Abstract
While inhibition of T cell co-inhibitory receptors has revolutionized cancer therapy, the mechanisms governing their expression on human T cells have not been elucidated. Here we show that type 1 interferon (IFN-I) regulates co-inhibitory receptor expression on human T cells, inducing PD-1/TIM-3/LAG-3 while inhibiting TIGIT expression. High-temporal-resolution mRNA profiling of IFN-I responses established the dynamic regulatory networks uncovering three temporal transcriptional waves. Perturbation of key transcription factors (TFs) and TF footprint analysis revealed two regulator modules with different temporal kinetics that control expression of co-inhibitory receptors and IFN-I response genes with SP140 highlighted as one of the key regulators that differentiates LAG-3 and TIGIT expression. Finally, we found that the dynamic IFN-I response in vitro closely mirrored T cell features in acute SARS-CoV-2 infection. The identification of unique TFs controlling co-inhibitory receptor expression under IFN-I response may provide targets for enhancement of immunotherapy in cancer, infectious diseases, and autoimmunity.
Introduction
Immune checkpoint blockade targeting T cell co-inhibitory receptors has revolutionized cancer treatment. While signals such as IL-27 are associated with T cell expression of co-inhibitory receptors in mice1, 2, the regulatory mechanisms of co-inhibitory receptor induction in human T cells are still unknown. Type 1 IFN (IFN-I) serves as a first line defense system towards viral infection and its effect on T cells has been extensively studied in the context of T cell activation3, survival4, and effector functions5. IFN-I is also induced during chronic viral infection, autoimmunity, and cancer6, 7, 8, and accumulating evidence suggests that IFN-I may have immunomodulatory functions beyond their conventional role on T cells in acute viral infection9, 10, 11, 12. Notably, continuous exposure to IFN-I is implicated to promote T cell exhaustion which is marked by aberrant expression of co-inhibitory receptors (i.e., PD-1, TIM-3, LAG-3, and TIGIT) in chronic viral infection and cancer12, 13, 14, 15, 16. While IFN-I has a central role in regulating immune responses and murine models have enabled the elucidation of a functional role of IFN-I on T cells, it has become clear that this response varies greatly between rodents and human17. Thus, it is of critical importance to elucidate the biological consequences of IFN-I on human T cells and in the context of human diseases.
Here, we constructed a dynamic gene regulatory network of human primary T cell response to IFN-I stimulation using high-resolution gene expression profiling, chromatin accessibility analysis and perturbation of key network regulators. This regulatory network revealed both canonical and non-canonical IFN-I transcriptional regulators and identified unique regulators that control the expression of co-inhibitory receptors. To provide direct in vivo evidence for the role of IFN-I on co-inhibitory receptors, we then performed single-cell RNA-sequencing in subjects infected with SARS-CoV-2, where viral load was strongly associated with T cell IFN-I signatures. We found that the dynamic IFN-I response in vitro closely mirrored T cell features with acute IFN-I linked viral infection, with high LAG3 and decreased TIGIT expression. Finally, our gene regulatory network identified SP140 as a key regulator for differential LAG3 and TIGIT expression. The construction of co-inhibitory regulatory networks induced by IFN-I and unique transcription factors controlling their expression provides further resource for the identification of targets for enhancement of immunotherapy in cancer, infectious diseases, and autoimmunity.
Results
The impact of IFN-β on co-inhibitory receptors in human T cells
IL-27 has been identified as a crucial cytokine that promotes the induction of a co-inhibitory receptor module, with T cell exhaustion in murine tumor models1. Several reports have suggested that IL-27 functions downstream of IFN-β18, an important member of the IFN-I family. Thus, we hypothesized that IFN-β facilitates the induction of co-inhibitory receptors in humans. We first assessed the effect of IL-27 and IFN-β on the induction of core co-inhibitory receptors (TIM-3, LAG-3, PD-1, and TIGIT) in vitro using human primary naïve CD4+ and CD8+ T cells (Extended Data 1). Both IL-27 and IFN-β promoted significantly higher TIM-3 expression as compared to the expression of control groups without addition of exogenous cytokines. Of note, IFN-β induced more LAG-3 and TIM-3 expression compared to IL-27 in both CD4+ and CD8+ T cells (Figure 1a, Extended Data 2a). Both IL-27 and IFN-β suppressed the expression of TIGIT in CD4+ and CD8+ T cells (Extended Data 2b). We also observed increased production of IL-10 induced by IFN-β; however, IL-10 induction by IL-27 was modest in our in vitro culture settings which may reflect the difference between mouse and human T cell responses toward IL-27 stimulation (Extended Data 2c). Next, we further determined the impact of IFN-β treatment on gene expression kinetics for co-inhibitory receptors by qPCR. Gene expression dynamics for core co-inhibitory receptors (HAVCR2, LAG3, PDCD1) were upregulated by IFN-β for most time points. In contrast, TIGIT was downregulated, which was confirmed by protein expression using flow cytometry (Figure 1b, c). We examined the expression of other co-inhibitory receptors and found IFN-β induced co-expression of multiple co-inhibitory receptors (e.g., HAVCR2, PDCD1, LAG3) but inhibited expression of others (e.g., TIGIT, CD160, BTLA) (Figure 1d, Extended Data 2d). As T cell activation and entry into cell cycle could be confounding factors on the measurement of co-inhibitory receptor expression, we performed proliferation assays using cell trace violet dye and examined the level of T cell activation markers (CD25, CD44, and CD69) with IFN-β treatment. There was no statistical difference in cellular division or degree of T cell activation between control and the IFN-β condition after four days of human primary T cell culture. Additionally, there was even less proliferation of memory CD4+ T cells with IFN-β, indicating that the induction of co-inhibitory receptors by IFN-β is not affected by T cell activation state (Extended Data 3a-c), consistent with previous studies 19, 20. Furthermore, we confirmed a dose dependent effect of IFN-β treatment on co-inhibitory receptor expressions (Extended Data 3d). The effects of IFN-β on the level of LAG-3, TIM-3, PD-1 and TIGIT were consistently observed across each cellular division status, strongly suggesting that IFN-β induced changes on co-inhibitory receptors were not biased by T cell proliferation (Extended Data 3e). Collectively, these data elucidate a role for IFN-β as a cytokine that can directly control multiple co-inhibitory receptors in both human CD4+ and CD8+ T cells in vitro.
Figure 1. IFN-β differently regulates LAG-3, TIM-3, PD-1 and TIGIT in human T cells.
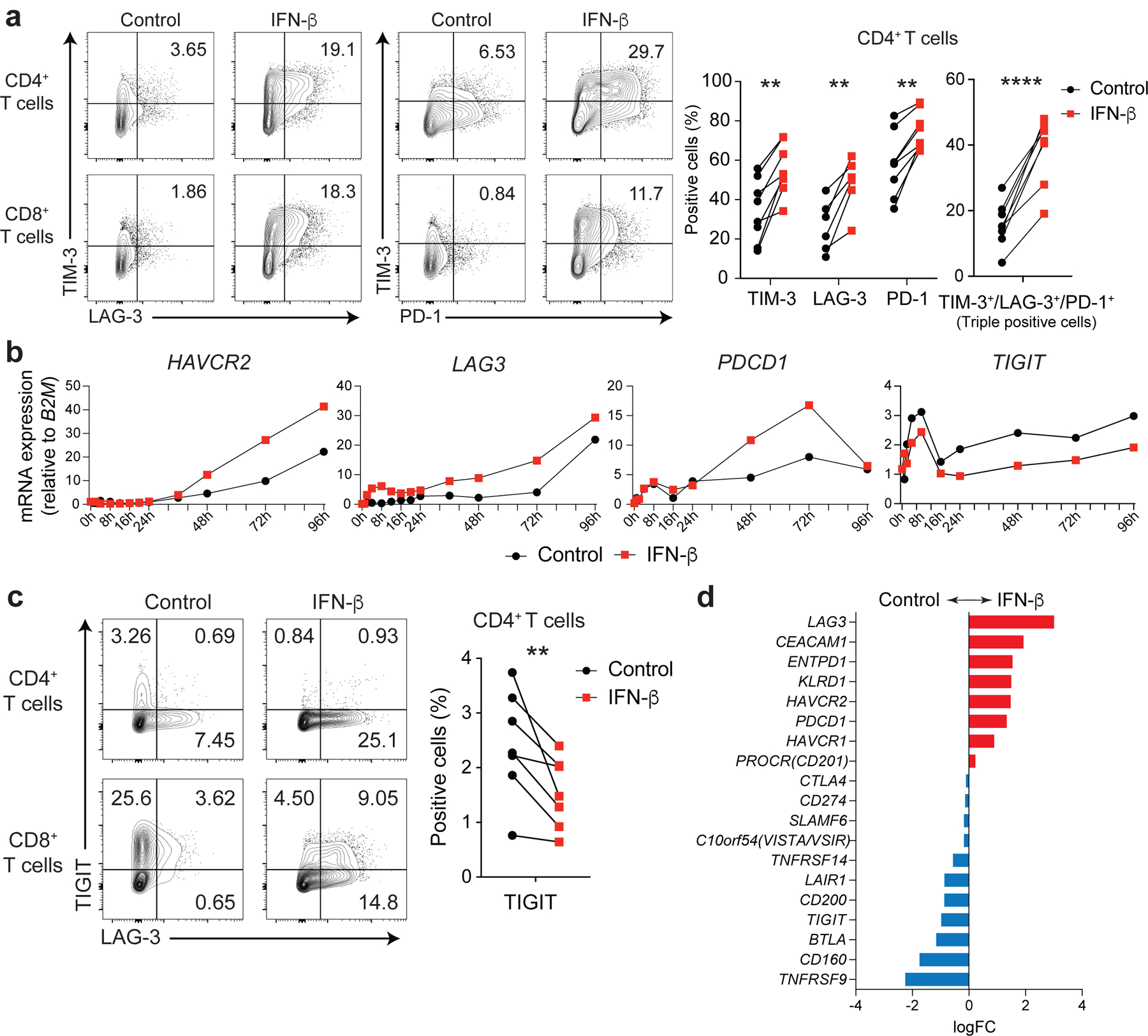
Effects of IFN-β on LAG-3, TIM-3, PD-1, and TIGIT expression on human naïve CD4+ and CD8+ T cells cultured with anti-CD3/CD28 for 96h in the absence (Control) or with 500 U/ml IFN-β (IFN-β). a, Representative contour plots of flow cytometry analysis on surface LAG-3, TIM-3, and PD-1 (left), quantitative expression for LAG-3, TIM-3, and PD-1 expression on naïve CD4+ T cells (n = 6; biologically independent samples) (middle), quantitative analysis for triple-positive (LAG-3, TIM-3, and PD-1) cells in naïve CD4+ T cells (n = 6; biologically independent samples) (right). b, Gene expression kinetics of LAG3, HAVCR2, PDCD1, and TIGIT quantified by qPCR with 13 timepoints in naïve CD4+ T cells. Average expression values from two subjects are plotted. c, IFN-β induces LAG-3 but suppresses TIGIT expression on human naïve CD4+ and CD8+ T cells. Representative contour plots of flow cytometry analysis (left), quantitative analysis for TIGIT positive cells in naïve CD4+ T cells (n = 8; biologically independent samples) (right). d, Co-inhibitory receptors expression pattern under IFN-β treatment in naïve CD4+ T cells by qPCR (n = 4; biologically independent samples). Red and blue bars represent higher expression in IFN-β treatment and Control conditions, respectively. Data was represented as mean +/− SD. **p < 0.01, ****p < 0.0001. Paired Student’s t test.
High resolution transcriptomic dynamics of IFN-β response
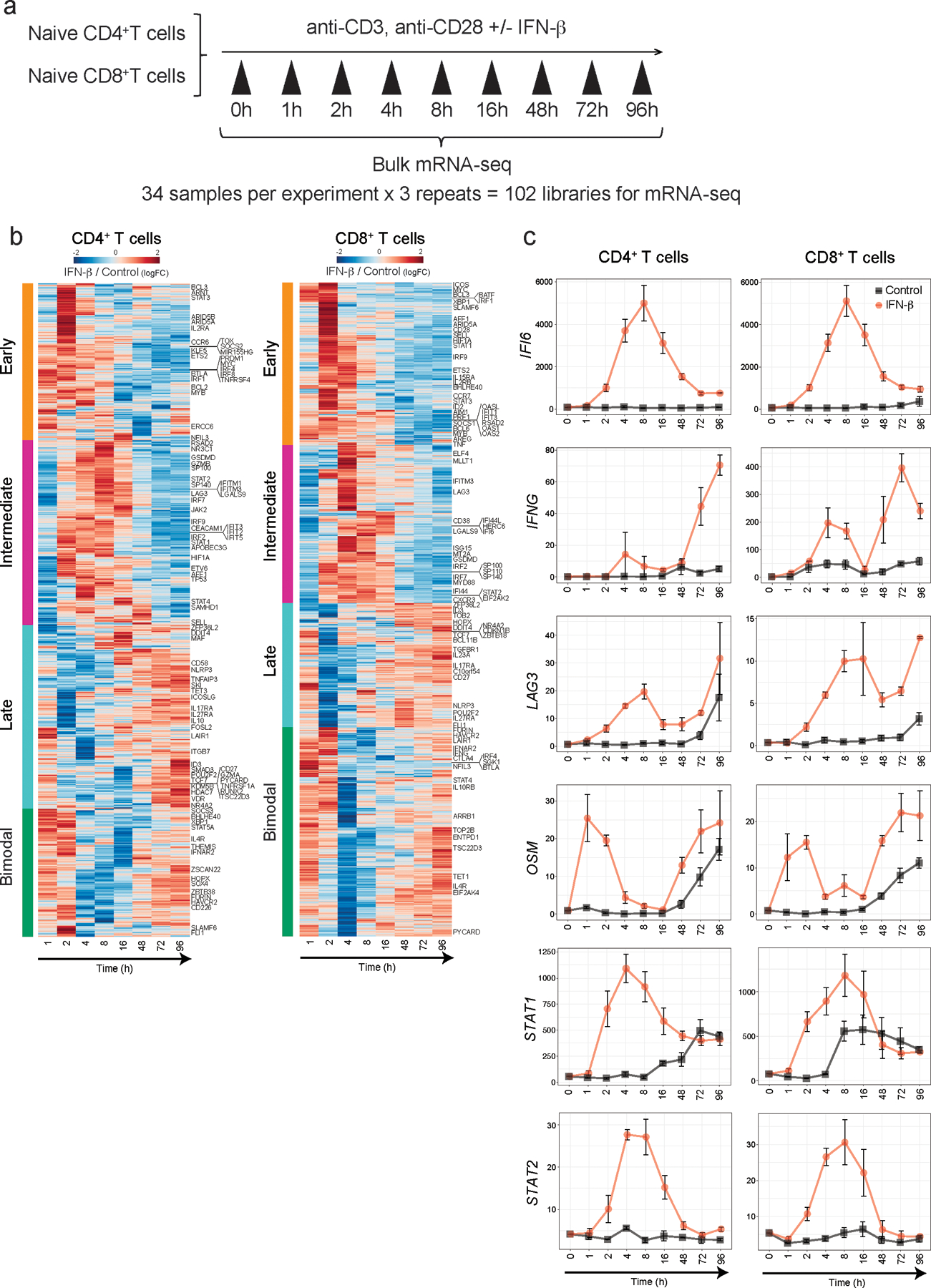
To uncover the regulatory mechanisms underlying the IFN-β response in human primary T cells, we generated a transcriptional profile at high temporal resolution. We used bulk mRNA-seq at ten time points along a 96-hour time course with and without IFN-β treatment (Extended Data 4a). To avoid inter-individual genetic variation, we selected one healthy subject whose T cells exhibited a stable response to IFN-β, and repeated the experiment three times at a two-week interval for each experiment, analogous to studying one mouse strain. We identified 1,831 differentially expressed genes (DEGs) for CD4+ T cells and 1,571 DEGs for CD8+ T cells across time points with IFN-β treatment, revealing a temporal shift of gene expression patterns in both CD4+ and CD8+ T cells (Figure 2a, Extended Data 4b, Methods). The genome-wide transcriptional profiles from three independent experiments demonstrated highly consistent results across time points (Figure 2b). Specifically, three transcriptional waves were observed during 96 hours of IFN-β response: early phase (1–2 h), intermediate phase (4–16 h), and late phase (48–96 h). As expected, we observed abundant induction of classical IFN stimulated genes (ISGs) (i.e., IFI6, MX1/2, RSAD2, STAT1/2/3, SP100/110/140), which peaked at the early-intermediate phase. IFN-I induced cytokines produced by T cells (IFNG, IL10, GZMB, PRF1) were also upregulated at intermediate-late phase (Figure 2c, Extended Data 4c). OSM (Oncostatin M), which is reported to amplify IFN-β response and suppress Th17 differentiation20, was significantly induced by IFN-β from the early phase and maintained induction at all time points (Extended Data 4c). Among DEGs, we identified dynamic expression of 134 transcription factors (TFs) for CD4+ T cells and 100 TFs for CD8+ T cells, which were both up- and down-regulated over the course of differentiation (Figure 2c). We further explored the dynamic changes at the level of chromatin accessibility by using an assay for transposase accessible chromatin sequencing (ATAC-seq)21 for CD4+ and CD8+ T cells over three different time points corresponding to three transcriptional waves (2, 8, and 72 h) (Extended Data 5a). Principal component analysis (PCA) highlighted the differential chromatin accessibility at the late phase (72 h) compared to the other phases (Extended Data 5b). Differential chromatin accessibility analysis at each time point identified temporal changes of chromatin accessibility; in the control the chromatin accessibility was reduced at the early time point (2 h) and then became more accessible over time (Extended Data 5c). This temporal change was altered by IFN-β treatment with more chromatin accessibility compared to control condition at 8 h, which was further enhanced at 72 h (Extended Data 5d). Taken together, these high temporal resolution transcriptional and chromatin accessibility data highlight the impact of IFN-β treatment on both the dynamics of T cell activation and their final differentiation state.
