Abstract
Purpose
Deploying external artificial intelligence (AI) models locally can be logistically challenging. We aimed to use the ACR AI-LAB software platform for local testing of a chest radiograph (CXR) algorithm for COVID-19 lung disease severity assessment.
Methods
An externally developed deep learning model for COVID-19 radiographic lung disease severity assessment was loaded into the AI-LAB platform at an independent academic medical center, which was separate from the institution in which the model was trained. The data set consisted of CXR images from 141 patients with reverse transcription-polymerase chain reaction–confirmed COVID-19, which were routed to AI-LAB for model inference. The model calculated a Pulmonary X-ray Severity (PXS) score for each image. This score was correlated with the average of a radiologist-based assessment of severity, the modified Radiographic Assessment of Lung Edema score, independently interpreted by three radiologists. The associations between the PXS score and patient admission and intubation or death were assessed.
Results
The PXS score deployed in AI-LAB correlated with the radiologist-determined modified Radiographic Assessment of Lung Edema score (r = 0.80). PXS score was significantly higher in patients who were admitted (4.0 versus 1.3, P < .001) or intubated or died within 3 days (5.5 versus 3.3, P = .001).
Conclusions
AI-LAB was successfully used to test an external COVID-19 CXR AI algorithm on local data with relative ease, showing generalizability of the PXS score model. For AI models to scale and be clinically useful, software tools that facilitate the local testing process, like the freely available AI-LAB, will be important to cross the AI implementation gap in health care systems.
Key Words: ACR AI-LAB, AI, chest radiograph, COVID-19, local testing
Visual Abstract
O wonder! How many goodly creatures are there here! How beauteous mankind is! O brave new world, that has such people in’t.
—William Shakespeare, The Tempest [1]
Background
A vigorous debate has emerged how to best bring the benefits of the brave new world of artificial intelligence to bear in our clinical enterprises. When individual practices deploy artificial intelligence (AI) today, most contract with individual commercial companies to deploy their clinical solutions, or they use a “platform vendor” to choose from algorithms available on those aggregated marketplaces. In either scenario, validation of these algorithms with site-specific data is recommended to ensure that performance of the algorithm with local data conforms to stand-alone performance testing. In a broader sense, and during the development of algorithms, AI algorithms need to be validated using real-world data that reflect the spectrum of disease in a range of practice types with variable imaging devices, before commercial clinical deployment. The ability of radiology practices to participate in such algorithm validation is hampered by their rightful reluctance to release their (anonymized) patient data beyond their institution for commercial use. Algorithm developers, on the other hands, are concerned with protecting the proprietary nature of their trained algorithms. Therefore, a need exists for solutions, and serves as an intermediary, bringing together practices and their data with developers to train, test, validate, and assess AI algorithms. Multiple approaches are emerging that address this need in different ways, with or without the need to move source data. As an example, the Medical Imaging and Data Resource (MIDRC), funded by the National Institute of Biomedical Imaging and Bioengineering and implemented by a consortium of professional societies and academic resources, facilitates central data collection and AI research on various entities. Platform and marketplace vendors are beginning to incorporate tools for acceptance testing into their commercial offerings. Early-stage commercial offerings are emerging, which promise to enable interaction of research or commercial algorithms with local data without the need to share the data.
The ACR AI-LAB application has been developed by the ACR Data Science Institute as a platform to lower the barrier to entry for radiologists to engage with AI algorithms under development by external entities, without the need to share patient data externally [2]. This platform aims to democratize participation in AI algorithm development and evaluation. One should remember that even when using an AI algorithm intermediary platform, a practice may need additional resources to participate in these activities. Practices need to have the ability to identify suitable patient cohorts, or build examination-specific filters, and identify suitable images from each examination to present to the AI algorithm. This may be viewed as an insurmountable hurdle by some, particularly in the case of small and midsized practices with limited informatics resources.
