Abstract
We examined antibody and memory B cell responses longitudinally for ∼9–10 months after primary 2-dose SARS-CoV-2 mRNA vaccination and 3 months after a 3rd dose. Antibody decay stabilized between 6 and 9 months, and antibody quality continued to improve for at least 9 months after 2-dose vaccination. Spike- and RBD-specific memory B cells remained durable over time, and 40%–50% of RBD-specific memory B cells simultaneously bound the Alpha, Beta, Delta, and Omicron variants. Omicron-binding memory B cells were efficiently reactivated by a 3rd dose of wild-type vaccine and correlated with the corresponding increase in neutralizing antibody titers. In contrast, pre-3rd dose antibody titers inversely correlated with the fold-change of antibody boosting, suggesting that high levels of circulating antibodies may limit the added protection afforded by repeat short interval boosting. These data provide insight into the quantity and quality of mRNA-vaccine-induced immunity over time through 3 or more antigen exposures.
Keywords: SARS-CoV-2, COVID-19, mRNA, vaccine, booster, Omicron, variants of concern, immune memory, antibody, memory B cell
Graphical abstract

Immunization with 2 doses of mRNA vaccine encoding the ancestral SARS-CoV-2 spike protein induces a population of durable memory B cells with broad reactivity to viral variants including Omicron. Boosting with a 3rd dose of ancestral vaccine increases variant-neutralizing antibody levels, highlighting the significance of vaccine-induced B cell memory.
Introduction
SARS-CoV-2 infections continue to cause significant morbidity and mortality worldwide (Carvalho et al., 2021). Since the virus was identified in late 2019, several SARS-CoV-2 variants of concern (VOC) have emerged. Mutations found in SARS-CoV-2 variants, particularly those in the spike glycoprotein, can alter viral transmission and immune recognition (Garcia-Beltran et al., 2021; Greaney et al., 2021a, 2021b). Of these VOC, the Delta (B.1.617.2) variant had considerable impact due to its increased infectivity and partial escape from neutralizing antibodies (Mlcochova et al., 2021; Planas et al., 2021). In November 2021, scientists in South Africa identified and characterized the Omicron (B.1.1.529) variant (Viana et al., 2022). In the weeks following identification, Omicron spread rapidly, outcompeting Delta to become the dominant variant in the United States and many parts of the world.
A major concern about Omicron is the large number of mutations in the spike protein, including ∼15 amino acid changes in the spike receptor-binding domain (RBD). In vitro data indicate that these mutations have a substantial effect on evading antibody responses in convalescent or mRNA-vaccinated (Pfizer BNT162b2 or Moderna mRNA-1273) individuals. This effect is more pronounced than that of other VOC, with a ∼10-fold to ∼40-fold reduction in neutralization capacity compared with wild-type virus using either pseudovirus or live virus neutralization assays, and little-to-no neutralizing activity against Omicron detected at >6 months after the primary 2-dose vaccine series (Cameroni et al., 2022; Cele et al., 2021; Garcia-Beltran et al., 2022; Schmidt et al., 2021a).
In addition to circulating antibodies, memory B cells represent an important source of long-term immunity (Sette and Crotty, 2021; Victora and Nussenzweig, 2012). In contrast to antibodies that decline over the first 3–6 months postvaccination (Levin et al., 2021), antigen-specific memory B cells appear highly stable over time (Goel et al., 2021a). Upon re-exposure to antigen, either through vaccination or infection, these memory B cells can differentiate into antibody-secreting cells (ASCs) and rapidly produce new antibodies (Laidlaw and Ellebedy, 2021). Indeed, recent non-human primate studies of mRNA vaccination highlight recall antibody responses from memory B cells as a key factor in protection from severe COVID-19 pathology in the lungs (Gagne et al., 2022a). Previous work has shown that mRNA vaccines induce robust memory B cell responses that continue to evolve via germinal center reactions for months after primary vaccination (Goel et al., 2021b, 2021a; Kim et al., 2021; Röltgen et al., 2022; Turner et al., 2021). As a result, immunization with mRNA vaccines encoding the original Wuhan spike protein generates a population of high-affinity memory B cells that can bind the Alpha, Beta, and Delta variants and produce neutralizing antibodies upon restimulation.
Serologic data indicate that antibody responses to Omicron can be at least partially boosted in the short term (up to ∼1 month) after a 3rd vaccine dose (Muecksch et al., 2022; Muik et al., 2022; Schmidt et al., 2021b; Xia et al., 2022), suggesting that immunological memory generated by 2-dose vaccination has some reactivity against the Omicron spike protein. A 3rd vaccine dose also provides increased protection from Omicron variant infection (Shrestha et al., 2022). However, it is unclear how long these boosted antibody responses to Omicron may last and what percent of memory B cells retain binding to Omicron and other variants. Moreover, the dynamics of memory B cell responses in humans are poorly understood, and whether boosting with the original Wuhan spike can overcome antigenic changes by efficiently reactivating Omicron-binding memory B cells is unknown (Kotaki et al., 2022; Wang et al., 2022). Finally, it remains unclear what features of immunity induced by 2-dose vaccination determine optimal boosting following a 3rd vaccine dose, and how immune responses are affected by additional antigen encounters beyond a 3-dose vaccine schedule. The answers to these questions should inform how to optimize the use of additional vaccine doses for protection against Omicron and future VOC.
Results
Study design
We examined antibody and memory B cell responses to SARS-CoV-2 in a longitudinal cohort of 61 individuals receiving mRNA vaccines (Pfizer BNT162b2 or Moderna mRNA-1273). This cohort has been previously described through 6-months post-2 doses of mRNA vaccine (Goel et al., 2021b, 2021a; Painter et al., 2021). A total of 45 individuals were infection naive, and 16 had recovered from a prior SARS-CoV-2 infection. Paired serum and peripheral blood mononuclear cell (PBMC) samples were collected at 10 different time points, ranging from prevaccine baseline to ∼9–10 months postprimary 2-dose vaccination, as well as prior to a 3rd vaccine dose, ∼2 weeks post-3rd dose, and ∼3 months post-3rd dose (Figure 1 A). Nine individuals had a confirmed postvaccination (commonly referred to as “breakthrough”) infection during the study period and are indicated in all analyses. Additional cohort information is provided in Tables S1 and S2.
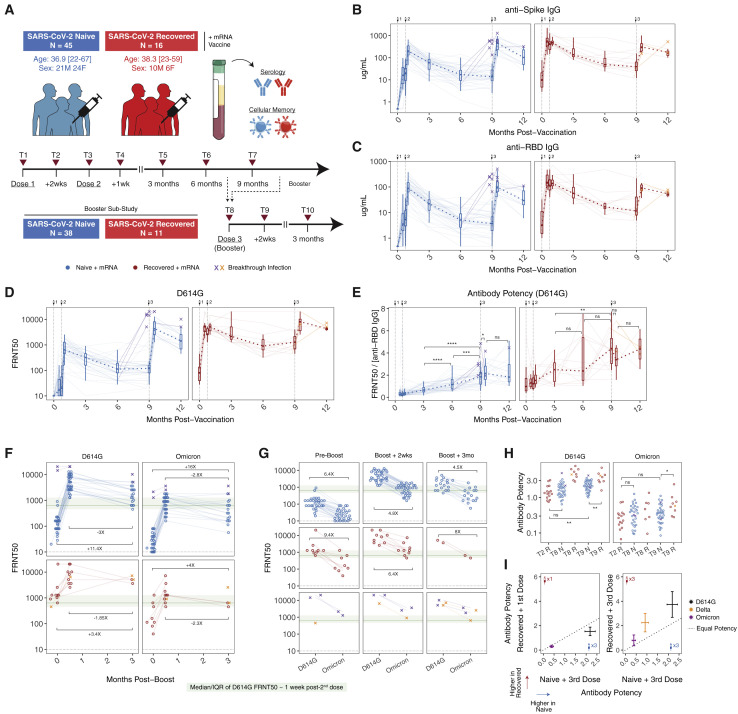
Figure 1.
Antibody responses after 2 and 3 doses of mRNA vaccine
(A) Study design and cohort characteristics.
(B and C) (B) Anti-spike and (C) anti-RBD IgG concentrations over time in plasma samples from vaccinated individuals.
(D) Pseudovirus (PSV) neutralization titers against wild-type D614G spike protein over time in plasma samples from vaccinated individuals. Data are represented as focus reduction neutralization titer 50% (FRNT50) values.
(E) Antibody neutralization potency against D614G over time. Potency was calculated as neutralizing titer (FRNT50) divided by the paired concentration of anti-RBD IgG.
(F and G) Plasma neutralizing activity against D614G and Omicron before and after booster vaccination. Dotted lines indicate limit of detection for the assay. Green boxes and lines indicate interquartile range (IQR) and median of D614G neutralizing titers ∼1 week following the second vaccine dose in SARS-CoV-2-naive subjects.
(H and I) Comparison of antibody potency against D614G, Delta, and Omicron between SARS-CoV-2-naive and previously infected vaccinees. For (I), bars indicate mean with 95% confidence intervals. Statistics were calculated using unpaired nonparametric Wilcoxon test with Benjamini-Hochberg correction for multiple comparisons. Breakthrough infection samples were excluded from statistical comparisons. Median fold-changes for selected comparisons are indicated. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant. Binding antibody and D614G pseudovirus neutralization data from prevaccine baseline through 6 months postprimary vaccination were described previously (Goel et al., 2021a).
