2% Hellmanex into each well. Let sit for 1h.
Wash 3 × with distilled water. Remove Hellmanex, and pipette in 100 μL of dH2O. Repeat three times with fresh water. Remove excess water.
Pipette 20 μL 1 M NaOH into each well. Let sit for 30min. Do not let sit longer, as it might begin to degrade the plastic.
Wash 3 × with distilled water, as in step 2.
Let sit in hood until residual water evaporates.
Dissolve PEG silane in 95% ethanol to a concentration of 20 mg/mL. Pipette 20 μL into each well, cover with parafilm. Let the PEG silane react with the silica overnight (10–14h) in the dark.
Next day wash 3 × with distilled water, as in step 2.
Dry the wells, as in step 5.
4.1.2. Chamber Protocol
Solutions and Reagents
Glass slides 75 × 25mm, 1 mm thickness
Cover slips 25 × 25mm, .15 mm thickness
2% Hellmanex detergent
1 M NaOH
Distilled water
95% Ethanol
PEG silane powder
Procedure
Immerse glass slides and coverslips in a beaker or slide holder with 2% Hellmanex. Let sit for 1h.
Wash 3 × with distilled water. Hellmanex can be removed and water directly added to slide holders. Repeat three times with fresh water and remove excess water.
Immerse glass slides in 1 M NaOH. Let sit for 30 min.
Wash 3 × with distilled water, as in step 2.
Dry slides with nitrogen gas dispensed from a standard nitrogen gas tank. If nitrogen tank is not available the slides can be air-dried in a chemical fume hood for 2–3h.
Dissolve PEG silane in 95% ethanol to a concentration of 20 mg/mL. Pipette 50 μL of solution onto slide and cover with cover slip. Press down to remove any air bubbles. Let the PEG silane react with the silica overnight (10–14h) in the dark. During this time we prevent the solution from drying out by placing the slides on the top rack of an empty tip box with half an inch of water in the bottom.
Wash 3 × with distilled water while keeping track of which side was exposed to PEG silane.
Dry slides as in step 5. Drying with nitrogen is preferred as it avoids residue forming on glass surface.
4.2. Creating Chambers
Once glass slides and coverslips are clean, they can be assembled with double-sided tape into chambers with a volume of ~10 μL (Fig. 2A).
Solutions and Reagents
PEG silane-coated glass slides and cover slips
Double sided tape, 0.5 mm thick
Procedure
Creating chambers
- Cut out strips of tape and adhere to the PEG silane-coated side of the slide in parallel fashion, leaving ~0.5 mm in between each strip (Fig. 2).
- The length between two strips of tape and the height of tape will determine the volume of sample needed to fill chamber.
- Depending on width of tape and channel, you can assemble between 1 and 5 chambers per slide.
Adhere cover slip to taped slide. Apply pressure with a pipette tip to remove any pockets of air.
4.3. Imaging Droplets
After preparing chambers or the 384-well plates, the nPhos-HP1α sample can be inserted and visualized with brightfield microscopy.
Notes
Droplet formation and size is often time sensitive. It is therefore important to measure and standardize the time spent between preparing solutions and imaging on the microscope, especially when comparing different conditions and protein mutants.
Solutions and Reagents
384-Well plates OR chambers
Vacuum grease (optional)
Final Buffer Components
20 mM HEPES pH 7.5
75 mM KCl
1 mM DTT
Final protein concentration, 300 μM nPhos-HP1α
Imaging With 384-Well Plate
Pipette 20 μL nPhos-HP1α sample into well of plate.
Image with a 40 × objective on a brightfield microscope (Fig. 1B).
Imaging With Chambers
Pipette nPhos-HP1α sample into space between cover slip and slide. Volume of sample will depend on channel width. Capillary action will pull your sample further into the chamber. If you want to avoid movement of the sample you can seal each side of the chamber with vacuum grease.
Image with a 40 × objective on a brightfield microscope.
5. QUANTIFYING PHASE SEPARATION
We utilize two methods to quantitate the phase separation properties of nPhos-HP1α, a spin down assay and a turbidity assay. These two methods quantify two different, though related, properties associated with phase separation. These quantitative measures allow for systematic comparison between mutants and a means to compare the potencies of external ligands that regulate droplet formation.