Figure 2. Three waves of dynamic transcriptomic changes by IFN-β in human T cells.
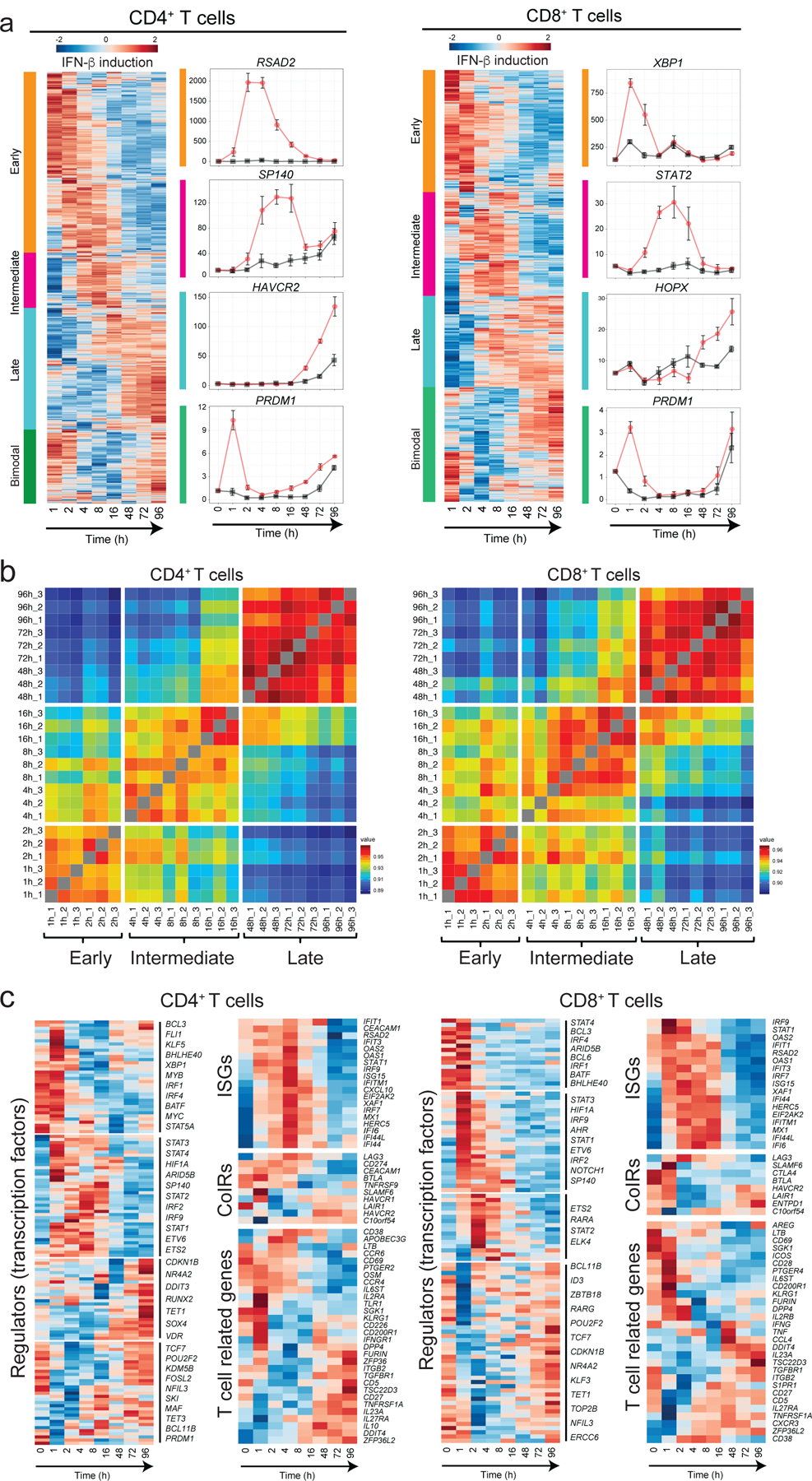
a, Gene expression profiles under IFN-β treatment in naïve CD4+ and CD8+ T cells. Differential expression of gene levels for eight time points with IFN-β stimulation (log2(expression)) are shown in heatmap. Based on the expression kinetics, the genes are clustered into four categories: early, intermediate, late, and bimodal (up regulated at early and late phase). Representative individual gene expression kinetics from each cluster are shown (mean+/− SD). b, Correlation matrix of global gene expression representing three transcriptional waves on CD4+ (left) and CD8+ T cells: early (1–2h), intermediate (4–16h), and late (48–96h). Eight timepoints with three replicates are shown. c, Temporal transcriptional profiles of differentially expressed genes for four categories are shown; transcriptional regulators (transcription factors), ISGs, co-inhibitory receptors, and key T cell associated factors for CD4+ (left) and CD8+ T cells.
Genetic perturbation identifies two IFN-I regulatory modules
Genetic perturbation of key transcription factors allows inference of the gene regulatory network with our high temporal resolution transcriptional data, leading to the elucidation of the regulatory mechanisms by which IFN-I response controls co-inhibitory receptor expressions. To narrow the list of TFs for perturbation, we prioritized TFs that are differentially expressed in both CD4+ and CD8+ T cells. We then further selected TFs associated with T cell exhaustion: 1) human tumor infiltrating T cells (TILs) 22, 23, 24, 25; 2) HIV specific T cell signature in progressive patients 26; and 3) IL-27 driven co-inhibitory regulators 1 (see Methods). We confirmed that these TFs are identified as ISGs in human immune cells by the Interferome dataset27 (Figure 3a). In total, 31 TFs were listed as candidates based on the overlap between ISG, TIL and IL-27 signatures and we chose 19 of them for perturbation. Since TCF-1 (encoded by TCF7) and Blimp-1 (encoded by PRDM1) are known to express functionally distinct isoforms, we also targeted a unique sequence for the long isoforms (TCF7L and PRDM1L, respectively), resulting in perturbation of 21 different targets in total.
Figure 3. Perturbation of key transcription factors in quiescent human T cells.
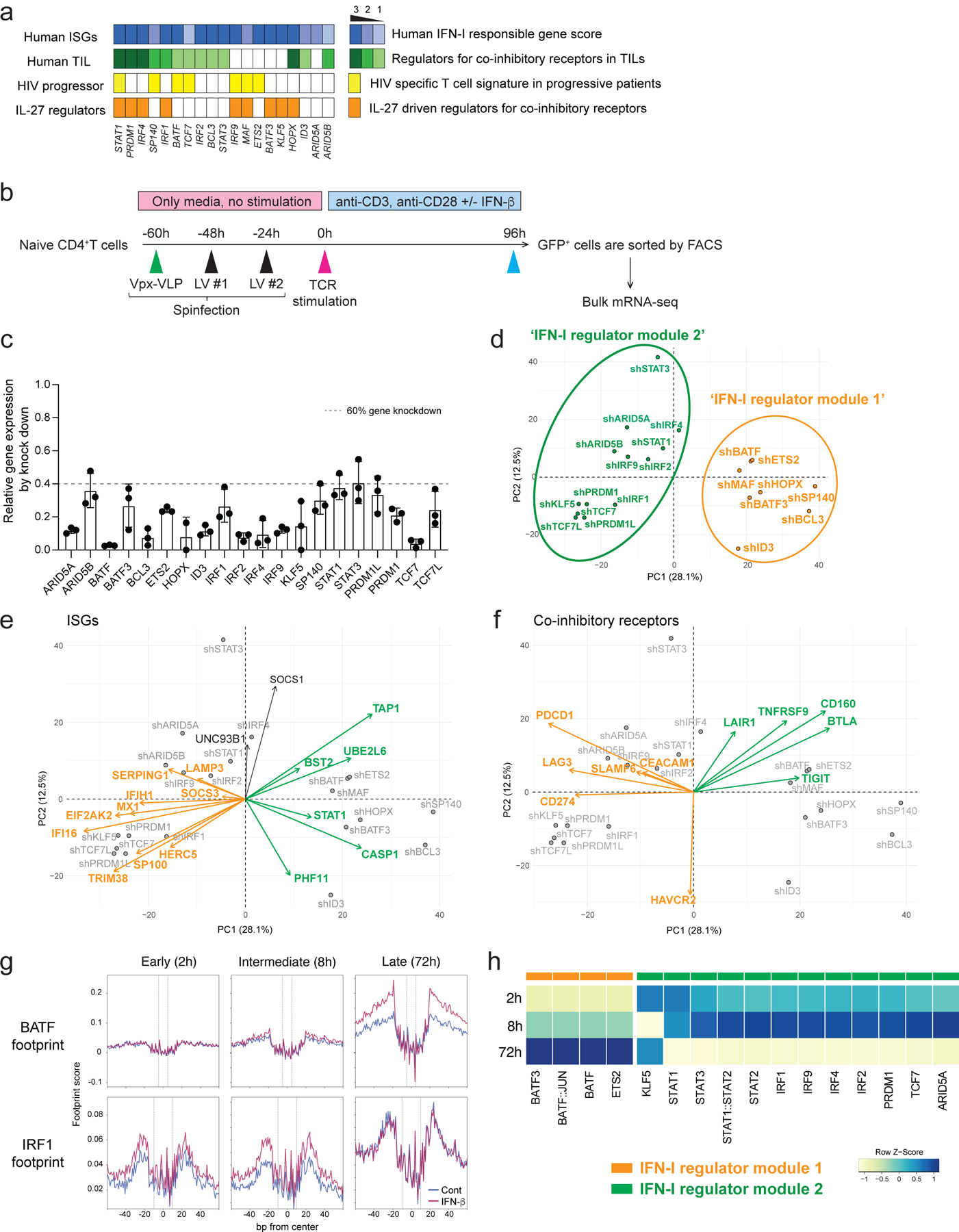
a, Characterization of candidate TFs for perturbation. Perturbed TFs are listed based on the overlap between differentially expressed TFs of CD4+ T cells and CD8+ T cells. Human ISG score (top; blue), human TIL co-inhibitory receptors score (green), HIV specific T cell signature genes in progressive patients (yellow), and IL-27 driven co-inhibitory receptor regulators (orange) are shown for each TFs. b, Experimental workflow of Vpx-VLP supported lentiviral shRNA perturbation. Ex vivo isolated naïve CD4 T cells were transduced with Vpx-VLPs, followed by two times of lentiviral particle transduction before starting T cell activation. T cells were stimulated with anti-CD3/CD28 in the absence or presence of IFN-β (500 U/ml) for 96h and GFP positive cells were sorted by FACS. RNAs were extracted from sorted cells and applied for mRNA-seq. Perturbations for all 21 shRNAs are performed with human CD4+ T cells isolated from the same individual as in Figure 2. c, Gene knockdown efficiency is shown as relative expression over scramble shRNA transduced controls. Dotted line represents 60% of gene knockdown. d-f, PCA plots and biplots based on differentially expressed genes by perturbation. d, PCA plot demonstrating the two modules of TF regulators on perturbation with 21 TFs. Characterization of shRNA-based gene knockdown for each TF being plotted. Labels represent perturbed TF gene names. ‘IFN-I regulator module 1’ is colored in green and ‘IFN-I regulator module 2’ is in orange. e, f, PCA biplot showing differential regulation by modules of regulator TFs; e, for ISGs and f, for co-inhibitory receptors. Orange and green arrows (vectors) are highlighting two groups of genes affected inversely by the different modules of TFs. g, Footprint plots for BATF and IRF1 at three time points (2, 8, 72 h). The dashed lines represent the edges of each TF motif. h, A heatmap showing the TF footprint activity of perturbed TFs across three phases.
Considering the majority of these regulators are induced at the early (1–2 h) and intermediate (4–16 h) phases, it was important to perform gene deletion prior to T cell receptor activation. For human primary T cells gene knockdown, we adopted lentiviral delivery of shRNAs with lentiviral gene product X (Vpx) containing virus-like particles (VLPs) to efficiently transduce lentivirus into unstimulated primary human naïve T cells28, 29 (Figure 3b). Spinoculation with Vpx-VLPs significantly increased the number of GFP expressing T cells (~30–60 %) compared to normal LV particle transduction without Vpx-VLPs (~1–5 %) and resulted in successful transduction of lentiviral vectors into non-blasting/quiescent cells (Extended Data 6a). We achieved efficient knockdown of at least 60 % gene expression for 21 target TFs in human naïve CD4+ T cells (Figure 3c).
To identify the effect of perturbation for each regulator, PCA was applied to determine the changes in RNA expression associated with each transcription factor knockdown (Figure 3d). PC1 divided the impact of perturbation into two modules of regulators: BATF, MAF, ETS2, HOPX, SP140, BCL3, ID3, and BATF3 constituted ‘IFN-I regulator module 1’, and IRF1, IRF2, IRF4, STAT1, STAT3, ARID5A, ARID5B, TCF7, PRDM1, PRDM1L, KLF5, and TCF7L constituted a distinct ‘IFN-I regulator module 2’. To visualize the contribution of the selected genes to the PCs and the directionality of the contribution, a PCA biplot analysis was adopted. In these biplots, the loadings of each variable (gene) are represented as arrows and the changes of gene expression by perturbation are represented as points, providing a faithful representation of the relationships among each gene (arrows) based on the contribution to the effect of each perturbation. We found that ISGs are divided into two groups: classical ISGs that are correlated with ‘IFN-I regulator module 1’ (depicted in orange arrows in Figure 3e), and the other ISGs that are correlated with ‘IFN-I regulator module 2’ (depicted in green arrows in Figure 3e, Extended Data 6b), which is predominantly explained by PC1. These results suggest that ISGs are bi-directionally regulated by different modules of TFs. Furthermore, these modules contributed differently to the regulation of co-inhibitory receptors; TIGIT, CD160, BTLA as one module and LAG3, HAVCR2, PDCD1 as another module (Figure 3f, Extended Data 6c). Within ‘IFN-I regulator module 1’, STAT3 positively regulated HAVCR2 but not LAG3 and PDCD1 expression, which is predominantly contributed by PC2 in the biplot (Figure 3f). Taken together, our perturbation with the Vpx-VLP system elucidated two distinct TF modules simultaneously regulating opposing IFN-I target genes, which sheds light on the central roles of non-canonical IFN-I induced regulators in human T cells.
