Abstract
Despite the success of the BNT162b2 mRNA vaccine, the immunological mechanisms that underlie its efficacy are poorly understood. Here we analyzed the innate and adaptive responses to BNT162b2 in mice, and show that immunization stimulated potent antibody and antigen-specific T cell responses, as well as strikingly enhanced innate responses after secondary immunization, which was concurrent with enhanced serum IFNγ levels one day following secondary immunization. Notably, we found that natural killer cells and CD8 T cells in the dLNs are the major producers of this circulating IFNγ. Analysis of knockout mice revealed that induction of antibody and T cell responses to BNT162b2 was not dependent on signaling via TLRs 2, 3, 4, 5, 7, nor inflammasome activation, nor the necroptosis or pyroptosis cell death pathways. Rather, the CD8 T cell response induced by BNT162b2 was dependent on type I IFN-dependent MDA5 signaling. These results provide insights into the molecular mechanisms by which the BNT162b2 vaccine stimulates immune responses.
The BNT162b2 mRNA vaccine developed by Pfizer and BioNTech is the first FDA-approved mRNA vaccine in history. The vaccine demonstrated 95% efficacy against symptomatic COVID-19 disease caused by SARS-CoV-2 infection in a phase-II/III trial 1, 2. However, despite its widespread use, the mechanisms by which BNT162b2 stimulates protective immunity remain largely unknown. BNT162b2 comprises N1-methyl-pseudouridine (m1Ψ) nucleoside-modified mRNA encapsulated in a lipid nanoparticle (LNP). Following administration in muscle, the mRNA is translated, resulting in the expression of the spike protein and spike-specific B and T cell responses 3. Kariko et al. found that the m1Ψ-modification on RNA dampens the inflammatory response mediated by TLR3, TLR7, or RIG-I, and increases translational efficiency and biological stability 3, 4. mRNA vaccines encapsulated in LNPs were also found to induce a high magnitude of germinal center B (GC B) cell and follicular helper T cells (TFH) responses after a single immunization 5, 6, 7, 8.
Several studies have analyzed immune responses to the BNT162b2 vaccine in humans and revealed insights about the nature of the antibody and T cell responses to vaccination 1, 2, 9, 10, 11, 12, 13. Despite our emerging understanding of immune responses to BNT162b2, there are major knowledge gaps on its mechanisms of action. For example, there is a paucity of understanding on the nature of the innate response to BNT162b2 and, in particular, how BNT162b2 is sensed by the host’s innate immune system. Studies of mRNA vaccines against cancer demonstrate that they can trigger TLR4 14, TLR7 15, or STING 16 signaling pathways, but such knowledge is lacking in the context of BNT162b2. Arunachalam et al used a systems vaccinology approach to analyze the innate and adaptive immune responses to vaccination with BNT162b2 in humans 9. Our analysis demonstrated detectable neutralization antibody titers after primary immunization, which was enhanced considerably upon secondary immunization. Furthermore, there was only a modest innate immune response, at 1 day or 7 days after primary immunization. Surprisingly however, secondary immunization with BNT162b2 induced a much higher magnitude of innate immune response than after primary vaccination 9. This enhanced “secondary innate response,” was characterized by increased levels of IFNγ in the plasma, enhanced transcriptional signatures of innate immunity and antiviral immunity in myeloid cells. However, the cellular sources of the rapidly produced IFNγ and its role in the enhanced innate immune response after secondary vaccination remains unclear. Furthermore, although vaccination with BNT162b2 has been shown to induce substantial frequencies of antigen-specific CD8+ T cell responses in the blood 9, 17, there is currently no data on the nature of immune responses in tissues, as most studies have focused on analyzing immune responses in the blood. In addition, the in vivo distribution of the mRNA and the spike protein that it encodes, after intramuscular vaccination, remains poorly understood. To address these knowledge gaps, we performed a detailed analysis of innate and adaptive immune responses to BNT162b2 vaccination in mice. In particular, we analyzed: (i) the adaptive immune response induced by BNT162b2 after both primary and booster vaccines in organs including the lung and spleen; (ii) the innate immune response in dLNs, non-draining lymph nodes and other organs at the single-cell level, using systems immunology approaches; and (iii) the innate sensing mechanisms that regulate antigen-specific antibody and T cell responses to BNT162b2 vaccination.
Results
BNT162b2 immunization stimulates robust germinal center responses
To evaluate immune responses to BNT162b2, we immunized C57BL/6 mice on days 0 and 21 with 5 μg BNT162b2 by intramuscular injection (Fig. 1a). Anti-spike binding antibody (bAb) immunoglobulin G (IgG) responses, comprised of both IgG1 and IgG2c, increased significantly at 14 days post-primary immunization and persisted until day 21, while the secondary immunization increased the response more than 10-fold (Fig. 1b). Consistent with binding antibody responses, neutralizing antibody (nAb) responses against the SARS-CoV-2 wild type (WT) strain and variants of concern including B1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), and B.1.429 (Epsilon) were detected on day 21 and increased significantly following secondary immunization (Fig. 1c). While the neutralizing titers against the Alpha and Epsilon variants did not differ significantly from that of the wild type (WT) strain, nAb titers at day 21 against the Beta, Gamma, and Delta variants were lower by 8.6-, 4.3-, and 6.5-fold respectively relative to the titers against the WT strain (Fig. 1c), consistent with findings in humans 18. In addition to serological responses, we examined the responses of GC B cells (CD19+CD38-CD95+) and TFH cells (CD3+CD4+PD1+CXCR5+) as well as plasma cells (CD44+CD138+). BNT162b2 immunization induced a strikingly high magnitude of GC B cells, TFH cells, and plasma cells in dLNs, which peaked on day 7 and decreased by day 21 (Fig. 1d–f). Collectively, these data demonstrate the potent stimulation of humoral immune responses by BNT162b2. In addition, we tested antibody responses in mice immunized with 0.2 μg of the vaccine, which is an equivalent dose to that used in humans based on body weight19, 20. The antibody kinetics mirrored the response induced by the higher dose, but the magnitude was approximately 10-fold lower (Extended Data Fig.1a).
Figure 1. BNT162b2 vaccine induces robust germinal center B cell and TFH response in mice.
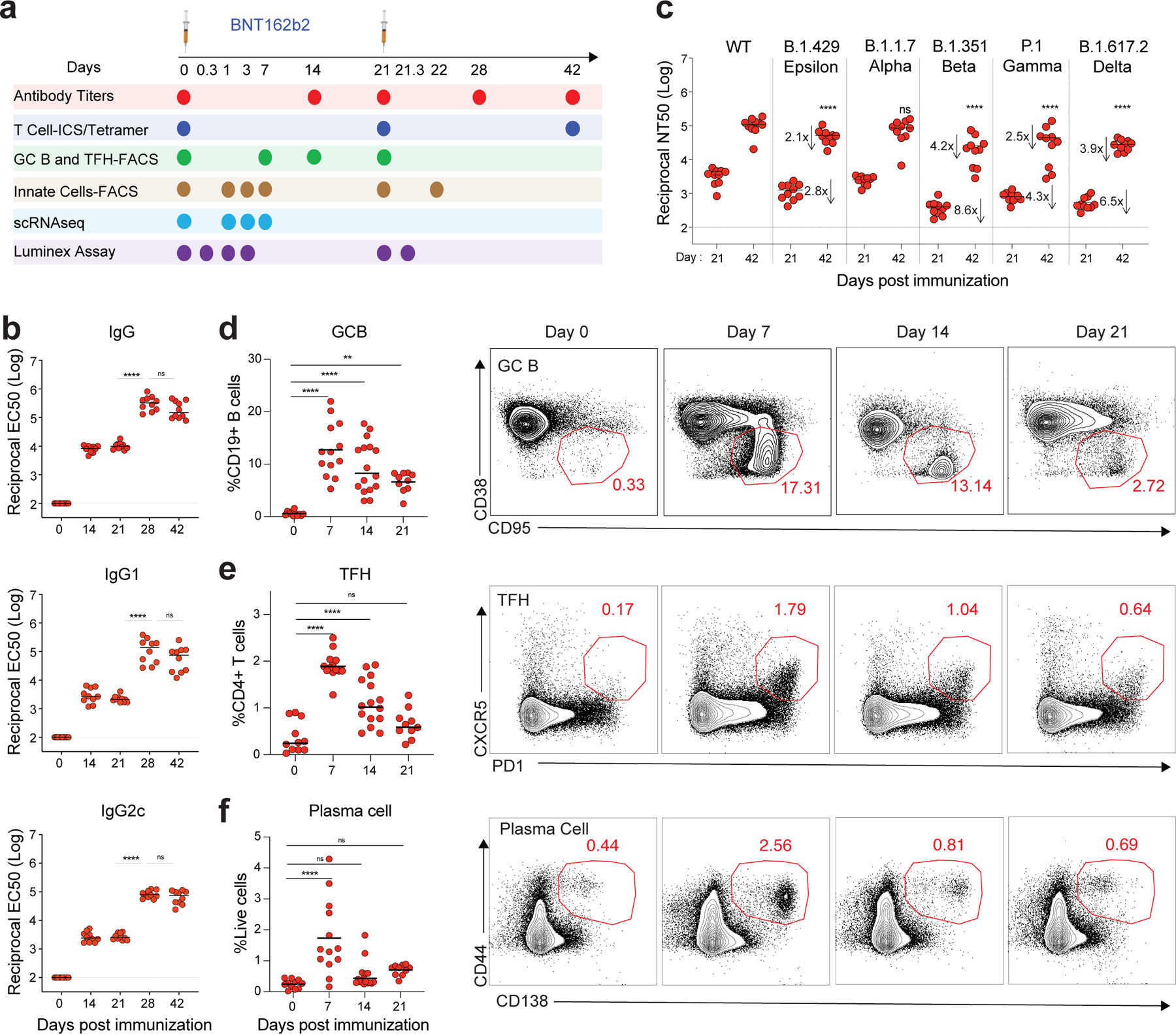
a, Experimental timeline and tests. b, Serum anti-S IgG, IgG1a, and IgG2c binding titers detected by ELISA. c, Pseudo-typed lentivirus neutralization antibody titers against SARS-CoV-2 WT and variants of concern (B.1.429, B.1.1.7, B.1.351, P.1 and B.1.617.2) measured at days 21 and 42. n=10 mice per time point in b-c. d-f, Germinal center B cell (d), T follicular helper cell (e), and plasma cell (f) response after BNT162b2 immunization. n=11, 13, 16, 10 for day 0, 7, 14, 21, respectively. Data were combined from two or more independent experiments. One-Way ANOVA followed by Tukey’s test was applied in (b-f). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Differences between groups were considered significant for P-values < 0.05.
T cell response induced by BNT162b2 vaccination
Next, we measured antigen-specific T cell responses in spleen and lung tissue on days 21 and 42 post-vaccination following the prime-boost immunization strategy presented in Fig. 1a. While the primary immunization elicited detectable antigen-specific CD4 and CD8 T cell responses in both tissues consistent with previous reports8, 21, there was a striking increase in the frequency of antigen-specific CD8 T cells after secondary immunization, especially in the lung, where we observed a median of 10% MHC-I tetramer+ CD8 T cells on day 42 (Fig. 2a). There was a robust induction of spike-specific CD8 T cells secreting IFNγ, TNF, and IL-2 following in vitro stimulation with an overlapping peptide pool (Fig. 2b). The CD4 T cell responses were primarily Th1-type with little induction of IL-4 response (Fig. 2c), consistent with previous reports8, 21. As tissue resident memory T cells (TRMs) play an essential role in preventing local infection22, 23, we measured TRM (CD3+CD8+CD44+CD69+CD103+) levels in the lung at day 21 and day 42 after BNT162b2 immunization. The data demonstrated that BNT162b2 immunization did not induce TRMs in the lung (Fig. 2d), consistent with previous findings24. The low dose (0.2 μg) immunization resulted in ~5-fold lower magnitude of T cell responses (Extended Data Fig.1b–d).
Figure 2. T cell response induced by BNT162b2 vaccination in mice.
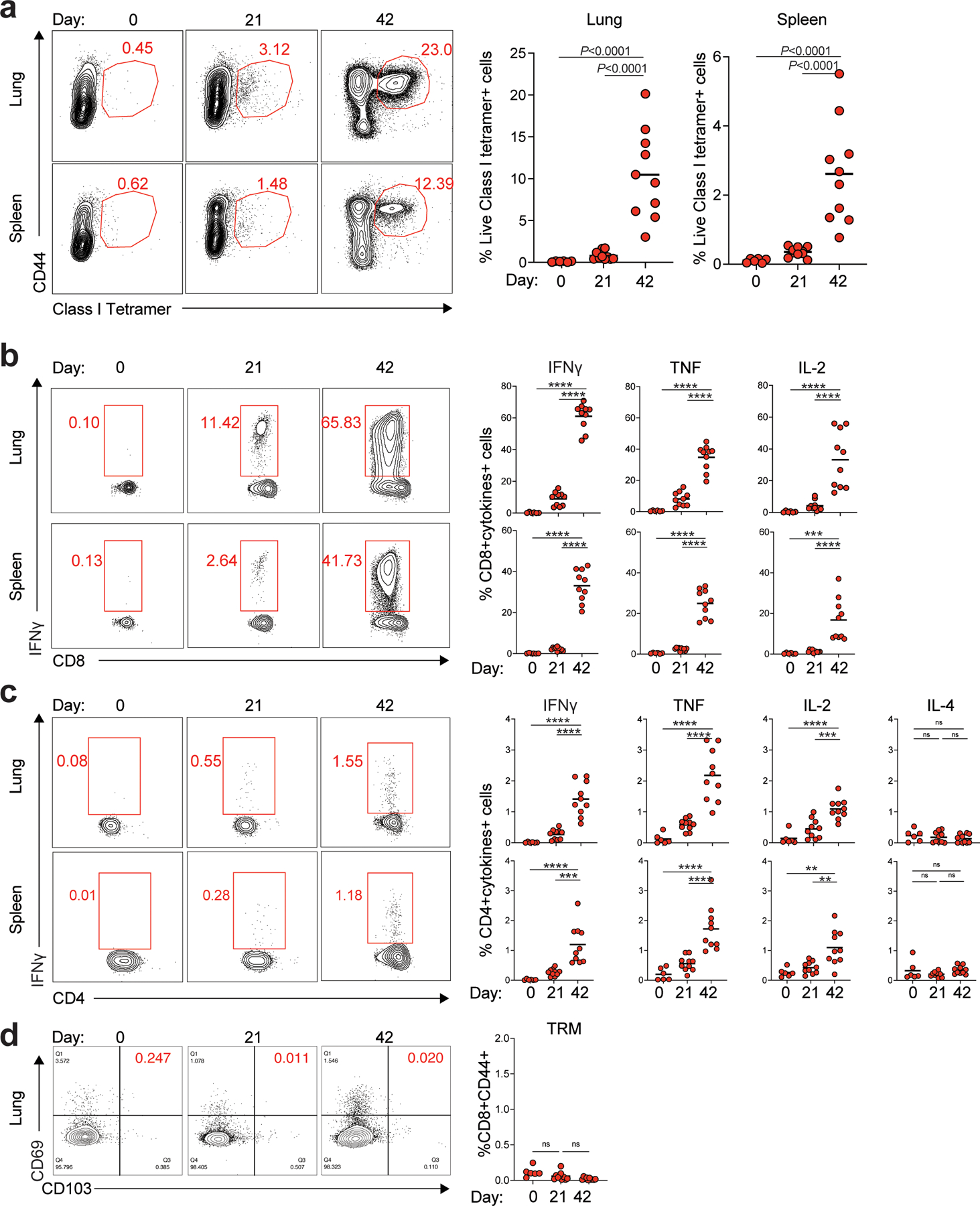
a, Class I tetramer specific CD8 T cell responses in lung and spleen at days 21 and 42 after BNT162b2 immunization. b-c, Antigen-specific CD8+ T (b) and CD4+ T (c) cell response in lung and spleen at days 21 and 42 measured by FACS intracellular cytokine staining assay. d, Tissue-resident memory T cells in lung at days 21 and 42. Data were combined from two independent experiments. One-Way ANOVA followed by Tukey’s test was applied in (a-d). n=6 for day 0, n=10 for day 21 and day 42, respectively. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
BNT162b2 induces robust innate immune responses
To profile the innate immune response induced by BNT162b2, we first analyzed the dLNs by multiparametric flow cytometry (Extended Data Fig.2a). Frequencies of monocytes (CD11b+Ly6Chi), pDCs (CD11cloPDCA1+), and CD103+migratory DCs increased significantly on day 1 and returned to baseline levels on day 7 (Extended Data Fig.2b). In contrast, resident DCs (both CD8a+rDC and CD11b+rDC) decreased in frequency on day 1 but increased significantly on day 7 (Extended Data Fig.2b). Further, multiple innate immune cells including monocytes, macrophages (CD11b+Ly6CloF4/80+), pDCs, and DC subsets (CD8a+rDC, CD11b+rDC, CD103+mDC, and CD11b+mDC) were highly activated, as indicated by the enhanced expression of the activation marker CD86 (Fig. 3a). Low dose (0.2 μg) induced lower levels of innate cell activation (Extended Data Fig.1e). These innate cell types remained activated until day 3 post-immunization (Fig. 3b). In comparison, the innate cells were moderately activated in the contralateral lymph nodes (non-dLNs), lung, and spleen (Fig. 3c, d).