Antibody responses
As we have previously described for this cohort, 2-dose mRNA vaccination in previously uninfected individuals induced high titers of binding and neutralizing antibodies, whereas vaccination in individuals with a prior SARS-CoV-2 infection (commonly referred to as “hybrid immunity”) resulted in higher antibody titers, consistent with an anamnestic response from prevaccination immunological memory (Goel et al., 2021b, 2021a). Although these previous studies documented a decline in antibodies from their peak ∼1 month postvaccination to 6 months, here we extended our analysis of this cohort to later time points. These data revealed a stabilization of antibody titers between 6 and 9 months postvaccination for both individuals with and without previous SARS-CoV-2 infection, with little-to-no decrease in neutralizing antibody titers after 6 months (Figures 1B–1D). These findings are consistent with ongoing antibody production from long-lived plasma cells in the later phases of immune memory after vaccination.
To evaluate the quality of antibody responses, we calculated an antibody potency index. Although antibody-mediated protection can be influenced by functions other than neutralization (Bournazos and Ravetch, 2017; Lu et al., 2017), we defined a potency index based on the ratio of neutralization titers to the total concentration of anti-RBD-binding IgG. Antibody potency increased significantly over time after the 2nd vaccine dose, with a continued increase in potency from 6 to 9 months postvaccination in the infection-naive group as antibody concentrations began to plateau (Figure 1E). These observations suggest decay of lower quality antibody from short-lived ASCs, as well as continued emergence of postgerminal center affinity-matured plasma cells over time that produce higher quality antibody later in the response. This improvement in the quality of antibody for at least 9 months is also consistent with a recent report demonstrating the continued presence of spike-binding germinal center B cells in axillary lymph nodes at 29 weeks postvaccination (Kim et al., 2021).
In addition to the primary 2-dose vaccine series, most of our cohort went on to receive a 3rd dose of mRNA vaccine. A 3rd dose of vaccine in infection-naive individuals significantly increased binding and neutralizing antibodies, with both reaching a similar level to that observed in previously infected individuals after the 2-dose vaccine series (Figures 1B–1D). A 3rd dose of mRNA vaccine in these COVID-recovered individuals (i.e., a 4th exposure) also significantly boosted antibody responses; however, the relative magnitude of this increase was less than observed in the recall response after the initial 2-dose vaccine series (Figures 1B–1D). Several individuals in this cohort also experienced breakthrough infections after 2 or 3 doses of vaccine. Although the sample size was limited (n = 3), infection following 2-dose vaccination appeared to boost antibodies to similar levels compared with previous infection with 2 doses of vaccine (Figures 1B–1D), suggesting that the total number of antigen exposures may be as important as the relative order of exposure to infection and vaccination.
To quantify neutralizing capacity of vaccine-induced antibody responses against VOC, we generated pseudotyped viruses encoding the Delta and Omicron spike proteins. Consistent with previous reports, neutralizing titers against Omicron were significantly reduced relative to D614G, with ∼20% of individuals having Omicron neutralization titers below the limit of detection at ∼9 months postprimary vaccination (Figures 1F and 1G). Following a 3rd dose, neutralizing titers to Omicron increased by a median of ∼45-fold in COVID-naive individuals, with similar kinetics and magnitude of boosting as neutralizing antibodies against D614G (Figures 1F and 1G). Although neutralization against Omicron declined 2.8-fold from peak levels between 2 weeks post-3rd dose and 3 months post-3rd dose, titers remained 16-fold above pre-3rd dose baseline in COVID-naive individuals (Figure 1F), indicating that an additional vaccine dose has a lasting benefit for antibodies against Omicron. In paired comparisons for individuals, Omicron-neutralizing antibodies had a 4.8-fold lower median neutralizing titer compared with D614G at the peak response after the 3rd vaccine dose and 4.5-fold lower titer 3 months later. Despite the relative loss of neutralizing activity, peak Omicron neutralizing titers after the 3rd dose were comparable with neutralizing titers against D614G ∼1 week after the 2nd vaccine dose, where clinical efficacy has previously been defined (Figures 1F and 1G) (Khoury et al., 2021). Moreover, Omicron neutralizing titers at 3 months post-3rd dose were higher than pre-3rd dose D614G neutralizing titers (Figures 1F and 1G).
Finally, to investigate potential differences in the quality of antibody recall responses, we compared antibody potency 2 weeks after the first dose of mRNA vaccine in individuals with pre-existing immune memory from infection to antibody potency 2 weeks after a 3rd dose of mRNA vaccine in previously uninfected individuals. Previous reports have suggested that infection generates greater antibody potency and breadth than 2-dose mRNA vaccination alone (Cho et al., 2021), but little is known about the impact of a 3rd vaccine dose on antibody potency. In this cohort, antibody potency against both D614G and Omicron was slightly higher following a 3rd dose of mRNA vaccine compared with recall responses following the 1st dose in SARS-CoV-2-recovered individuals (Figures 1H and 1I), suggesting that a 3rd dose of mRNA vaccine drives antibody potency to similar levels to “hybrid immunity.” Of note, potency continued to increase in SARS-CoV-2-recovered individuals between the initial recall response to vaccine and ∼9 months postvaccination (Figures 1E, 1H, and 1I). This finding indicates that there may be ongoing evolution after a vaccine-induced recall response that can result in further improvement of antibody potency. Taken together, these data demonstrate that antibody responses, including neutralizing antibodies to Omicron, are effectively boosted by a 3rd dose of mRNA vaccine with sustained benefit at ∼3 months post-3rd dose.
Memory B cell responses
We next investigated B cell responses to mRNA vaccination. Antigen-specific B cell responses were quantified from bulk PBMCs by flow cytometry using fluorescently labeled SARS-CoV-2 spike and RBD probes as previously described (Dan et al., 2021; Goel et al., 2021b, 2021a). Influenza hemagglutinin (HA) was used as a historical antigen for a specificity control. Plasmablasts were identified as CD20− CD38++ non-naive B cells. Memory B cells were identified as CD20+ CD38lo/int non-naive B cells. Full gating strategy is shown in Figure S1 .
Figure S1.
Gating strategy for identifying SARS-CoV-2-specific B cell responses, related to Figure 2
Single cells were identified based on FSC-A and FSC-H. Live cells were identified based on negative staining with Ghost 510 viability dye. Lymphocytes were identified from bulk PBMCs based on FSC-A and SSC-A. Total B cells were then identified from live lymphocytes as CD3− CD19+ cells. Memory B cell subsets were identified based on expression of IgD, CD20, CD27, and CD38. IgD+ CD27− naive B cells were excluded from all analysis. From non-naive B cells, memory B cells were identified as CD20+ CD38lo/intermediate and plasmablasts were identified as CD20lo CD38+. Antigen-specificity of memory B cells and plasmablasts was determined based on binding to fluorescently labeled antigen probes. First, a decoy probe (BV711-streptavidin) was used to exclude cells that nonspecifically bind streptavidin. Decoy negative cells were then assessed for binding to full-length SARS-CoV-2 spike protein or influenza hemagglutinin (HA) from the 2019 flu vaccine season. Spike+ B cells were subsequently analyzed for cobinding to a receptor-binding domain (RBD) probe. Regarding phenotype, immunoglobulin isotype (IgG, IgM, and IgA) was measured on all antigen-binding populations. CD71 was used as a marker of activated B cells.
Consistent with our plasma antibody data, re-exposure to SARS-CoV-2 antigen, either through a 3rd mRNA vaccine dose or breakthrough infection, resulted in a significant expansion of spike-binding plasmablasts ∼1 week after antigen encounter (Figures 2A and 2B). Overall, the rapid emergence of antibody-secreting cells following antigen re-encounter is consistent with recall from a pool of memory B cells. These data are also consistent with findings after viral challenge in SARS-CoV-2 mRNA-vaccinated monkeys, where anamnestic antibody responses from memory B cells were identified as a major protective mechanism (Gagne et al., 2022a).
Figure 2.
Memory B cell responses after 2 and 3 doses of mRNA vaccine
(A) Flow cytometry gating strategy for SARS-CoV-2-specific plasmablasts.
(B) Frequency of spike+ plasmablasts ∼1 week after booster vaccination or postvaccine breakthrough infection. Data are represented as a percentage of total B cells.
(C) Flow cytometry gating strategy for SARS-CoV-2-specific memory B cells.
(D and E) (D) Frequency of spike+ and (E) spike+ RBD+ memory B cells over time in PBMCs from vaccinated individuals. Data are represented as a percentage of total B cells.
(F and G) (F) Fold-change in the frequency of spike+ and (G) spike+ RBD+ memory B cells after booster vaccination relative to paired preboost samples. Median fold-change is indicated in dashed blue or red lines. Dashed black lines at fold-change = 1 indicate no change in frequency compared with preboost samples.
(H) Isotype composition of spike+ memory B cells in vaccinated individuals pre and postboost.
(I) Activation status of spike+ memory B cells over time in vaccinated individuals following booster vaccination. Statistics were calculated using unpaired nonparametric Wilcoxon test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant. Memory B cell responses from prevaccine baseline through 6 months postprimary vaccination were reanalyzed from a previous dataset (Goel et al., 2021a).
See also Figure S1.