Our institution sought to participate in AI algorithm testing using the AI-LAB to better understand the process. During the first half of 2020, all hospitals in our metropolitan area were heavily affected by the first wave of COVID-19 infections; for this reason, there was a particular interest for testing a COVID-19 chest radiology (CXR) algorithm trained to assess disease severity. Multiple AI algorithms have been developed for detection of COVID-19 on CXR [3, 4, 5, 6]. However, since the radiographic findings of the COVID-19 infection are nonspecific and both Centers for Disease Control and ACR do not currently recommend CXR or CT for the primary diagnosis of COVID-19, there is limited clinical value for diagnostic AI in this regard [7]. We hypothesized that we could use AI-LAB, in the absence of local data science infrastructure or expertise, to deploy an already trained AI model to reliably and repeatedly assess the severity of COVID-19 lung disease across many patients at our institution. Multiple research groups have developed different AI models that can predict the radiographic severity of lung involvement based on lung opacities [8, 9, 10].
In this study, we evaluated the feasibility of deploying and testing such an AI model developed at another institution on our local institutional data using AI-LAB. We used the previously published Pulmonary X-ray Severity (PXS) score model, a convolutional Siamese neural network-based model for continuous disease severity evaluation [8,11,12]. Model outputs were correlated with manual lung disease severity assessments by radiologists and associated with clinical outcomes at our institution.
We describe our experience conducting an applied clinical data science research project using the AI-LAB platform, including site requirements for data preparation, ground truth annotation, validation, and testing AI algorithms. We tested a chest radiograph algorithm to assess lung disease severity among patients with COVID-19 during the first pandemic surge.
Methods
Institution
The midsized academic radiology practice located in the Northeastern United States is a 335-bed hospital serving a suburban population of a metropolitan area in the United States with minimal data science infrastructure and no internal access to data scientists, no high-performance graphics processing units (GPU)-based computers or designated general purpose AI software prior to the activity reported here.
Clinical Scenario
Our radiology group partnered with data scientists at another academic medical center in our metropolitan area to use a COVID-19 CXR-based lung disease severity quantification algorithm, which had been trained at that other institution.
Infrastructure Setup and Institutional Review Board
As an early adopter, pilot site, we had joined an ACR-facilitated research consortium for the purpose of AI model testing and exchange. At the outset, we internally assessed our data science infrastructure to conduct the proposed AI algorithm testing and consulted with the AI-LAB developer team to obtain recommendations on the necessary computing capability to implement a local installation of CONNECT/AI-LAB.
This HIPAA-compliant study was performed with approval from the Lahey Hospital & Medical Center Institutional Review Board with a waiver of informed consent. The study was performed by our radiology group in our radiology department, which is an official participating pilot site for the AI-LAB platform.
Study Cohort
We used a combined query of an imaging and laboratory database (Primordial RadMetrix, Nuance Communications Inc, Burlington, Massachusetts) and our electronic health records (Epic, Verona, Wisconsin) to identify the patient cohort. The query retrospectively identified consecutive patients with positive COVID-19 reverse transcription-polymerase chain reaction tests who also had a CXR on clinical presentation (to the emergency room, outpatient clinics, and inpatient wards) performed between March 16, 2020, and April 18, 2020. Since hospital admission was one of the primary outcomes and the presentation CXR was not available for 20 transfer patients, these patients were excluded. Admission, intubation, and death dates were recorded for each patient. Admission, intubation, and death within 3 days of the presentation CXR were calculated and recorded as primary clinical outcomes. Due to low incidence of death within 3 days of admission, a combined outcome of death or intubation within 3 days was used.
Manual Radiologist Assessment of Pulmonary Disease Severity
The Radiographic Assessment of Lung Edema (RALE) score was initially devised to assess lung edema based on degree and extent of lung opacity in patients with acute respiratory distress syndrome [13]. A modified version of this score (mRALE) was used in our study. Each lung was assigned an mRALE score for the extent of involvement by consolidation or ground glass opacities (0 = none, 1 = <25%, 2 = 25%-50%, 3 = 50%-75%, 4 = >75% involvement) [7]. Each lung score was then multiplied by an overall lung density score (1 = hazy, 2 = moderate, 3 = dense). The scores from each lung are added together to form the patient-level mRALE score. Examples of this scoring are demonstrated in Figure 1 .