Given that mRNA expression of TFs is not necessarily linked with the regulatory function of TFs, we performed footprint analysis on our ATAC-seq data by using TOBIAS30. IFN-β treatment enhanced the enrichment of IRFs and STATs footprints at the early and intermediate phases, in contrast, AP-1 footprints were highly enriched at the late phase by IFN-β (Extended Data 6d). This dynamic switch of TF footprints from IRFs/STATs to AP-1 family under IFN-I response represents the two differential regulatory modules indicated as ‘IFN-I regulator module 1’ (containing IRF1/2/4/9 and STAT1–3) and ‘IFN-I regulator module 2’ (containing AP-1 family TFs: BATF and BATF3) (Figure 3g, h). In light of the role of AP-1 facilitating human T cell exhaustion with higher PD-1 and LAG-3 expression31, the enhanced AP-1 footprints at the later phase of IFN-β treatment supports the role of IFN-I on induction of T cell exhaustion in human T cells11, 12. Taken together, the perturbation of key regulatory factors with ATAC-seq footprint analysis revealed two different regulator modules that have different temporal characteristics of activation and differentially control ISGs and co-inhibitory receptors under IFN-I response.
Dynamic regulatory network reveals central IFN-I regulators
To characterize the impact of differentially expressed transcription factors (DETFs) in response to IFN-β, we generated transcriptional regulatory networks describing TFs and their target genes for each of the transcriptional waves identified (Figure 4a, Methods). When comparing the three regulatory networks, early and late networks had similar numbers of TFs (46 and 42 TFs, respectively), although the intermediate network contained 73 TFs (Figure 4b, top). The ratio between up and down-regulated TFs differs between the three regulatory waves. The early and intermediate networks contained more up-regulated TFs than down-regulated TFs; in contrast, the late network had more down-regulated than up-regulated TFs. Thus, IFN-I induced differentiation involves dominance of up-regulated TFs in the first 16 hours, replaced by the dominance of down-regulated TFs after 48 hours. We next ranked the TFs based on the enrichment of their target genes and their centrality in the networks (Methods), highlighting the significance of each TF to the network (Figure 4b, bottom). STAT1/2, which are well established as being directly downstream of IFN-I signaling, were highlighted as one of the top ranked up-regulators in the early and intermediate wave networks (Figure 4b, Extended Data 4c). Of note, STAT1 and STAT2 showed contrastive scores with Hypergeometric test (HG) and Centrality (Cent) 32; STAT1 was higher in Centrality and lower in Hypergeometric scores. In contrast, STAT2 was higher in Hypergeometric and lower in Centrality scores. These data suggest that STAT2 is specialized to control IFN-I regulatory TFs at early and intermediate phases, while STAT1 is less specific to IFN-I signaling but plays a more general role in T cell survival and memory formation 26, 33. In the late network, effector function related regulators were upregulated (TBX21, HIF1A, FOSL1/2, MAF); in contrast, the TFs associated with Treg differentiation and maintenance (IKZF2, MYB, FOXP1, STAT5A) were down-regulated, suggesting the skewed differentiation toward an effector-like signature.
Figure 4. Transcriptional regulatory network under IFN-I response.
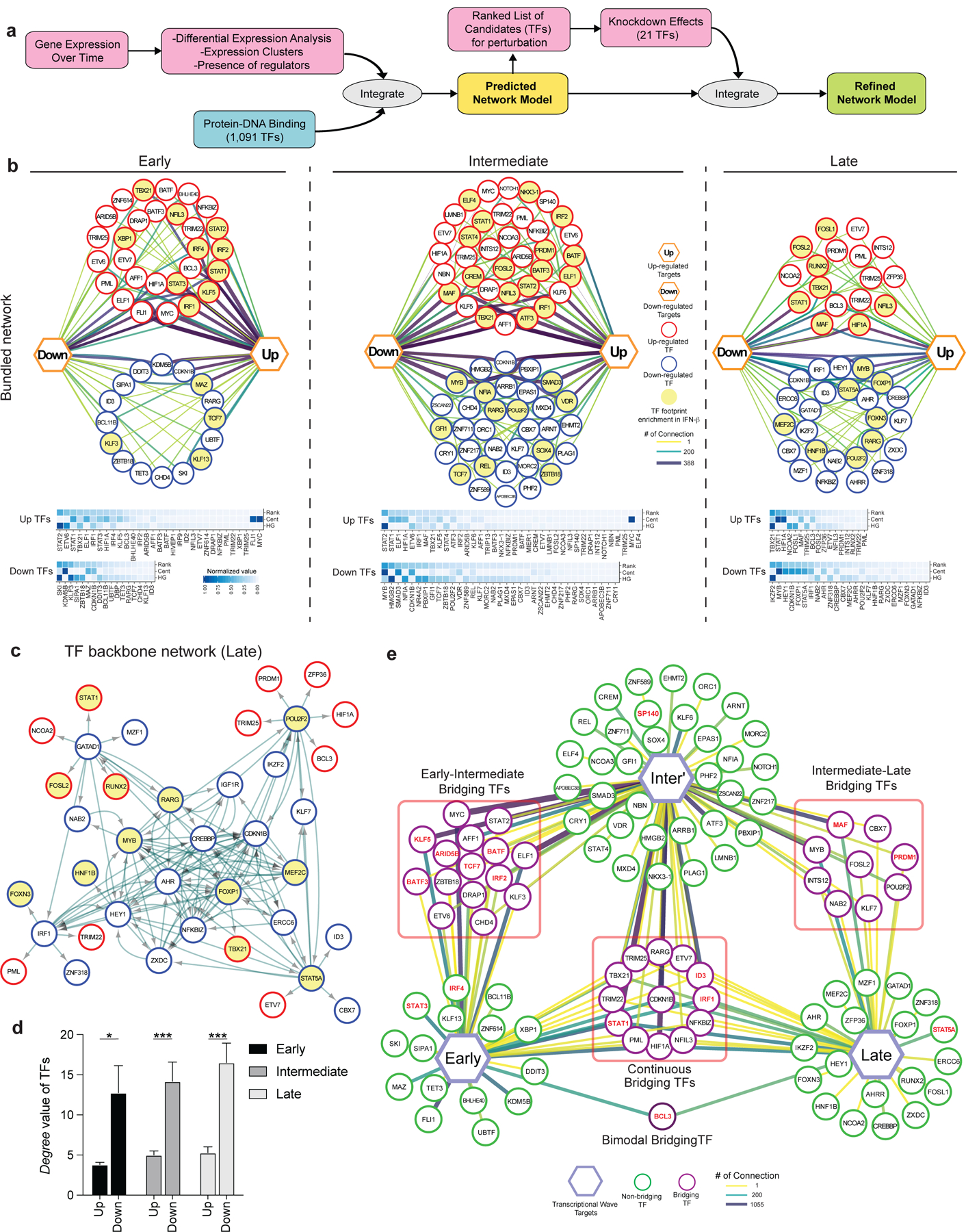
a, Overview of regulatory network generation. b, In depth view of the transcriptional regulation at each transcriptional wave. Top row; the representation of regulatory networks highlighting TFs interaction. The thicker and darker an edge is the more TF-target connections it represents. Target genes are represented by up and down hexagons, according to their response to IFN-β. Middle row: heatmaps representing a ranking of the TFs based on their centrality, connectivity and gene-target enrichment in the corresponding regulatory network. ‘Cent’ stands for centrality, which is a parameter that is given to each node, based on the shortest path from the node to the other nodes in the network. It represents how central and connected a node is to the rest of the network. ‘HG’ stands for hyper-geometric, the value in the heatmap is the of a hypergeometric enrichment test of target genes of each TF in the network. The rank column is an average of both HG and Cent values, after score rescaling (0–1). c, Directed TF backbone networks at the late wave. The regulation and interaction of each DE TFs at each transcriptional wave were depicted. b,c, Red circles represent up-regulated TFs, blue circles represent down-regulated TFs, and the arrows represent the direction of regulation from TF to TF. TF footprints enriched in IFN-β condition were highlighted in yellow. d, Bar plot show the mean level of the degree of connectivity values for either up or down regulated DE TFs. p values were calculated by an independent two-way t-test. *p < 0.05, ***p < 0.001. e, Dynamics of TFs regulation across the transcriptional waves. Each hexagon represents targets from each transcriptional wave. Green circles represent regulatory TFs which are differentially expressed only in one transcriptional wave they are connected to, while purple circles represent bridging TFs, which are differentially expressed in all transcriptional waves they are connected to. Perturbed TFs in Figure 3 were highlighted in red. The thicker and darker an edge is, the more TF-target connections it represents.
To further study the relationships between these IFN-I driven transcriptional regulators, we generated directed backbone networks that displayed the TFs that participate in each transcriptional time wave and visualized the interactions among them using the ECOplusMaST algorithm32 (Figure 4c, Extended Data 7a, Methods). This analysis emphasizes that much of the signaling involves cross regulation between the TFs themselves. The central regulatory TFs, which are represented as more connected with the other TFs, were dominated by down-regulated TFs in response to IFN-β (Figure 4d) in all temporal transcriptional networks. In contrast, TFs less central were primarily up-regulated. Interestingly, we found TGF-β associated TFs (i.e. SKI, SMAD3, IKZF2) and gene signatures enriched in the downregulated genes at each time wave (Figure 4b; bottom heatmap, Extended Data 7b), indicating counter regulatory mechanisms between IFN-I response and TGF-β signaling in human T cells 34, 35. These results suggest that a loss of suppression by the key TFs that configure the backbone network may trigger the activation of downstream effector TFs under IFN-I response.
Although T cell differentiation under IFN-β is characterized by three major transcriptional waves, we hypothesized that there are key TFs that bridge each wave to the next. To this end, we specifically identified TFs that participate in more than one of these transcriptional waves, and termed these ‘Bridging TFs’ (Figure 4e). Examples of dominant Bridging TFs between early and intermediate waves include KLF5 and STAT2. Examples of intermediate to late waves include MAF, PRDM1, and MYB. Finally, there are TFs that were upregulated throughout the entire differentiation; such as STAT1, HIF1A, and TBX21. Generally, Bridging TFs tend to be more dominant than other TFs; thus, it is possible that Bridging TFs play an important role in the transition between different transcriptional waves. Indeed, our perturbation experiment demonstrated the critical roles of those Bridging TFs in the regulation of ISGs and co-inhibitory receptors (Figure 4e). Our computational analysis revealed the temporal dynamics of complex regulatory interactions during the IFN-I response and highlighted the usefulness of our approach in discovering this new aspect of IFN-I induced transcriptional regulation.
In vivo link of IFN-I regulators and co-inhibitory receptors
To provide direct in vivo evidence for the role of IFN-I on T cell co-inhibitory receptor expression, we sought to validate our regulatory network in humans where the IFN-I response of T cells is induced acutely. As acute viral infections are strongly associated with IFN-I responses, we examined a number of clinical models where viral infection is closely linked to IFN-I T cell response. Our analysis of single cell RNA seq (scRNA-seq) data of T cells in COVID-19 patients revealed an extremely high correlation between viral load and IFN-I score (r=0.8), and time difference between paired samples and the respective change in IFN-I score (r=0.97) 36, providing a unique opportunity to generate a rich dataset to determine whether the in vitro T cell response to IFN-I can be validated during an acute viral human infection strongly associated with a IFN-I signal.
By using our scRNA-seq data, we subclustered T cell populations into 13 subpopulations and identified five CD4+ T cell and five CD8+ T cell subsets (Figure 5a, Extended Data 8a, b). We first focused on total CD4+ T cells and CD8+ T cells and confirmed that the IFN-I response signature is higher in progressive patients who required admission to the ICU and eventually succumbed to the disease (Figure 5b). Expression of co-inhibitory receptors differed across disease conditions, but the trend was conserved between CD4+ and CD8+ T cells. We observed a strikingly similar pattern of co-inhibitory receptor expression with IFN-I stimulation in vitro and in vivo. Indeed, we observed the upregulation of ‘IFN-I up co-inhibitory receptors’ (LAG3/HAVCR2) and the downregulation of ‘IFN-I down co-inhibitory receptors’ (TIGIT/LAIR1/SLAMF6) in T cells from COVID-19 patients (Figure 5c, d). As expected, ‘IFN-I up co-inhibitory receptors’ were positively correlated with expression of canonical ISGs, but ‘IFN-I down co-inhibitory receptors’ were not, suggesting that different regulatory mechanisms are dictating co-inhibitory receptor expression patterns (Figure 5e). Next, we investigated which subpopulation of CD4+ and CD8+ T cells is more affected by the IFN-I response and computed the IFN-I score across subpopulations for each T cell subtype in COVID-19 patients (Figure 5f). Within the subpopulations that exhibited higher IFN-I scores, dividing CD4+/CD8+ T cells and ISG+ CD8+ T cells were increased in COVID-19 patients, particularly in severe patients 36 37. Moreover, these subpopulations and effector T cells expressed higher level of co-inhibitory receptors compared to the other subpopulations (Figure 6a, Extended Data 8c). Of note, the ‘IFN-I regulator module-1’ score was higher in these T cell subsets than the ‘IFN-I regulator module-2’ score (Extended Data 8d), implying a more profound role of ‘IFN-I regulator module 1’ on controlling T cell signature in COVID-19.
Figure 5. Linked IFN-I response and co-inhibitory receptor expression in T cells in COVID-19.
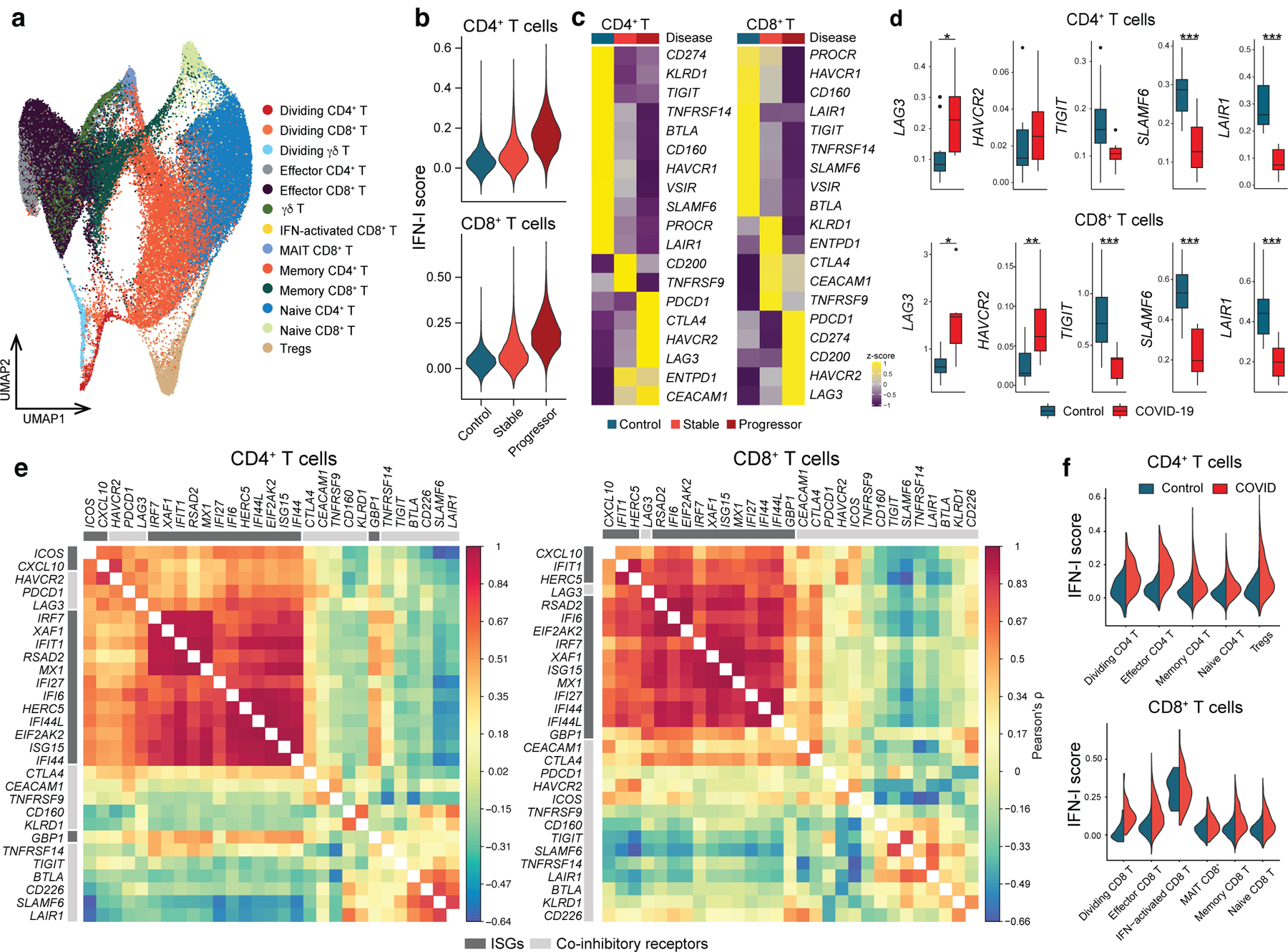
a, UMAP representation of T cells from healthy control samples (n = 13; biologically independent samples) and COVID-19 samples (n = 18; biologically independent samples). 13 subcluster were identified. b, IFN-I score for CD4+ and CD8+ T cells across the three disease conditions. c, Heatmaps for co-inhibitory receptors expression in CD4+ and CD8+ T cells across the three disease conditions. d, Expression of key co-inhibitory receptors between control vs COVID-19 for CD4+ and CD8+ T cells. Average expression per subject for each gene is shown. *p < 0.05, **p < 0.01, ***p < 0.001. Two-sided Kruskal-Wallis test. e, Correlation matrix of ISGs (dark gray) and co-inhibitory receptors (light gray) in CD4+ and CD8+ T cells in COVID-19 patients. f, IFN-I score for subsets of CD4+ and CD8+ T cells between control vs COVID-19.