We previously demonstrated that mRNA vaccines induce durable and functional memory B cells to SARS-CoV-2 that are stable for at least 6 months after vaccination (Goel et al., 2021a). Here, we extended these observations by tracking responses further into the memory phase. Spike- and RBD-specific memory B cell numbers continued to remain highly stable through at least 9 months postvaccination in both SARS-CoV-2-naive and previously infected individuals with no evidence of decline in numbers from 6 to 9 months postprimary vaccination (Figures 2C–2E). Notably, 34/35 SARS-CoV-2-naive individuals had spike- and RBD-specific memory B cell frequencies above their prevaccine baseline at the 9-month time point, highlighting the continued durability of mRNA vaccine-induced cellular immunity.
Upon receipt of a 3rd mRNA vaccine dose, these memory B cells expanded in number. At ∼2 weeks postboost, there was a median of 2.2-fold increase in spike-specific and 3.6-fold increase in RBD-specific memory B cells in infection-naive individuals, with similar boosting for spike- and RBD-specific memory B cells in COVID-recovered vaccinees (Figures 2F and 2G). By 3 months post-3rd vaccine dose in infection-naive subjects, memory B cells had declined from peak levels but still remained ∼1.5-fold more abundant than before boosting (Figures 2F and 2G). A 3rd vaccination did not markedly change the isotype composition of the response from what was reported previously in this cohort after two doses, with a majority of memory B cells remaining IgG+ (Figure 2H). A 3rd dose of vaccine also induced a population of CD71+ activated B cells, consistent with reactivation of memory B cells (Figure 2I). This activation status, however, transitioned back to a resting memory (RM) phenotype by 3 months post-3rd dose (Figure 2I). Thus, memory B cells were rapidly reactivated by re-exposure to antigen, through either infection or vaccination, and this reactivation was associated with the induction of plasmablasts, numerical expansion of memory B cells, and re-establishment of B cell memory.
Memory B cell responses to Omicron and other variants
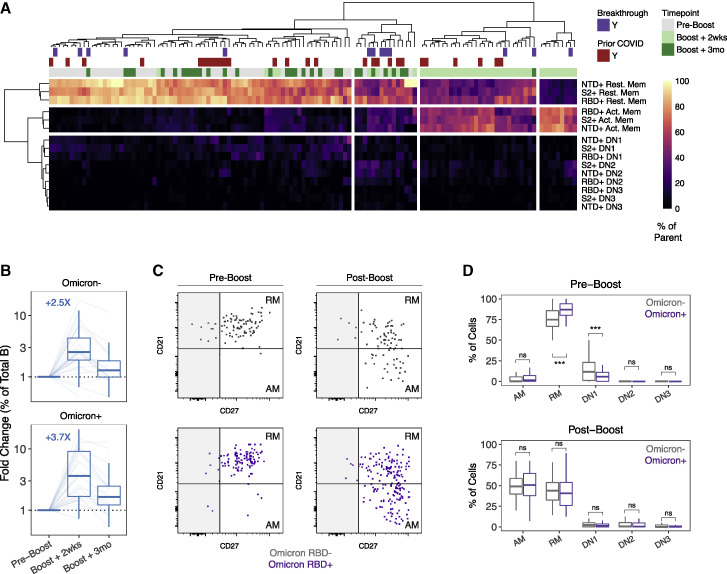
A major question is whether vaccination with the original Wuhan spike protein induces effective immunological memory to VOC including Omicron, and if so, whether Omicron-reactive memory B cells can be efficiently boosted by subsequently revaccinating with wild-type vaccine. To investigate if mRNA vaccine-induced memory B cells were capable of recognizing the Omicron variant and determine how Omicron binding related to specificity for other VOC, we designed a modified flow cytometry panel with antigen probes for 9 SARS-CoV-2 antigens. This panel included full-length spike, N-terminal domain (NTD), S2 domain, wild-type (WT, Wuhan-Hu-1) RBD, and 4 variant RBDs (Alpha, Beta, Delta, and Omicron). Nucleocapsid (N) was included for a response indicative of infection but not vaccination. For this analysis, B cells were enriched from the total PBMCs by negative selection at 3 time points: pre-3rd dose, ∼2 weeks after the 3rd dose, and ∼3 months after the 3rd dose. Representative plots and gating strategy are shown in Figures 3 A, 3B, and S2 . Briefly, antigen specificity and phenotype were identified according to the following gating strategy: spike+ memory B cells were first identified from total memory B cells as described above. spike+ memory B cells were subsequently gated based on cobinding to NTD or S2 probes. Memory B cells that were spike+ but did not bind NTD or S2 were then examined for binding to the WT RBD probe, as well as variant RBD probes.
Figure 3.
Variant-reactive memory B cell responses after 2 and 3 doses of mRNA vaccine
(A and B) (A) Experimental design and (B) flow cytometry gating strategy for SARS-CoV-2 variant-reactive memory B cells.
(C) Frequency of NTD+, WT RBD+, all variant RBD+, and S2+ memory B cells in vaccinated individuals pre- and post-boost.
(D) Fold-change in the frequency of antigen-specific memory B cells after booster vaccination relative to paired preboost samples. Median fold-change for each variable is indicated in dashed blue or red lines. Dashed black lines at fold-change = 1 indicate no change in frequency compared with preboost samples.
(E) Variant cross-binding of RBD-specific memory B cells in vaccinated individuals. Data are represented as a percentage of WT RBD+ cells.
(F) Boolean analysis of variant cross-binding memory B cell populations in vaccinated individuals ∼2 weeks after 3rd vaccination or at a cross-sectional time point in individuals with a postvaccine breakthrough infection. Pie charts indicate the fraction of WT RBD+ memory B cells that cross-bind zero, one, two, three, or four variant RBDs. Colored arcs indicate cross-binding to specific variants.
(G) Comparison of RBD variant cross-binding between SARS-CoV-2-naive and previously infected vaccinees before and ∼2 weeks after 3rd vaccination. For (G), bars indicate mean with 95% confidence intervals. Statistics were calculated using unpaired nonparametric Wilcoxon test with Benjamini-Hochberg correction for multiple comparisons. Breakthrough infection samples were excluded from statistical comparisons. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
See also Figure S2.
Figure S2.
Gating strategy for identifying SARS-CoV-2 variant-reactive memory B cell populations, related to Figures 3 and 4
B cells were pre-enriched from total PBMCs by negative selection with a STEMCELL isolation kit. Lymphocytes were identified based on FSC-A and SSC-A. Single cells were identified based on FSC-A and FSC-H. Live cells were identified based on negative staining with Ghost 510 viability dye. Total B cells were then identified from live lymphocytes as CD3− CD19+ cells. IgD+ naive B cells were excluded from all analysis. From non-naive B cells, plasmablasts were identified as CD27++ CD38+ and excluded. Antigen-specificity of memory B cells was determined based on binding to fluorescently labeled antigen probes. First, a decoy probe (BUV615-streptavidin) was used to exclude cells that nonspecifically bind streptavidin. Decoy negative cells were then assessed for binding to full-length SARS-CoV-2 spike protein or SARS-CoV-2-nucleocapsid protein. Spike+ B cells were subsequently analyzed for cobinding to NTD and S2 probes. Spike+ B cells that did not bind NTD or S2 were analyzed for binding to wild-type receptor-binding domain (RBD) probe. Cross-binding to Alpha, Beta, Delta, and Omicron variant RBD probes was measured on WT RBD-specific B cells. Regarding phenotype, memory B cell subsets were identified based on CD11c, CD21, and CD27 staining. CD21+ CD27+ B cells were defined as resting memory and CD21− CD27+ B cells were defined as activated memory. CD27− memory B cells were split into double-negative (DN) subsets based on CD11c and CD21 staining.
As previously described, 2-dose vaccination induced memory B cells specific for all domains of the spike protein, with S2 representing the immunodominant part of the response (Goel et al., 2021a). A 3rd dose of mRNA vaccine boosted NTD-, WT RBD-, and S2-specific memory B cells with a 2.4-fold increase in NTD-specific memory B cells, 3.2-fold increase in WT RBD-specific memory B cells, and a 1.9-fold increase in S2-specific memory B cells (Figures 3C and 3D). Notably, memory B cells that recognized all RBD variants (Alpha, Beta, Delta, and Omicron) simultaneously had the greatest fold-change after the boost (3.8-fold; Figures 3C and 3D). All RBD-binding memory B cells, regardless of cross-binding specificity, declined in frequency from peak levels to 3 months after the 3rd dose, but these memory B cells remained above preboost levels in most individuals (Figure 3D). Taken together, these data indicate that a 3rd exposure to wild-type spike was sufficient to expand memory B cells targeting multiple different VOC.
To further investigate variant reactivity within the memory B cell compartment, we quantified cross-binding to different VOC probes as a percentage of WT RBD-binding cells. Nine months after primary vaccination, >90% of memory B cells that bound WT RBD also bound Alpha RBD containing a single N501Y substitution (Figure 3E). The L452R and T478K mutations found in Delta resulted in a moderate loss of binding, with ∼80% of WT RBD+ cells still able to cross-bind Delta RBD (Figure 3E). The K417N, E484K, and N501Y mutations found in Beta were slightly more immune evasive than Delta, with ∼70% of WT RBD+ memory B cells able to cross-bind Beta RBD (Figure 3E). Notably, ∼55% of WT RBD-binding memory B cells after 2 doses of vaccine were still able to cross-bind Omicron RBD (Figure 3E). Most Omicron RBD-specific memory B cells were also capable of recognizing Alpha, Beta, and Delta RBDs, although binding overlap was less complete for Omicron and Delta compared with other combinations (Figure 3F), likely due to the L452R mutation that is found in Delta but not in Omicron. As a result, a considerable fraction (∼40%–50%) of RBD-specific memory B cells could bind all 4 VOC RBDs simultaneously (Figures 3E and 3F). These “All Variant+” memory B cells may represent a source of broad protection that is resilient to future VOC. Boosting appeared to slightly increase memory B cell cross-binding to Beta, Delta, and Omicron RBDs (Figure 3E), and the overall VOC binding profiles were similar in both COVID-naive and prior COVID vaccinees pre- and post-boost (Figure 3G).