Fig. 1.
Representative example images of modified Radiographic Assessment of Lung Edema (mRALE) and Pulmonary X-ray Severity (PXS) scores in chest radiographs of patients with COVID-19.
For the purpose of this study, two staff radiologists and a radiology fellow were trained to visually assess CXR and assign mRALE scores by first assessing a training set of 10 sample CXRs with feedback on how their scores correlated with the group. Then, each radiologist independently assigned an mRALE score for each frontal CXR image from the study cohort. The average mRALE score across all readers was imported to the AI-LAB as the reference standard (Fig. 2 ).
Fig. 2.
Data processing. CXR = chest radiograph; mRALE = modified Radiographic Assessment of Lung Edema; PXS = Pulmonary X-ray Severity.
AI Model Sharing in AI-LAB
The PXS score model was previously developed at Massachusetts General Hospital, a large tertiary care hospital, initially using CXRs from patients admitted with COVID-19 and was further fine-tuned using outpatient clinic CXRs at that institution [11,12]. The model takes a CXR image of interest and compares it with a pool of normal CXR images. A continuous disease severity score is calculated as the median of the Euclidean distances between the image of interest and each image in the pool of normal studies, as it passes through twinned neural networks. Please see the cited work for details on the design and implementation of this neural network architecture. The model was packaged into a Docker file (Docker Inc, Palo Alto, California), which could then be loaded onto the AI-LAB platform and imported locally at our institution.
Statistical Analysis
Pearson correlation was used to evaluate the correlation between mRALE assessments by different radiologist raters and the correlation between the average mRALE and PXS scores. The Mann-Whitney U test was used for comparison of the PXS score between groups. Bootstrap 95% confidence intervals (CIs) were calculated for the correlation between the average mRALE and PXS scores and for the area under the curve of the receiver operating characteristic (AUROC) curves.
Results
Implementation Process Outcomes
Infrastructure
After engaging our radiology and enterprise informatics teams, we identified a need for a dedicated, high-performance GPU-based server in our institution. Although only basic GPU based hardware is required to run pretrained models (also known as model inference), we preferred to “future-proof” our investment in computational resources and opted to acquire a high-performance GPU server, which would equip us to retrain and optimize models locally, if desired. To obtain the server, we worked with the customary hospital hardware supplier to ensure the physical server had sufficient motherboard power supply to support the graphics card(s) of choice. After determination of the hardware specifications in collaboration with the AI-LAB team and the vendor, we ultimately decided on a rack-based Dell R740XD PowerEdge Server (Dell Technologies, Austin, TX), with 3x Nvidia Tesla T4 GPUs (16 GB memory each; Nvidia, Santa Clara, CA) and 4 TB of SAS-based Solid State Drives for data storage needs.
The installation process, including setting up the AI-LAB software, and configuration of all required docker containers was performed. At the time we first implemented this software, some of the installation process required more manual steps, since we were the first institution in the United States to adopt this platform. Since then, the installation process has been streamlined with the development of a new installer software that requires less manual input.
COVID-19 CXR AI Algorithm Access
The AI-LAB team assisted with the upload of the COVID-19 CXR AI model. During the experiment, the authoring institution made improvements to the algorithm. The new model was packaged using AI-LAB Inference Model Standards [14]. Because the algorithm was packaged using the appropriate model standards, the AI-LAB platform was able to receive the updated docker container and make it available for subscription in its cloud. We subsequently downloaded and used the updated model on our prepared data, running it on our local instance of AI-LAB. Total time spent by our informatics analyst in this step of the collaboration was approximately 2 hours.
We imported the frontal view DICOM files for each of these CXRs from our institution into AI-LAB. For patients with more than one frontal view CXR image associated with the study accession (eg, large body habitus or difficult positioning requiring multiple attempts at image acquisition), we manually selected the image that contained the most lung in the image. All clinical data were used for testing, with no retraining or model tuning of the algorithm using our institution’s data.