Figure 6. Integration of IFN-I regulatory network with T cell signature in COVID-19.
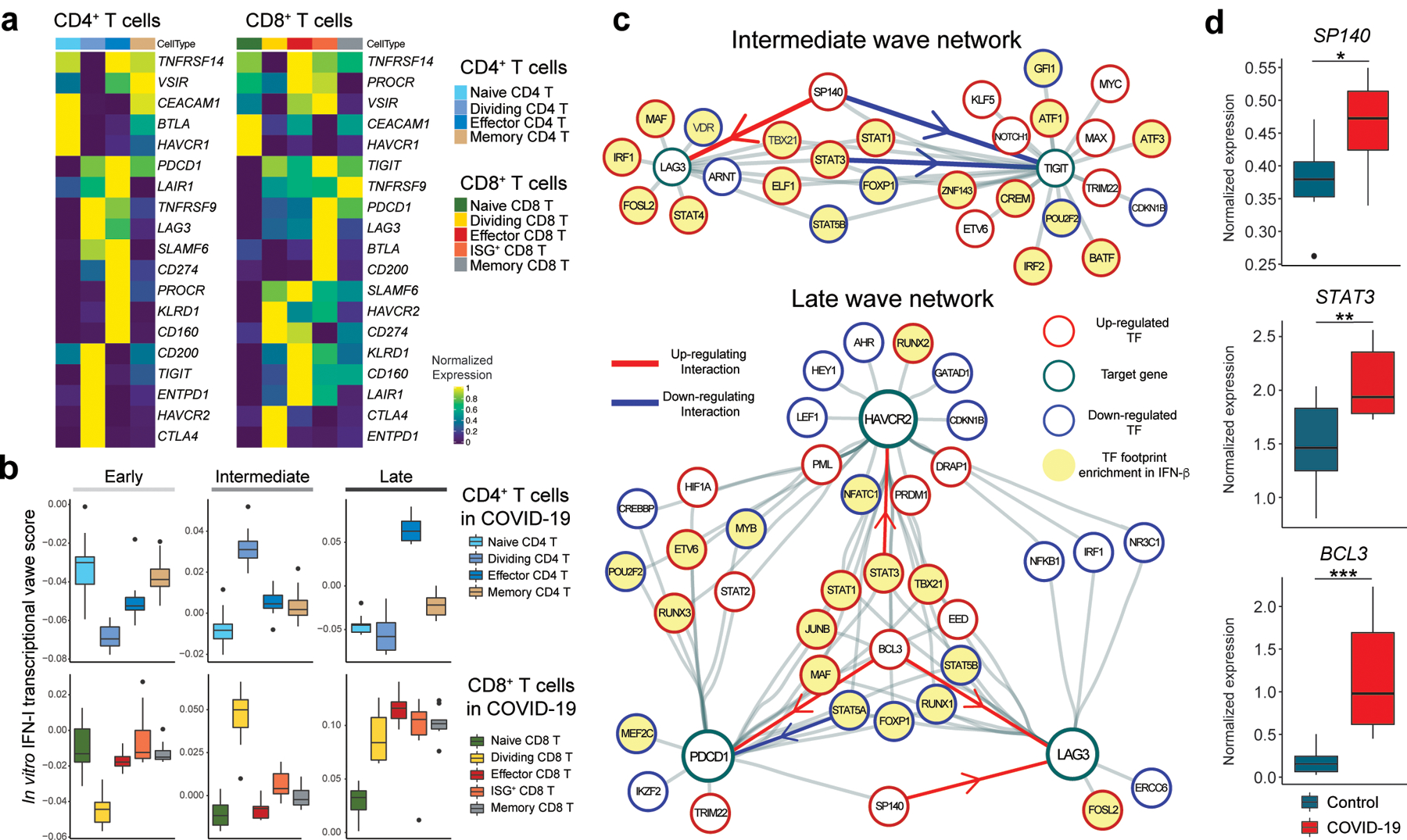
a, Heatmap showing co-inhibitory receptors expression for subsets of CD4+ and CD8+ T cells in COVID-19. b, Computed three transcriptional waves (early, intermediate, and late) score for the subsets of CD4+ and CD8+ T cells in COVID-19 patients. Scores were calculated based on upregulated DEGs of CD4+ and CD8+ T cells for each transcriptional wave. c, Regulatory relationship between regulators in intermediate phase network for LAG3 and TIGIT (top) and late phase network for LAG3, HAVCR2, and PDCD1 (bottom) are shown. Positive regulation (TF to target) is highlighted in red and negative regulations in blue. TF footprints enriched in IFN-β condition were highlighted in yellow. d, Box plots showing expression of key regulators between control vs COVID-19 for CD4+ T cells. Average expression per subject for each gene is shown. *p < 0.05, **p < 0.01, ***p < 0.001. Two-sided Kruskal-Wallis test.
We then examined which subpopulations were more enriched in the three transcriptional waves of IFN-I response. DEGs specific for each wave were used to compute the scores for the CD4+ and CD8+ T cell subpopulations in COVID-19 patients (Supplementary Table 1, Methods). We found that T cells induced in vitro with IFN-I strongly mirrored the intermediate wave score on dividing CD4+ and CD8+ T cells, and the late wave score on effector CD4+ T cells and ISG+ CD8+/effector CD8+ T cells (Figure 6b) in COVID-19 patients. Given that expansion of dividing CD4+/CD8+ T cells are a unique characteristic of acute viral infections, including COVID-19, we applied the intermediate phase IFN-I regulatory network with the dividing CD4+ T cell gene expression signature to examine the relationships between regulators and target genes in this subpopulation. This analysis highlights the regulators that function in establishing the characteristics of the dividing CD4+ T cell population under IFN-I response in vivo (Extended Data 8e). As LAG-3 is the most upregulated co-inhibitory receptor in the dividing CD4+ T cell population, we utilized the network analysis to elucidate the specific regulation of LAG3 and TIGIT that were regulated in an opposing manner under IFN-I response both in vivo (Figure 5c, d) and in vitro (Figure 1). Analysis of the intermediate wave gene regulatory network demonstrated that SP140 is a bi-directional regulator for LAG3 and TIGIT under IFN-I response, which is supported by the observation in COVID-19 patients, where increased SP140 and LAG3 but decreased TIGIT expression were demonstrated (Figure 5c, d, Figure 6c, d, Extended Data 8c). In addition, the late wave network demonstrated the complex interaction of regulators for LAG3, HAVCR2, and PDCD1, in which BCL3 and STAT3 are highlighted as positive regulators on LAG3 and HAVCR2 respectively (Figure 6c). Importantly, both BCL3 and STAT3 were significantly elevated in T cells in COVID-19 patients (Figure 6d). This can be highly relevant to effector T cell development under acute viral infection in which these co-inhibitory receptors play a critical role in regulating effector function38 and, of note, the late wave signature was enriched in effector T cells in COVID-19 patients (Figure 6b). Finally, we validated the actions of these key regulators on major co-inhibitory receptor expressions (LAG-3, PD-1, TIM-3, and TIGIT) at the protein level with our in vitro system, where T cell proliferation was further controlled by determining T cell division state in parallel with gene knockdown (Extended Data 9a, b). Additionally, STAT5A overexpression suppressed PD-1 induction under IFN-β treatment in human primary CD4+ T cells, which validated the observation that STAT5A negatively regulates co-inhibitory receptors, and its downregulation is necessary to elicit IFN-I mediated PD-1 expression (Figure 6e, Extended Data 9c). We also confirmed that T cell activation was not significantly altered by gene perturbation across conditions (Extended Data 9d), suggesting that our in vitro perturbation system with shRNA-mediated gene knockdown was not confounded by T cell activation or proliferation, two well-known factors linked with co-inhibitory receptor expression. Taken together, these findings strongly suggest that in vitro regulatory networks can be utilized as a powerful tool to explore the human acute viral response in vivo.
Discussion
Here, our systematic, computational and biological approach identifies IFN-I as a major driver of co-inhibitory receptor regulation in human T cells. While classical ISG induction has been extensively studied, those investigations have focused primarily on the canonical JAK-STAT pathway downstream of IFN-I receptor. Given that IFN-I exhibits multiple functions in context-dependent roles, a more complex understanding of the IFN-I response beyond this canonical pathway with a more extensive analysis of ISG transcriptional regulation in T cells is critical for elucidating the mechanism of co-inhibitory receptor regulation.
In this study, we comprehensively explored the induction of co-inhibitory receptor expression with IFN-I, elucidating previously unrecognized patterns of T cell co-inhibitory receptors. Although previous studies have shown that PD-1 is induced by IFN-I in T cells using mouse models of viral infection39 and the blockade of IFN-I restores T cell effector capacity with reduction of PD-1 and TIM-3 in the chronic HIV infection model16, the full spectrum of co-inhibitory receptor expression controlled by IFN-I in the context of human diseases has not been investigated. Here, we build a dynamic gene regulatory network that controls IFN-I response, and identify key regulatory modules of ISG transcription in T cell responses to IFN-β. Our approach revealed two mutually antagonistic modules of ISG regulators, which exhibit opposite temporal kinetics under IFN-β treatment, may explain how the harmonized IFN-I induced T cell response is achieved. Within the two modules, we highlighted SP140 as a potential regulator that controls LAG-3 and TIGIT in an opposing manner, and STAT3 as a positive regulator for TIM-3. SP140 might have a DNA binding motif similar to AP-140, which supports our findings of SP140 as one of ‘IFN-I regulatory module 1’ TFs that contains AP-1 family TFs. These findings provide novel insight into the landscape of the ISG transcriptional network, and sheds light on the large contribution of the noncanonical IFN-I pathway during IFN-I response in T cells 37, 41, 42. Although the newly identified regulators (e.g., SP140, BCL3) in this study are not necessarily directly downstream of the conventional JAK/STAT pathway and may act differently depending on the context, they are nevertheless attractive targets for manipulation of specific downstream functional molecules such as co-inhibitory receptors in T cells. One of the limitations in this study is that although our regulatory network takes into account all differentially expressed genes by IFN-β treatment, which were further ranked the key regulatory factors for the enrichment score under treatment with IFN-β, it does not exclude the possibility that they play an important role regulating certain genes or perhaps the same genes under other conditions.
We demonstrate the relevance of our in vitro T cell IFN-I response to humans by integrating scRNA-seq data from COVID-19 patients, where a predominant T cell IFN-I response was observed. Intriguingly, the expression pattern of co-inhibitory receptors on T cells in vitro are highly replicated in severe COVID-19 cases, and classical ISGs were well correlated with one module of co-inhibitory receptors (LAG3/PDCD1/HAVCR2), but not with the other modules (TIGIT/CD160/BTLA/LAIR1). While dynamics of IFN-I on T cells from COVID-19 patients should be considered with higher temporal profiling, we confirmed that the IFN-I response was clearly reduced at a later point; thus, our data based on an earlier collection of blood should reflect the active ISG transcriptomics during human acute viral response. Given the IFN-I response has been shown to contribute to chronic viral infection and cancer, the novel regulators that we identified can be examined in these diseases, as in vitro T cell culture systems only partially model chronic infection or cancer milieu in vivo. Further investigations of factors that characterize acute viral infections are likely to differ from chronic viral infections and will be of interest to explore in the context of long-term human infections not well modeled by COVID-19. Finally, we have previously shown that TIM-3 is decreased on T cells in patients with multiple sclerosis. As predicted based on our investigations, TIM-3 expression increased on T cells in MS patients after IFN-β treatment43. Further studies in COVID-19 patients who are treated with IFN-β may provide further insight into the functional relevance of T cell IFN-I response in COVID-19 pathogenesis44, 45.
In conclusion, our systems biology approach identified cytokine signals and regulatory mechanisms that drive the expression of co-inhibitory receptors in humans, and provided a pathway to comprehensively capture the dynamics of their expression in humans. Our results will advance the understanding of the host immune response to a variety of viral infections and could serve as a resource for mining of existing datasets. Uncovering novel ISG regulators controlling co-inhibitory receptors will create a foundation for further development of new therapeutics for a multitude of different malignant and infectious diseases.
Methods
Study subjects
Peripheral blood was drawn from healthy controls who were recruited as part of an Institutional Review Board (IRB)-approved study at Yale University, and written consent was obtained. All experiments conformed to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
Human T cell isolation and culture
Peripheral blood mononuclear cells (PBMCs) were prepared from whole blood by Ficoll gradient centrifugation (Lymphoprep, STEMCELL Technologies) and used directly for total T cell enrichment by negative magnetic selection using Easysep magnetic separation kits (STEMCELL Technologies). Cell suspension was stained with anti-CD4 (RPA-T4), anti-CD8 (RPA-T8), anti-CD25 (clone 2A3), anti-CD45RO (UCHL1), anti-CD45RA (HI100) and anti-CD127 (hIL-7R-M21, all from BD Biosciences) for 30 minutes at 4°C. Naïve CD4+ T cells (CD4+/CD25neg/CD127+/CD45ROneg/CD45RA+) and naïve CD8+ T cells (CD8++/CD25neg/CD127+/CD45ROneg/CD45RA+) were sorted on a FACSAria (BD Biosciences). Sorted cells were plated in 96-well round-bottom plates (Corning) and cultured in RPMI 1640 medium supplemented with 5 % Human serum, 2 nM L-glutamine, 5 mM HEPES, and 100 U/ml penicillin, 100 μg/ml streptomycin, 0.5 mM sodium pyruvate, 0.05 mM nonessential amino acids, and 5% human AB serum (Gemini Bio-Products). Cells were seeded (30,000–50,000/wells) into wells pre-coated with anti-human CD3 (2 μg/ml, clone UCHT1, BD Biosciences) along with soluble anti-human CD28 (1μg/ml, clone 28.2, BD Biosciences) in the presence or absence of human IFN-β (500 U/ml: Pestka Biomedical Laboratories) or IL-27 (100 ng/ml: BioLegend) without adding IL-2. To avoid interindividual batch effects, T cells used for RNA-seq and ATAC-seq were isolated from one individual (37-year-old, male, healthy individual with no evidence of viral infection, cancer, and autoimmune diseases at the time of sample collection).