To investigate how memory B cells with different antigen specificities were impacted by a 3rd dose of vaccine containing Wuhan spike, we examined the activation phenotype of these cells ∼2 weeks after the 3rd vaccine dose. Memory B cell activation state was defined based on the expressions of CD21, CD27, and CD11c (Figure 3B). CD21− CD27+ B cells were identified as activated memory (AM) and CD21+ CD27+ cells were identified as RM (Lau et al., 2017). CD27− cells were split into double-negative (DN) 1, 2, and 3 subsets based on CD21 and CD11c staining (Figure 3B). As expected, revaccination with a 3rd dose induced a clear transition from a RM to an AM B cell phenotype in spike-specific cells (Figure 4 A). DN1, DN2, and DN3 cells only represented a small fraction of the overall response. NTD+, RBD+, and S2+ memory B cell activation states clustered together (Figure 4A), indicating that antigen re-exposure activates B cells specific for all parts of the spike protein.
Figure 4.
Activation of Omicron-reactive B cell memory after a 3rd dose of mRNA vaccine
(A) Heatmap and hierarchal clustering of memory B cell activation status by antigen specificity at pre- and post-3rd dose time points. Prior COVID infection and/or postvaccine breakthrough infection are indicated.
(B) Median fold-change in the frequency of Omicron RBD-binding versus nonbinding memory B cells after booster vaccination relative to paired preboost samples. Dashed black lines at fold-change = 1 indicate no change in frequency compared with preboost samples.
(C) Representative flow cytometry plots for activation phenotype of Omicron RBD-binding versus Omicron RBD nonbinding (but still wild-type RBD binding) memory B cells.
(D) Frequency of activated memory (AM), resting memory (RM), or double-negative (DN) subsets in Omicron RBD-binding versus nonbinding memory B cells before and ∼2 weeks after a 3rd vaccination. For (B)–(D), analysis was restricted to SARS-CoV-2-naive vaccinees with no breakthrough infection. Statistics were calculated using paired nonparametric Wilcoxon test with Benjamini-Hochberg correction for multiple comparisons. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
See also Figure S2.
Within the RBD-specific memory B cell population, it has been unclear if variant-reactive memory B cells could be activated by a 3rd dose of vaccine, and if so, would there be a preference for VOC nonbinders over VOC binders. Here, a 3rd dose of mRNA vaccine encoding the original Wuhan spike boosted Omicron RBD-binding memory B cells by 3.7-fold compared with 2.5-fold for memory B cells that did not bind Omicron RBD (Figure 4B). Moreover, a similar fraction of these Omicron RBD-binding memory B cells had an activated phenotype compared with non-Omicron RBD-binding memory B cells (Figures 4C and 4D). Although there were no obvious differences in the recruitment of Omicron RBD-binding versus nonbinding memory B cells in the recall response to a 3rd dose of wild-type spike protein, it will be important to determine if heterologous boosting with variant-specific vaccines or variant infection can preferentially recruit cross-reactive or variant-specific memory B cells.
Taken together, these data indicate that mRNA vaccines encoding the original Wuhan spike protein generated memory B cells that bind Omicron and other variant RBDs. These memory B cells were maintained without decline for at least 9–10 months after the primary 2-dose vaccine series. A 3rd dose of mRNA vaccination activated Omicron-reactive memory B cells at a similar proportion to Omicron RBD nonbinding memory B cells. Thus, mRNA vaccination generates a robust population of memory B cells that maintain reactivity against multiple SARS-CoV-2 VOC, including Omicron, and these cells are efficiently re-engaged by a 3rd vaccine dose.
Immune relationships and predictors of boosted responses
Having quantified antibody and memory B cell responses individually, we next evaluated relationships between different antigen-specific antibody and memory B cell parameters over the course of primary 2-dose vaccination and after a 3rd vaccine dose. To visualize the trajectory of vaccine-induced immunity over time, we clustered samples based on antibody and memory B cell responses using uniform manifold approximation and projection (UMAP) (Figure 5 A). Infection-naive and COVID-recovered individuals clustered apart from each other at the prevaccination baseline time point, as well as at early time points following the primary 2-dose vaccine series (Figure 5B). Notably, these two groups began to converge in UMAP space at later memory time points and were indistinguishable after the 3rd vaccine dose. This was also true in an additional UMAP generated using a larger set of parameters at pre- and post-boost time points, including Omicron-neutralizing antibodies, variant-reactive memory B cells, and memory B cell phenotype (Figures S3 A and S3B). We also examined correlations between antibody and memory B cell responses over time. Correlation analysis was restricted to individuals without prior COVID or postvaccine infection. Antibody and memory B cell responses became more tightly correlated at memory time points before the 3rd dose (Figures 5C and S3C), supporting other findings that mRNA vaccines generate coordinated germinal center responses that ultimately result in the export of both long-lived plasma cells and memory B cells (Kim et al., 2021).
Figure 5.
Immune relationships after 2 and 3 doses of mRNA vaccine
(A) UMAP of antibody and memory B cell responses to mRNA vaccination. Data points represent individual participants and are colored by time point relative to primary vaccine.
(B) UMAP coordinates of SARS-CoV-2-naive and SARS-CoV-2-recovered subjects over time. Labels indicate centroids for each group at the indicated time point. Breakthrough infection samples were excluded from calculations of group centroids.
(C) Correlation matrix of antibody and memory B cell responses over time in SARS-CoV-2-naive subjects.
(D) Correlation of preboost RBD+ memory B cell frequencies with neutralizing antibody recall responses to D614G and Omicron. Recall responses were calculated as the difference between pre- and post-boost titers ∼2 weeks after the 3rd vaccine dose.
(E) Change in binding and neutralizing antibody responses after a 3rd vaccine dose in individuals with 3 versus 4 exposures to SARS-CoV-2 antigen (∼2 weeks post-3rd dose in SARS-CoV-2-naive and SARS-CoV-2-recovered individuals), calculated as in (D). Dotted lines indicate no change in antibodies.
(F) Correlation of preboost binding antibody responses with change in antibody responses after boost, calculated as in (D).
(G) Peak binding and neutralizing antibody levels after 3 versus 4 exposures to SARS-CoV-2 antigen. Dotted lines indicate the limit of detection for the assay.
(H) Correlation of preboost binding antibody levels with peak postboost antibody levels.
(I) Fold-change in antibody responses after 3 versus 4 exposures to SARS-CoV-2 antigen. Dotted lines indicate no change in antibodies.
(J) Correlation of fold-change in antibody responses after boosting with pre-3rd dose antibody levels. Statistics were calculated using unpaired nonparametric Wilcoxon tests with Benjamini-Hochberg correction for multiple comparisons. All correlations were calculated using nonparametric Spearman rank correlation. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
See also Figure S3.
Figure S3.
Expanded analysis of immune relationships and correlation of antibody boosting with time since primary vaccination, related to Figure 5
(A) UMAP of antibody and memory B cell responses to mRNA vaccination, including Omicron neutralization titers, variant-binding memory B cell frequencies, and memory B cell phenotypes. Data points represent individual participants and are colored by time point: 9 months, pre-boost; 9.5 months, boost + 2 weeks; 12 months, boost + 3 months.
(B) Kernel density plots of Omicron neutralizing antibodies and all variant+ memory B cells. Red contours represent areas of UMAP space that are enriched for indicated immune components.
(C) Correlation matrix of antibody and memory B cell responses over time in SARS-CoV-2-naive subjects.
(D) Correlation of preboost wild-type and variant-reactive RBD+ memory B cell frequencies with neutralizing antibody recall responses to D614G and Omicron. Recall responses were calculated as the difference between pre and post-boost titers ∼2 weeks after the 3rd vaccine dose.
(E) Correlation of peak postboost antibody levels (∼2 weeks after the 3rd dose) with days since primary vaccination.
(F) Correlation of fold-change in antibody responses after boosting with days since primary vaccination. All correlations were calculated using nonparametric Spearman rank correlation. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant.
Finally, we investigated how different immune features and sequential exposures to SARS-CoV-2 spike protein affected the absolute and relative magnitudes of recall responses. The pre-3rd dose frequency of RBD+ memory B cells correlated with the absolute change in neutralizing antibody titers (pre-3rd dose subtracted from post-3rd dose) against D614G and Omicron ∼2 weeks after revaccination (Figures 5D and S3D). This observation is consistent with the notion that memory B cells generated after 2 doses of vaccine are an important predictor of subsequent recall responses to SARS-CoV-2 antigens. When comparing individuals with 3 exposures with SARS-CoV-2-spike antigen (three vaccine doses) versus 4 (infection plus three vaccine doses), both groups showed a similar increase in antibody levels after the 3rd vaccine dose (Figure 5E). The absolute increase in antibody titer after the 3rd vaccine dose was not obviously correlated with antibody titers prior to the 3rd vaccine dose, suggesting that the frequency of RBD-binding memory B cells prior to boosting is the primary determinant of the magnitude of new antibody production (Figure 5F). Individuals reached similar peak anti-spike- and RBD-binding antibody titers after the 3rd vaccine dose, regardless of whether it was their 3rd or 4th exposure (Figure 5G). Despite similar binding antibody levels, individuals with 4 total exposures reached slightly higher neutralizing titers against D614G and Omicron (Figure 5G). Pre-3rd dose antibody titers were moderately correlated with peak post-3rd dose antibody titers (Figures 5H and S3C), suggesting that antibody production after boosting is additive across the full range of preboost antibody levels.