5.1. Spin Down Assay
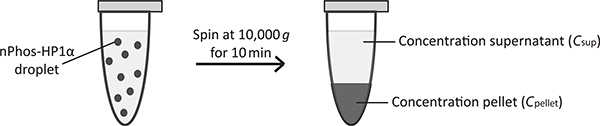
This is a simple experiment that utilizes the separation of the solution into two distinct phases. The basis for this assay is akin to a solubility measurement. If the concentration of the protein is greater than can remain soluble, the excess protein separates out in the form of droplets. The droplets, which are denser, will spin down to the bottom of a test tube with centrifugation. The concentration of the supernatant and the pellet can then be determined by measuring absorbance at 280 nm (A280) (Fig. 3). This is a rapid assay, which provides reproducible measurement of the protein concentration that remains in the supernatant. However, the supernatant concentration is dependent on both the density of the droplets and the speed of the centrifuge. Care must be taken to ensure the same rotor speed is maintained across different measurements. Further, it is best practice to visualize the supernatant on a brightfield microscope after centrifugation to see if any droplets remain in the supernatant.
Fig. 3.
Spin down assay. nPhos-HP1α forms phase-separated droplets under the conditions listed in Section 4.1. After spinning, the droplets pellet to the bottom and A280 reads can be taken to calculate the protein concentration in the supernatant and pellet.
Solutions and Reagents
Microcentrifuge
NanoDrop Spectrophotometer
nPhos-HP1α dialyzed as in Section 4.1
Buffer Components
20 mM HEPES pH 7.5
75 mM KCl
1 mM DTT
Procedure
Pipette 10 μL of nPhos-HP1α prepared in Section 4.1 into a new 1.5 mL tube. Let incubate at room temperature for 5min.
Spin solution down at 10,000 × g for 5 min in a table top microcentrifuge.
Remove 4 μL of supernatant. Take an A280 measurement on the NanoDrop Spectrophotometer in triplicate. The average of the three is the supernatant concentration (Csup).
The remainder of supernatant can be removed, and A280 measurement taken to determine the protein concentration within the phase. However, we typically do not measure the pellet concentration as removing the supernatant can be technically challenging if the pellet is small.
5.2. Turbidity Assay
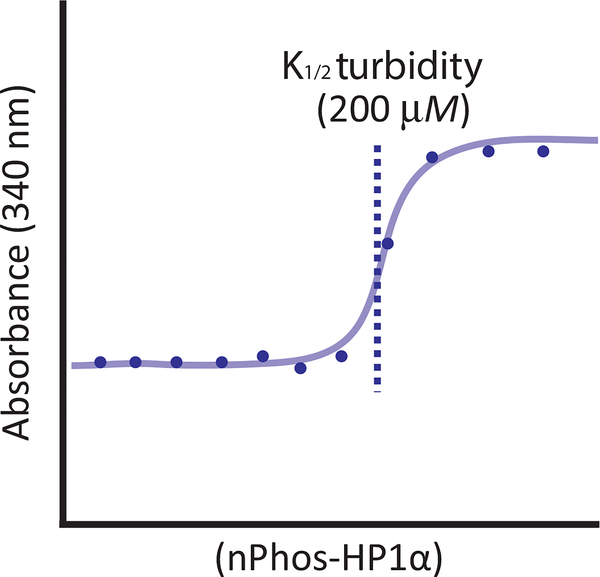
A classic example of turbidity in biochemistry is the transformation of a solution of tubulin molecules from clear to cloudy upon the induction of polymerization. Longer polymers of tubulin scatter more light than dimers, and this output has often been measured by the absorbance of the solution at 350nm (Gaskin, Cantor, & Shelanski, 1974; Mirigian, Mukherjee, Bane, & Sackett, 2013). Similarly, nPhos-HP1α is a monomer or dimer at low concentrations but when phase separation occurs, droplets form with much larger radii than an individual dimer. These droplets will scatter light more effectively. We define the K1/2 turbidity as the concentration of protein necessary to reach half-maximal turbidity (Fig. 4). Assaying for turbidity allows for reproducible, quantitative measurements of phase separation. However, due to the dependence of the scattering signal on droplet size, smaller droplets may be underrepresented. This assay requires a dilution series of nPhos-HP1 and a plate reader that can measure absorbance at 340 nm. If needed the assay can also be adapted to be carried out using a cuvette instead of a plate but this requires larger sample volumes.
Fig. 4.
Turbidity assay. Schematic of turbidity data for nPhos-HP1α, with K1/2 turbidity referring to the half-maximal protein concentration.
Notes
This procedure can be done with glass 384-well plates treated with PEG silane. After quantitation of 340 absorption the same plate can be taken to the microscope and imaged. Subsequent imaging is best practice, as light scattering at 340 nm is dependent on the size of the droplets.