Figure 3. BNT162b2 induces robust innate immune response in draining lymph nodes.
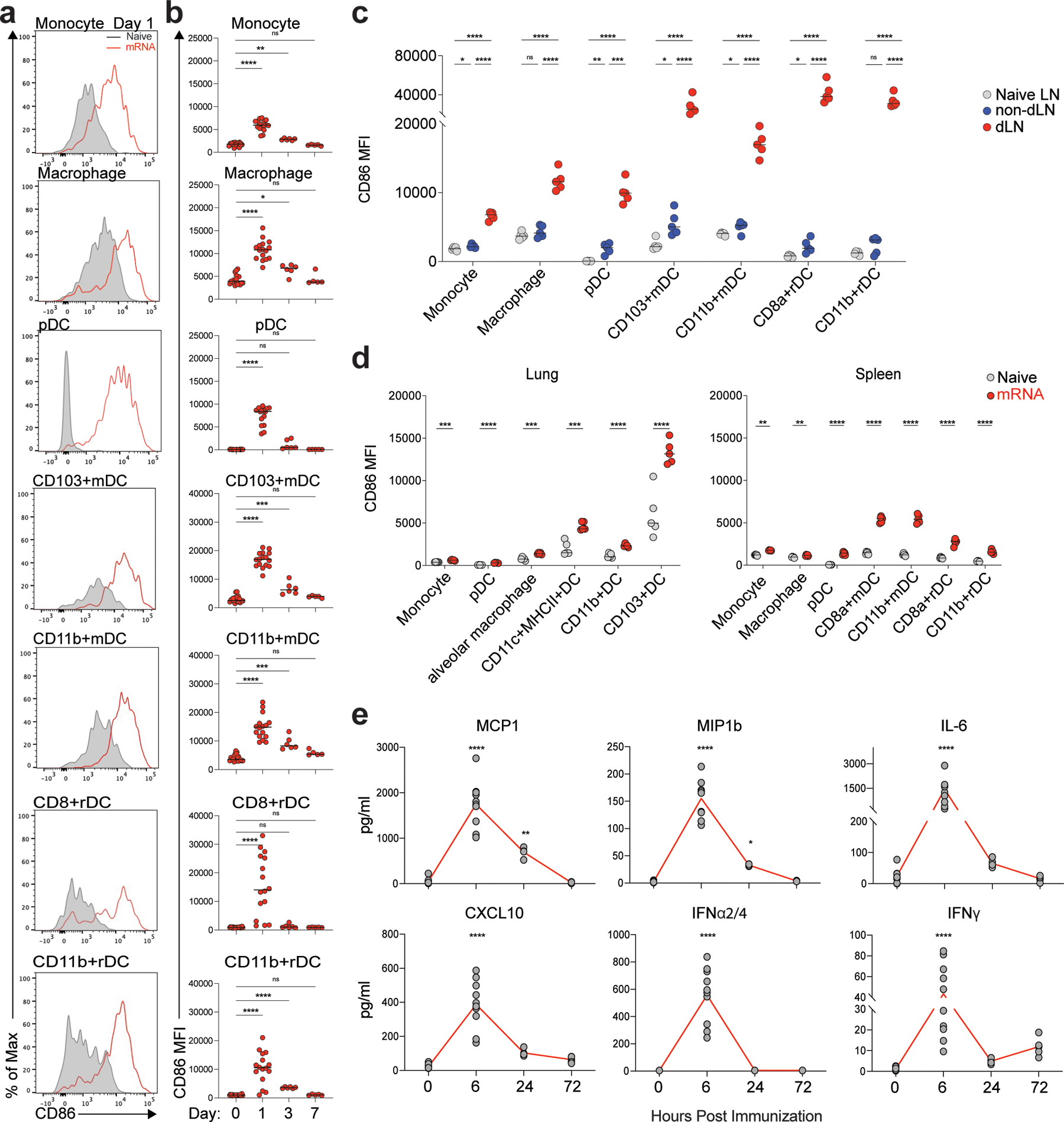
a, Activation of innate cells at day 1 after BNT162b2 immunization in dLNs indicated by activation marker CD86. b, Dynamics of innate cell activation at days 1, 3, and 7 after BNT162b2 immunization. n=20, 16, 7, 5 for day 0, 1, 3, and 7, respectively. c-d, Innate cell activation in contralateral lymph nodes (non-dLN) compared to that in dLNs (c), and in lung and spleen (d) at day 1 post-prime. n=5 for each group. e, Dynamics of cytokines in serum at 0, 6, 24 and 72 h after BNT162b2 immunization. n=10 for 0 and 6h, n=5 for 24h and 72h. Data were combined from at least two independent experiments (a, b, e) or one representative of two independent experiments (c, d). One-Way ANOVA followed by Tukey’s test was applied in (b-e). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Next, we analyzed serum cytokine responses using a Luminex assay. BNT162b2 immunization induced the release of many cytokines, including MCP1, MIP1b, IL-6, and CXCL10, as well as IFNγ, that peaked at 6 h post-immunization (Fig. 3e) in contrast to the significantly elevated levels of only two cytokines/chemokines (IFNγ and CXCL10) in human volunteers after receiving the same vaccine9. It should, however, be noted that in the study in humans, Arunachalam et al only measured plasma cytokine levels at 1 and 7 days post vaccination, and not at the earlier time point of 6 h examined in the current mouse study 9. All the cytokines, except IFNγ, returned to normal levels at day 3 post-immunization. Strikingly, there was a significant induction of IFNα ( IFNα2 and IFNα4) in mice at 6 h, which was not observed in humans at the 1-day time point we examined9. Of note, the lower dose (0.2 μg) immunization did not induce IFNα as seen in humans; however, there was also an absence of IFNγ response (Extended Data Fig.1f), suggesting that there was not a human equivalent dose of BNT162b2 in mice.
Single-cell transcriptomics of innate immunity to BNT162b2
To further understand the cellular and molecular mechanisms involved in the immunogenicity of BNT162b2, we performed single-cell transcriptional profiling of cells in the dLNs 1, 3 and 7 days post-immunization (Fig. 4a). As a benchmark, we compared the responses to BNT162b2 with those of the live attenuated yellow fever vaccine YF-17D, one of the most effective vaccines ever developed in humans 25. Our previous work showed that YF-17D activates multiple subsets of DCs via several different Toll-like receptors to stimulate potent adaptive immune responses 26, 27. We employed an “enrich-mix” strategy in which CD11b+ myeloid cells, pDC (CD11b-CD11clo PDCA-1+), and DCs (CD11c+ MHC-II+) were enriched by cell sorting and then mixed with total cell suspension at a ratio of 1:1 (Fig. 4a). The resulting cell suspension was subjected to droplet-based single-cell gene expression profiling. After quality control, we obtained 52,788 high-quality transcriptomes (Fig. 4b). Using dimensionality reduction via UMAP and graph-based clustering, we identified 20 cell clusters containing all major innate and adaptive immune cell types (Fig. 4b; Extended Data Fig.3a). First, we analyzed the number of differentially expressed genes (DEGs) within each cluster. The YF-17D vaccine stimulated a profound DEG response in multiple cell types, as seen in previous study 28, which peaked at day 1 and returned to baseline by day 7. Interestingly, the mRNA vaccine stimulated a larger number of DEGs, especially in monocyte/macrophage, DCs, and natural killer (NK) cells on day 1 that persisted until day 7 (Fig. 4c). Gene-set enrichment analysis using blood transcriptional modules (BTMs) revealed activation of type I IFN and antiviral responses in multiple cell types, which peaked at day 1 and returned to baseline levels by day 7 (Fig. 4d). In contrast, cell-cycle and transcription associated genes increased at day 1 and persisted to day 7, especially in NK cells and T cells (Fig. 4d). The overall responses were comparable to that of the YF-17D immunization (Extended Data Fig.3b–c).
Figure 4. Single-cell transcriptional response induced by BNT162b2 and YF-17D immunization in dLNs.
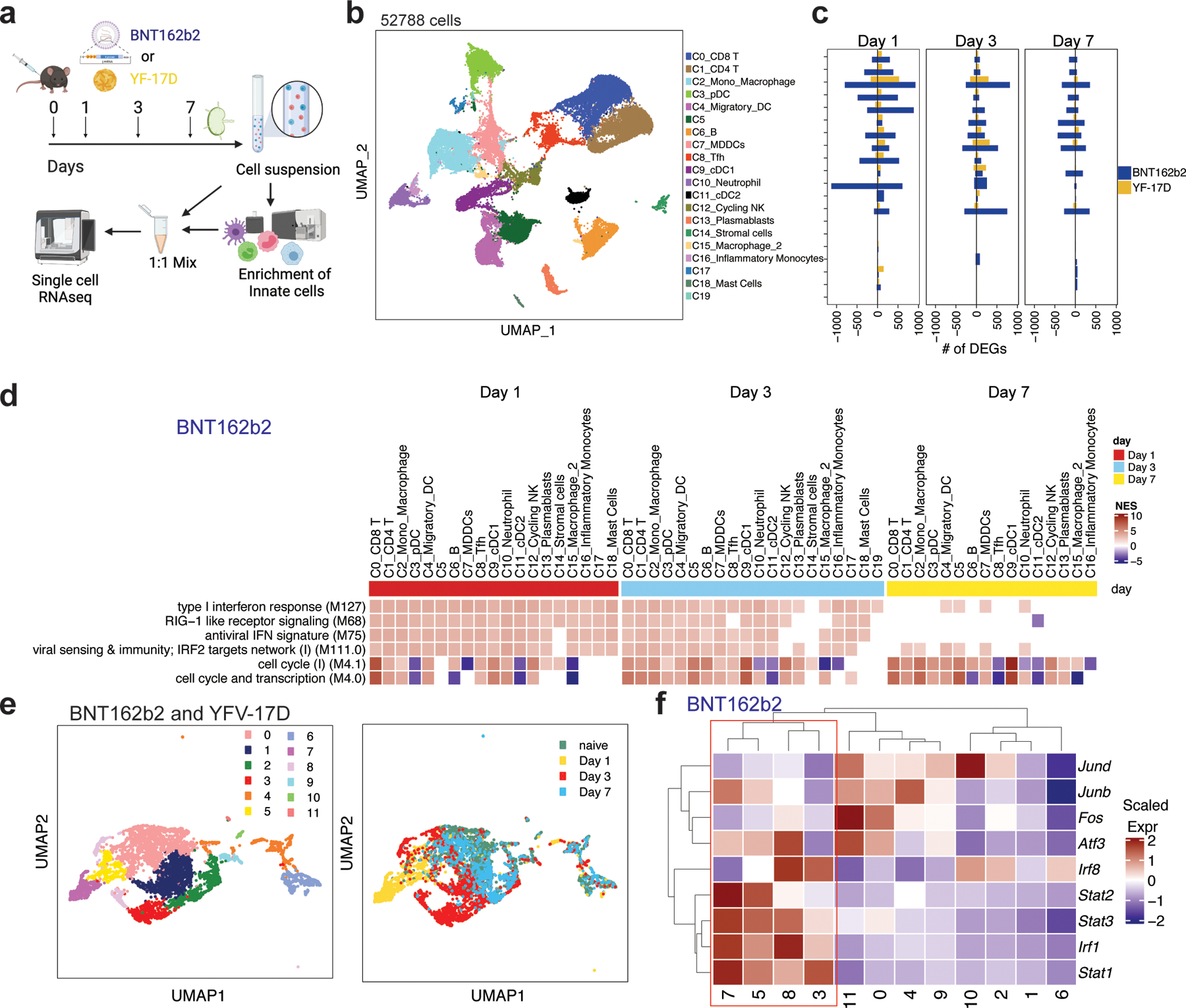
a, Experimental design and timeline. b, UMAP of cell types clustered by single-cell transcriptional analysis of 52,788 cells after quality control. c, DEGs induced by BNT162b2 and YF-17D immunization at days 1, 3, and 7. d, Significantly enriched interferon and cell cycle BTMs (false discovery rate [FDR]< 0.05, absolute normalized enrichment score [NES]> 2) across all clusters from days 1 to 7 after BNT162b2 immunization. Only clusters with a significantly modulated pathway are shown. e, UMAP of subclusters in C2 (Mono_Macrophage) and C15 (Macrophage) after BNT162b2 and YF-17D immunization, respectively. f, Heatmap of key interferon response and AP-1 transcription factors after BNT162b2 immunization. Samples used for scRNAseq were pooled from three independent experiments containing 8–10 mice.
Arunachalam et al demonstrated that the BNT162b2 vaccination in humans results in an enhanced innate immune response following the secondary vaccination in comparison with the primary vaccination 9. A crucial aspect of that study was the discovery of an innate cell cluster (C8) using single-cell transcriptional profiling that emerged 1 day after secondary vaccination, a ~100-fold increase in comparison to the first day after primary vaccination. We determined that these cells were a transcriptional counterpart of an epigenetically programmed monocyte population that Wimmers et al described in humans vaccinated with the H5N1 avian influenza subunit vaccine adjuvated with AS03 H5N1+AS03 29. The core transcriptional signature of these cells was an increased expression of interferon-stimulated genes (ISGs) (ISG15, GBP1, IFITM3, IFIH1, ANKRD22, IFI35, MX1, IRF1, IRF7, IRF8, STAT1) and decreased expression of AP-1 transcription factors FOS and JUN 9, 29. Therefore, we asked if a similar cell population would also emerge in response to mRNA vaccination in mice. We re-embedded C2_Mono_Macrophage and C15_Macrophage clusters using genes that characterized the epigenetically remodeled monocyte population enriched in humans 21 days post-vaccination with two doses of H5N1/AS03 29. Principal components were calculated from the 2000 most variable genes within the cells originally part of macrophage clusters C2 and C15. Applying Louvain community detection to the nearest-neighbor graph structure derived from PCA embeddings with a resolution of 0.2, the monocytes/macrophages were segregated into 12 subclusters (Fig. 4e). Cluster membership differences between cells derived from mRNA and YF-17D treated mice were mostly due to cells profiled on day 1 and day 3 (Fig. 4e). Strikingly, we found subclusters (3, 5, 7, and 8) that appeared within 1 and 3 days post-immunization with increased levels of Irf8, Stat1, Stat2, Irf1, and Stat3, and decreased levels of Atf3, Fos, Junb, and Jund, mirroring the innate immune cell populations described above (Fig. 4f; Extended Data Fig.3d).
BNT162b2 stimulates enhanced secondary innate immune response
A hallmark of the immune response to BNT162b2 vaccination in humans is the striking increase in the innate immune response following secondary vaccination, relative to the response after primary vaccination 9. To determine whether a similar effect was observed in mice, we measured serum cytokine/chemokine responses after the primary and secondary immunizations. Consistent with observation in humans 9, median IFNγ response increased 8.6-fold, from 44.5 pg/mL at 6 h after immunization (Day 0.3) to 383.1 pg/mL at 6 h after the secondary immunization (Day 21.3) (Fig. 5a). The data generated with the Luminex assay provided evidence that IFNγ and other cytokines like IL-2, CCL2, CCL4, and CCL5 were enhanced at 6 h after the secondary compared with those after the primary immunization, while there was not much difference between prime and boost with a 0.2 μg dose (Extended Data Fig.4a). Consistent with an increased IFNγ level after secondary immunization, innate cells, including monocytes, macrophages and DCs, were more activated after secondary than after primary immunization, as judged by the level of CD86 expression (Fig. 5b).
Figure 5. Enhanced innate immune response after boost is mediated by IFNγ production.