To determine whether residual antibody levels affected the relative benefit of a 3rd vaccine dose, we calculated the fold-change in binding and neutralizing antibodies at 2 weeks after revaccination compared with paired pre-3rd dose samples. A 3rd vaccine dose induced a 10-fold to 100-fold increase in antibody titers for individuals with a total of 3 immune exposures to SARS-CoV-2 spike (Figure 5I). By contrast, the fold increase in antibodies following a 3rd vaccine dose in those with previous COVID (i.e., 4 total exposures) was significantly lower with only a 5-fold to 10-fold boost (Figure 5I). Notably, the pre-3rd dose concentration of anti-spike or anti-RBD IgG was strongly negatively correlated with the corresponding fold-change in antibody after the 3rd vaccine dose, regardless of exposure history (Figure 5J). Accordingly, the relative benefit of boosting with an additional vaccine dose may be greatest for individuals with lower levels of preboost antibody.
One concern that has arisen is that vaccinating too soon after a previous exposure might lead to limited boosting. Existing data on this topic for SARS-CoV-2 are so far limited to the time interval between the 1st and 2nd vaccine doses (Chatterjee et al., 2022). Here, we did not observe any significant association between peak antibody titers post-3rd dose and time since primary vaccination (Figure S3E; range = 206–372 days), although there was a weak positive association between the fold-change in antibody responses after a 3rd dose and the time since primary vaccination (Figure S3F). Taken together, these data suggest that boosting with a 3rd dose of mRNA vaccine efficiently re-engages memory B cells to produce new antibodies. Moreover, despite an increase in antibody titers in all subjects after the 3rd vaccine dose, the relative benefit of this increase, measured by fold-change in antibody titers, was greatest in those with lower preboost antibody levels.
Discussion
mRNA vaccination generates protective immunity against SARS-CoV-2 by inducing potent antibody responses as well as memory B cells that can rapidly respond and produce new antibodies upon antigen re-exposure. The establishment of immunity after 2 doses of mRNA vaccine has been well characterized (Cho et al., 2021; Goel et al., 2021a; Mateus et al., 2021; Rodda et al., 2022; Tarke et al., 2022). However, it remains unclear how additional vaccine doses and combinations of vaccination and infection affect the magnitude and quality of immune responses, particularly against immune-evasive SARS-CoV-2 variants like Omicron. In this study, we examined antibody and memory B cell responses through ∼9 months following the primary 2-dose vaccine series as well as up to ∼3 months following a 3rd vaccine dose. In particular, the longitudinal nature of this study enabled detailed analysis of the magnitude, durability, and quality of SARS-CoV-2 vaccine-induced immunity over ∼1 year and multiple antigen exposures.
This study provides several key pieces of data relevant to SARS-CoV-2 and mRNA vaccine immunobiology. Although antibody titers declined from peak levels observed ∼1 week after the second dose to ∼6 months postprimary vaccination, these antibody titers then stabilized between 6 and 9 months postvaccination with continued improvement of neutralization potency over this period. A 3rd vaccine dose at ∼9 months postprimary vaccination increased antibody responses ∼10-fold to 100-fold, including boosting neutralizing antibodies against the Omicron variant. Moreover, the antibody titers achieved after the 3rd dose were similar to those observed in SARS-CoV-2 recovered individuals after 2 doses of mRNA vaccine, commonly referred to as hybrid immunity. Breakthrough infection after 2 doses of mRNA vaccine also appeared to produce similar increases in antibody to a 3rd vaccine dose. These boosted antibody responses subsequently declined over time but still remained significantly above preboost levels at 3 months post-3rd dose.
In contrast to antibodies, which decayed over time following vaccination, memory B cell numbers remained highly stable in the blood with no evidence of decay at ∼9 months postprimary vaccination. Notably, 2-dose vaccination generated a robust memory B cell response against the Omicron variant, with ∼40%–50% of RBD-binding memory B cells able to cross-bind Alpha, Beta, Delta, and Omicron. A 3rd vaccine dose efficiently recruited memory B cells with cross-reactivity to multiple VOC, resulting in amplification of antibody responses capable of neutralizing spike proteins from immune-evasive SARS-CoV-2 VOC including Omicron. The ability of approximately half of the memory B cell pool to bind multiple variants indicates that the antibodies encoded by these memory B cells are targeting more conserved epitopes of RBD or are of higher quality and able to overcome epitope changes associated with mutations in VOC. It will be important to determine how resilient these memory B cell responses are to emerging variants. For example, BA.2 shares many features with previous VOC but also contains additional mutations not observed in the BA.1 Omicron sublineage (Yamasoba et al., 2022). Regardless, mRNA vaccines encoding the original Wuhan spike are capable of generating and boosting memory B cell responses and associated antibodies with the capacity to recognize major current SARS-CoV-2 VOC and may provide lasting protection against future variants.
Our data also identify several factors that predict the absolute magnitude and relative benefit of boosting. Boosting with a 3rd vaccine dose universally increased binding and neutralizing antibody responses compared with preboost levels, and preboost memory B cell frequency was the best predictor of the increase in antibody levels after boosting. Binding antibody titers achieved after a 3rd dose were similar in individuals with or without a prior SARS-CoV-2 infection. However, infected-then-vaccinated individuals with a total of 4 immune exposures reached slightly higher neutralizing titers against D614G and Omicron than SARS-CoV-2-naive individuals who received 3 doses of vaccine, suggesting that additional vaccination or infection following a 3-dose vaccine regimen may still have a quantitative benefit to antibody responses. Despite similar peak antibody titers after a booster dose, a 3rd exposure to SARS-CoV-2-Spike antigen increased antibodies 10-fold to 100-fold, whereas a 4th exposure (i.e., in individuals with prior infection) resulted in a lower fold-change (5-fold to 10-fold). A major factor influencing this difference in the relative magnitude of the boost was pre-3rd vaccine dose antibody concentrations, which were inversely correlated with the fold-change of antibody boosting. This is consistent with previous data from mouse models showing that high levels of immunoglobulin may attenuate recall responses (Pape et al., 2011). Accordingly, the relative value of additional vaccine doses will likely be greatest for individuals with lower preboost antibody titers, including immunocompromised or older populations.
Overall, this study supports the utility of a 3rd vaccine dose to recall immunological memory and boost circulating antibody levels. These recall responses were resilient to VOC, as a 3rd dose of mRNA vaccine encoding the original Wuhan spike efficiently re-engaged Omicron-reactive memory B cells generated by the first two doses. Our results also highlight immunological factors that may constrain the added protection afforded by repeated short interval boosting. Additional work is required to evaluate variant-specific vaccines and how boosting with a modified antigen may augment recall responses compared with boosting with the original Wuhan strain sequence (Gagne et al., 2022b). Nevertheless, these data highlight the urgent need to better understand what antibody titers are necessary for protection against infection and/or severe disease (Khoury et al., 2021). Should this threshold be defined, it may be useful to implement serologic testing to maximize the benefit and equity of additional vaccine doses moving forward.