Lentiviral and Vpx-VPLs production
Lentiviral plasmids encoding shRNA for gene knockdown or open reading frame (ORF) of STAT5A for overexpression were obtained from Sigma-Aldrich (MISSION shRNA, Sigma) and Horizon Discovery Biosciences (Precision LentiORF), respectively. Scramble shRNA (Sigma; for perturbation experiment) or GFP control vectors (Origene; for protein validation experiment, Horizon Discovery Bioscience; for overexpression experiment) are used as the controls for lentiviral transduction. Each plasmid was transformed into One Shot Stbl3 chemically competent cells (Invitrogen) and purified by ZymoPURE plasmid Maxiprep kit (Zymo research). Lentiviral pseudoparticles were obtained after plasmid transfection of 293T cells using Lipofectamine 2000 (Invitrogen) or TurboFectin 8.0 Transfection Reagent (Origene). To prepare Vpx-VLPs, 293T cells were co-transfected by Lipofectamine 2000 or TurboFectin 8.0 Transfection Reagent with the 5 μg pMDL-X, 2.5 μg pcRSV-Rev, 3.5 μg X4-tropic HIV Env, and 1 μg pcVpx/myc, as described previously with some modifications37,38. The medium was replaced after 6–12 h with fresh media with 1X Viral boost (Alstem). The lentivirus or Vpx-VLPs containing media was harvested 72 h after transfection and concentrated 80 times using Lenti-X concentrator (Takara Clontech) or Lenti Concentrator (Origene). LV particles were then resuspended in RPMI 1640 media without serum and stored at −80°C before use. Virus titer was determined by using Jurkat T cells and Lenti-X GoStix Plus (Takara Clontech).
Lentiviral transduction with Vpx-VPLs
Two step Vpx-VLP and LV transduction was performed as described previously with some modiciations36. Vpx are pseudotyped with X4-tropic HIV Env to promote efficient entry of Vpx-VLPs into quiescent human T cells37. FACS-sorted naïve CD4+ T cells were plated at 50,000 cells/well in round bottom 96 well plate and chilled on ice for 15 min. 25–50 μl of Vpx-VLPs were added to each well and mixed with cold cells for an additional 15 min, then spinfected with high-speed centrifugation (1200 g) for 2 hour at 4 °C. Immediately after centrifugation, cells are cultured overnight at 37 °C. Vpx-transduced cells are spinoculated again with LV particles containing shRNAs or ORF with high-speed centrifugation (1000 g) for 1.5 hours at RT. After 24 hours of incubation, the second transduction of LV particles with shRNAs or ORF were performed as well as the first time spinoculation. After a second LV transduction, cells were washed and plated into 96-well round-bottom plates pre-coated anti-human CD3 (2 μg/ml) with soluble anti-human CD28 (1 μg/ml), in the presence or absence of human IFN-β (500 U/ml). Cells are collected at day 4–6 after anti-CD3/CD28 stimulation and GFP positive cells were sorted by FACSAria or analyzed by Fortessa.
Real time quantitative PCR
Total RNA was extracted using RNeasy Micro Kit (QIAGEN), or ZR-96 Quick-RNA kit (Zymo Research), according to the manufacturer’s instructions. RNA was treated with DNase and reverse transcribed using TaqMan Reverse Transcription Reagents (Applied Biosystems) or SuperScript IV VILO Master Mix (Invitrogen). cDNAs were amplified with Taqman probes (Supplementary Table 2) and TaqMan Fast Advanced Master Mix on a StepOne Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. Relative mRNA expression was evaluated after normalization with B2M expression.
Flow cytometry analysis
Cells were stained with LIVE/DEAD Fixable Near-IR Dead Cell Stain kit (Invitrogen) and surface antibodies for 30 min at 4 °C. For intracellular cytokine staining, cells were treated with 50 nM phorbol-12-myristate-13-acetate (MilliporeSigma) and 250 nM ionomycin (MilliporeSigma) for 4 hours in the presence of Brefeldin A (BD Biosciences) before harvesting. Cells were washed and fixed with BD Cytofix™ Fixation Buffer (BD Biosciences) for 10 min at RT, then washed with PBS. Intracellular cytokines were stained in permeabilization buffer (eBioscience) for 30 min at 4 °C. Antibody details are provided in Supplementary Table 3. Cells were acquired on a BD Fortessa flow cytometer with FACSDiva (BD Pharmingen) and data was analyzed with FlowJo software v10 (Threestar).
RNA-seq library preparation and data analysis
10,000 cells per condition were subjected to RNA-seq. cDNAs were generated from isolated RNAs using SMART-Seq v4 Ultra Low Input RNA Kit for sequencing (Takara/Clontech). Barcoded libraries were generated by the Nextera XT DNA Library Preparation kit (Illumina) and sequenced with a 2×100 bp paired-end protocol on the HiSeq 4000 Sequencing System (Illumina).
After sequencing, adapter sequences and poor-quality bases (quality score < 3) were trimmed with Trimmomatic. Remaining bases were trimmed if their average quality score in a 4 bp sliding window fell below 5. FastQC was used to obtain quality control metrics before and after trimming. Remaining reads were aligned to the GRCh38 human genome with STAR 2.5.246. We used Picard to remove optical duplicates and to compile alignment summary statistics and RNA-seq summary statistics. After alignment, reads were quantitated to gene level with RSEM 47 using the Ensembl annotation. Normalized Fragments Per Kilobase Million (FPKM) expression values of the three replicates were used for the analysis below.
Identification of three transcriptional waves
The correlation matrix is created by Pearson correlating 48 the IFN-β expression profile of each time point with all the other time points, creating a symmetric matrix of Pearson correlation coefficients.
Differential expression calculation
The differential expression (DE)49 for each timepoint was calculated using the DEseq2 50 R package. Three separate testing methods were used for calculating DE genes: Wald 51, likelihood ratio test (LRT) 52, and time-course 53. For each of the three methods, genes with false discovery rate (FDR) adjusted P.value bellow 0.05, were regarded as DE. More specifically, for CD4+ T cells, if TFs appeared in two out of the three calculating methods (agree by two), they were regarded as DE. For CD8+ T cells, if TFs appeared in any of the methods above, they were regarded as DE.
Selection of regulators for perturbation
The list of TFs for perturbation was selected based on following criteria: 1) overlapped differentially expressed TFs across the time points between CD4+ and CD8+ T cells based on the above-mentioned method. Intersection of DETFs in our in vitro data were chosen; 2) differentially expressed TFs in human tumor infiltrated T cells22, 23, 24, 25 that were significantly correlated with exhausted T cell cluster (defined by the upregulation of LAG-3/PD-1/TIM-3). ‘Human TIL score’ – defined as the number of times a gene was shared between the four different human cancer TIL datasets; 3) HIV specific T cell signature upregulated in progressive patients compared to stable patients26; 4) TFs that were induced by IL-27 and categorized as IL-27 driven co-inhibitory receptor modules1. ‘Human ISG score’ – defined as the number of times it was shared between the three different categories (T cells, PBMCs, and all immune cells) of human ISGs identified by Interferome database. All perturbed TFs were confirmed as IFN-I induced genes and showed ‘human ISG score’ greater than 1.
Expression data of all time points under IFN-β treatment expression was divided by the control expression data, creating fold-change ratio data of treatment over control. For visualization purposes, the fold change data in figure 2a was then shown for the DE genes only, logged with a base of 10, and scaled per row. The results were clustered hierarchically. In Figure 2c specific genes of interest were picked, such as ISGs, co-inhibitory receptors, T cell related genes, and transcription factors.
Heatmap of perturbed TFs
21 TFs were perturbed using lentiviral shRNA, together with a scramble shRNA (SCR) as control. ISGs (IFN Score B54) and co-inhibitory receptors were selected to visualize the expression differences in the two modules, in Extended Data 5b and c, respectively. The effect of TF perturbation on IFN-β response for each gene of interest (GOI) was calculated as follows. For each shRNA perturbation, the effect of IFN-β treatment over control condition was calculated for GOIs. This fold change value was then logged with a base of 2. To normalize the effect of the shRNA protocol, we further normalized the data by the scrambled shRNA (SCR), in an identical manner to the shRNA perturbation described above. Finally, the log2 transformed shRNA values were subtracted by the log2 transformed SCR values as shown in the equation bellow:
For the heatmap visualization, the normalized values calculated by the equation above were clustered hierarchically.
Differential expression analysis for each TF perturbation was conducted as follows: Perturbed TF+IFN-β treatment expression vs Perturbed TF untreated expression. DE genes in this analysis that were regarded as significant (FDR adjusted P.value lower than 0.05) were marked with a white plus sign on their tile.
PCA and PCA biplot analysis
The PCA was applied to the DE genes (defined above) as variables, and perturbed genes as observations. The data was normalized by the scramble shRNA control, in the same manner as the Heatmap of perturbed TFs. Although PDCD1 was not defined as a DE gene across the time course, it was differentially expressed at later time points in mRNA-seq and qPCR, thus we manually added it to Figure 5f. Genes of IFN Score B54 (see Supplementary Table 1) were represented as ISGs in Figure 3e. The PCA and biplot analysis were calculated and visualized using the R package FactoMineR55.
Regulatory networks
Following the DE analysis in each transcriptional wave, the DE genes were defined as TFs and their targets (from->to). The targets of all DE TFs were determined using ChIP-seq data from the database GTRD56. TFs and target genes defined as DE were added to the network as nodes (circles), and edges (lines) were added between them. The network plots were created using the Cytoscape57 software.
Top Regulatory TFs Heatmaps
We ranked the DE TFs of each transcriptional time wave to identify the dominant regulators in each stage of the differentiation process using two methods. Hyper-geometric (HG), were the values in the heatmap are the log P. value of the hypergeometric enrichment test . The HG calculation was conducted using the python SciPy package58. The second method was network centrality (Cent), which is a parameter that is given to each node, based on the shortest path from the node to the other nodes in the network. It represents how central and connected a node is to the rest of the network59. The centrality calculation was conducted using the python NetworkX60 package. The rank column is an average of both HG and Cent values, after scaling to the range of 0 to 1.
Integration of Perturbation Data to Regulatory Networks
Following the generation of the regulatory network mentioned above, we further refined the network by integrating the TF perturbation data. Genes that were significantly affected by a TF perturbation were added as a “validated” edges between the perturbed TF and the target gene. If a gene was up-regulated by a TF perturbation, the interaction between them is registered as down-regulation. If a gene was down-regulated by a TF perturbation, the interaction between them is registered as up-regulation.
Reduced TF networks
For visualization purposes we used a network reduction algorithm. The algorithm presents all the TFs in the network (nodes) but reduces some of the connections between them (edges). The reduced TF networks were calculated using the NetworkToolbox61 package in Rstudio. The method of the reduction calculation was ECOplusMaST32, which applies the efficiency cost optimization (ECO) neural network filtering method combined, with the Maximum Spanning Tree (MaST)62 filtering method.
The bar plots depicted at the bottom represent the mean level of the degree63 value for either up or down regulated TFs, in the complete networks (not reduced ones). The degree of a node is the number of connections it has to other nodes. An independent two-way t-test was conducted between the degree values of the up and down TFs for each time wave. The P value of each test is presented in the caption of each bar plot.
Bridging TFs Network
DE TFs and their target genes from the three transcriptional waves were combined to create a comprehensive network emphasizing the dynamics between transcriptional waves. In this network TFs and their target genes were annotated by the transcriptional wave in which they appear as DE. TFs that participate in more than one transcriptional wave are regarded as bridging TFs.
ATAC-seq library preparation and data analysis
Cells were split into RNA-seq (10,000 cells) and ATAC-seq (15,000 cells) at the time of collection. Cells for ATAC-seq were cryopreserved by using Recovery Cell Culture Freezing Medium (Gibco) with concentration of 15,000 cells/100 μl. Cryopreserved cells were thawed and immediately proceeded to Tn5 digestion (50 μl of transposase mixture: 25 μl of 2x TD buffer, 2.5 μl of TDE1 (both from Illumina), 0.5 μl of 1% digitonin (Promega), 22 μl of nuclease-free water)64. Transposition reactions were incubated at 37°C for 30 minutes in a thermal shaker with agitation at 300 RPM. Transposed DNA was purified using a Zymo DNA Clean and Concentrator-5 Kit (Zymo research) and purified DNA was eluted in 10 μl elution buffer. Transposed fragments were amplified and purified as described previously64 with modified primers65. Libraries were quantified using qPCR (KAPA Library Quantification Kit) prior to sequencing. All ATAC libraries were sequenced using a 2×150 bp paired-end protocol on the Nova-seq Sequencing System (Illumina).
ATAC-seq Alignment, Feature Calling and Quantitation
We trimmed adapter sequences before aligning reads to the GRCh38 human genome assembly with Bowtie 2. We used FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to obtain sequence quality control metrics both before and after read trimming. We obtained alignment summary metrics with Picard tools (http://broadinstitute.github.io/picard). We retained reads that mapped uniquely to the primary assembly with MAPQ >= 30 and re- moved optical duplicates. We shifted cut sites by +4bp and −5bp on positive and negative strands, respectively.
We used MACS266 to call ATAC-seq peaks within individual samples using a q < 0.2 threshold. For each library, we use ataqv67 to calculate TSS enrichment and the fraction of reads in peaks (FRiP). We excluded peaks within ENCODE blacklist regions.
We used Markov clustering68 to identify a preliminary set of consensus features across libraries. Edges were weighted by the proportion of overlapping bases between each pair of peaks. Peaks that did not overlap with at least one peak from another library were excluded from further analysis. After clustering, we defined preliminary clusters to be the maximal (union) coordinates of all constituent peaks.
We used a two-state Hidden Markov Model to identify the central, maximally replicated portion of each preliminary feature. Consensus features were defined as the maximal extent of all “open” segments within each cluster.
We used featureCounts69 to quantitate the number of reads within each peak for each library. Outliers were identified through principal component analysis.
ATAC-seq Samples PCA
The PCA was conducted on ATAC-seq peaks from 19 CD4+ T cell samples, and from 21 CD8+ T cell samples. Two samples were removed from the CD4+ samples due to large variability relative to the other replicates.
Differential Chromatin Accessibility
Differentially accessible ATAC peaks were calculated using DEseq2 R package. Peaks with FDR adjusted P.value below 0.05, and fold change of ±1 are regarded as differentially accessible peaks.
Transcription Factor footprint analysis
We used TOBIAS30 to identify transcription factor footprints within accessible chromatin features. We pooled libraries by cell type (CD4+ and CD8+ T cells) and treatment status (control and IFN-β) at 2 h, 8 h and 72 h time points. Within each pool, we corrected for Tn5 cut site bias and calculated footprint scores with TOBIAS. We used TOBIAS to identify TF motifs differentially footprinted at each time point.