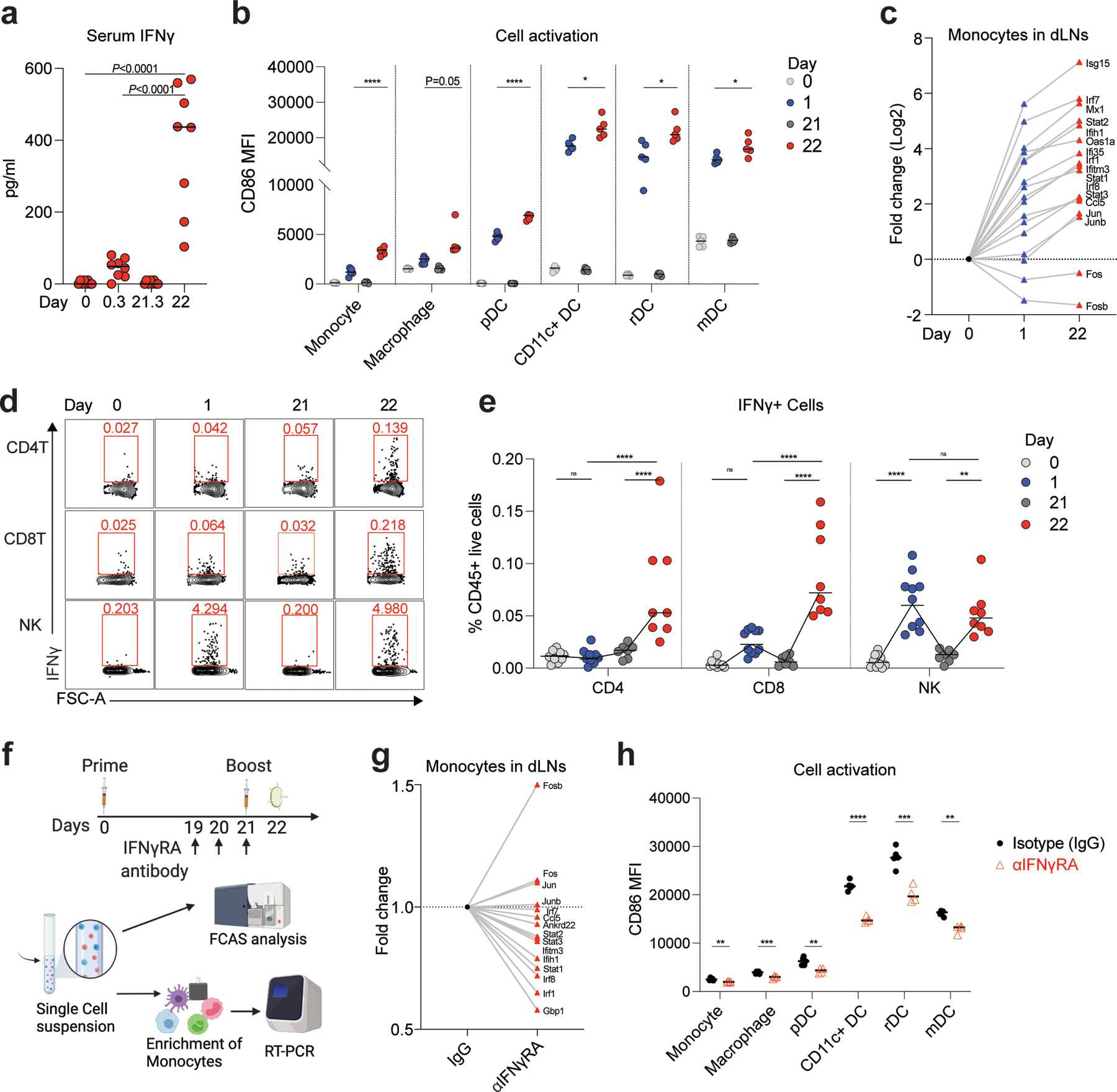
a, Serum IFNγ measured by ELISA at days 0, 0.3 (6 h post-prime), 21, and 21.3 (6 h post-boost). n=8 for each time point. b, Innate cell activation at days 0, 1, 21, and 22 after BNT162b2 immunization. n=5. c, Fold change of key interferon response genes and AP-1 transcription factors in monocytes at days 0, 1, and 22 after BNT162b2 immunization. d-e, Flow cytometry of cells producing IFNγ in dLNs at the indicated time. n=10 for day 0 and 1, n=7, 8 for day 21 and 22, respectively. f, Experimental design and timeline. g, Fold change of key interferon response genes and AP-1 transcription factors in IFNγRA neutralized group (αIFNγRA) against isotype (IgG) control. h, Innate cell activation at day 22 after IFNγRA neutralization. n=5 for IgG group, n=4 for αIFNγRA treated group. Data were combined from two independent experiments (a, e) or one representative of two independent experiments (b, c, g, h). One-Way ANOVA followed by Tukey’s test was applied in (a). P-values in (b, h) were determined using Student’s t-tests (two-tailed). Two-Way ANOVA followed by Tukey’s test was applied in (e). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
In addition, we purified monocytes and quantified the expression of key ISGs and AP-1 transcription factors that distinguished Cluster 8 from the other myeloid cells, defined in the human study with BNT162b2 9. All the key ISGs (Isg15, Mx1, Ifih1, Oas1a, Ifi35, Ifitm3, Ccl5, Irf1, Irf7, Irf8, Stat1, Stat2, and Stat3) were induced after the primary immunization, and importantly, the expression was increased further after the secondary immunization (Fig. 5c). Further, the AP-1 transcription factors Fos and Fosb were down-regulated (Fig. 5c), as in previous studies 9, 29. Next, in order to identify the cellular origin of plasma IFNγ, we performed ex vivo flow cytometry (Extended Data Fig.4b). Notably, while the NK cells produced IFNγ after both primary and secondary immunizations, CD4 and CD8 T cells contained the highest frequency of IFNγ-expressing cells on day 22 (Fig. 5d, e). To determine whether the IFNγ produced by T cells and NK cells within one day of secondary immunization acted on myeloid cells to induce the expression of ISGs, we blocked the activity of IFNγ in vivo with an IFNγ receptor neutralizing antibody against the IFNγ receptor, before secondary immunization on day 21 (Fig. 5f) and measured gene expression on day 22. We observed a significant reduction in the expression of all ISGs (Gbp1, Ifih1, Ifitm3, Ankrd22, Ccl5, Irf1, Irf7, Irf8, Stat1, Stat2, and Stat3) in monocytes of anti-IFNγ receptor antibody-treated mice (αIFNγRA) versus mice treated with an isotype control (IgG), while the AP-1 TFs like Fos, Fosb, Jun, and Junb were up-regulated (Fig. 5g). Furthermore, activation of monocytes, macrophages, and DCs was significantly reduced in anti-IFNγ receptor antibody-treated mice (Fig. 5h), suggesting that IFNγ signaling plays an important role in the innate immune activation induced by BNT162b2. We also measured spike-specific T cell and antibody responses on day 42 after IFNγ receptor blockade. We found that the antigen-specific CD4 and CD8 T cell responses did not differ significantly in the lung; nor was there a difference in antibody response (Extended Data Fig.4c–e). However, there was a ~2-fold reduction in the spike-specific CD8 T cells producing IFNγ in the spleen (Extended Data Fig.4c). To further delineate the relative importance of T cells versus NK cells in producing the burst of IFNγ on day 22, we depleted NK or T cells (CD4 and CD8 T cells) before boost using NK1.1 or CD4/CD8 depletion antibodies. The T cell depletion, but not NK cell depletion abrogated serum IFNγ on day 22 (Extended Data Fig.4f), 1 day after the boost demonstrating that the CD4 and CD8 T cells were the primary source of the circulating IFNγ. Consistent with this, the activation of innate immune cells measured by CD86 MFI was more substantially reduced in T cell-depleted mice than in the naïve mice or mice depleted of NK cells (Extended Data Fig.4g).
mRNA uptake by lymph node DCs and macrophages
Many characteristics of LNP-mRNA vaccines, including size of the LNP, pKa of the ionizable lipid, and lipid gradients, affect the tissue and cell specificity of the mRNA vaccine 15, 30. Using the scRNAseq data, we investigated the cells containing spike mRNA (S mRNA) in the dLNs. We observed high levels of spike mRNA reads on day 1 that decreased sharply by day 3 (Fig. 6a). The reads were primarily restricted to Clusters 2 and 4, which represent monocyte/macrophages and migratory DCs, respectively (Fig. 6a). Quantification of cells with at least one read per cell demonstrated that 40% of monocyte/macrophages and 20% of migratory DCs contained spike mRNA on day 1 (Fig. 6b). The frequency of these cells increased after vaccination (Fig. 6c). We also measured mRNA in various tissues using real-time PCR and found that the dLNs contained the highest concentration of mRNA (Fig. 6d). Interestingly, the spleen tissue also contained mRNA but at much lower concentration (Fig. 6d). Consistent with the transcriptional data, serum concentration of spike protein increased on day 1 and decreased by day 7 (Fig. 6e). There was a small amount of mRNA in other tissues such as muscle, liver, lung, and non-dLNs (Fig. 6e), consistent with previous reports 31. Finally, we analyzed the gene expression profile associated with the spike mRNA signals in macrophages and migratory DCs (Fig. 6f). In both cell clusters, the spike mRNA signal was associated strongly with IFN and innate immune response-related BTMs including type I interferon response (M127), activated DCs (M64 and M67) in C2, chemokine cluster (II) (M27.1), TLR and inflammatory signaling (M16) in C4, and RIG−I like receptor signaling (M68) and antiviral IFN signature (M75) in both C2 and C4 (Fig. 6f), suggesting direct activation of innate immune signaling by the mRNA. Notably, Ifih1, the gene encoding MDA5, and downstream targets of MDA5 signaling were highly correlated with the spike mRNA signal in monocytes and macrophages (Fig. 6g). On the other hand, expression of TLRs and MyD88 was highly associated with the spike mRNA levels in the migratory DCs (Fig. 6g).
Figure 6. Uptake of BNT162b2 by DCs and macrophages in dLNs.
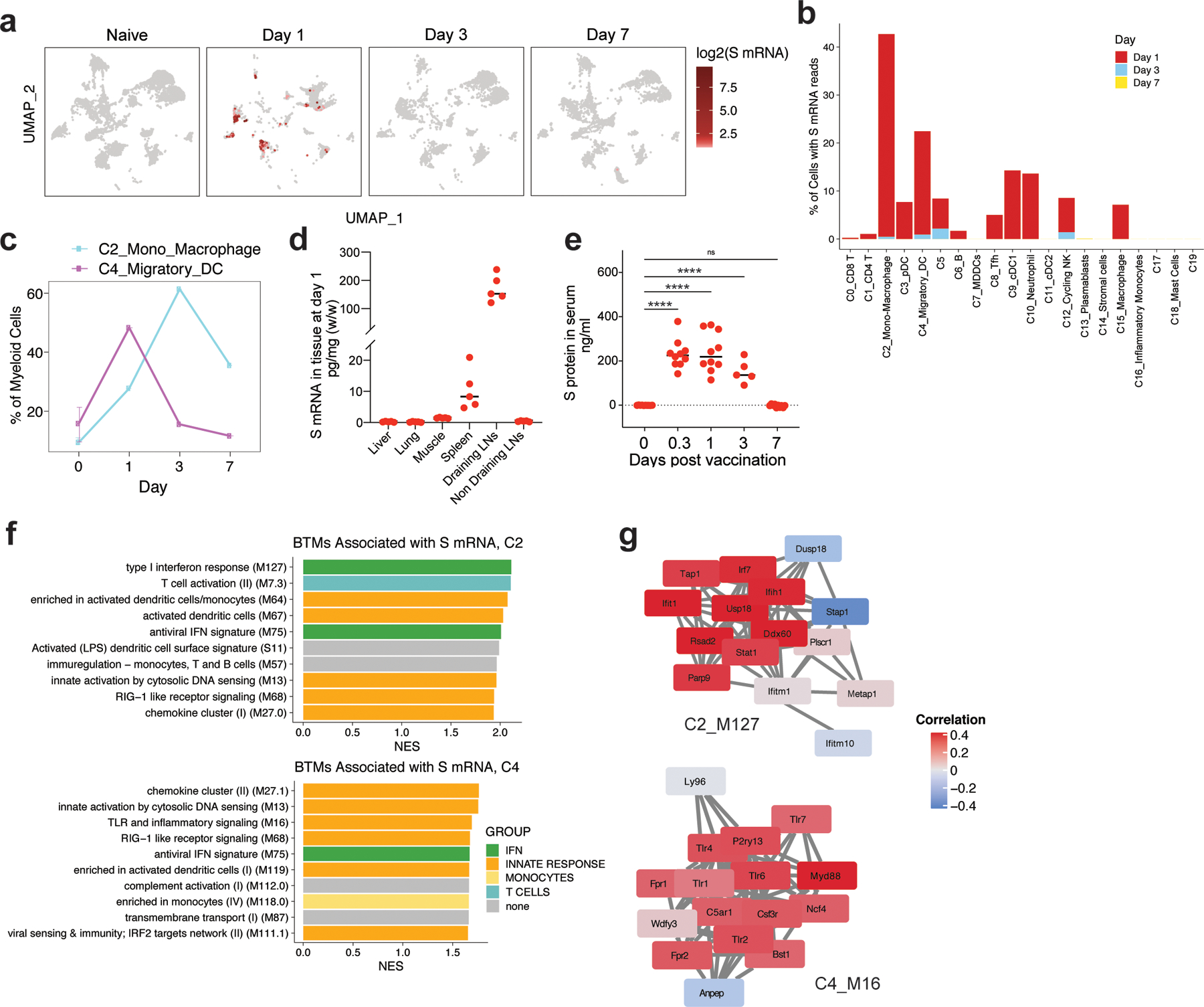
A, Distribution of S mRNA at the single-cell level (read >1/cell) from days 1 to 7 after BNT162b2 immunization. Cell clusters of UMAP are as shown in Fig. 4b. b, Percentage of cells with S mRNA reads in each cluster over time. c, Percentage of C2 and C4 in myeloid cells over time. d, Distribution of S mRNA in tissues at day 1 after BNT162b2 immunization measured by TaqMan based RT-PCR. n=5 for each tissue. e, S protein in mouse serum detected by ELISA. n=10 f, The BTMs associated with S mRNA signal in C2_Mono_Macrophage and C4_Migratory DC. g, The correlation between S mRNA signal and gene expression in BTM module M127 (Type I interferon response) in C2_Mono_Macrophage, and M16 (TLR and inflammatory signaling) in C4_Migratory DC, respectively. Samples used for scRNAseq were pooled from three independent experiments containing 8–10 mice. Data were one representative of two independent experiments (d) or combined from two independent experiments (e). One-Way ANOVA followed by Tukey’s test was applied in (e). ****P<0.0001.
BNT162b2 stimulates CD8 T cell responses via MDA5-IFNα axis
TLRs are the most studied sensors of pathogen-associated molecular patterns 32. TLR3 and TLR7 recognize double- and single-stranded RNA respectively 33, 34, 35. Therefore, we first examined whether deficiency of these receptors results in a diminished immune response to BNT162b2. However, immunization of Tlr3−/− or Tlr7−/− mice did not cause a reduction of antibody or T cell responses (Extended Data Fig.5a–c), suggesting that they are not the primary sensors of BNT162b2 in vivo. Previous evidence indicates that the SARS-CoV-2 spike protein binds to and stimulates TLR2 and TLR4 signaling 36, 37. Further, we have shown that the immune responses to the seasonal flu and other unadjuvanted vaccines are controlled by microbiota via the TLR5 pathway 38. Therefore, in addition to examining TLR3 and 7, we also measured antibody and T cell responses in Tlr2−/−, Tlr4−/−, and Tlr5−/− mice. None of the mice demonstrated an impaired immune response to BNT162b2 vaccination (Extended Data Fig.5d–i), providing evidence that these receptors are also not the primary sensors of BNT162b2.
Inflammasomes are critical regulators of innate immunity that regulate adaptive immunity to vaccines and adjuvants 39, 40. We examined the role of the inflammasome pathway using Asc−/− and Nlrp3−/− mice and found no significant difference in antibody or T cell responses in comparison to WT mice (Extended Data Fig.6a–c). Next, we examined the role of the cGAS-STING pathway, a cytoplasmic nucleic acid-sensing mechanism 41, 42. cGas−/− and Sting−/− mice did not display a reduced response (Extended Data Fig.7a–c), suggesting that these proteins do not play a role in immunogenicity of BTN162b2. Kim et al reported that Damage-Associated Molecular Patterns (DAMPs), released during cell death pathways such as necroptosis, regulate immunity to adjuvants 43. Like the adjuvant MF59 43, BNT162b2 induced DAMP signals, including double-stranded DNA and HMGB1 peaking at 24 h post-immunization, and uric acid peaking at 2 days post-immunization (Extended Data Fig.8a–c). However, Ripk3−/− and Gsdmd−/− mice displayed no effect of BNT162b2-mediated T cell and antibody responses compared with littermate controls (Extended Data Fig.8d–i).