“Data Wrangling” Challenges
AI-LAB has the ability to bulk upload ground truth information (eg, radiologist-generated labels of disease) and imaging studies. However, other important data set curation functions are still to be developed. Importantly, almost any imaging-based data science project requires selection of the appropriate series of an imaging examination for input into the AI model. Most digital radiography devices and PACS designate each exposure or image as a separate series within a single examination. For our own experiment, we needed to upload the optimal frontal CXR image and ensure that it was also the one the readers in this study had based their evaluation on. In 90% of our patients there was only a single image, but in 10% there was more than one image. This was due to acquisition challenges in often critically ill patients. Lacking a universal series selection tool at our institution, we retrieved studies of interest from PACS, batch anonymized them, manually selected the image series of interest, which, together with activities such as meetings with readers (to ensure reads matched key series) and ACR team members, required approximately 12 hours of analyst time for the entire cohort. We used a shared anonymized spreadsheet to ensure that the series of interest was communicated unambiguously to the readers.
AI Algorithm Assessment
Cohort Characteristics
One hundred forty-one patients positive for COVID-19 by reverse transcription-polymerase chain reaction who had CXRs were included in the study cohort. Patient demographics are summarized in Table 1 . Most patients (n = 130, 92%) were imaged in the emergency room setting. Most patients (n = 120, 85%) required hospital admission. A subset of patients (n = 14, 10%) were intubated within 3 days of CXR acquisition. Six patients (4%) died within 3 days of CXR acquisition.
Table 1.
Patient demographics
| Variable | Data |
|---|---|
| Median age (y) (Q1-Q3) | 73 (63-80) |
| Female, N (%) | 60 (43%) |
| Median BMI (kg/m2) (Q1-Q3) | 27 (23-31) |
| Patient type/Imaging Setting, n (%) | |
| Outpatient | 1 (1) |
| ED | 130 (92) |
| Inpatient | 10 (7) |
| Median mRALE (Q1-Q3) | 3 (1-5) |
| mRALE, n (%) | |
| mRALE=0 | 12 (9) |
| 0< mRALE≤4 | 92 (65) |
| 4< mRALE≤10 | 34 (24) |
| mRALE>10 | 3 (2) |
Manual Assessment of Lung Disease Severity
The correlation between the mRALE scores assigned by the three radiologists who independently assessed each image varied (r = 0.71, 0.78, 0.82). The average of the assigned mRALE were used as the reference standard for the deep learning PXS score. The median of the reference standard mRALE scores in this cohort was 2.7 (interquartile range = 1.3-5).
Testing PXS Score in AI-LAB
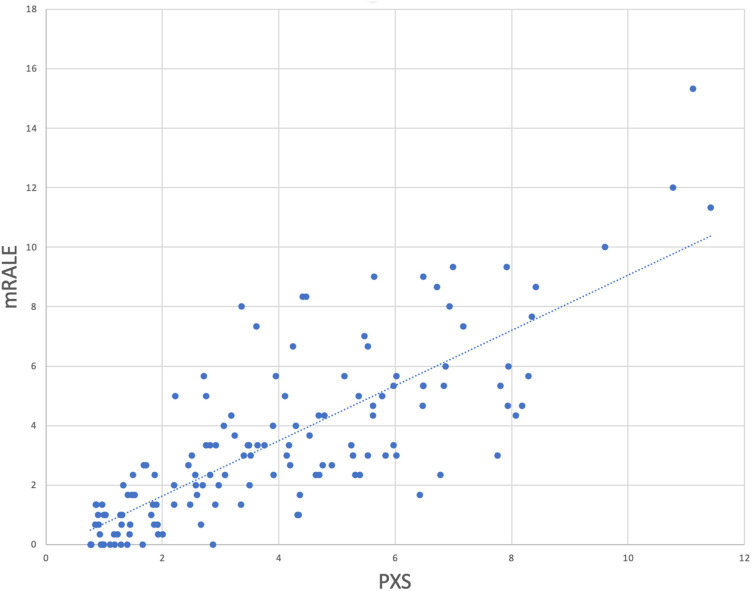
The PXS score deployed in AI-LAB correlated with the mRALE score assigned by the radiologist readers (r = 0.80) (Fig. 3 ).