Reanalysis of COVID-19 single-cell RNA sequencing data
A PBMC single-cell RNA seq data set of 10 COVID-19 patients and 13 matched controls was reanalyzed which had been previously performed and reported by us36. We have described the full cohort and detailed methods elsewhere. From eight of the ten COVID-19 samples, PBMCs from two different time points had been analyzed. Four of the COVID-19 patients had been classified as progressive, the other six COVID-19 patients as stable. Briefly, single cell barcoding of PBMCs and library construction had been performed using the 10x Chromium NextGEM 5prime kit. Libraries had been sequenced on an Illumina Novaseq 6000 platform. Raw reads had been demultiplexed and processed using Cell Ranger (v3.1) mapping to the GRCh38 (Ensembl 93) reference genome. Resulting gene-cell matrices had been analyzed using the package Seurat63 in the software R including integration of data, clustering, multiplet identification and cell type annotation. The final annotated R object was used and re-analyzed in Seurat with default settings - unless otherwise specified - as follows:
The three cell populations “Dividing T & NK”, “Effector T” and “Memory CD4 & MAIT” were each subsetted and reclustered to obtain a finer cell type granularity as they included a mix of CD4, CD8, MAIT and gamma delta T cells. Per subset, the top 500 variable genes were determined by the “FindVariableFeatures” function using the “vst” method. Data was scaled using the “ScaleData” function regressing out the total number of UMI and the percentage of UMIs arising from the mitochondrial genome. After Principal Component (PC) Analysis, the first 10 Principal Components (PCs) were utilized to detect the nearest neighbors using the “FindNeighbors” function and clustered by Seurat’s Louvain algorithm implementation “FindClusters” using a resolution of 0.2 for “Dividing T & NK”, of 0.3 for “Effector T” and of 0.1 for “Memory CD4 & MAIT” subsets. Cluster-specific gene expression profiles were established using the “FindAllMarkers” per cluster and per subset to annotate the clusters. New cell type annotations were then transferred back to the full dataset.
A new Uniform Approximation and Projection (UMAP) embedding was created by integrating the datasets on a subject level as follows: A subset containing all T cells was generated, which was then split by subject. For each subject, the top 2000 variable genes were selected, then integration anchors determined by “FindIntegrationAnchors” (with k.filter = 150). These anchors were used to integrate the data using the “IntegrateData” function with top 30 dimensions. The integrated data was scaled, subjected to a PC analysis and the top 13 PCs used as input for the “RunUMAP” function on 75 nearest neighbors.
Module scores were calculated using the “AddModuleScore” function using a) all genes within the GO list “RESPONSE TO TYPE I INTERFERON” (GO:0034340)64 and b) all genes significantly associated with either of the three waves in our in vitro perturbation experiments (see Supplementary Table 1). Differential gene expression was established using Seurat’s implementation of the Wilcoxon Rank Sum test within the “FindMarkers” function with a Bonferroni correction for multiple testing.
Statistical analysis
Detailed information about statistical analysis, including tests and values used, is provided in the figure legends. P-values of 0.05 or less were considered significant.
Extended Data
Extended Data Fig. 1. FACS gating strategy for isolating naïve CD4+ and CD8+ T cells and schematic experimental workflow.
Representative gating strategy for sorting naïve CD4+ and CD8+ T cells are shown. FACS isolated cells were immediately plated on 96 well round bottom plates coated with anti-CD3 (2 μg/ml) and soluble anti-CD28 (1 μg/ml) in the absence or presence of human IL-27 (100 ng/ml) or IFN-β (500 U/ml).
Extended Data Fig. 2. IFN-β differently regulates co-inhibitory receptors in human T cells.
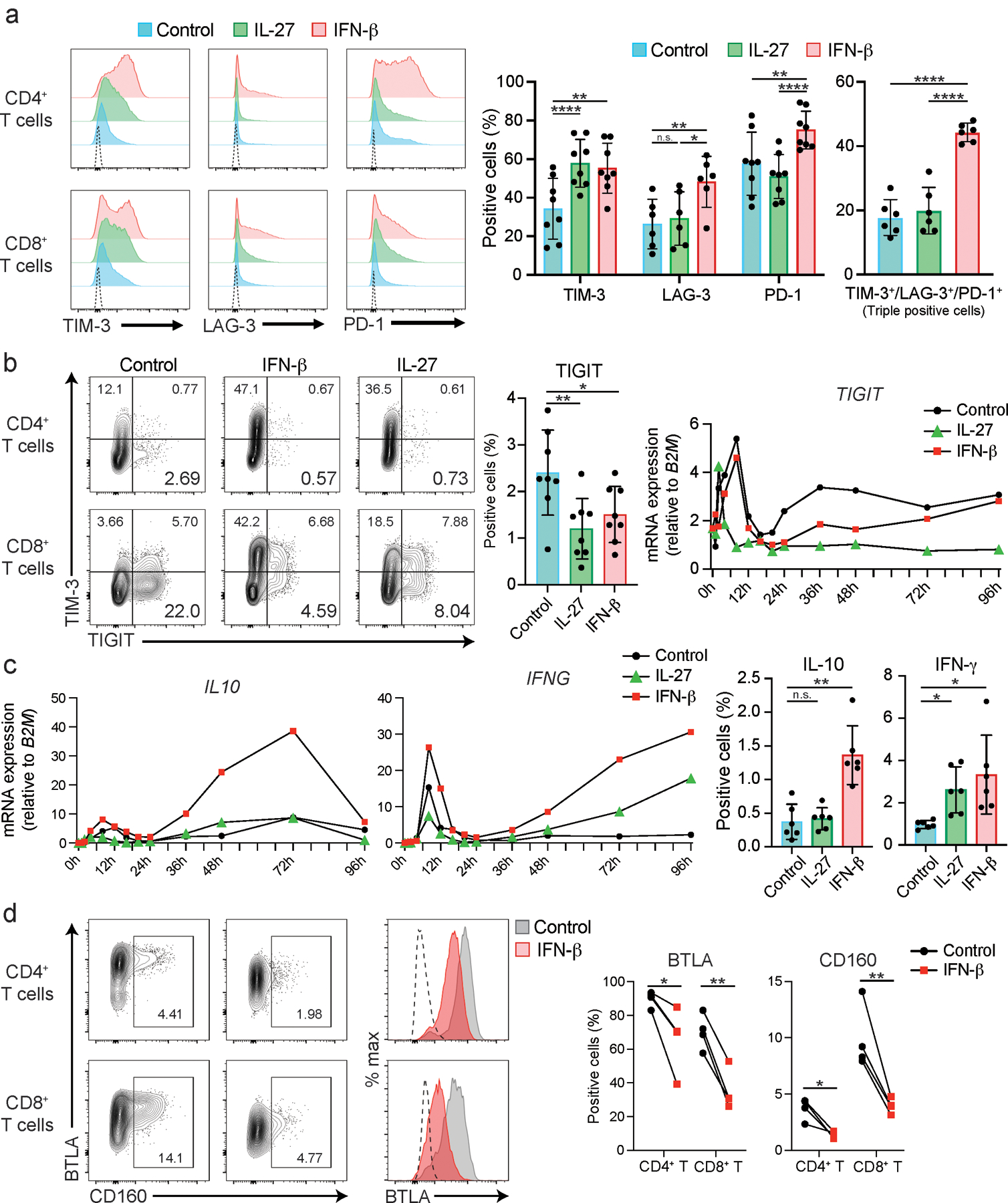
a, Representative histograms of surface expression of TIM-3, LAG-3, and PD-1 assessed by flow cytometry at 72–96 hours after stimulation. Dotted lines represent isotype control staining. Percent single positive cells for TIM-3, LAG-3, and PD-1 (middle) and triple positive cells (right) are shown (n = 6–8; biologically independent samples). *p < 0.05, **p < 0.01, ****p < 0.0001. b, Representative contour plots of flow cytometry analysis for TIM-3 and TIGIT expression in naïve CD4+ and CD8+ T cells (left). Cells were treated as Extended Data 1 and analyzed at 72 hours of culture. Percent TIGIT positive cells in naïve CD4+ T are shown (n = 8; biologically independent samples). *p < 0.05, **p < 0.01. (middle). qPCR analysis of TIGIT expression over the time course (13 time points from 0 to 96 hours). Each dot represents the average expression of two independent individuals’ data. ****p < 0.0001. c, qPCR analysis of IL10 and IFNG expression over the time course (13 time points from 0 to 96 hours). Each dot represents the average expression of two independent individuals’ data (left). IL-10 and IFN-γ production assessed by intracellular staining (right). Cells are treated as in a, and cytokines are stained intracellularly. Cytokine positive cells are detected by flow cytometry (n = 6; biologically independent samples). d, Representative contour plots of flow cytometry analysis for CD160 and BTLA expression and overlayed histogram for BTLA expression in naïve CD4+ and CD8+ T cells (left). Cells were treated as outlined in b and analyzed at 72 hours of culture. Percent positive cells for CD160 and BTLA in naïve CD4+ T are shown (n = 8; biologically independent samples) (right). *p < 0.05, **p < 0.01. Repeated-measures one-way ANOVA with Tukey’s multiple comparisons test (a-c), and paired student’s t-test (d).
Extended Data Fig. 3. The impact of IFN-β on co-inhibitory receptors is not associated with T cell activation.
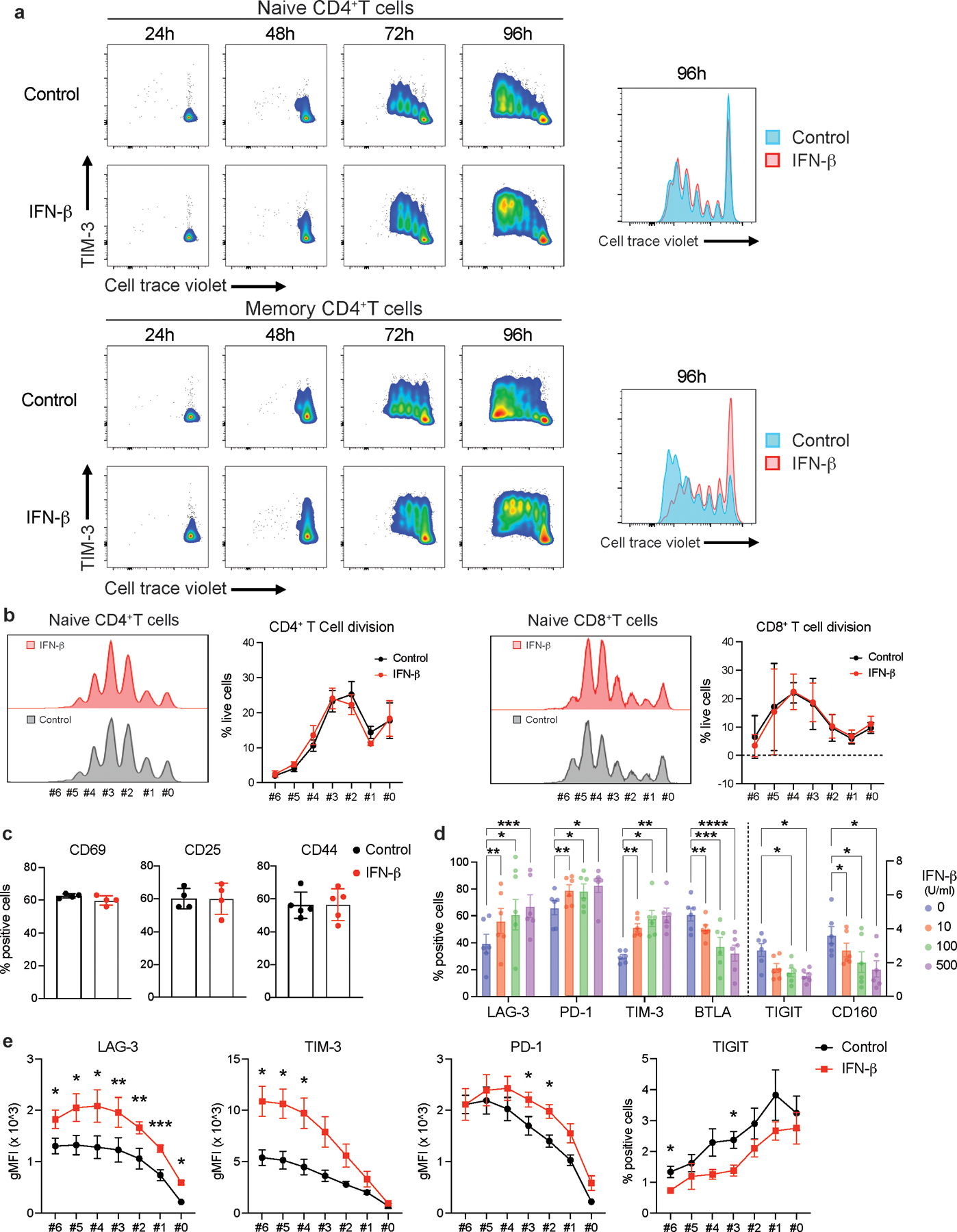
a, Representative plots for T cell proliferation assay using cell trace violet dye. Naive and memory CD4+ T cells were stimulated with anti-CD3 and anti-CD28 in the absence or presence of IFN-β. TIM-3 expression and cellular proliferation were assessed at 24, 48, 72, and 96 hours after stimulation. Overlayed histogram for control and IFN-β condition were shown at right. b, Frequency of living cells at each cellular division state was calculated in naïve CD4+ T cells (left) and naïve CD8+ T cells (right) at 96 hours. There is no statistical difference observed between control vs IFN-β condition (n=5; biologically independent samples). c, T cell activation markers (CD44, CD25, CD69) were quantified by flow cytometry in naïve CD4+ T cells at 96 hours. There is no statistical difference observed between control vs IFN-β condition (n=4–5; biologically independent samples). d, Dose dependent effects of IFN-β on co-inhibitory receptors assessed by flow cytometry (n=6; biologically independent samples). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Two-way ANOVA with Dunnett’s multiple comparisons test. e, LAG-3, PD-1, TIM-3, and TIGIT expressions were assessed by flow cytometry at each cellular division state (n=4; biologically independent samples). *p < 0.05, **p < 0.01, ***p < 0.001. Paired t test at each time point. Data was represented with mean +/− SEM.
Extended Data Fig. 4. Dynamic transcriptomic changes by IFN-β in human T cells.
a, Schematic experimental setup for high temporal resolution transcriptional profiling. b, Heatmap showing log fold change of DE gene expression between IFN-β and control Th0 condition at each timepoints for naive CD4+ (left) and CD8+ T cells (right). Genes are clustered based on the three transcriptional wave or bi-modal pattern. c, Line plots for IFI6, IFNG, LAG3, OSM, STAT1, and STAT2 expression in naive CD4+ (left) and CD8+ T cells (right).
Extended Data Fig. 5. Dynamic changes of chromatin accessibility by IFN-β in human T cells.