Limitations of the study
There are several limitations to this study. Although we studied immunological responses to mRNA vaccination, the precise correlates of vaccine efficacy are still being defined. Thus, these data on antibody and memory B cell responses cannot be directly translated to levels of clinical protection. In addition, we measured antibody quality using an index dependent on neutralization but did not measure other non-neutralizing functions of antibodies that could have additional roles in antiviral immunity. Regarding our investigation of memory B cells, our strategy for examining Omicron RBD-reactive cells required simultaneous binding to WT RBD. It remains possible that there are also populations of Omicron RBD-specific memory B cells that do not bind WT RBD. Finally, this study examined predominantly young, healthy subjects, and the immunological responses observed may not be entirely representative of those observed in older individuals, those with compromised immune systems, or other demographically distinct cohorts.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BUV563 anti-CD3 | BD Biosciences | Cat#748569; RRID: AB_2872978 |
| BV750 anti-CD19 | Biolegend | Cat#302262; RRID: AB_2810434 |
| BUV805 anti-CD20 | BD Biosciences | Cat#612905; RRID: AB_2870192 |
| BUV395 anti-CD27 | BD Biosciences | Cat#563815; RRID: AB_2744349 |
| BUV661 anti-CD38 | BD Biosciences | Cat#612969; RRID: AB_2870242 |
| APC-H7 anti-CD71 | BD Biosciences | Cat#563671; RRID: AB_2738364 |
| AF700 anti-CD11c | Biolegend | Cat#337220; RRID: AB_2561503 |
| FITC anti-IgA | Miltenyi | Cat#130-113-475; RRID: AB_2726166 |
| BV480 anti-IgD | BD Biosciences | Cat#566138; RRID: AB_2739536 |
| PE/Dazzle 594 anti-CD21 | Biolegend | Cat#354922; RRID: AB_2750243 |
| PE-Cy7 anti-IgG | Biolegend | Cat#410722; RRID: AB_2750227 |
| PerCP/Cy5.5 anti-IgM | Biolegend | Cat#314512; RRID: AB_2076098 |
| mouse anti-VSV Indiana G, 1E9F9 | Absolute Antibody | Cat#Ab01402-2.0 |
| goat anti-human IgG-HRP | Jackson Immuno Research Laboratories | RRID: AB_2337596 |
| goat anti-human IgM-HRP | SouthernBiotech | RRID: AB_2795603 |
| Bacterial and virus strains | ||
| SARS-CoV-2 VSV pseudotypes | Generated for this paper | N/A |
| Biological samples | ||
| Human peripheral blood samples from SARS-CoV-2 mRNA vaccine recipients | Collected at the University of Pennsylvania | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 Biotyinlated Full Length Spike | R&D Systems | Cat#AVI10549-050 |
| SARS-CoV-2 Biotyinlated Full Length Spike | R&D Systems | Cat#BT10500-050 |
| HA(ΔTM)(A/Brisbane/02/2018)(H1N1) | Immune Tech | Cat#IT-003-00110ΔTMp |
| HA(ΔTM)(B/Colorado/06/2017) | Immune Tech | Cat#IT-003-B21ΔTMp |
| SARS-CoV-2 Biotyinlated RBD | Acro Biosystems | Cat#SPD-C82E9-25ug |
| SARS-CoV-2 Biotyinlated RBD (N501Y) | Acro Biosystems | Cat#SPD-C82E6-25ug |
| SARS-CoV-2 Biotyinlated RBD (K417N/E484K/N501Y) | Acro Biosystems | Cat#SPD-C82E5-25ug |
| SARS-CoV-2 Biotyinlated RBD (L452R/K478N) | Acro Biosystems | Cat#SPD-C82Ed-25ug |
| SARS-CoV-2 Biotyinlated RBD (Omicron) | Acro Biosystems | Cat#SPD-C82E4-25ug |
| SARS-CoV-2 Biotyinlated N-Terminal Domain | Sino Biological | Cat#40591-V49H-B |
| SARS-CoV-2 Biotyinlated S2 | Acro Biosystems | Cat#S2N-C52E8-25ug |
| SARS-CoV-2 Biotyinlated Nucleocapsid | R&D Systems | Cat#BT10474-050 |
| BV421 Streptavidin | Biolegend | Cat#405226 |
| BV605 Streptavidin | Biolegend | Cat#405229 |
| BV711 Streptavidin | BD Biosciences | Cat#563262 |
| BV786 Streptavidin | BD Biosciences | Cat#563858 |
| BUV615 Streptavidin | BD Biosciences | Cat#613013 |
| BUV737 Streptavidin | BD Biosciences | Cat#612775 |
| BB515 Streptavidin | BD Biosciences | Cat#564453 |
| PE Streptavidin | Biolegend | Cat#405203 |
| PE-Cy7 Streptavidin | Biolegend | Cat#405206 |
| APC Streptavidin | Biolegend | Cat#405207 |
| Ghost Viability Dye Violet 510 | Tonbo | Cat#13-0870-T100 |
| Human TruStain FcX (Fc Receptor Blocking Solution) | Biolegend | Cat#422302 |
| EZ-Link Micro NHS-PEG4 Biotinylation Kit | Thermo Fisher | Cat#21955 |
| Zeba Spin Desalting Columns 7K MWCO | Thermo Fisher | Cat#89894 |
| Experimental models: Cell lines | ||
| 293T | ATCC | RRID: CVCL_0063 |
| VeroE6/TMPRSS | Stefan Pohlman | N/A |
| Recombinant DNA | ||
| Plasmid: pCAGGS SARS-CoV-2 spike | Florian Krammer | N/A |
| Plasmid: pCAGGS SARS-CoV-2 RBD | Florian Krammer | N/A |
| Plasmid: pCG1 SARS-CoV-2 D614G delta18 | Paul Bates Lab | N/A |
| Plasmid: pCG1 SARS-CoV-2 B.1.617.2 delta18 | Paul Bates Lab | N/A |
| Plasmid: pCG1 SARS-CoV-2 B.1.1.529 delta18 | Paul Bates Lab | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, E. John Wherry (wherry@pennmedicine.upenn.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subject recruitment and sampling
Sixty-one individuals were enrolled in the longitudinal vaccine study with informed consent and approval from the University of Pennsylvania Institutional Review Board (IRB# 844642). Of the 61 individuals, 45 were SARS-CoV-2 naïve prior to receiving their first vaccine dose and 16 had recovered from prior SARS-CoV-2 infection. All participants were otherwise healthy, with no self-reported history of chronic health conditions. Prior SARS-CoV-2 infection status was determined by a combination of self-reporting and laboratory detection of pre-existing immune responses. All subjects received mRNA vaccines, either Pfizer (BNT162b2) or Moderna (mRNA-1273). Samples were collected at 10 timepoints: baseline (T1), 2 weeks post-1st dose (T2), day of 2nd dose (T3), 1 week post-2nd dose (T4), 3 months post-primary immunization (T5), 6 months post-primary immunization (T6), 9-10 months post-primary immunization (T7), pre 3rd dose (T8), 2 weeks post 3rd dose (T9), and 3 months post 3rd dose (T10). Individuals received booster doses between 6 and 12 months post-primary vaccination. For clarity, we assigned booster doses as taking place at 9 months on longitudinal graphs. For the subset of individuals who received a booster vaccination after collection of T7, an additional sample was collected when they were transferred to the booster sub-study (T8). For those who received a booster dose before the T7 timepoint was collected, they were transferred to the booster sub-study for collection of T8 without collection of T7. For simplicity, T7 and T8 were visualized as a single timepoint at ∼9 months in most figures. Peripheral blood samples (80-100mL) and clinical questionnaire data were collected at each of the timepoints listed above. Full cohort and demographic information is provided in Table S1. Additional healthy donor PBMC samples were collected with approval from the University of Pennsylvania Institutional Review Board (IRB# 845061).
Method details
Processing of peripheral blood
Venous blood was collected into sodium heparin and EDTA tubes by standard phlebotomy. Plasma was separated by centrifuging blood tubes at 3000rpm for 15 minutes and aliquots were stored at -80°C for subsequent antibody analyses. The remaining cellular fraction was diluted with an equal volume of RPMI + 1% FBS + 2mM L-Glutamine + 100 U Penicillin/Streptomycin (R10 medium) and layered above a lymphoprep gradient (STEMCELL Technologies) in SEPMATE tubes (STEMCELL Technologies). SEPMATE tubes were centrifuged at 1200g for 10 minutes and the PBMC fraction was decanted into fresh tubes, washed once with R10, and treated with ACK lysis buffer (Thermo Fisher) for 5 minutes. The ACK reaction was stopped with an additional wash, and cells were then resuspended in R10, filtered with a 70μm cell strainer, and counted using a Countess automated cell counter (Thermo Fisher). PBMCs were aliquoted and cryopreserved in 90% FBS 10% DMSO.
Measuring SARS-CoV-2 binding antibodies
SARS-CoV-2-specific antibody levels in plasma were measured by enzyme-linked immunosorbent assay (ELISA) as previously described (Flannery et al., 2020; Goel et al., 2021b, 2021a). Plasmids encoding recombinant full-length SARS-CoV-2 Spike protein and RBD were provided by F. Krammer (Mt. Sinai) and purified by nickel-nitrilotriacetic acid resin (Qiagen). ELISA plates (Immulon 4 HBX, Thermo Fisher Scientific) were coated overnight at 4°C with 2 μg/mL recombinant protein or PBS. After overnight incubation, the plates were washed with phosphate-buffered saline containing 0.1% Tween-20 (PBS-T) and then blocked for 1 hour with PBS-T supplemented with 3% non-fat milk powder. Plasma samples were heat-inactivated for 1 hour at 56°C and diluted in PBS-T supplemented with 1% non-fat milk powder. After washing the plates with PBS-T, 50 μL of diluted sample was added to each well and plates were incubated for 2 hours. Plates were washed again with PBS-T and then incubated for 1 hour with 50 μL of 1:5000 diluted goat anti-human IgG-HRP (Jackson ImmunoResearch Laboratories) or 1:1000 diluted goat anti-human IgM-HRP (SouthernBiotech). After secondary antibody, plates were washed again with PBS-T. 50 μL SureBlue 3,3’,5,5’-tetramethylbenzidine substrate (KPL) was added to each well and plates were incubated for 5 minutes. The reaction was then stopped by adding 25 μL of 250 mM hydrochloric acid to each well. Plates were read with a SpectraMax 190 microplate reader (Molecular Devices) at an optical density (OD) of 450 nm. Monoclonal antibody CR3022 was included on each plate to convert OD values into relative antibody concentrations. Plasmids to express CR3022 were provided by I. Wilson (Scripps).