Fig. 3.
Modified Radiographic Assessment of Lung Edema (mRALE) and Pulmonary X-ray Severity (PXS) correlation.
PXS Association With Clinical Outcomes
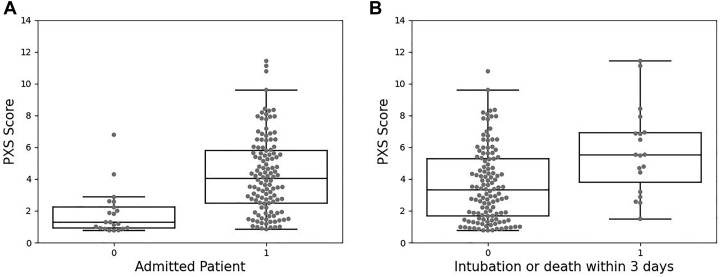
The PXS score was significantly higher in patients admitted to the hospital within 3 days of CXR acquisition than for those patients who did not require admission (4.0 versus 1.3, P < .001) (Fig. 4 a). The PXS score was also significantly higher in patients requiring intubation or death within 3 days (5.5 versus 3.3, P = .001) (Fig. 4b).
Fig. 4.
Pulmonary X-ray Severity (PXS) score comparison between (a) patients who were admitted within 3 days (1) and those who were not (0) and (b) patients who were intubated or died within 3 days (1) and those who survived and were not intubated within 3 days (0).
The AUROC was 0.84 (bootstrap 95% CI 0.73-0.93) for identifying patients who were admitted to the hospital (Fig. 5 a). The AUROC was 0.73 (bootstrap 95% CI 0.61-0.84) for the composite outcome of intubation or death within 3 days (Fig. 5b).
Fig. 5.
Receiver operating characteristic (ROC) curves for (a) area under the curve (AUC) of the ROC (AUROC) for detection of hospital admission within 3 days and (b) AUROC for the composite outcome of intubation or death within 3 days. Solid lines show the ROC curve from the 50th percentile bootstrap of the AUROC, and the dotted lines show the ROC curves from the 2.5th percentile bootstrap and 97.5th percentile bootstrap of the AUROC. CI = confidence interval.
Discussion
This study represents the first description of the use of the AI-LAB, a computational platform to facilitate evaluation of an external proprietary AI algorithm using institution-specific patient-level data without the need to export patient data beyond the firewall of the health care system. The aggregate setup time blocks comprised 3 to 6 months for hardware and software infrastructure, followed by 3 months for this first experiment. Infrastructure planning and setup consumed approximately 30 hours of radiology IT analyst time, about 5 to 10 hours of administrative time (eg, for contracts, institutional review board) and an estimated 20 hours of radiologist time for internal and external meetings. The capital investment for the hardware did not exceed $20,000. The AI-LAB software and the AI algorithms were obtained free of charge. The experiment itself required approximately 12 hours of radiology IT analyst and 2 hours of Epic Data analyst time and 40 hours of radiologist time between three readers and external assistance from the academic center that developed the AI model. After migrating the data set from the PACS, a script was used to curate the data based on the series description. We identified that some examinations had multiple instances of the same projection image do to clipped anatomy, position error, or exposure. Those examinations were manually reviewed by a radiologist to determine the best image to be used for the AI.
We demonstrate with this effort that it is feasible to set up and successfully use clinical data science infrastructure without any previous institutional history or dedicated personnel in this field. The overall investment of time and resources was deemed reasonable in return for the outcome we achieved. The interinstitutional effort created learning about data science workflow steps for all stakeholders and provides further evidence of the potential of an external platform to facilitate radiology practice participation in AI algorithm assessment. This platform offers an opportunity for successful engagement of clinical radiologists in the absence of on-site data scientists and more robust on-site data science infrastructure.