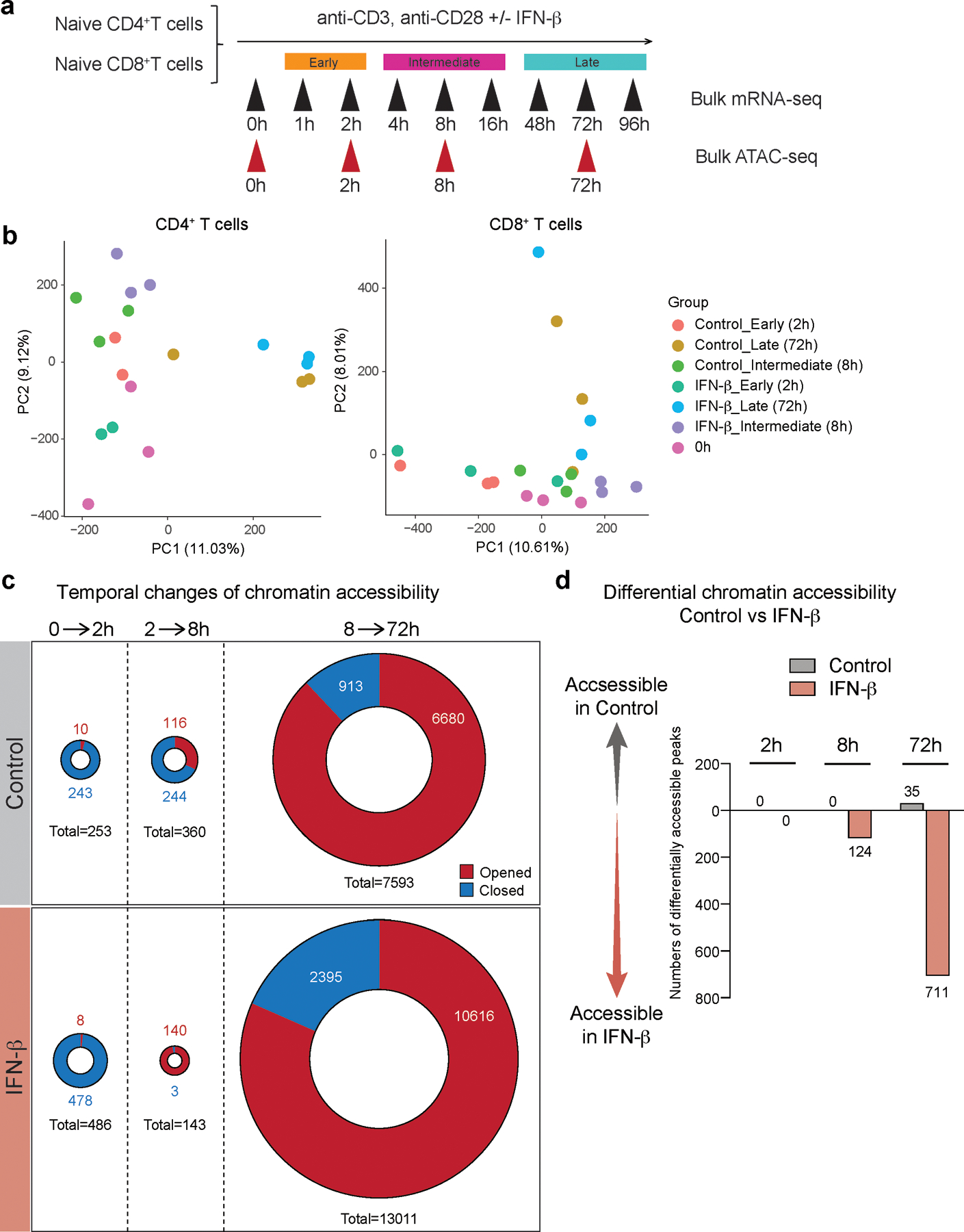
a, Schematic experimental setup for ATAC-seq experiment. b, PCA plots based on ATAC-seq peaks. Each dot represents one sample demonstrating the difference between the control and IFN-β groups, and the four different time points (0h, 2h, 8h, 72h). c, Pie charts depicting the differential chromatin accessibility between two time points, for both control and IFN-β treatment. The red or blue colored areas represent the numbers of opened or closed ATAC peaks compared to earlier time point, respectively. d, Bar plots describing the differential accessibility between IFN-β treatment and control at each time points.
Extended Data Fig. 6. Two IFN-I regulatory modules govern both transcriptional and epigenetic changes during IFN-I response.
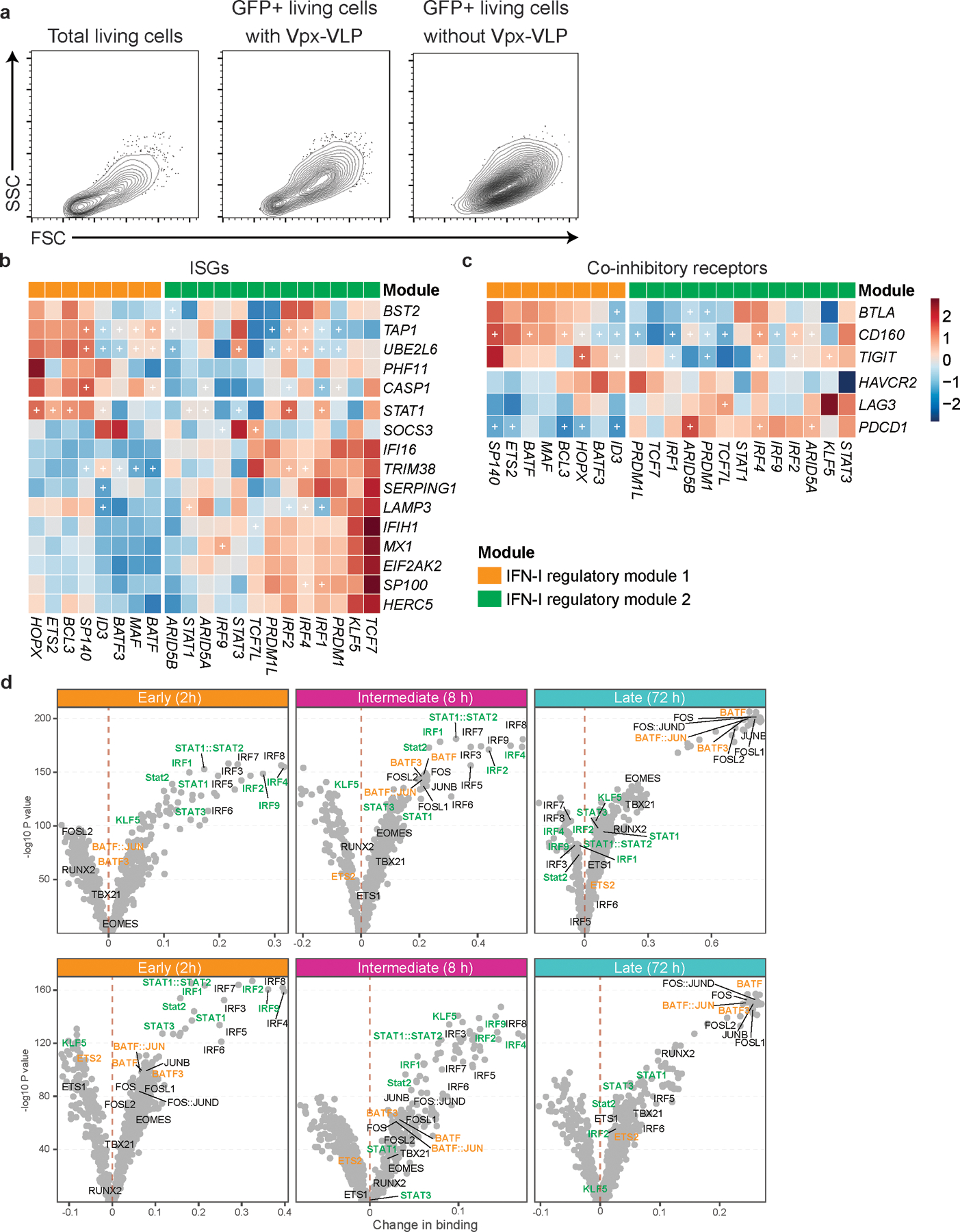
a, Contour plots for total living cells and backgating analysis for GFP positive cells. Primary naïve CD4+ T cells were transduced with scramble shRNA control LV with or without Vpx-VLPs pre-transduction. Cells are collected at 96 hours after starting stimulation and analyzed by flow cytometry. b, c, Heatmaps showing the effect of TFs perturbation under IFN-β stimulation on ISGs (b) and co-inhibitory receptors (c). Values in the heatmap were normalized by subtractions of log10 fold change of scramble shRNA control over perturbed expression. The “+” sign indicates a statistically significant effect with an adjusted P.value < 0.05 (details in Methods). d, Volcano plots depicting differential TF binding activity against the −log10(p value) (both provided by TOBIAS) of all investigated TF motifs; each dot represents one motif. Positive binding activity represents more enrichment in IFN-β treatment compared to control. The motifs for IFN-I regulator module 1 and IFN-I regulator module 2 were highlighted in orange and green, respectively.
Extended Data Fig. 7. Backbone network analysis identify downregulated TGF-β signature during IFN-I response.
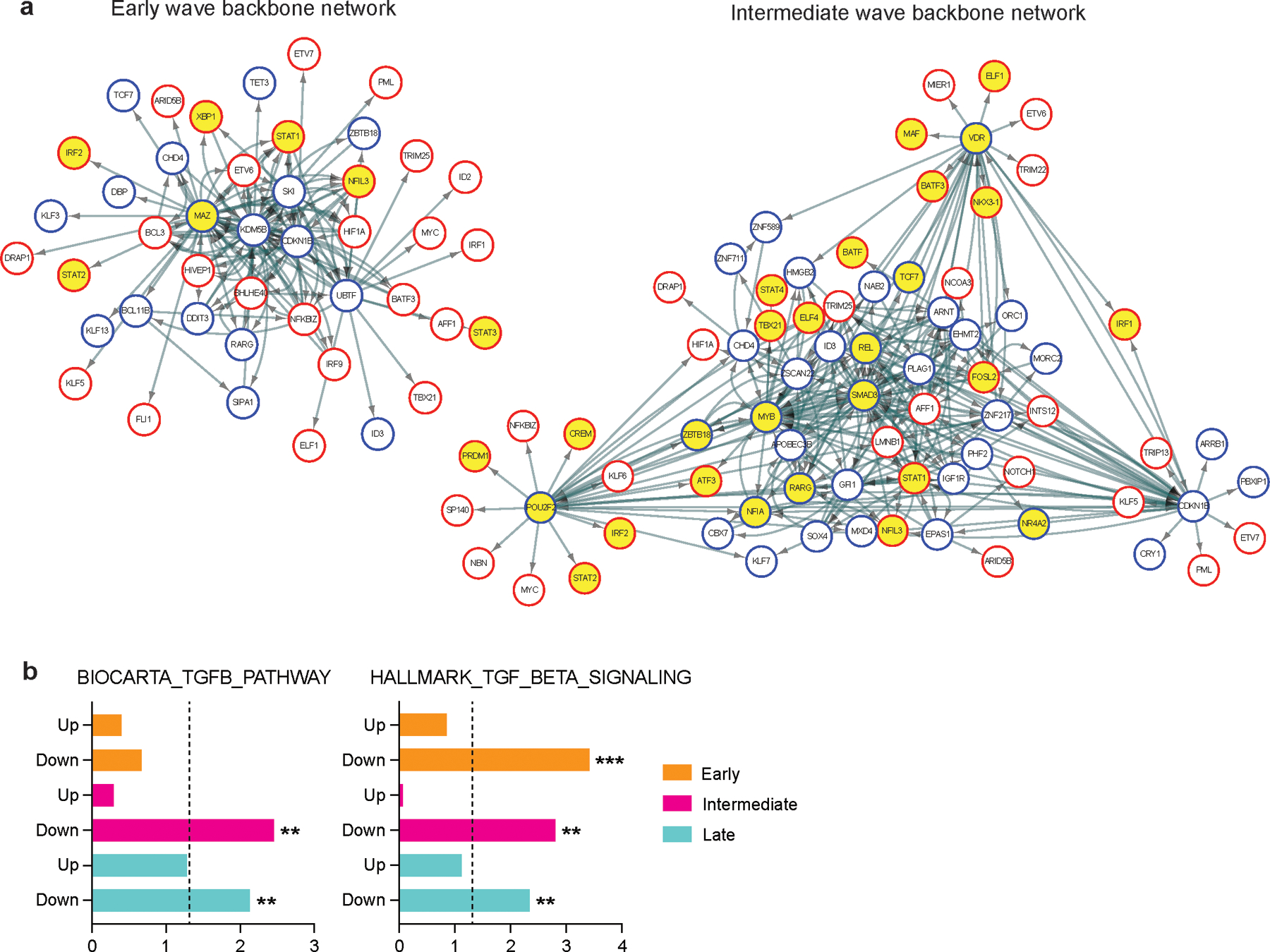
a, Directed TF backbone networks at the early and intermediate waves. The regulation and interaction of each DETFs at each transcriptional wave were depicted. TF footprints enriched in IFN-β condition were highlighted in yellow. b, ssGSEA for TGF-β signaling pathways with upregulated TFs (Up) and downregulated TFs (Down) at each transcriptional wave. Adjusted p values are shown and the dotted line indicates a threshold of significance defined by adjusted p value = 0.05. **p < 0.01, ***p < 0.001. Data was represented with mean +/− SEM.
Extended Data Fig. 8. IFN-I regulator expression profiles in T cells from COVID-19.
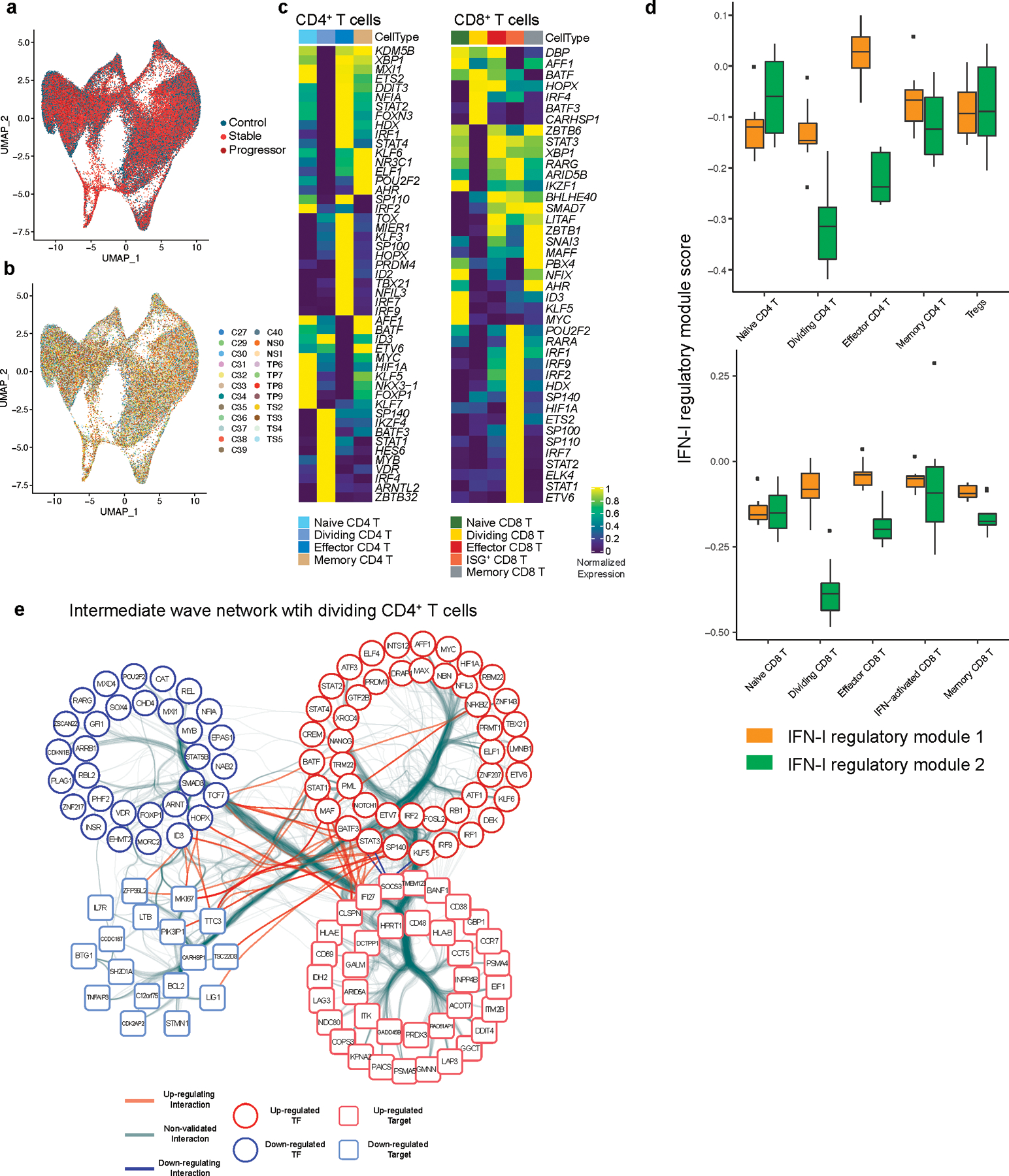
a, b, UMAP representation of T cells from healthy control samples (n = 13; biologically independent samples) and COVID-19 samples (n = 18; biologically independent samples) color coded by a, disease conditions and b, each individual. Cells from the same individual were labeled as one subject code, which resulted in 10 individual codes shown in b. c, Heatmap showing the expression of DETFs for CD4+ and CD8+ T cells in each T cell subset. d, IFN-I regulator module 1 and 2 scores for CD4+ and CD8+ T cells across sub cell types. e, Bundled regulatory network showing interactions between regulators at intermediate phase and transcriptional signature of dividing CD4+ T cells in COVID-19. Regulators at the intermediate phase are marked with circles (red; upregulated TFs, blue; downregulated TFs), and genes that are differentially expressed in dividing CD4+ T cells in COVID-19 were marked with squares (light red; upregulated DEGs, light blue; downregulated DEGs).