The RIG-I-like receptor (RLR) family of proteins sense cytosolic RNA and are a second family of putative sensors of BNT162b244, 45. To evaluate whether RLRs are involved in mRNA recognition, we immunized mice deficient of MDA5. There was no reduction in the antibody responses (Fig. 7a, b). Interestingly, Mda5−/− mice had a striking reduction in the frequency of antigen-specific CD8 T cells as evidenced by fewer class I tetramer specific CD8 T cells and IFNγ-producing CD8+ T cells after primary (Extended Data Fig.9) and secondary immunizations (Fig. 7c, d) suggesting that the MDA5 sensing of BNT162b2 is responsible for the induction of spike-specific CD8 T cell responses. We also observed that the concentration of serum total IFNα was below the detection limit in Mda5−/− mice, while the WT B6 mice produced as much as 2 ng/mL of IFNα at 6 h measured by ELISA (Fig. 7e). Further, the activation of innate immune cells measured by CD86 expression was significantly downregulated in these mice (Fig. 7f). Since type I IFNs are known to directly stimulate clonal expansion of CD8 T cells46, 47, we examined whether mice deficient in IFNAR1 are likewise deficient in CD8 T cell responses. Consistent with our hypothesis, there was a striking reduction in spike-specific CD8 T cell frequencies enumerated by tetramer staining in the lung and spleen of Ifnar1−/− mice (Fig. 7g). In addition, there was also a significant but incomplete down-regulation in the frequency of spike-specific T cells secreting IFNγ (Fig. 7h); however, there was only a negligible effect on the antibody response (Fig. 7i). Moreover, the serum cytokine/chemokine responses (Fig. 7j, k) and the activation of innate immune cells measured by CD86 expression were also reduced (Fig. 7l). Collectively, these data show that MDA5 senses BNT162b2 to induce expression of IFNα, which stimulates antigen-specific CD8 T cell expansion via IFNAR1, to promote high magnitude of antigen-specific CD8 T cells. Thus, MDA5 deficiency leads to IFNα deficiency, which in turn leads to reduced CD8 T cell response after primary vaccination, which then leads to reduced recall responses. Finally, we also detected a reduced frequency of CD8 T cells, and negligible reduction of IgG titers in Batf3−/− mice (Extended Data Fig.10), which lack cross-presenting CD8α+ DCs48, suggesting the cross-presentation is important for BNT162b2 induced T cell responses.
Figure 7. MDA5-IFNAR1 axis is important for BNT162b2 induced CD8 T cell response.
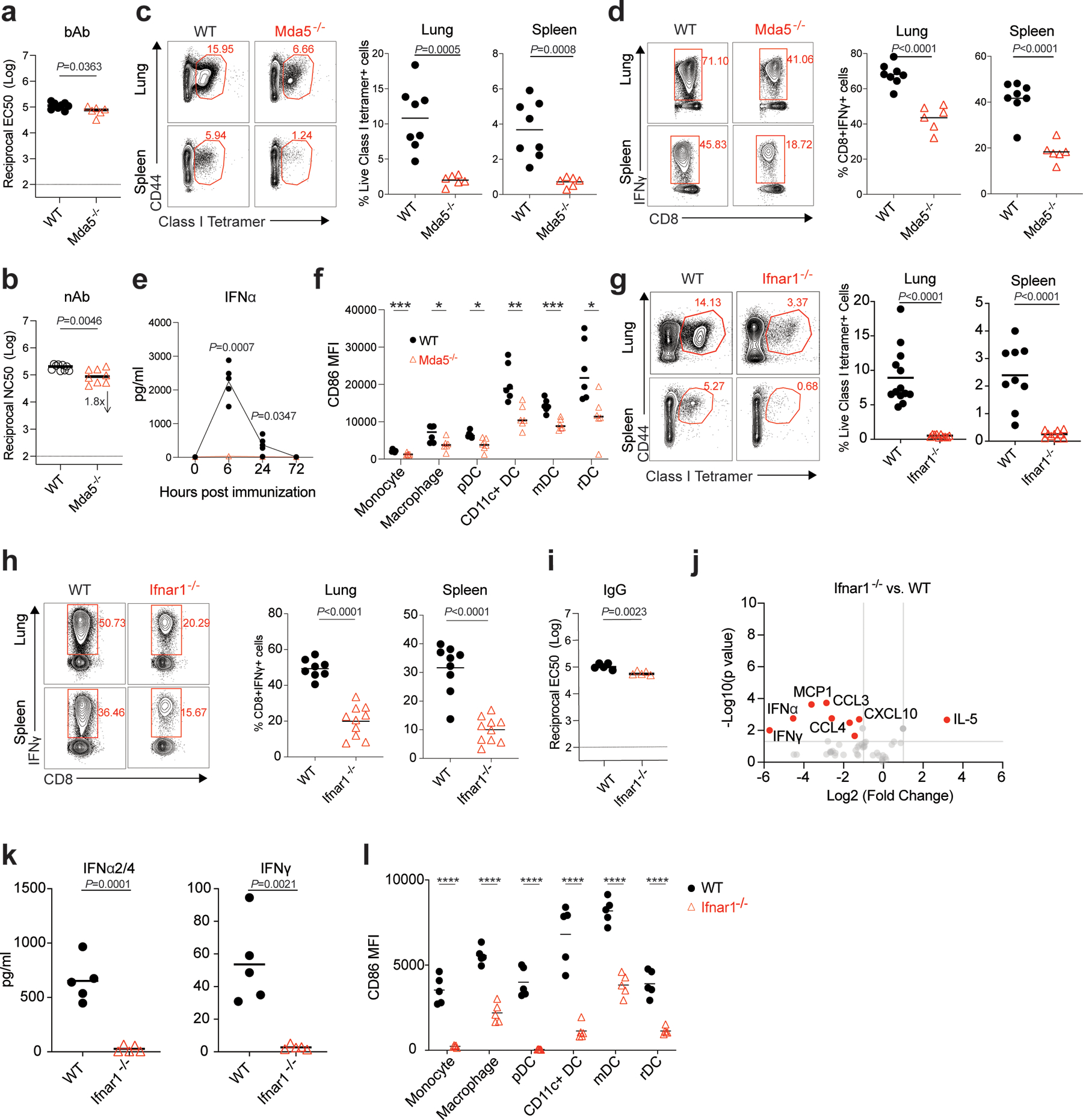
a-b, bAb (a) and nAb (b) titers of WT and Mda5−/− mice at day 42. In (a), n=8 for WT, n=6 for Mda5−/−. In (b), n=9 for WT, n=8 for Mda5−/−. c, Class I tetramer specific CD8 T cell response in the lung and spleen of WT and Mda5−/− mice measured at Day 42 after BNT162b2 immunization as shown in Fig. 1a. d, Antigen-specific CD8 T cell response in lung and spleen of WT and Mda5−/− mice detected at day 42 by intracellular cytokine staining (ICS) assay. In (c-d), n=8 for WT, n=6 for Mda5−/−. e, Serum total IFNα levels in WT (n=5) and Mda5−/− (n=4) mice at 0, 6, 24, and 72 h measured by ELISA. f, Activation of innate cells in WT and Mda5−/− mice at day 1 post-prime. n=6 for each group. g, Class I tetramer specific CD8 T cell response in lung and spleen tissue of WT and Ifnar1−/− mice measured at day 42. For Lung, n=13 for WT, n=10 for Ifnar1−/−; for spleen, n=9 for WT, n=10 for Ifnar1−/−. h, Antigen-specific CD8 T cell response in lung and spleen of WT and Ifnar1−/− mice detected at day 42 by ICS assay. For Lung, n=8 for WT, n=10 for Ifnar1−/−; for spleen, n=9 for WT, n=10 for Ifnar1−/−. i, IgG titers of WT and Ifnar1−/− mice at day 42. N=5 for each group. j, Cytokines/chemokines in serum of WT and Ifnar1−/− mice measured by Luminex at 6 h measured by Luminex. k, Serum IFNα2/4 (IFNα2 and IFNα4) and IFNγ levels in WT and Ifnar1−/− mice at 6 h. n=5 for each group. l, Activation of innate cells in WT and Ifnar1−/− mice at day 1 post-prime. n=5 for each group. Data were combined from at least two independent experiments (b-d, g-h) or one representative experiment (a, e-f, i-l). P-values were determined using Student’s t-tests (two-tailed). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Discussion
The current study, involving a detailed analysis of immune responses to vaccination of mice with the BNT162b2 vaccine, provided several new mechanistic insights (Fig. 8). As is the case in humans, after secondary immunization of mice, BNT162b2 induced a 10-fold greater magnitude of antigen-specific binding and neutralizing antibodies, and potent GC B cell and TFH responses. Surprisingly, there was a strikingly high magnitude of antigen-specific CD8+ T cells in the lung and spleen, after the secondary immunization. The higher magnitude of T cell response in lung than in spleen is not unique to the mRNA vaccine, as we have observed this in mice immunized with YF-17D49, and with AddaVax or MF59 plus Ova43. Experiments using a low dose (0.2 μg) of vaccine also resulted in similar B and T cell responses, but the magnitude was 5–10-fold lower than the high dose. Consistent with findings in humans9, we observed in mice enhanced innate immune responses, including enhanced plasma IFNγ concentrations, enhanced activation of DCs and myeloid cells in the dLNs, with a potent transcriptional signature of ISGs, and diminished expression of AP-1 transcription factor genes, after the secondary immunization with BNT162b2. We extended these findings to show that the CD4 and CD8 T cells were the primary source of serum IFNγ 1-day after secondary immunization, which results in the enhanced myeloid cell activation after the secondary immunization. The heightened IFNγ following secondary immunization could conceivably enhance innate antiviral immunity during the first few days or weeks of vaccination, a period during which antigen specific T and B cell responses are still nascent. In line with this notion, Wimmers et al showed that vaccination with H5N1+AS03 induces epigenetic changes in monocytes and myeloid DCs associated with enhanced IRF accessibility and heightened resistance to infection with unrelated blood-borne viruses, such as dengue virus and Zika virus29. Whether BNT162b2 can also cause the epigenetic changes on a genomic level to provide broader protection against other infectious diseases remains an open question in both humans and mice.
Figure 8. Cartoon of mode of action of BNT162b2 in vivo.
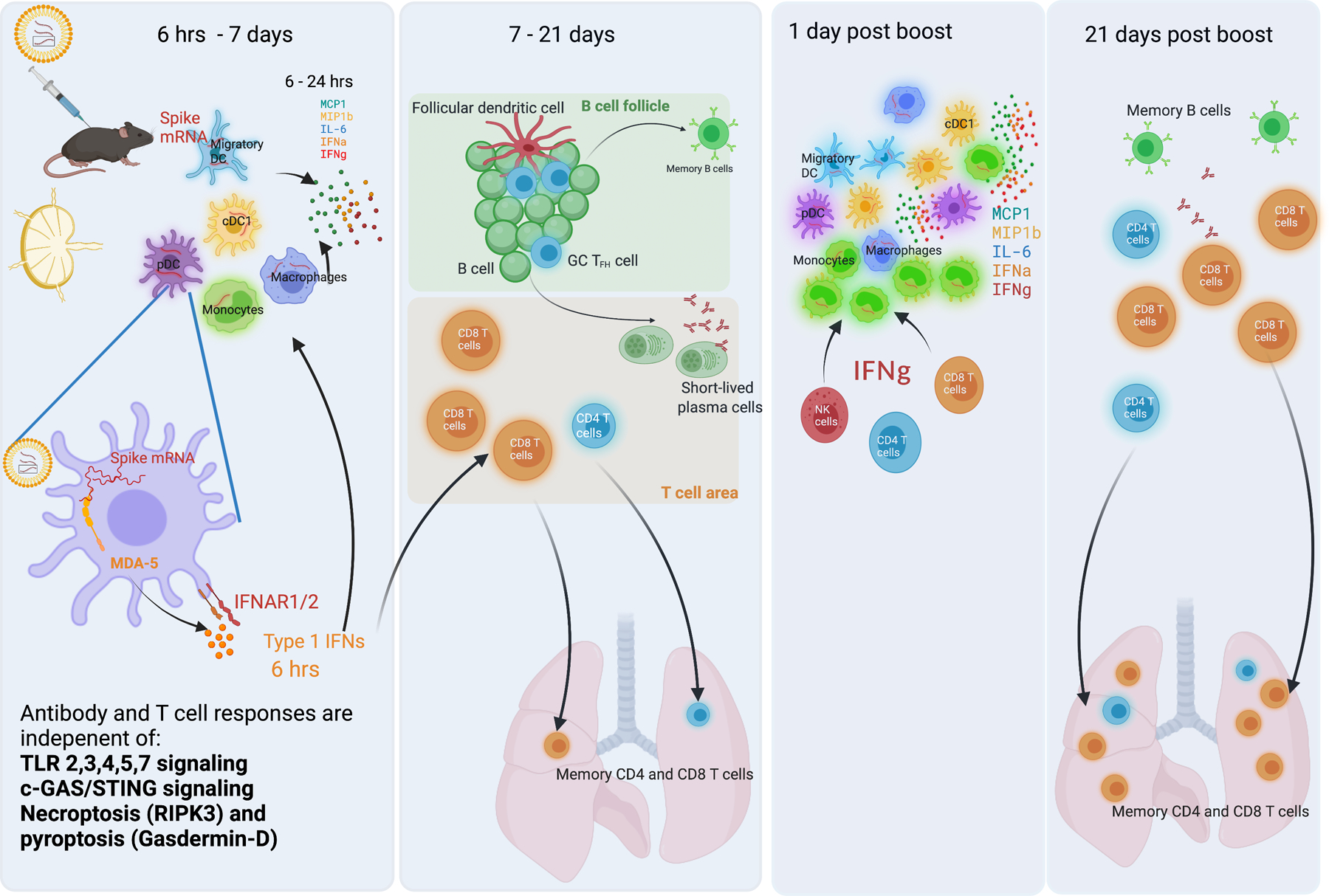
a. At 6 hrs-7 days post-BNT162b2 prime, DCs and macrophages in dLNs take up the vaccine and express spike proteins. These innate cells are highly activated and produce cytokines, including IL-6, IFNα, IFNγ, MCP1, and MIP1b. In this process, the MDA5-IFNAR1 signaling pathway is essential for IFNα production and innate cell activation. b, At days 7 to 21, strong GC B and TFH responses are induced in B cell follicle. CD8 T cell response is increased mildly in both spleen and lung tissue. c, At days 1 to 3 days post-boost, much more IFNγ is produced by NK, CD4, and CD8 T cells, contributing to the enhanced innate cell activation after boost. d, At 21 days post-boost, the antibody response, CD4, and CD8 T cell responses increase significantly. The MDA5-IFNAR1 signaling pathway is essential for CD8 T cell response in both spleen and lung tissue.
The distribution and dynamics of the BNT162b2 mRNA and the spike protein encoded by it, in tissues and cell types in vivo remains unclear. In this study we found that BNT162b2 mRNA could be detected in migratory DCs and monocyte/macrophage subsets in the dLNs for at least 7 days following intramuscular immunization. Furthermore, mRNA could be detected in the spleen, and the spike protein itself was detectable in the serum, for up to 7 days post immunization.
Another insight to emerge from this study is how that innate immune system senses the BNT162b2 mRNA vaccine and modulates adaptive immunity. By analyzing responses in various strains of knockout mice, we found that induction of antigen-specific T and antibody responses was independent of TLRs 2,3,4,5 and 7, or the STING-cGAS DNA sensing pathway, and independent of NLRP3 dependent ASC inflammasome activation. Furthermore, mice deficient in RIP3 kinase and Gasdermin D, the central mediators of necroptosis 50, 51 and pyroptosis 52 respectively, mounted normal T and antibody responses to BNT162b2, arguing that these cell death pathways were not essential for the immunogenicity of this vaccine. However, we found that the MDA5-IFNAR1 signaling pathway is critical for the CD8+ T cell response induced by BNT162b2. A previous study showed that MDA5 deficiency resulted in a modest impairment of antigen-specific CD8 T response following primary immunization with OVA+polyI:C, but a more pronounced impairment of secondary T cell responses53. MDA5 deficiency affects clonal expansion of CD8 T cells after primary as well as secondary immunization with BNT162b2 via impaired production of IFNα, within the first few hours of primary vaccination. These data are consistent with studies showing a critical role for type I IFN responses in the clonal expansion of antigen-specific CD8 T cells46. mRNA is known to spontaneously form dsRNA structures, and it is likely that MDA5 recognizes these double stranded elements54 in the target cells.