Solutions and Reagents
Temperature controlled plate reader that can read absorbance at 340 nm. We utilize the Spectramax M5 plate reader.
96- or 384-well plate with clear bottom
nPhos-HP1α dialyzed in Section 4.1
Buffer Components
20 mM HEPES pH 7.5
75 mM KCl
1 mM DTT
Procedure
Beginning at 400 μM, perform a twofold serial dilution using the same buffer as above in a 12-tube PCR strip with a final volume of 25 μL for each sample.
Transfer 20 μL of each sample to clear bottom 384-well plate.
Let incubate for 5 min.
Read absorbance at 340 nm at room temperature on plate reader.
Analyzing Data
The data are plotted and fit with the following sigmoid function, as shown in Fig. 4 schematic.
A = absorbance value at max concentration; B = absorbance value at min concentration; Xc = inflection point, half-maximal concentration; k = slope factor of sigmoid curve; Y = absorbance; x = protein concentration.
6. CHARACTERIZING PHASE SEPARATION OF OTHER PROTEINS
If it is speculated that a protein of interest phase separates, the first hurdle is a successful protein purification, as droplets stick to many common purification tools such as columns and spin concentrators. Analysis of the protein sequence can facilitate this process, and the first relevant question is whether there are sequence stretches predicted to be disordered. If the predicted disordered regions are enriched for charged residues, then purification in a high salt regime (200 mM KCl or greater) is recommended as low salt promotes electrostatic interactions. If instead these regions contain an abundance of hydrophobic residues, purification in a lower salt regime (30–100 mM KCl) may be preferable as increasing salt concentration promotes hydrophobic interactions. Once purification is achieved, some relevant parameters to consider are protein concentration, salt concentration, pH, and crowding conditions. In some experiments researchers have added crowding agents such as PEG and Ficoll, to mimic the macromolecular crowding within cells (Lin, Protter, Rosen, & Parker, 2015). After dialyzing the protein into a buffer space where phase separation is suspected to occur, titrations of the protein and salt concentrations can be performed and imaged as in Section 4. This will allow for visualization of any turbidity and droplet formation, and subsequent quantitation of the two phase separation properties described in Section 5. Care should be taken to match the in vitro phase separation reaction conditions as much as possible to those expected in the cell. Correspondingly, interpretation of the physiological significance of phase separation observed in vitro should take into account potential differences between the in vitro and cellular conditions.
7. CONCLUSIONS
Within this chapter we define our protocol for imaging nPhos-HP1α droplets and two methods to quantitate phase separation properties. The emergence of phase-separated droplets is only one of many causes for the emergence of a turbid solution. Classic examples include protein denaturation and precipitation or tubulin polymerization. Therefore, if presented with turbidity it is essential to image the solution to determine the cause.
Phase separation of proteins in vitro is strongly linked to the ionic strength of the solvent, the temperature and protein concentration used. A thorough investigation of additional buffer parameters like mono or divalent salts and pH while using the above techniques can facilitate teasing apart the mechanisms of multivalency. To assess the physiological relevance of droplet formation in vitro, it is important to (i) match as best as possible the in vitro buffer conditions to those in the corresponding cellular context and (ii) determine how the cellular concentration of a protein compares with the Csup and K1/2 turbidity values.
REFERENCES
- Banani SF, Lee HO, Hyman AA, & Rosen MK (2017). Biomolecular condensates: Organizers of cellular biochemistry. Nature Reviews. Molecular Cell Biology, 18(5), 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskin F, Cantor C, & Shelanski ML (1974). Turbidimetric studies of the in vitro assembly and disassembly of porcine neurotubules. Journal of Molecular Biology, 89, 737–758. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, & Jülicher F (2014). Liquid-liquid phase separation in biology. Annual Review of Cell and Developmental Biology, 30(1), 39–58. 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, et al. (2017). Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature, 547(7662), 236–240. 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, & Narlikar GJ (2018). The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry, 57(17), 2540–2548. 10.1021/acs.biochem.8b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature, 483(7389), 336–340. 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, & Parker R (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding protein. Molecular Cell, 60(2), 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirigian M, Mukherjee K, Bane SL, & Sackett DL (2013). Measurement of in vitro microtubule polymerization by turbidity and fluorescence. Microtubules, in vitro: Vol. 115. (pp. 215–229). Elsevier. 10.1016/B978-0-12-407757-7.00014-1. [DOI] [PubMed] [Google Scholar]
- Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, & Karpen GH (2017). Phase separation drives heterochromatin domain formation. Nature, 547(7662), 241–245. 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]