Extended Data Fig. 9. Validation of key IFN-I regulators controlling co-inhibitory receptors in human T cells.
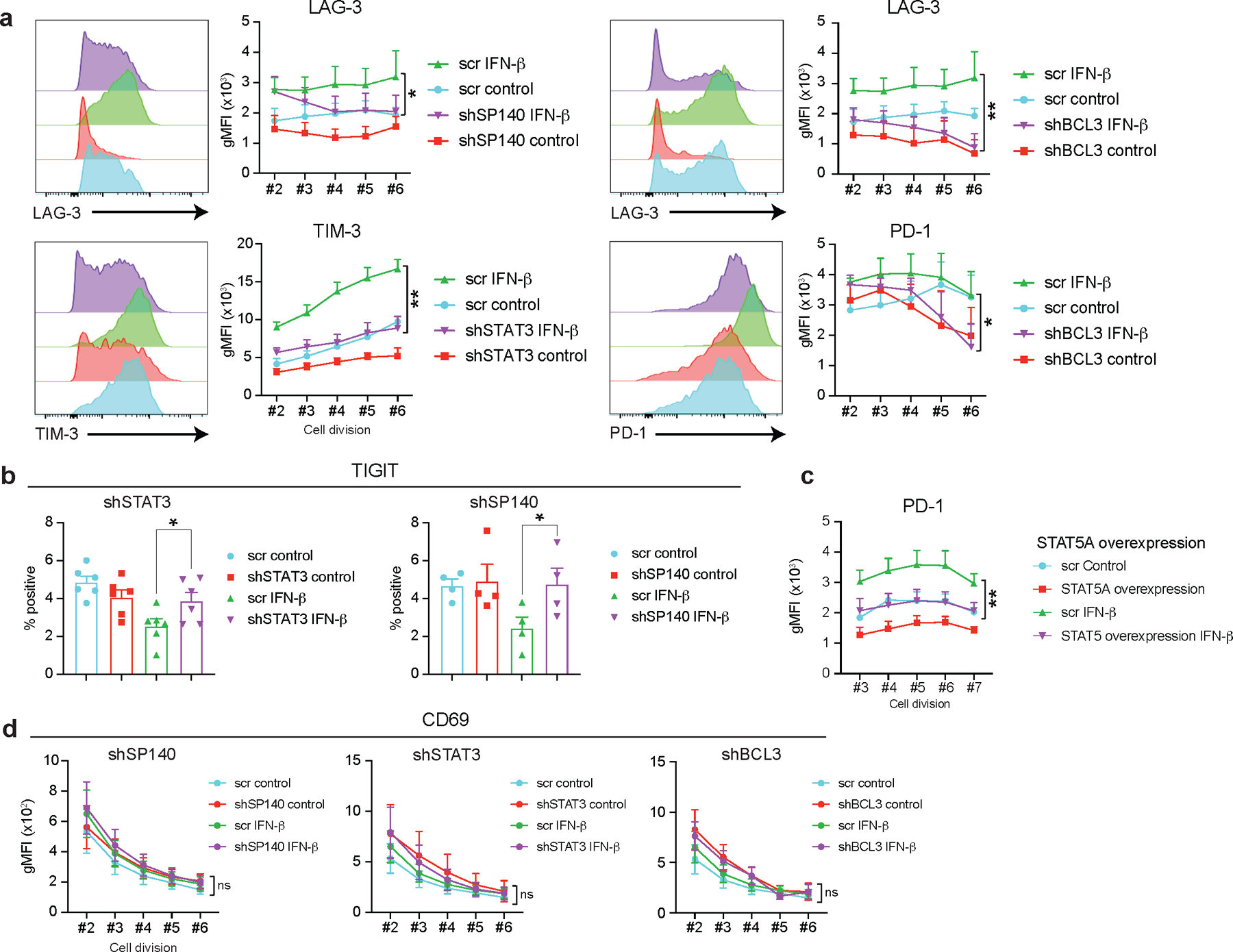
a, Co-inhibitory receptor expression was assessed at each cellular division state (division #2–6) under each TF gene knockdown performed as well as in Figure 3. The effects of each gene perturbation over GFP control vector (GFP) treated with IFN-β were determined by repeated-measures two-way ANOVA with Fisher’s LSD test. The statistical significance between GFP control vector with IFN-β (GFP + IFN-β) vs gene knockdown with IFN-β (shRNA + IFN-β) condition was shown. *p < 0.05, **p < 0.01. b, Frequency of TIGIT positive cells was determined in perturbation for SP140 and STAT3 (n=4; biologically independent samples). *p < 0.05. c, STAT5A was overexpressed on human primary naïve CD4+ T cells and PD-1 expression was shown at each cellular division state (division #3–7). Repeated-measures two-way ANOVA with Fisher’s LSD test was applied and the comparison between GFP control vector with IFN-β (GFP + IFN-β) vs STAT5A overexpression with IFN-β (STAT5A overexpression + IFN-β) condition was shown (n=6; biologically independent samples). **p < 0.01. d, CD69 expression was assessed at each cellular division state with three TF perturbation experiments (SP140, STAT3, and BCL3) (n=4; biologically independent samples). Statistics were determined as well as in a. ns; not significant. Data was represented with mean +/− SEM.
Supplementary Material
Acknowledgements
We thank L. Devine and C. Wang for assistance with FACS based cell sorting; G. Wang, M. Machado, and C. Castaldi at Yale Center for Genome Analysis for support with 10x Genomics library and ATAC-seq library preparation, and sequencing; P. Coish for his diligent proofreading of this paper; K. Raddassi and L. Zhang for processing scRNA-seq samples; L. Geng and M. Zhang for preparation of the bulk RNA-seq libraries, and sequencing; M. Taura for assistance developing Vpx-VLP based lenti viral transduction; N. Chihara and H. Asashima for useful discussions and input on the manuscript. This work was supported by grants to T.S.S. from Race to Erase MS; M.R.L. from the National MS Society and the Consortium of MS Centers (Career Transition Award); D.A.H. from the National Institute of Health (NIH) (U19 AI089992, R25 NS079193, P01 AI073748, P01 AI039671, and P50 CA121974), the National Multiple Sclerosis Society (NMSS) (CA 1061-A-18 and RG-1802–30153), the Nancy Taylor Foundation for Chronic Diseases, and Erase MS; V.K.K. from NIH (R01 NS045937, R01 NS030843, R01 AI144166, P01 AI073748, P01 AI039671 and P01 AI056299) and the Klarman Cell Observatory; N.K. from NIH (R01HL127349, R01HL141852 and U01HL145567); A.M. from The Alon fellowship for outstanding young scientists, Israel Council for Higher Education, from the Israel Science Foundation (1700/21), from the Israel Cancer Association (ICA, 01028753) and from the Israel Cancer Research Fund (ICRF) Research Career Development Awards; J.C.S from DoD (W81XWH-19–1-0131). RNA sequencing service was conducted at Yale Center for Genome Analysis and Yale Stem Cell Center Genomics Core facility, the latter supported by the Connecticut Regenerative Medicine Research Fund and the Li Ka Shing Foundation.
Footnotes
Competing interests
D.A.H. has received research funding from Bristol-Myers Squibb, Sanofi, and Genentech. He has been a consultant for Bristol Myers Squibb, Compass Therapeutics, EMD Serono, Genentech, and Sanofi Genzyme over the last three years. Further information regarding funding is available on: https://openpaymentsdata.cms.gov/physician/166753/. V.K.K. has an ownership interest and is a member of the SAB for Tizona Therapeutics. V.K.K. is also a co-founder and has an ownership interest and a member of SAB in Celsius Therapeutics and Bicara Therapeutics. V.K.K.’s interests were reviewed and managed by the Brigham and Women’s Hospital and Partners Healthcare in accordance with their conflict of interest policies. N.K. served as a consultant to Biogen Idec, Boehringer Ingelheim, Third Rock, Pliant, Samumed, NuMedii, Theravance, LifeMax, Three Lake Partners, Optikira, Astra Zeneca over the last 3 years, reports Equity in Pliant and a grant from Veracyte and non-financial support from MiRagen and Astra Zeneca. Has IP on novel biomarkers and therapeutics in IPF licensed to Biotech. The remaining authors declare no competing interests.
Data availability
The sequence data generated in this study are available under accession number GSE195543. Single-cell RNA-seq data presented in this study is available in GSE155223.
References
- 1.Chihara N et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558, 454–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeLong JH et al. IL-27 and TCR Stimulation Promote T Cell Expression of Multiple Inhibitory Receptors. Immunohorizons 3, 13–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtsinger JM, Valenzuela JO, Agarwal P, Lins D & Mescher MF Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol 174, 4465–4469 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Marrack P, Kappler J & Mitchell T Type I interferons keep activated T cells alive. J Exp Med 189, 521–530 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Bon A et al. Direct stimulation of T cells by type I IFN enhances the CD8+ T cell response during cross-priming. J Immunol 176, 4682–4689 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez-Navajas JM, Lee J, David M & Raz E Immunomodulatory functions of type I interferons. Nat Rev Immunol 12, 125–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crow MK & Ronnblom L Type I interferons in host defence and inflammatory diseases. Lupus Sci Med 6, e000336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Musella M, Manic G, De Maria R, Vitale I & Sistigu A Type-I-interferons in infection and cancer: Unanticipated dynamics with therapeutic implications. Oncoimmunology 6, e1314424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welsh RM, Bahl K, Marshall HD & Urban SL Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog 8, e1002352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Axtell RC, Raman C & Steinman L Type I interferons: beneficial in Th1 and detrimental in Th17 autoimmunity. Clin Rev Allergy Immunol 44, 114–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teijaro JR et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340, 207–211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson EB et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340, 202–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumeister SH, Freeman GJ, Dranoff G & Sharpe AH Coinhibitory Pathways in Immunotherapy for Cancer. Annu Rev Immunol 34, 539–573 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Anderson AC, Joller N & Kuchroo VK Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity 44, 989–1004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford A et al. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity 40, 289–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhen A et al. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J Clin Invest 127, 260–268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mestas J & Hughes CC Of mice and not men: differences between mouse and human immunology. J Immunol 172, 2731–2738 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Molle C et al. IL-27 synthesis induced by TLR ligation critically depends on IFN regulatory factor 3. J Immunol 178, 7607–7615 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Petricoin EF 3rd et al. Antiproliferative action of interferon-alpha requires components of T-cell-receptor signalling. Nature 390, 629–632 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Marshall HD, Urban SL & Welsh RM Virus-induced transient immune suppression and the inhibition of T cell proliferation by type I interferon. J Virol 85, 5929–5939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luoma AM et al. Molecular Pathways of Colon Inflammation Induced by Cancer Immunotherapy. Cell 182, 655–671 e622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sade-Feldman M et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013 e1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh DY et al. Intratumoral CD4(+) T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 181, 1612–1625 e1613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789 e718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quigley M et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med 16, 1147–1151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusinova I et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 41, D1040–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laguette N et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldauf HM et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18, 1682–1687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bentsen M et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun 11, 4267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gennert DG et al. Dynamic chromatin regulatory landscape of human CAR T cell exhaustion. Proc Natl Acad Sci U S A 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Vico Fallani F, Latora V & Chavez M A Topological Criterion for Filtering Information in Complex Brain Networks. PLoS Comput Biol 13, e1005305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofer MJ et al. Mice deficient in STAT1 but not STAT2 or IRF9 develop a lethal CD4+ T-cell-mediated disease following infection with lymphocytic choriomeningitis virus. J Virol 86, 6932–6946 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grunwell JR et al. TGF-beta1 Suppresses the Type I IFN Response and Induces Mitochondrial Dysfunction in Alveolar Macrophages. J Immunol 200, 2115–2128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas DA & Massague J TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Unterman A et al. Single-Cell Omics Reveals Dyssynchrony of the Innate and Adaptive Immune System in Progressive COVID-19. medRxiv, 2020.2007.2016.20153437 (2020). [DOI] [PMC free article] [PubMed]
- 37.Mostafavi S et al. Parsing the Interferon Transcriptional Network and Its Disease Associations. Cell 164, 564–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grosso JF et al. LAG-3 regulates CD8+ T cell accumulation and effector function in murine self- and tumor-tolerance systems. J Clin Invest 117, 3383–3392 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terawaki S et al. IFN-alpha directly promotes programmed cell death-1 transcription and limits the duration of T cell-mediated immunity. J Immunol 186, 2772–2779 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Ghiboub M et al. Modulation of macrophage inflammatory function through selective inhibition of the epigenetic reader protein SP140. bioRxiv, 2020.2008.2010.239475 (2020). [DOI] [PMC free article] [PubMed]
- 41.Gil MP et al. Biologic consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci U S A 98, 6680–6685 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Xu L, Su J, Peppelenbosch MP & Pan Q Transcriptional Regulation of Antiviral Interferon-Stimulated Genes. Trends Microbiol 25, 573–584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang L, Anderson DE, Kuchroo J & Hafler DA Lack of TIM-3 immunoregulation in multiple sclerosis. J Immunol 180, 4409–4414 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Monk PD et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med 9, 196–206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosi E et al. Interferon beta-1a (IFNbeta-1a) in COVID-19 patients (INTERCOP): study protocol for a randomized controlled trial. Trials 21, 939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
References associated with the Methods section
- 46.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ly A, Marsman M & Wagenmakers EJ Analytic posteriors for Pearson’s correlation coefficient. Stat Neerl 72, 4–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bojnordi MN et al. Differentiation of Spermatogonia Stem Cells into Functional Mature Neurons Characterized with Differential Gene Expression. Mol Neurobiol 54, 5676–5682 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen YM, Weng YT, Dong X & Tsong Y Wald tests for variance-adjusted equivalence assessment with normal endpoints. J Biopharm Stat 27, 308–316 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Rubinstein ML, Kraft CS & Parrott JS Determining qualitative effect size ratings using a likelihood ratio scatter matrix in diagnostic test accuracy systematic reviews. Diagnosis (Berl) 5, 205–214 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Leong HS et al. A global non-coding RNA system modulates fission yeast protein levels in response to stress. Nat Commun 5, 3947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Sherbiny YM et al. A novel two-score system for interferon status segregates autoimmune diseases and correlates with clinical features. Sci Rep 8, 5793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lê S, Josse J & Husson F FactoMineR: An R Package for Multivariate Analysis. 2008 25, 18 (2008). [Google Scholar]
- 56.Yevshin I, Sharipov R, Kolmykov S, Kondrakhin Y & Kolpakov F GTRD: a database on gene transcription regulation-2019 update. Nucleic Acids Res 47, D100–D105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Virtanen P et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piraveenan M, Prokopenko M & Hossain L Percolation centrality: quantifying graph-theoretic impact of nodes during percolation in networks. PLoS One 8, e53095 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hagberg A, Swart P & Chult D Exploring Network Structure, Dynamics, and Function Using NetworkX, 2008.
- 61.Christensen AP NetworkToolbox: Methods and Measures for Brain, Cognitive, and Psychometric Network Analysis in R. The R Journal 10, 422–439 (2018). [Google Scholar]
- 62.Szalkai B, Varga B & Grolmusz V Mapping correlations of psychological and structural connectome properties of the dataset of the human connectome project with the maximum spanning tree method. Brain Imaging Behav 13, 1185–1192 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Rubinov M & Sporns O Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Corces MR et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orchard P, Kyono Y, Hensley J, Kitzman JO & Parker SCJ Quantification, Dynamic Visualization, and Validation of Bias in ATAC-Seq Data with ataqv. Cell Syst 10, 298–306 e294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Enright AJ, Van Dongen S & Ouzounis CA An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data generated in this study are available under accession number GSE195543. Single-cell RNA-seq data presented in this study is available in GSE155223.