Whether these findings with BNT162b2 are applicable to other mRNA vaccines, such as mRNA-1273 from Moderna 55 and MRT5500 from Sanofi 56, remains to be determined. Although our data provides no evidence for a role for TLRs 2, 3, 4, 5 and 7, Alameh et al showed that GCB and Tfh responses induced by a different mRNA vaccine (a nucleoside-modified HA mRNA-LNP vaccine) is dependent on MyD88 but not MAVS57. Whether this difference is due to synergistic triggering through multiple TLRs, or differences between the BNT162b2 vaccine from Pfizer-BioNTech versus the vaccine used by Alameh et al remains to be determined.
As will be appreciated, the aforementioned experiments in mice have revealed several new mechanistic insights about the innate and antigen-specific T and B cell responses induced by the BNT162b2 mRNA vaccine. A key question relates to relevance of these various responses to protection against infection and disease caused by SARS-CoV-2. Israelow et al. found that mRNA-mediated protection from SARS-CoV-2 infection in mouse models is primarily mediated by humoral responses while the cell-mediated immune responses are efficient in clearing infection24. The extent to which the immunological defects observed in the MDA5−/− and IFNAR−/− mice, as well the defects in secondary innate immunity caused by blockade of IFNγ activity prior to secondary vaccination, remains to be determined. Finally, it should be remembered that whilst the immune systems of mice and humans are broadly similar, nearly 80 million years of evolutionary divergence of these species have resulted in many differences in the details of how their immune systems work 58. Therefore, the results obtained in mice should be interpreted in the context of similar studies in humans, in order to arrive at an integrated understanding of how our immune systems sense and respond to mRNA vaccines 58, 59, 60. In summary, our study provides rich insights into the mechanism of BNT162b2’s effects in vivo. These results not only help us better understand the mechanism of the BNT162b2 vaccine but will also help in the design of improved vaccines in the future.
Methods and Reagents Details
Mice and immunization
C57BL/6, B6129SF2/J, Tlr2−/−, Tlr3−/−, Tlr4−/−, Tlr5−/−, and Tlr7−/− mice were purchased from Jackson Laboratories. Ripk3−/−, Gsdmd−/−, and littermates were gifts from Genentech Inc (Alex Gitlin). Ifnar1−/−, Mda5−/−, Sting−/−, cGas−/−, and Batf3−/− mice were bred in our animal facility at Stanford University. Asc−/− and Nlrp3−/− mice were initially obtained from Genentech Inc (Vishva Dixit) and maintained in our private colony at Jackson Laboratories. Tlr3−/− mice are on B6129SF2/J background, other strains are on C57BL/6 background. Mice were sex-matched and aged between 8 and 14 weeks.
Discarded remnant material (Pfizer/BioNTech) was used within manufacturer’s guidelines for stability at 50 μL of BNT162b2 mRNA vaccine (5 μg/mouse or 0.2 μg/mouse), as described in previous study 61. In addition, since this was not available for purchase and since only remnant (otherwise to be discarded) material could be used at the time (since the product was approved for EUA only), we obtained approval from the Dean of Research at Stanford, Office for General Counsel at Stanford, and the FDA-CBER Vaccines Division). For immunization, mice were injected intramuscularly with 50 μL of BNT162b2 mRNA vaccine (5 μg/mouse or 0.2 μg/mouse) or subcutaneously at the base of the tail with 100 μL of 106 PFU YF-17D. All mice in this study were maintained under specific pathogen-free conditions, 12 light/12 dark cycle, temperatures of ~18–23°C with 40–60% humidity, and handled according to the protocol approved by the Institutional Animal Care and Use Committee (IACUC) of Stanford University.
Flow cytometry analysis of innate cells
Draining iliac lymph nodes from BNT162b2 immunized mice, draining inguinal lymph nodes from YF-17D immunized mice, along with whole lung or spleen, were harvested and digested with 1 mg/mL collagenase type IV (Worthington) for 20 min at 37 °C, followed by smashing with a 100 μm strainer to make a single-cell suspension. Red blood cells from the lung and spleen were lysed before staining. Single-cell samples were then stained with Zombie UV™ (1:300, BUV496; BioLegend #423107), anti-Ly6C (1:500, BV780; BioLegend #128041), anti-Ly6G (1:400, APC-Cy7; BioLegend #127624), anti-CD19 (1:100, BB700; BD #566411), anti-CD3 (1:100, BB700; BD #742175), anti-MHCII (1:400, AF700; eBioscience #56-5321-82), anti-CD11b (1:300, BV650; BioLegend #101239), anti-CD11c (1:400, BV421; BioLegend #117330), anti-CD86 (1:300, A647; BioLegend #105020), anti-Siglec-F (1:400, PE-CF594; BD #562757), anti-CD45 (1:200, BV610; BioLegend #103140), anti-CD169 (1:200, PE-Cy7; BioLegend #142412), anti-PDCA-1 (1:200, BUV563; BD #749275), anti-CD8a (1:200, BUV805; BD #612898), anti-CD103 (1:100, PE; eBioscience #12-1031-82), anti-NK1.1 (1:200, BV510; BioLegend #108738), and anti-F4/80 (1:100, BUV737; BD #749283). Data was collected on a BD FACSymphony analyzer with BD FACSDiva (v8.0.1).
Flow cytometry of GC B, TFH, and plasma cells in lymph nodes
dLNs were harvested and smashed with a 100 μm strainer to make a single-cell suspension followed by staining for viability with Ghost Dye Violet 510 (Tonbo Biosciences) for 5 min on ice in 1x PBS-2mM EDTA. After washing out viability dye, cells were blocked with Fc receptor antibody α-CD16/32 (clone: 2.4G2, BD) for 5 min on ice prior to staining with fluorochrome-conjugated antibodies in FACS staining buffer (1x PBS, 3% FBS, 1mM EDTA, 0.02% sodium azide): CD3 (1:100, clone: 17A2, BioLegend), CD4 (1:200, clone: GK1.5, BioLegend), CXCR5 (1:50, clone: L138D7, BioLegend), PD1 (1:200, clone: 29F.1A12, BioLegend), CD19 (1:200, clone:6D5/CD19, BioLegend), CD95 (1:200, clone: Jo2, BD Biosciences), CD38 (1:200, clone: 90, BioLegend), CD44 (1:300, clone: IM7, BioLegend), CD138 (1:200, clone: 281–2; BD Biosciences). Surface staining was carried out for 30 min on ice followed by fixation for 10 min in room temperature. Data were acquired in LSR-II and analyzed using FlowJo analysis software v10.
Intracellular cytokine staining assay
Whole Spleen and lung were harvested at 42 days post-prime (21 days post-boost). Briefly, mononuclear populations from the lung were isolated from the interphase of a 70–40% Percoll gradient of single suspension prepared by enzymatic digestion with 1 mg/mL type IV collagenase and DNase I. Single-cell suspensions from the spleen were prepared without digestion. Cells were plated at ~2 × 106 cells/well in 96-well U-shaped plates and re-stimulated with S-specific overlapping peptide pools (1 μg/mL of each peptide) in complete RPMI1640 medium for overnight incubation at 37 °C in the presence of brefeldin-A (10 μg/mL). At day 2, cells were stained with Ghost Dye Violet 510 (Tonbo Biosciences) for 10 min on ice in 1x PBS-2mM EDTA. After washing out viability dye, cells were blocked with Fc receptor antibody α-CD16/32 (clone: 2.4G2, BD) for 5 min on ice prior to staining with fluorochrome-conjugated antibodies in FACS staining buffer (1x PBS, 2% FBS): CD3 (1:50, clone:145-2C11, BioLegend), CD8α (1:200, 53–6.7, BioLegend), CD4 (1:200, clone: RM4-5, BioLegend), CD44 (1:400, clone: IM7, BioLegend), CD69 (1:200, clone: H1.2F3; BioLegend), CD45 (1:200, Clone 30-F11, BioLegend). Cells were incubated for 30 min on ice for surface staining followed by fixation and permeabilization in BD fix perm buffer and then stained intracellularly with fluorochrome-conjugated antibodies in fix perm buffer: IFNγ (1:100, XMG1.2, BioLegend), TNF (1:100, clone: MP6-XT22, BioLegend), IL-2 (1:100, clone: JES6-5H4, BioLegend), IL-4 (1:100, clone:11B11, BioLegend) per the manufacturer’s recommendation. Data were acquired in LSR-II and analyzed using FlowJo v10.
Antigen-specific tetramer and tissue-resident memory T cell staining
After staining cells with viability dye, cells were surface stained with anti-CD3 (1:50, clone:145-2C11, BioLegend), CD8α (1:200, 53–6.7, BioLegend), CD4 (1:200, clone: RM4-5, BioLegend), CD44 (1:400, clone: IM7, BioLegend), CD45 (1:200, clone: 30-F11, BioLegend), CD69 (1:200, clone: H1.2F3, BioLegend), CD103 (1:200, clone: 2E7, BioLegend), Spike S specific Tetramer (1:200, residues 539–546, VNFNFNGL, H-2K(B)), followed by fixation. Data were acquired in LSR-II and analyzed with FlowJo v10.
Flow cytometry of IFNγ-producing cells in lymph nodes
After staining cells with viability dye, cells were surface stained with anti-CD3 (clone:145-2C11, BioLegend), CD8α (53–6.7, BioLegend), CD4 (clone: RM4-5, BioLegend), CD44 (clone: IM7, BioLegend), CD45 (Clone 30-F11, BioLegend), NK1.1 (clone: PK136, BioLegend), and TCR delta (1:100, clone: Gl3, BioLegend), fixed and permeabilized in BD fix perm buffer, and stained intracellularly with anti-IFNγ (XMG1.2, BioLegend) per manufacturer’s the recommendation. Data were acquired in LSR-II and analyzed with FlowJo v10. To block IFNγ receptor, 1mg/mouse of anti IFNγ receptor neutralizing antibody (clone:2E2, Bioxcell, BE0287) or isotype control (IgG, Bioxcell, BE0091) were given by intraperitoneal at day 19, 20, and 21 after prime. Then mice were boosted at day 21 after prime, and lymph nodes were harvested at day 1 post boost. To delete T cells or NK cells, 0.3mg/mouse of anti-mouse CD8 (clone:YTS 169.4, Bioxcell, BE0117), anti-mouse CD4 (clone: GK1.5, Bioxcell, BE0003-1), or anti-mouse NK1.1 (clone:PK136, Bioxcell, BE0036) or isotype control (clone:LTF-2, Bioxcell, BE0090 for anti-CD4/CD8, or clone:C1.18.4, Bioxcell, BE0085 for anti-NK1.1) were given by intraperitoneal at day 20 post prime. Mice were boosted at day 21. Serum IFNγ and innate cells activation in spleen were checked at day 22.
Luminex assay
The assay was performed by the Human Immune Monitoring Center at Stanford University. Mouse 48-plex Procarta kits (EPX480-20834-901) were purchased from Thermo Fisher and used according to the manufacturer’s recommendations with modifications as described below. Beads were added to a 96-well plate and washed in a BioTek ELx405 washer. Samples were added to the plate containing the mixed antibody-linked beads and incubated overnight at 4 °C with shaking. Cold (4 °C) and room temperature incubation steps were performed on an orbital shaker at 500–600 rpm. Following the overnight incubation, plates were washed in a BioTek ELx405 washer and biotinylated detection antibody was added for 60 min at room temperature with shaking. Plates were washed as described above and streptavidin-PE was added. After incubation for 30 min at room temperature, a wash was performed as above and reading buffer was added to the wells. Each sample was measured in singlets. Plates were read on a FM3D FlexMap instrument with a lower bound of 50 beads per sample per cytokine/chemokine. Custom Assay Chex control beads were purchased from Radix BioSolutions, and were added to all wells.
Anti-S binding ELISA
SARS-CoV-2 Spike protein was purchased from Sino Biologicals (40589-V08B1). High binding plates (96-well) were coated with 100 ng of S protein diluted at a concentration of 2 mg/mL in PBS. The plates were washed once and blocked with 3% non-fat milk for 1 h at room temperature. Sera samples serially diluted in 1% non-fat milk containing PBS were added to the plates and incubated at 37 °C for 1 h. The plates were washed 3X with PBS-T, and horseradish peroxidase conjugated goat anti-mouse IgG, IgG1, or IgG2c (SouthernBiotech, 1:6000 dilution) in PBS-T containing 1% non-fat milk was added and incubated for 1 h at room temperature. Wells were washed three times with PBS-T before addition of 3,3’,5,5’-tetramethylbenzidine substrate solution (Thermo Pierce). The reaction was stopped after 3 min by addition of 0.16 M sulfuric acid. The optical density at 450 nm was measured with a Bio-Rad microplate reader.
Pseudotyped lentivirus virus production
Viral transfections were done in HEK293T cells using Bio T transfection reagent. Five million cells were seeded in D10 medium (DMEM + 10% FBS, L-glutamate, penicillin, streptomycin, and 10 mM HEPES) in 10-cm plates one day prior to transfection. A five-plasmid system 62 was used for viral production. The Spike vector contained the 21 amino acid truncated form of the SARS-CoV-2 Spike sequence from the Wuhan-Hu-1 strain of SARS-CoV-2. Plasmids were added to D10 medium in the following ratios: 10 μg pHAGE-Luc2-IRS-ZsGreen, 3.4 μg FL Spike, 2.2 μg HDM-Hgpm2, 2.2 μg HDM-Tat1b, 2.2 μg pRC-CMV-Rev1b in a final volume of 1 mL; 30 μL of Bio T were then added. Transfection reactions were incubated for 10 min at room temperature, and then added up to 10 mL with D10 medium. This mixture was added slowly to plated cells. Culture medium was removed 24 h post-transfection and replaced with fresh D10 medium. Viral supernatants were harvested 72 h post-transfection by spinning at 300 × g for 5 min followed by filtering through a 0.45 μm filter. Viral stocks were aliquoted and stored at −80 °C until ready for use.
Neutralization assay
The target cells used for infection in viral neutralization assays were from a HeLa cell line stably overexpressing the SARS-CoV-2 receptor, ACE2, as well as the protease known to process SARS-CoV-2, TMPRSS2 63. ACE2/TMPRSS2/HeLa cells were plated one day prior to infection at 5,000 cells per well or two days prior to infection at 2,500 cells per well. White-walled, clear-bottom, 96-well plates were used for the assay (Thermo Fisher Scientific). On the day of the assay, dilutions of serum were made into sterile D10 medium to a final volume of 30 μL. Samples were run in technical duplicate in each experiment. All other wells contained only D10 medium. A virus mixture was made containing the virus of interest (for example, SARS-CoV-2 with a 21 amino acid deletion on the C terminus), D10 medium (DMEM + 10% FBS, L-glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, and 10 mM HEPES), and polybrene. Virus dilutions into medium were selected such that a suitable signal would be obtained in the virus-only wells. A suitable signal was selected such that the virus-only wells would achieve a luminescence of at least >10,000 RLU. Sixty microliters of this virus mixture were added to each of the serum dilutions to make a final volume of 120 μL in each well. Virus-only wells were made containing 60 μL D10 medium and 60 μL virus mixture. Cells-only wells were made containing 120 μL of D10 medium. The diluted serum/virus mixture was left to incubate for 1 h at 37 °C. Following incubation, the media was removed from the cells on the plates made one or two days prior, replaced with 100 μL of diluted serum/virus dilutions, and incubated at 37 °C for approximately 48 h. Infectivity readout was performed by measuring luciferase with a microplate reader (BioTek). Normalized values were fit with a three-parameter non-linear regression inhibitor curve in GraphPad Prism (v9.2.0) to obtain IC50 values.
S protein detection in serum
SARS-CoV-2 (2019-nCoV) Spike Detection ELISA Kit (Sino Biological, KIT40591) was used to measure S protein in mouse serum according to the manufacturer’s protocol. First, plates were washed three times followed by the addition of standard and diluted samples (2 μL mouse serum was added into 98 μL dilution buffer). Plates were then incubated for 2 h. After incubation, plates were washed three times and incubated with detection antibody for 1 h. Again, the plates were washed three times and 200 μL of substrate solution were added for 20 min. Finally, 50 μL stop solution were added and the optical density at 450 nm was measured with a microplate reader (Bio-Rad). All incubations were conducted at room temperature.
S mRNA vaccine detection by RT-PCR
Tissues were taken at day 1 post-immunization. Tissues were weighted and homogenized with a bead miller (Fisher). Total RNA was purified with a PureLink RNA mini kit (Invitrogen). RNA purified from BNT162b2 mRNA vaccine was diluted serially and served as the standard. A Luna universal probe one-step RT-PCR kit (NEB) was used to measure cycle threshold (Ct) of purified RNA with mRNA vaccine-specific primers (mVac-F: 5’-TACCAAGCTGAACGACCTGT, mVac-R: 5’-TTGCTGTTCCAGGCAATCAC, mVac-Probe: 5’-FAM-TGCCCGACGACTTCACCGGC-IBFQ).