Measuring SARS-CoV-2 neutralizing antibodies
HEK 293T cells were seeded at 5 X 106 cells per 10 cm dish, incubated for 24 hours, and transfected using calcium phosphate with 25 μg of pCG1 SARS-CoV-2 S D614G delta18, pCG1 SARS-CoV-2 S B.1.617.2 delta 18, or pCG1 SARS-CoV-2 S B.1.1.529 delta 18 expression plasmid encoding a codon optimized SARS-CoV-2 S gene with an 18-residue truncation in the cytoplasmic tail. Mutations present in the variant constructs are provided below. To increase the expression of transfected DNA, the supernatant was replaced with fresh media containing 5mM sodium butyrate after 12 hours. Twenty-four hours post-transfection, cells were infected for 2 hours with VSV-G pseudotyped VSVΔG-RFP at an MOI of ∼1-3. After the infection, the viral supernatant was replaced with fresh serum-free media. VSVΔG-RFP SARS-CoV-2 pseudotypes were harvested by collecting the culture media 28-30 hours after infection. These supernatants were clarified by centrifugation twice at 6000g and stored at -80°C for neutralization assays. Before performing the neutralization assay, serum samples were thawed and heat-inactivated for 30 minutes at 53 ⁰C. Vero E6 cells stably expressing TMPRSS2 were seeded at a density of 2.5x104 cells/well in 100 μl in a 96 well collagen-coated plate. VSVΔG-RFP SARS-CoV-2 pseudotype virus (100-300 focus forming units/well) was mixed with serum samples from a serial two-fold dilution and incubated for 1 hour at 37⁰C. To neutralize any potential VSV-G carryover virus, mouse anti-VSV Indiana G antibody (1E9F9) was also added at a concentration of 600 ng/ml (Absolute Antibody, Ab01402-2.0), and the VeroE6 TMPRSS2 cell culture media was replaced with this serum-virus mixture. After 21-22 hours, VeroE6 cells were washed and fixed with 4% paraformaldehyde before visualization on an S6 FluoroSpot Analyzer (CTL, Shaker Heights OH). Individual infected foci were enumerated and the values were compared to control wells without antibody. The focus reduction neutralization titer 50% (FRNT50) was measured as the greatest serum dilution at which focus count was reduced by at least 50% relative to control cells that were infected in the absence of human serum. The geometric mean FRNT50 titers for each sample were reported based on readings from at least two technical replicates.
| Variant | Mutations in Pseudovirus Construct |
|---|---|
| D614G (WT) | D614G |
| B.1.351 (Beta) | L18F, D80A, D215G, R246I, K417N, E484K, N501Y, D614G, A701V |
| B.1.617.2 (Delta) | T19R, G142D, del156-157, R158G, L452R, T478K, D614G, P681R, D950N |
| B.1.1.529 (Omicron) | A67V, del69-70, T95I, G142D, del143-145, N211I, del212, ins214 (EPE), G339D, S371L, S373P, S375F, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, G655Y, N679K, P681H, D796Y, N856K, Q954H, N969K, L981F |
WT SARS-CoV-2-specific memory B cell analyses
Antigen-specific B cells were detected as previously described (Goel et al., 2021b, 2021a). Biotinylated proteins were multimerized by mixing with fluorescently labeled streptavidin (SA) and incubated for 1 hour at 4°C as follows: full-length Spike protein and SA-BV421 at a mass ratio of 10:1 (200ng Spike with 20ng SA; ∼4:1 molar ratio); Spike RBD and SA-APC at a mass ratio of 2:1 (25ng RBD with 12.5ng SA; ∼4:1 molar ratio); biotinylated influenza HA pools and SA-PE at a mass ratio of 6.25:1 (100ng HA pool with 16ng SA; ∼6:1 molar ratio). Influenza HA antigens from the 2019 trivalent vaccine (A/Brisbane/02/2018/H1N1, B/Colorado/06/2017) were biotinylated using an EZ-Link Micro NHS-PEG4 Biotinylation Kit (Thermo Fisher) according to the manufacturer’s instructions and used as a control antigen. After removing excess biotin with Zeba Spin Desalting Columns 7K MWCO (Thermo Fisher) according to manufacturer's protocol, protein was quantified by Pierce BCA Assay (Thermo Fisher). SA-BV711 was not multimerized with biotinylated protein and was used as a decoy probe to gate out cells that non-specifically bind streptavidin. Cell staining was performed in a solution of PBS + 2% FBS and Brilliant Buffer (BD Bioscience) at a 1:1 ratio. Antigen probes for Spike, RBD, and HA were individually multimerized before each stain and combined after multimerization with 5μM free D-biotin (Avidity LLC) to minimize the risk of multi-antigen probe formation. 5x106 cryopreserved PBMCs were thawed and stained in a 96-well U-bottom plate as follows. First, cells were incubated with Human TruStain FcX (Fc receptor blocking solution, Biolegend, 1:200) and Ghost 510 Viability Dye (Tonbo, 1:600) for 15 minutes at 4°C. Cells were then washed once with PBS + 2% FBS and stained with 50μL antigen probe master mix containing 200ng Spike-BV421, 25ng RBD-APC, 100ng HA-PE, and 20ng SA-BV711 decoy for 1 hour at 4°C. Cells were then washed again and stained with anti-CD3, anti-CD19, anti-CD20, anti-CD27, anti-CD38, anti-CD71, anti-IgD, anti-IgM, anti-IgG, and anti-IgA for 30 minutes at 4°C. Cells were then washed again and fixed overnight at 4°C using 1% PFA in PBS. Gates were set using healthy donor samples stained without antigen probes and identical gates were used for all experimental runs.
Variant RBD, NTD, and S2-specific memory B cells
Variant RBD, NTD, and S2-specific memory B cells were measured using an approach similar to what was used to measure WT Spike antigens above. In this assay, SARS-CoV-2 nucleocapsid was used as a control antigen specific for infection but not vaccination. Probes were multimerized individually at 4°C for 1.5 hours by combing biotinylated proteins and fluorophore-conjugated SA at ∼4:1 molar ratios (moles of SA calculated without considering the fluorophore): 200ng full-length Spike protein was incubated with 20ng SA-BV421, 30ng N-terminal domain was incubated with 12ng SA-BV786, 25ng wild-type RBD was incubated with 12.5ng SA-BB515, 25ng Alpha RBD was incubated with 12.5ng SA-BV711, 25ng Beta RBD was incubated with 12.5ng SA-PE, 25ng Delta RBD was incubated with 12.5ng SA-APC, 25ng Omicron RBD was incubated with 12.5ng SA-PE-Cy7, 50ng S2 was incubated with 12ng SA-BUV737, 50ng nucleocapsid was incubated with 14ng SA-BV605. 12.5ng SA-BUV615 was used as a decoy probe. Antigen probes were prepared fresh before each stain and then pooled in together as an antigen probe master mix in a solution containing 5μM free D-biotin. Total B cells were enriched from 10-20x106 cryopreserved PBMCs by negative selection using an EasySep human B cell isolation kit (STEMCELL, #17954). B cells were then incubated in a 96-well U-bottom plate with Fc block and Ghost 510 Viability Dye for 15 minutes at 4°C. Cells were washed with PBS + 2% FBS and stained with 50μL antigen probe master mix for 1 hour at 4°C. After probe staining, cells were washed again and stained with anti-CD3, anti-CD11c, anti-CD19, anti-CD21, anti-CD27, anti-CD38, and anti-IgD for 30 minutes at 4°C. Cells were then washed and fixed in 1X Stabilizing Fixative (BD Biosciences) overnight at 4°C.
Flow cytometry
Flow cytometry data was collected on a BD Symphony A5 instrument. Standardized SPHERO rainbow beads (Spherotech) were used to track and adjust photomultiplier tubes over time. UltraComp eBeads (Thermo Fisher) were used for compensation. Up to 5x106 cells were acquired per sample. Data were analyzed using FlowJo v10 (BD Bioscience). For Boolean analysis of variant cross-binding, data were imported into SPICE 6 (NIH Vaccine Research Center (Roederer et al., 2011)).
Quantification and statistical analysis
High dimensional analysis and statistics
All data were analyzed in RStudio using custom scripts. Heatmaps were generated using the pheatmap package in R. Pairwise non-parametric correlations between variables were calculated and visualized as a correlogram using corrplot with FDR correction as described previously (Mathew et al., 2020). Five antigen-specific immune features were used to train the UMAP in Figure 5: anti-Spike IgG, anti-RBD IgG, D614G FRNT50, Spike+ memory B cell frequency, and RBD+ memory B cell frequency. Sixteen antigen-specific immune features were used to train the expanded UMAP in Figure S3, including binding and neutralizing antibodies (anti-Spike IgG, anti-RBD IgG, D614G FRNT50, and Omicron FRNT50), memory B cell frequencies (NTD+, wild-type RBD+, All Variant RBD+, S2+), % cross binding to variant RBDs (Alpha, Beta, Delta, Omicron, and All Variant), and memory B cell phenotype (% activated memory NTD+, RBD+, and S2+ cells). Antibody and cell frequency data were log10 transformed and then z-score normalized prior to generating UMAP coordinates using the umap package in R. Statistical tests are indicated in the corresponding figure legends. All tests were performed two-sided with a nominal significance threshold of p < 0.05. Benjamini-Hochberg (BH) correction was performed in all cases of multiple comparisons. As samples from some participants were missing at individual time points, unpaired tests were used unless otherwise indicated. ∗ indicates p < 0.05, ∗∗ indicates p < 0.01, ∗∗∗ indicates p < 0.001, ∗∗∗∗ indicates p < 0.0001. All data and code are available upon request.