IFNγ, IFNα, and DAMP signal detection
Serum IFNγ and IFNα levels were determined in serum samples by ELISA. Serum IFNγ was detected with the Quantikine ELISA kit (R&D Systems, MIF00). IFNα in serum was measured using ELISA sets (PBL, 421201). HMGB1 was detected with the Chemiluminescence ELISA kit (Novus Biologicals, NBP262782). ELISAs were performed according to the manufacturer’s instructions. Serum dsDNA concentrations were determined with a Quant-iT PicoGreen dsDNA Kit (Invitrogen, #P11496). Uric acid in serum was measured with a Uric Acid Assay Kit (abcam, #ab65344).
scRNAseq samples and preparation
Draining iliac lymph nodes from BNT162b2 immunized mice and draining inguinal lymph nodes from YF-17D immunized mice were harvested and digested into single-cell suspensions. One million total cells were set aside on ice. The rest of the cells were incubated with biotinylated CD3, CD19, and NK1.1 Ab on ice for 20 min. Streptavidin-labeled magnetic beads (BD IMag™ Streptavidin Particles Plus-DM) were added and incubated on ice for 30 min. Unbonded cells were collected and CD11b+ myeloid cells, pDCs (CD11clo, CD11b-PDCA-1+), and DCs (CD11c+MHC-II+) were sorted with flow cytometry. Sorted cells were combined with total lymphocytes at a 1:1 ratio and resuspended in cold PBS supplemented with 1% BSA (Miltenyi) and 0.5 U/μL RNase Inhibitor (Sigma Aldrich). Cells were partitioned into Gel Beads-in emulsion (GEMs) using the 10x Chromium 3’ V3 chemistry system (10x Genomics). The released RNA was reverse transcribed in the C1000 touch PCR instrument (Bio-Rad), in accordance with the manufacturer’s recommendations. Barcoded cDNA was extracted from the GEMs by post-GEM RT-cleanup and amplified for 12 cycles. Amplified cDNA was subjected to 0.6x SPRI beads cleanup (Beckman, B23318). Twenty five percent of the amplified cDNA was subjected to enzymatic fragmentation, end repair, A-tailing, adapter ligation, and 10X specific sample indexing as per the manufacturer’s protocol. Sequencing libraries were generated and the quality was assessed through Bioanalyzer (Agilent) analysis. Libraries were pooled and sequenced on the HiSeq 4000 instrument (Illumina) with a targeted read depth of 40,000 read pairs/cell.
sc-RNAseq data analysis
Cell Ranger v3.1.0 (10x Genomics) was used to quantitate transcript levels against the 10x Genomics GRCh38 reference (v3.0.0). Raw count data were filtered to remove cells with a mitochondrial RNA fraction >20% of total RNA counts per cell, cells with <100 unique features, or cells with <200 total reads. The filtered count matrix was used to create a Seurat 64 (v3.1.4) object. Filtered read counts were scaled by a factor of 10,000 and log transformed. Doublets were identified with scds 65 (v1.2.0); cells with a doublet score in the top decile were removed. The remaining 52,788 cells were processed with the default Seurat pipeline. Specifically, the most variable 2000 RNA features were used to perform principal component analysis on the log-transformed counts. The first 25 components were used for further downstream analyses, including clustering and UMAP projections. Clusters were identified with Seurat SNN graph construction followed by Louvain community detection on the resultant graph with a resolution of 0.2, yielding 20 clusters. Differential expression for each timepoint compared to the baseline was calculated with a Wilcox rank-sum test. Gene-set enrichment analysis was run using all genes ranked by Wald statistic.
Macrophage clusters C2 and C15 were re-embedded using genes that characterized an epigenetically remodeled monocyte population enriched in humans 21 days post-vaccination with two doses of H5N1/AS03 29. Principal components were calculated from the most variable genes within this set; Louvain community detection was again used with a resolution of 0.2 to determine cluster identity. Complex Heatmap (v2.2.0) was used for all heatmaps. All analysis was performed in R (v3.6.3).
Data availability
Single-Cell RNAseq data are publicly accessible in the GEO under accession numbers GSE179131. All analyses and visualizations were performed in R (V3.6.3).
Code availability
Computer code is available upon reasonable request.
Quantification and statistical analysis
scRNAseq statistical analysis was completed as described above. All other statistical analysis was performed with Prism (GraphPad Software v9.2.0). For comparing two groups, P-values were determined using Student’s t-tests (two-tailed). For comparing more than two groups, one-Way ANOVAs followed by Tukey’s test were applied. Differences between groups were considered significant for P-values < 0.05. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications 38, 43. Mice were assigned to the various experimental groups randomly. Data collection and analysis were not performed blind to the conditions of the experiments. Data distribution was assumed to be normal, but this was not formally tested. No data points were excluded from the analyses.
Extended Data
Extended Data Fig. 1. Immune response induced by low immunization dose in mice.
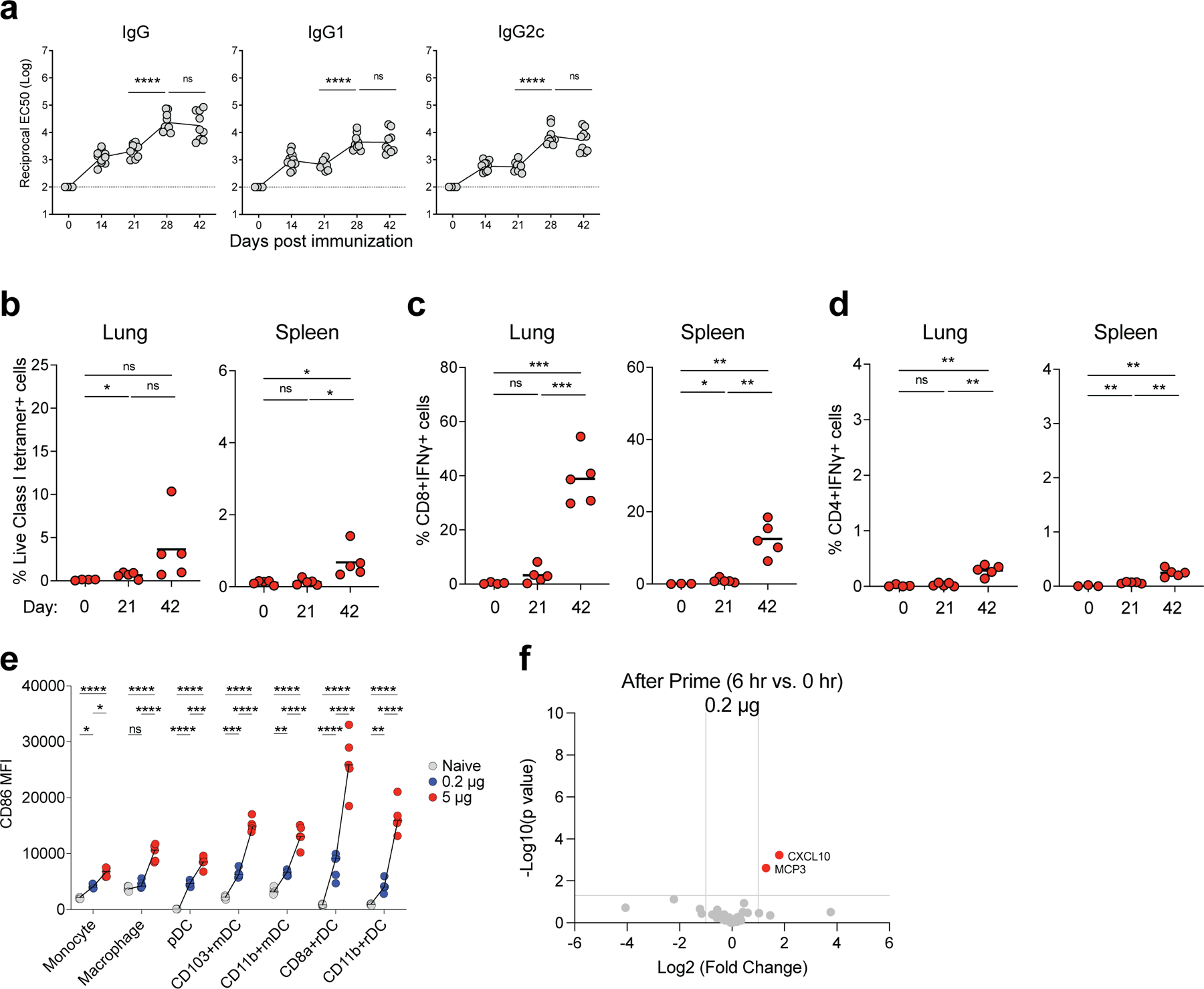
a, S binding titers induced by low dose (0.2 μg/mouse) of BNT162b2 immunization measured by ELISA. b, Class I tetramer specific CD8 T cell response in lung and spleen tissue of mice measured at days 21 and 42 after BNT162b2 immunization (0.2 μg/mouse). c-d, Antigen specific CD8 (c) and CD4 (d) T cell response in lung and spleen tissue of mice detected at days 21 and 42 by intracellular cytokine staining (ICS) assay. e, Comparison between the activation of innate cells in dLNs induced by low (0.2 μg/mouse) and high immunization doses (5 μg/mouse). f, Cytokines/chemokines produced at 6 h post prime with low (0.2 μg/mouse) dose detected by Luminex assay. Data were combined from two independent experiments (a) or one representative experiment (b-f). One-Way ANOVA followed by Tukey’s test was applied (a-e). P-values were determined using Student’s t-tests (two-tailed) (f). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Extended Data Fig. 2. Innate cells activated by BNT162b2.
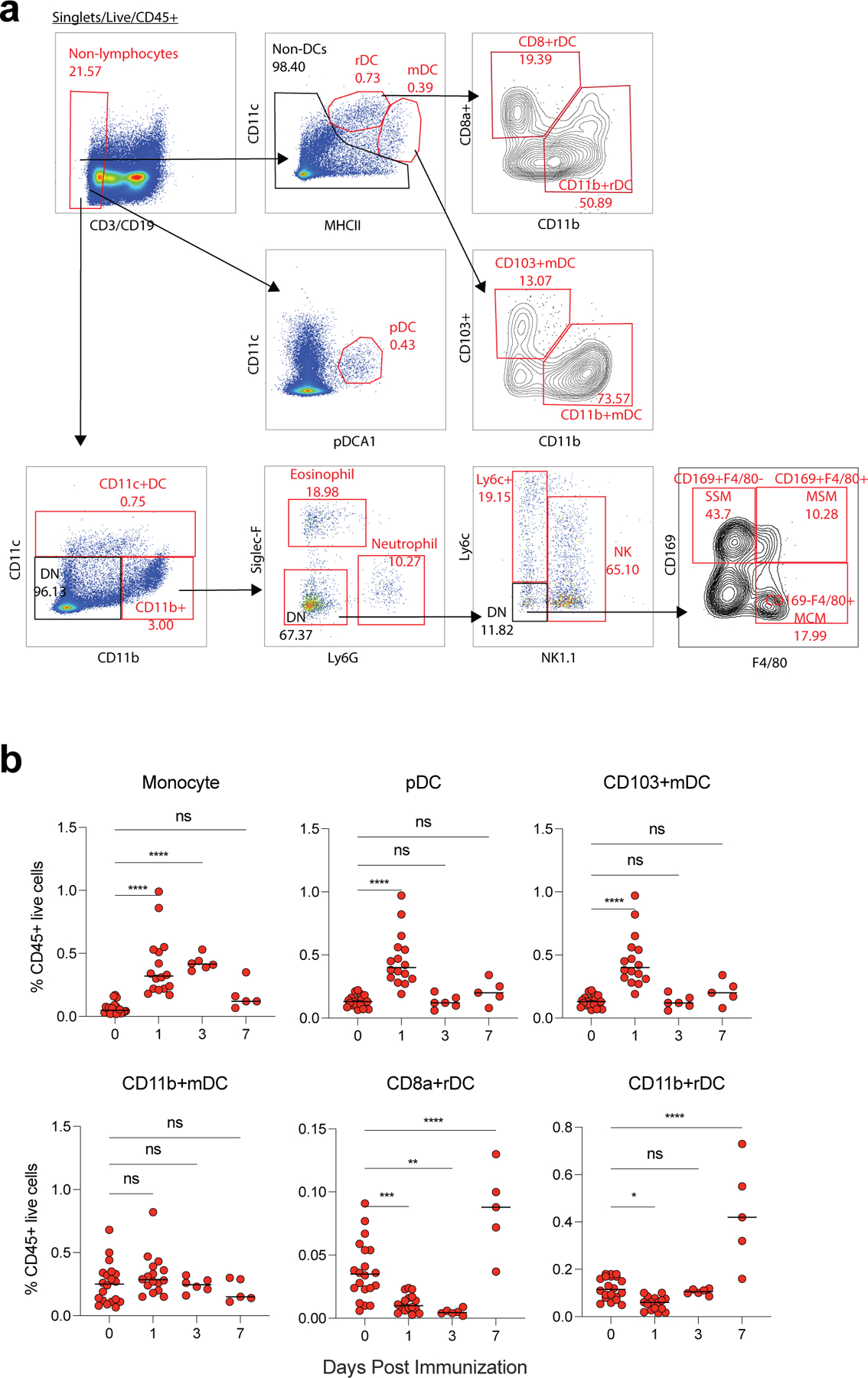
a, Gating strategy. b, Frequency of innate cells in live CD45+ cells after BNT162b2 immunization in dLNs. Data were combined from at least two independent experiments. One-Way ANOVA followed by Tukey’s test was applied in (b). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Extended Data Fig. 3. scRNAseq analysis of immune response induced by BNT162b2 and YF-17D.
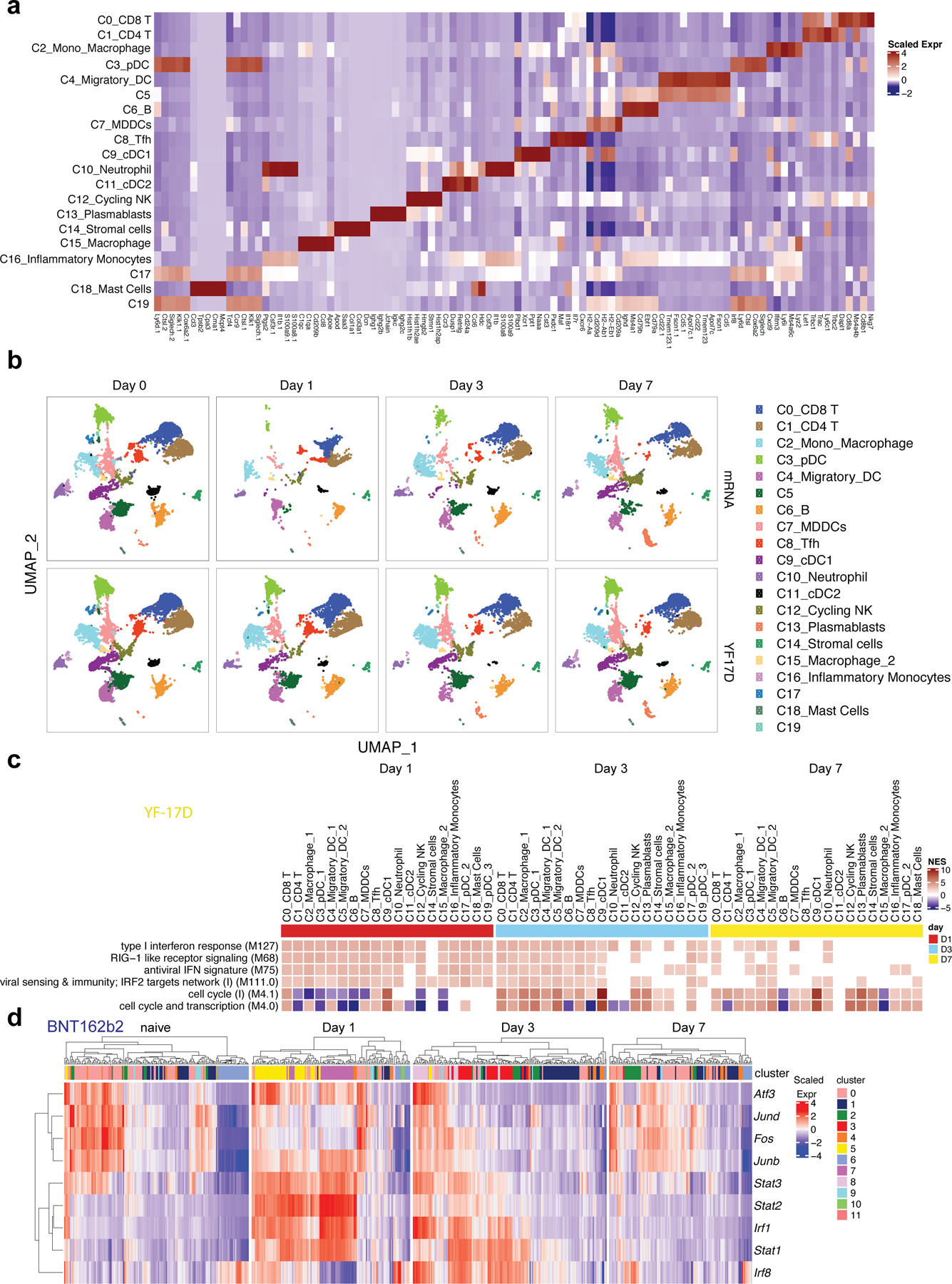
a, Clusters and their associated cluster-specific genes. b, UMAP of cell types clustered by single-cell transcriptional analysis at indicated time. c, Significantly enriched interferon BTMs (false discovery rate [FDR]< 0.05, absolute normalized enrichment score [NES]> 2) across all clusters from days 1 to 7 after YF-17D immunization. Only clusters with a significantly modulated pathway are shown. d, Heatmap of key interferon response and AP-1 transcription factors after BNT162b2 immunization over time. Samples used for scRNAseq were pooled from three independent experiments containing 8–10 mice.