Acknowledgments
We would like to thank the study participants for their generosity in making the study possible. We also thank Ali Ellebedy and Julian Zhou for helpful discussions and feedback, as well as the flow cytometry core at the University of Pennsylvania for technical support. Funding: This work was supported by NIH grants AI108545, AI155577, and AI149680 (to E.J.W.), AI152236 and AI142638 (to P.B.), HL143613 (to J.R.G.), R38 HL143613 (to D.A.O.), T32 AR076951-01 (to S.A.A.), T32 CA009140 (to J.R.G., D.A.O., and D.M.), T32 AI055400 (to P.H.), U19AI082630 (to S.E.H. and E.J.W.), P30CA016520, and NIH contract no. 75N93021C00015; Australian Government Medical Research Future Fund awards GNT2002073 (to M.P.D.), MRF2005544 (to M.P.D.), and MRF2005760 (to M.P.D.); an NHMRC program grant GNT1149990 (to M.P.D.); NHMRC Fellowship and Investigator grants (to D.S.K. and M.P.D.); funding from the National Health and Medical Research Council of Australia and the Australian Research Council (to D.S.K.); funding from the Allen Institute for Immunology (to S.A.A. and E.J.W.); a Cancer Research Institute-Mark Foundation Fellowship (to J.R.G.); the Chen Family Research Fund (to S.A.A.); the Parker Institute for Cancer Immunotherapy (to J.R.G. and E.J.W.); the Penn Center for Research on Coronavirus and Other Emerging Pathogens (to P.B.); a philanthropic gift from J. Lurie, J. Embiid, J. Harris, and D. Blitzer (to S.E.H.), the University of Pennsylvania Perelman School of Medicine COVID Fund (to R.R.G. and E.J.W.); the University of Pennsylvania Perelman School of Medicine 21st Century Scholar Fund (to R.R.G.); and the Paul and Daisy Soros Fellowship for New Americans (to R.R.G).
Author contributions
R.R.G., M.M.P., and E.J.W. designed the study. R.R.G., M.M.P., K.A.L., S.A.A., A.E.B., J.R.G., D.M., S.G., P.H., S.D., R.I.E., M.E.W., C.M.M., M.A., and N.T. performed experiments. A.R., D.S.K., W.K., D.A.O., and M.P.D. provided expertise on data analysis and methodology. R.R.G., M.M.P., S.A.A., A.P., A.H., H.S., S.H., S.K., J.D., S.L., K.D., J.T.H., M.M., J.C.W., S.A., O.K., and E.M.D. were involved in clinical recruitment and sample collection/processing. M.P.D., S.E.H., P.B., A.R.G., and E.J.W. supervised the study. All authors participated in data analysis and interpretation. R.R.G., M.M.P., and E.J.W. wrote the manuscript.
Declaration of interests
S.E.H. has received consultancy fees from Sanofi Pasteur, Lumen, Novavax, and Merck for work unrelated to this study. A.R.G. is a consultant for Relation Therapeutics. E.J.W. is consulting for or is an advisor for Merck, Marengo, Janssen, Related Sciences, Synthekine, and Surface Oncology. E.J.W. is a founder of Surface Oncology, Danger Bio, and Arsenal Biosciences.
Published: April 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.04.009.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bournazos S., Ravetch J.V. Fcγ receptor function and the design of vaccination strategies. Immunity. 2017;47:224–233. doi: 10.1016/j.immuni.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., de Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Krammer F., Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat. Rev. Immunol. 2021;21:245–256. doi: 10.1038/s41577-021-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D., Tauzin A., Marchitto L., Gong S.Y., Boutin M., Bourassa C., Beaudoin-Bussières G., Bo Y., Ding S., Laumaea A., et al. SARS-CoV-2 Omicron spike recognition by plasma from individuals receiving BNT162b2 mRNA vaccination with a 16-week interval between doses. Cell Rep. 2022;38:110429. doi: 10.1016/j.celrep.2022.110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery D.D., Gouma S., Dhudasia M.B., Mukhopadhyay S., Pfeifer M.R., Woodford E.C., Gerber J.S., Arevalo C.P., Bolton M.J., Weirick M.E., et al. SARS-CoV-2 seroprevalence among parturient women in Philadelphia. Sci. Immunol. 2020;5:eabd5709. doi: 10.1126/sciimmunol.abd5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne M., Corbett K.S., Flynn B.J., Foulds K.E., Wagner D.A., Andrew S.F., Todd J.M., Honeycutt C.C., McCormick L., Nurmukhambetova S.T., et al. Protection from SARS-CoV-2 Delta one year after mRNA-1273 vaccination in rhesus macaques coincides with anamnestic antibody response in the lung. Cell. 2022;185 doi: 10.1016/j.cell.2021.12.002. 113.e15–130.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne M., Moliva J.I., Foulds K.E., Andrew S.F., Flynn B.J., Werner A.P., Wagner D.A., Teng I.-T., Lin B.C., Moore C., et al. mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits comparable B cell expansion, neutralizing antibodies and protection against Omicron. Preprint at bioRxiv. 2022 doi: 10.1101/2022.02.03.479037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St. Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184 doi: 10.1016/j.cell.2021.04.006. 2372.e9–2383.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185 doi: 10.1016/j.cell.2021.12.033. 457.e4–466.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Apostolidis S.A., Painter M.M., Mathew D., Pattekar A., Kuthuru O., Gouma S., Hicks P., Meng W., Rosenfeld A.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naive and recovered individuals following mRNA vaccination. Sci. Immunol. 2021;6:eabi6950. doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29 doi: 10.1016/j.chom.2021.02.003. 463.e6–476.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:1–14. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Kim W., Zhou J.Q., Sturtz A.J., Horvath S.C., Schmitz A.J., Lei T., Kalaidina E., Thapa M., Alsoussi W.B., Haile A., et al. Germinal centre-driven maturation of B cell response to SARS-CoV-2 vaccination. Preprint at bioRxiv. 2021 doi: 10.1101/2021.10.31.466651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaki R., Adachi Y., Moriyama S., Onodera T., Fukushi S., Nagakura T., Tonouchi K., Terahara K., Sun L., Takano T., et al. SARS-CoV-2 Omicron-neutralizing memory B-cells are elicited by two doses of BNT162b2 mRNA vaccine. Sci. Immunol. 2022:eabn8590. doi: 10.1126/sciimmunol.abn8590. Published online February 3, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw B.J., Ellebedy A.H. The germinal centre B cell response to SARS-CoV-2. Nat. Rev. Immunol. 2021;22:7–18. doi: 10.1038/s41577-021-00657-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau D., Lan L.Y.L., Andrews S.F., Henry C., Rojas K.T., Neu K.E., Huang M., Huang Y., DeKosky B., Palm A.E., et al. Low CD21 expression defines a population of recent germinal center graduates primed for plasma cell differentiation. Sci. Immunol. 2017;2:eaai8153. doi: 10.1126/sciimmunol.aai8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2017;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Dan J.M., Zhang Z., Moderbacher C.R., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low dose mRNA-1273 COVID-19 vaccine generates durable T cell memory and antibodies enhanced by pre-existing cross-reactive T cell memory. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., Datir R., Collier D.A., Albecka A., Singh S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muecksch F., Wang Z., Cho A., Gaebler C., Tanfous T.B. ben, DaSilva J., Bednarski E., Ramos V., Zong S., Johnson B., et al. Increased potency and breadth of SARS-CoV-2 neutralizing antibodies After a third mRNA vaccine dose. Preprint at bioRxiv. 2022 doi: 10.1101/2022.02.14.480394. [DOI] [Google Scholar]
- Muik A., Lui B.G., Wallisch A.-K., Bacher M., Mühl J., Reinholz J., Ozhelvaci O., Beckmann N., Güimil Garcia R. de la C., Poran A., et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science. 2022;375:678–680. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immune responses to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54 doi: 10.1016/j.immuni.2021.08.001. 2133.e3–2142.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape K.A., Taylor J.J., Maul R.W., Gearhart P.J., Jenkins M.K. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Rodda L.B., Morawski P.A., Pruner K.B., Fahning M.L., Howard C.A., Franko N., Logue J., Eggenberger J., Stokes C., Golez I., et al. Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity. Cell. 2022 doi: 10.1016/j.cell.2022.03.018. Published online March 17, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Nozzi J.L., Nason M.C. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185 doi: 10.1016/j.cell.2022.01.018. 1025.e14–1040.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., et al. Plasma neutralization of the SARS-CoV-2 omicron variant. N. Engl. J. Med. 2021;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Rutkowska M., Poston D., DaSilva J., Zhang F., Bednarski E., Cho A., Schaefer-Babajew D.J., Gaebler C., et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature. 2021;600:512–516. doi: 10.1038/s41586-021-04005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha N.K., Shrestha P., Burke P.C., Nowacki A.S., Terpeluk P., Gordon S.M. Coronavirus disease 2019 (COVID-19) vaccine boosting in persons already protected by natural or vaccine-induced immunity. J. Neurol. Sci. 2022;428:117607. [Google Scholar]
- Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185 doi: 10.1016/j.cell.2022.01.015. 847.e11–859.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Nussenzweig M.C. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Wang K., Jia Z., Bao L., Wang L., Cao L., Chi H., Hu Y., Li Q., Jiang Y., Zhu Q., et al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature. 2022;603:919–925. doi: 10.1038/s41586-022-04466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Zou J., Kurhade C., Cai H., Yang Q., Cutler M., Cooper D., Muik A., Jansen K.U., Xie X., et al. Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.02.015. 485.e3–488.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., et al. Virological characteristics of SARS-CoV-2 BA.2 variant. Preprint at bioRxiv. 2022 doi: 10.1101/2022.02.14.480335. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.