Extended Data Fig. 4. IFNγ production after prime and boost.
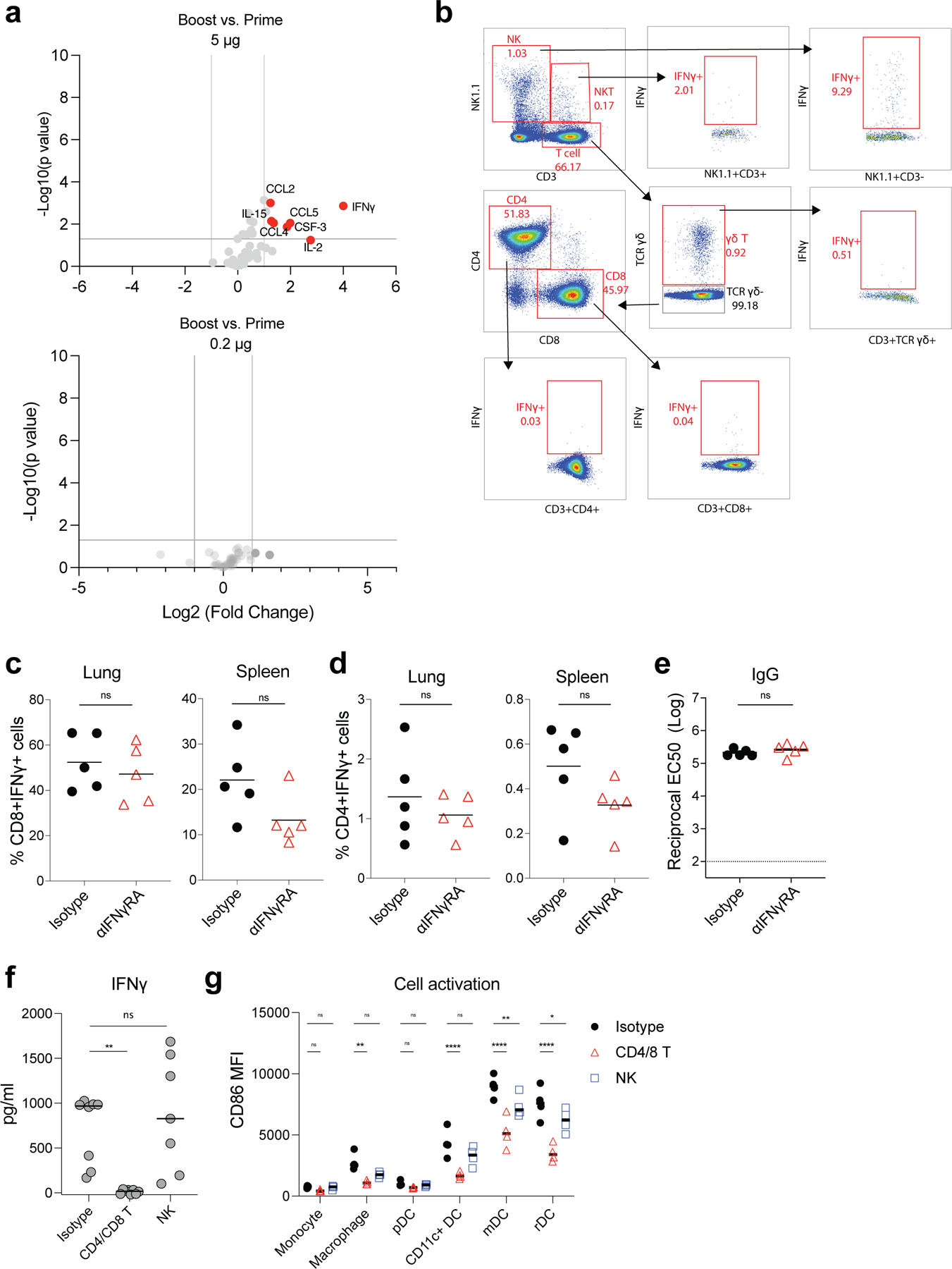
a, Luminex assay of serum cytokines/chemokines at 6 h post-prime and boost with 5 μg/mouse and 0.2 μg/mouse, respectively. Data were combined from five mice for each group. b, Gating strategy to analyze IFNγ producing cells in dLNs. c-e, CD8+ and CD4+ T cell responses, and IgG titer at day 42 after blocking of IFNγ receptor. Mice were treated with IFNγ receptor neutralizing antibody or isotype control before boost at day 21. f, Serum IFNγ level at day 22 (1 day after boost) after T cells (CD4 and CD8 T cells) or NK cells depletion at day 21. g, Innate cell activation in spleen at 1 day post boost after T or NK cell depletion. P-values were determined using Student’s t-tests (two-tailed) (a, c-e), One-Way ANOVA (f), or Two-way ANOVA (g). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
Extended Data Fig. 5. Roles of TLRs for BNT162b2-induced antibody and T cell response.
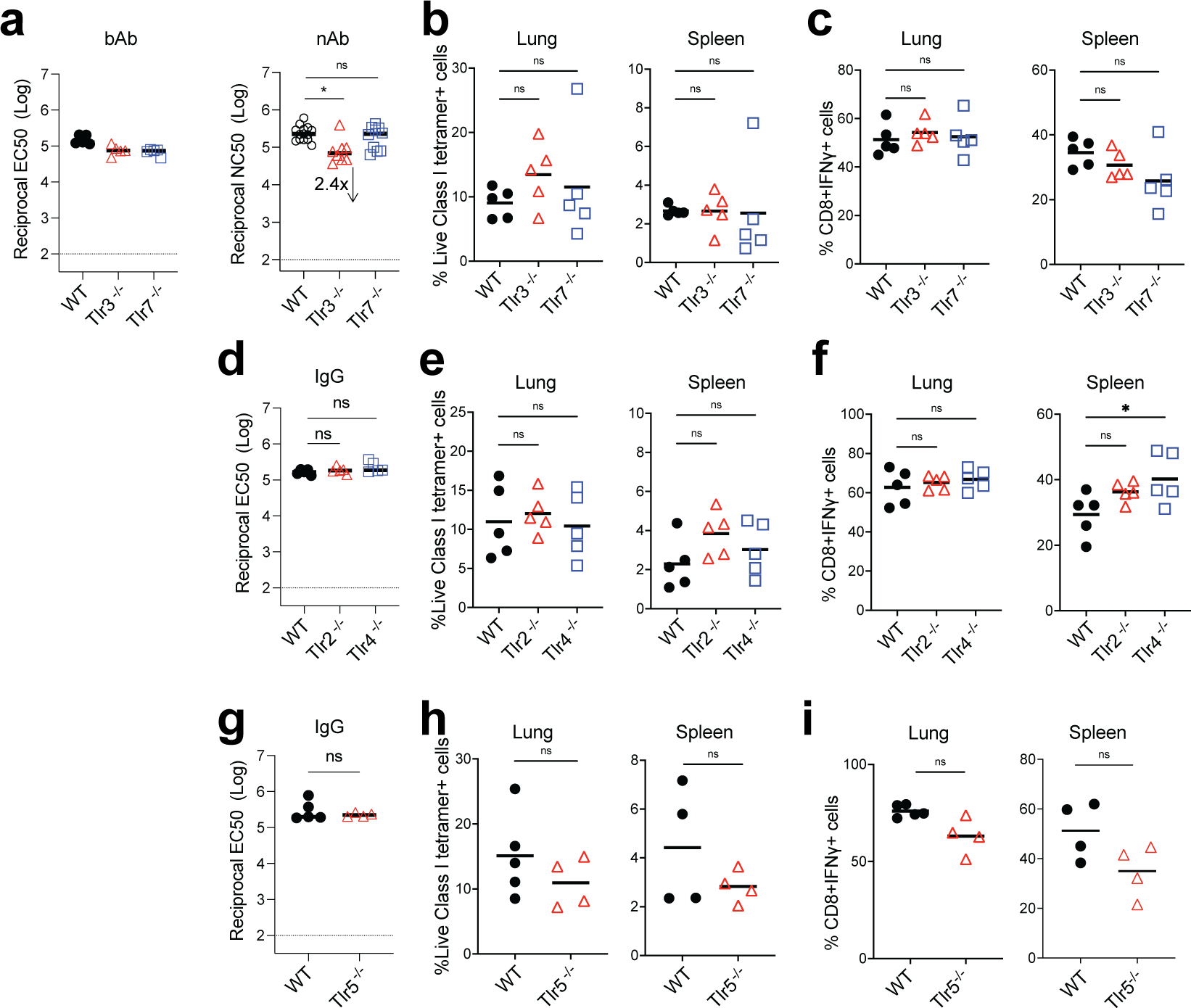
bAb and nAb titers of WT and Tlr3−/−, Tlr7−/− (a), bAb (IgG) titers of WT and Tlr2−/−, Tlr4−/− (d), Tlr5−/− (g) mice at day 42. CD8+ T cell response in WT and Tlr3−/−, Tlr7−/− (b-c), Tlr2−/−, Tlr4−/− (e-f), and Tlr5−/− (h-i) mice. Data were combined from two independent experiments. One-Way ANOVA followed by Tukey’s test was applied in (a-f). P-values in (g-i) were determined using Student’s t-tests (two-tailed). *P<0.05, **P<0.01.
Extended Data Fig. 6. Roles of inflammasome for BNT162b2-induced antibody and T cell response.
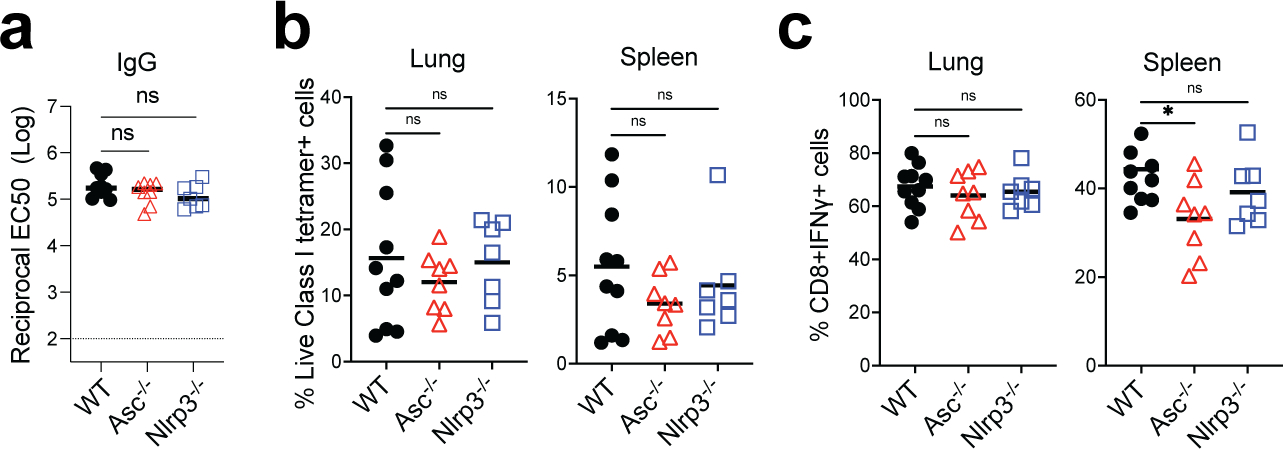
a, IgG titers of WT, Asc−/−, and Nlrp3−/− mice at day 42. (b-c), CD8+ T cell response in WT, Asc−/−, and Nlrp3−/− mice. Data were combined from two independent experiments. One-Way ANOVA followed by Tukey’s test was applied. *P<0.05.
Extended Data Fig. 7. Role of cGAS and STING for BNT162b2-induced antibody and T cell response.
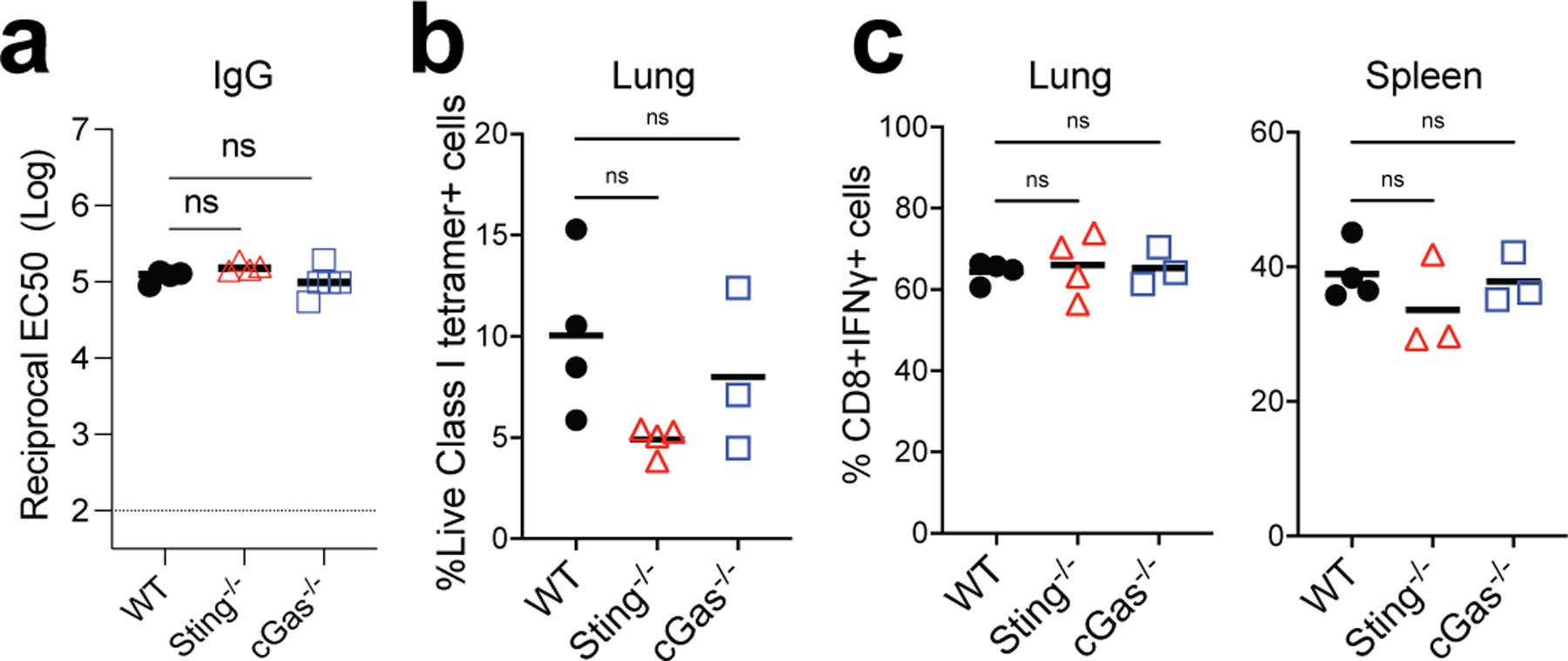
a, IgG titers of WT, cGas−/−, and Sting−/− mice at day 42. (b-c), CD8+ T cell response in WT, cGas−/−, and Sting−/− mice. Data are one representative of two independent experiments. One-Way ANOVA followed by Tukey’s test was applied.
Extended Data Fig. 8. Role of cell death in BNT162b2-induced antibody and T cell response.
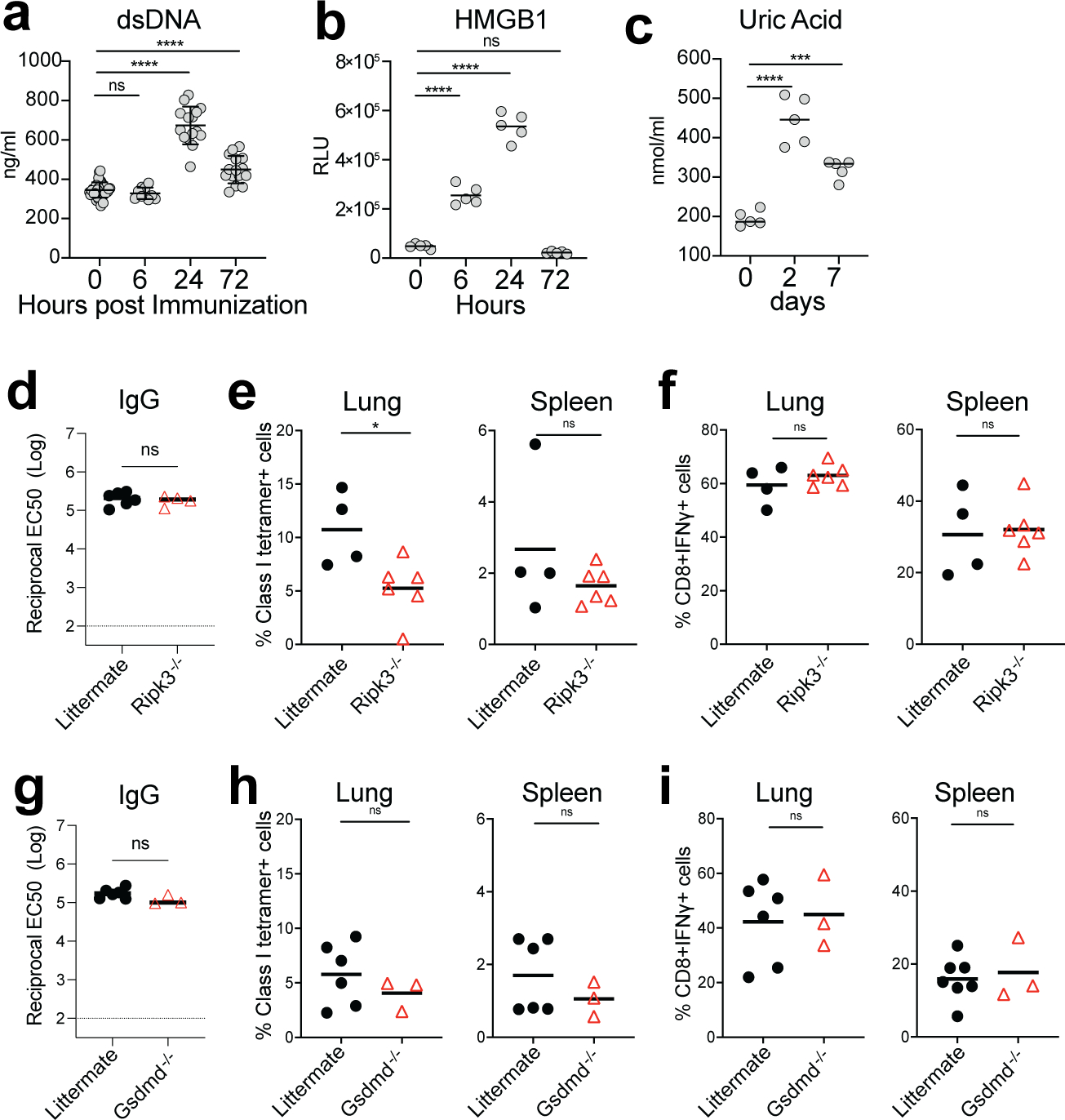
a-c, DAMP signals including dsDNA (a), HMGB1 (b), and uric acid (c) induced by BNT162b2 immunization at the indicated time. d, IgG titers of littermate and Ripk3−/− mice at day 42. e-f, CD8+ T cell response in littermate and Ripk3−/− mice. g, IgG titers of littermate and Gsdmd−/− mice at day 42. h-i, CD8+ T cell response in littermate and Gsdmd−/− mice. Data were one representative of at least two independent experiments (a), or one representative experiment (b-i). One-Way ANOVA followed by Tukey’s test was applied in (a-c). P-values in (d-i) were determined using Student’s t-tests (two-tailed). *P<0.05, ***P<0.001, ****P<0.0001.
Extended Data Fig. 9. Role of MDA5 in BNT162b2-induced T cell response after prime.
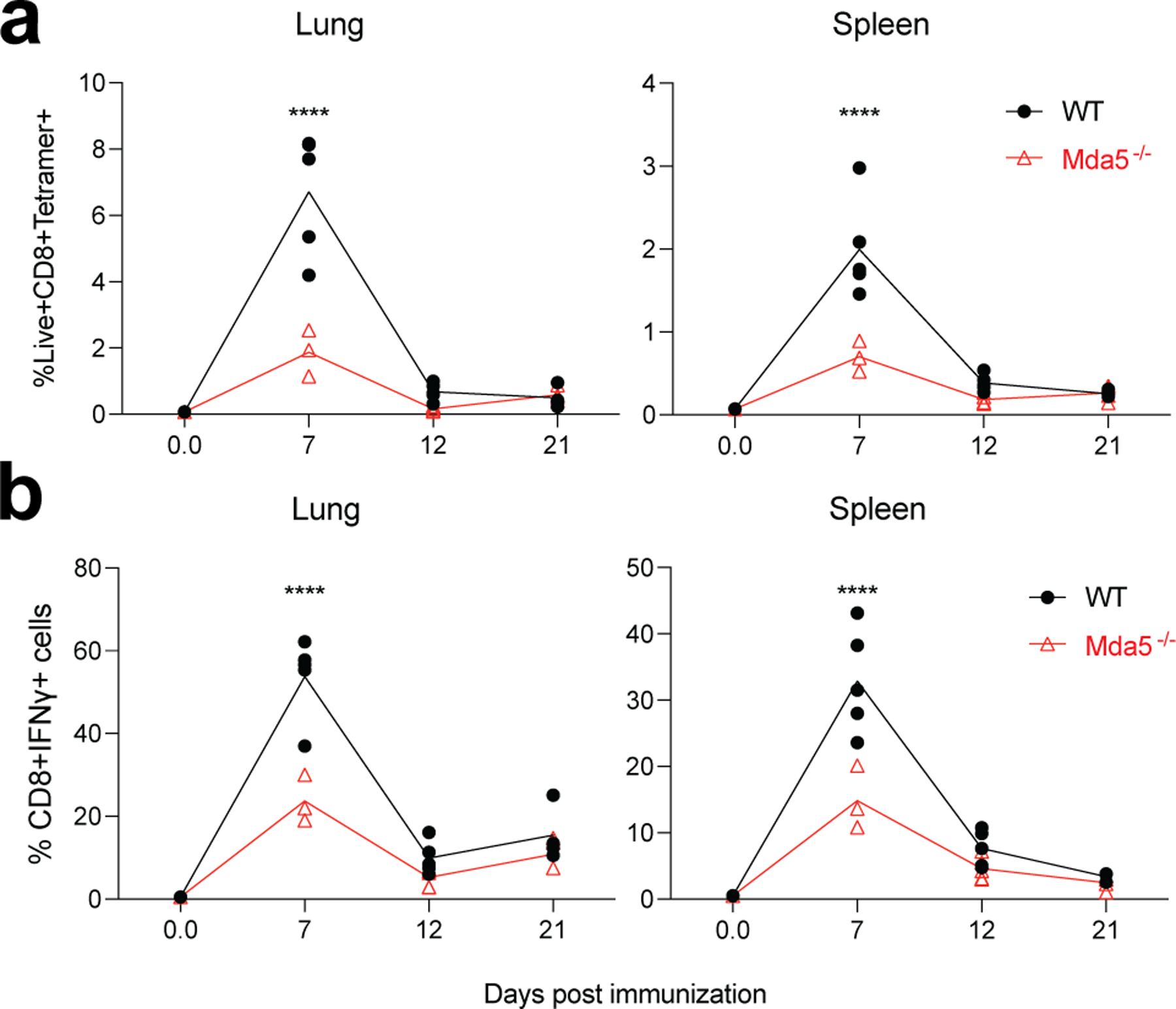
a-b, CD8+ T cell response in WT and Mda5−/− mice at indicated time post prime. P-values were determined using two-way ANOVA. ****P<0.0001.
Extended Data Fig. 10. Role of Batf3 in BNT162b2-induced antibody and T cell response.
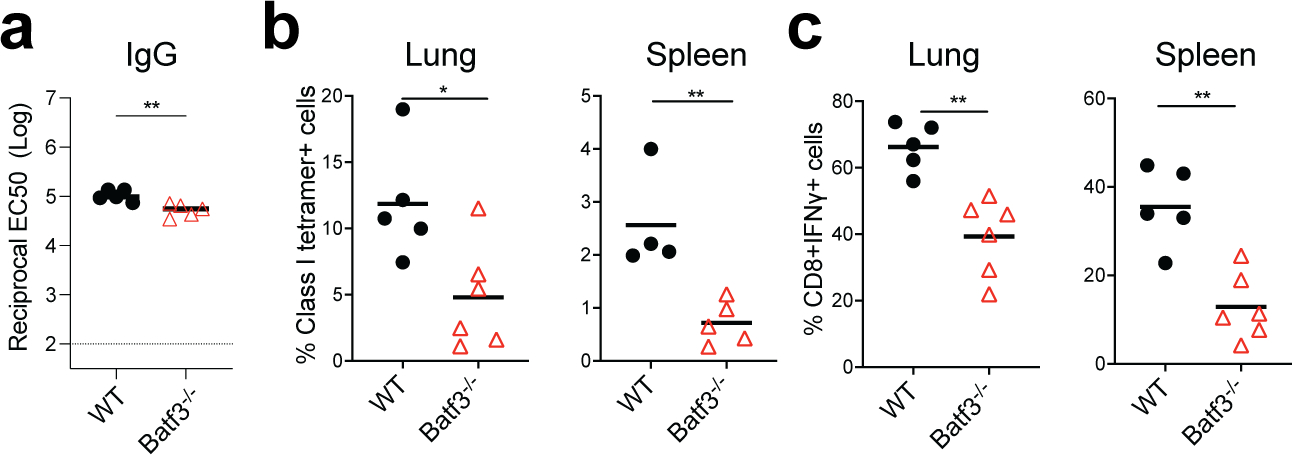
a, IgG titers of WT and Batf3−/− mice at day 42. b, CD8+ T cell response in WT and Batf3−/− mice. Data were one representative of three independent experiments. P-values were determined using Student’s t-tests (two-tailed). *P<0.05, **P<0.01.
Supplementary Material
Acknowledgments:
We thank the technical support from the Stanford Shared FACS Facility. Sorting was performed on an instrument in the Shared FACS Facility purchased by Parker Institute for Cancer Immunotherapy (PICI). We thank Dhananjay Wagh from Stanford Functional Genomics for preparing the single-cell library construction. We thank Lana Witt, Brooke Betts, and Tiffany Chang from Stanford Health care system for providing remnant vaccines. We thank the NIH tetramer core facility for providing the S specific tetramer for CD8+ T cells detection. We thank Ben Franco from Stanford Veterinary Service Center (VSC) for technical support.
The study was supported by NIH grants (R37 DK057665, R37 AI048638, U19 AI090023, U19 AI057266 (PI. Rafi Ahmed), U19 AI159840 (PI, Steven Foung)), the Bill and Melinda Gates Foundation, and the Soffer Fund endowment and Open Philanthropy to BP.
Footnotes
Competing Interests Statement
Bali Pulendran has served or is serving on the External Immunology Network of GSK, and on the scientific advisory board of Sanofi, Medicago, CircBio, Boehringer-Ingelheim. A.D.G. is a visiting scientist at Genentech, Inc. Other authors have declared no competing interests.
Reference:
- 1.Polack FP et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh EE et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N Engl J Med 383, 2439–2450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kariko K et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16, 1833–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kariko K, Buckstein M, Ni H & Weissman D Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Turner JS et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 596, 109–113 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lederer K et al. SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation. Immunity 53, 1281–1295 e1285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardi N et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med 215, 1571–1588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laczko D et al. A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity 53, 724–732 e727 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arunachalam PS et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature 596, 410–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarke A et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2, 100355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin U et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature 595, 572–577 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Liu J et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature 596, 273–275 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Painter MM et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity 54, 2133–2142 e2133 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H et al. Delivery of mRNA vaccine with a lipid-like material potentiates antitumor efficacy through Toll-like receptor 4 signaling. Proc Natl Acad Sci U S A 118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kranz LM et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Miao L et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol 37, 1174–1185 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Sahin U et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 586, 594–599 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Planas D et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596, 276–280 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Nair AB & Jacob S A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm 7, 27–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buschmann MD et al. Nanomaterial Delivery Systems for mRNA Vaccines. Vaccines (Basel) 9 (1), 65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel AB et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Arunachalam PS et al. T cell-inducing vaccine durably prevents mucosal SHIV infection even with lower neutralizing antibody titers. Nat Med 26, 932–940 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schenkel JM et al. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Israelow B et al. Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci Immunol 6, eabl4509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulendran B Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol 9, 741–747 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Querec T et al. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med 203, 413–424 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Querec TD et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 10, 116–125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee A et al. A molecular atlas of innate immunity to adjuvanted and live attenuated vaccines, in mice. Nat Commun 13, 549 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wimmers F et al. The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell 184, 3915–3935 e3921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardi N, Hogan MJ, Porter FW & Weissman D mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsay KE et al. Visualization of early events in mRNA vaccine delivery in non-human primates via PET-CT and near-infrared imaging. Nat Biomed Eng 3, 371–380 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Iwasaki A & Medzhitov R Toll-like receptor control of the adaptive immune responses. Nat Immunol 5, 987–995 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Alexopoulou L, Holt AC, Medzhitov R & Flavell RA Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Diebold SS, Kaisho T, Hemmi H, Akira S & Reis e Sousa C Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Heil F et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y et al. Publisher Correction: SARS-CoV-2 spike protein interacts with and activates TLR4. Cell Res 31, 825 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.;./uZheng M et al. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat Immunol 22, 829–838 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh JZ et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41, 478–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deets KA & Vance RE Inflammasomes and adaptive immune responses. Nat Immunol 22, 412–422 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Pulendran B, P SA & O’Hagan DT Emerging concepts in the science of vaccine adjuvants. Nat Rev Drug Discov 20, 454–475 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishikawa H & Barber GN STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim EH et al. Squalene emulsion-based vaccine adjuvants stimulate CD8 T cell, but not antibody responses, through a RIPK3-dependent pathway. Elife 9, e52687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehwinkel J & Gack MU RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20, 537–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato H et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Kolumam GA, Thomas S, Thompson LJ, Sprent J & Murali-Krishna K Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med 202, 637–650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welsh RM, Bahl K, Marshall HD & Urban SL Type 1 interferons and antiviral CD8 T-cell responses. PLoS Pathog 8, e1002352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hildner K et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravindran R et al. The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature 531, 523–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang DW et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Cho YS et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi J et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Cella M, Gilfillan S & Colonna M Cutting edge: polyinosinic:polycytidylic acid boosts the generation of memory CD8 T cells through melanoma differentiation-associated protein 5 expressed in stromal cells. J Immunol 184, 2751–2755 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Wu B et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152, 276–289 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Corbett KS et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalnin KV et al. Immunogenicity and efficacy of mRNA COVID-19 vaccine MRT5500 in preclinical animal models. NPJ Vaccines 6, 61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alameh MG et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 54, 2877–2892 e2877 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pulendran B & Davis MM The science and medicine of human immunology. Science 369 (6511):eaay4014. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis MM A prescription for human immunology. Immunity 29, 835–838 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mestas J & Hughes CC Of mice and not men: differences between mouse and human immunology. J Immunol 172, 2731–2738 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Warren CM et al. Assessment of Allergic and Anaphylactic Reactions to mRNA COVID-19 Vaccines With Confirmatory Testing in a US Regional Health System. JAMA Netw Open 4, e2125524 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crawford KHD et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 12 (5):513. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogers TF et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bais AS & Kostka D scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics 36, 1150–1158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-Cell RNAseq data are publicly accessible in the GEO under accession numbers GSE179131. All analyses and visualizations were performed in R (V3.6.3).