Many radiology-based AI models have been developed since the start of COVID-19 pandemic, with the hopes of improving diagnostic accuracy, speed, and risk assessment. However, for these algorithms to be safely used in clinical practice, they must be deployed and ideally tested locally before providing inferences on live patient data for use in clinical or operational decision making. AI-LAB enabled our practice to do just that. In this study, we successfully used AI-LAB to deploy and test a COVID-19 CXR AI algorithm that had been developed at another institution, showing generalizability of the previously developed external PXS score model on local data obtained at our own institution. The actual model deployment using AI-LAB was accomplished in a matter of days once the system setup had been completed. This demonstrates the feasibility of using AI-LAB to provide expedient solutions for assessment of algorithms across institutions, without the need to send actual source imaging or clinical data outside of the institution to test the model.
In general, lack of information technology and data science expertise at small and medium-sized institutions like ours might be considered as a major hurdle to participation in AI research or application of AI in radiology workflow. Platforms such as the AI-LAB, designed for federated algorithm use, can reduce the barriers to participation. MIDRC, launched in 2020, pursues a different approach by aggregating anonymized data in a central archive. This “centralized” approach also aims at achieving exposure of algorithms to a broader sample of data, through a different architecture that requires moving the data. MIDRC is currently geared toward the use case of COVID imaging, with the plan to expand into other disease entities in the future. Several commercial AI marketplaces exist with emerging assessment and analysis capabilities (examples include Blackford Analysis, Edinburgh, UK and Nuance Inc, Burlington, Massachusetts). A commercially available federated inference and training platform has also recently been launched (Rhino Health, Cambridge, Massachusetts). All these approaches provide options for imaging departments to engage with commercial and noncommercial AI offerings.
Clinical Utility of Model Validation With Own Institution Data
The COVID-19 deep learning model that we deployed and tested in AI-LAB in this study shows potential for predicting hospital admission or intubation or death within 3 days of presentation. This may become a useful tool for data-driven resource management within a health system. During the COVID-19 pandemic, many health systems allocated and moved precious resources (eg, ventilators) based on actual observed patient census. Similarly, ICU bed capacity was managed based on actual current capacity, initiating patient transfers as needed. One could envision a future state in which the repeated, at-scale use of the deep learning model–based prediction of near-term clinical prognosis of affected patients in a given health system could facilitate prospective, predictive management of resources and capacity. The correlation between mRALE score and PXS score in our study was 0.80 (95% CI 0.72-0.86), which is similar to the original study, which showed a PXS score of 0.86 (95% CI 0.80-0.90) in an internal test set and 0.86 (95% CI 0.79-0.90) in a different external hospital test set [8]. Although the 95% confidence intervals do overlap, the possible decrease in model performance could be related to differences in image acquisition technique and patient population.
Patients had less severe disease in our own study cohort (median mRALE 2.7) compared with the original study test sets (median mRALE 4.0 and 3.3). We also found in our study that PXS score can predict subsequent intubation or death within 3 days, with an AUROC of 0.73 (95% CI 0.61-0.84), which is less than the AUROC of 0.80 (95% CI 0.75-0.85) reported in the original study, though the bootstrap 95% confidence intervals overlap. The PXS score model was not trained to predict these outcomes (rather it was trained to evaluate lung disease severity), so it is not surprising that different patient populations may have different outcomes. Also, as new clinical management guidelines and therapeutic options arise, prediction of such outcomes may change. Thus, ongoing testing is needed to ensure that such predictions are updated, which AI-LAB can help to facilitate.
Many AI models, developed using curated institutional data, demonstrate high performance initially, but their performance not uncommonly degrades when deployed on data generated at a different institution [15]. This variability in generalization of the performance is a known issue, especially for models created based on single-institution data [16]. External platforms such as AI-LAB provide the opportunity for the developers to train and test their models on multiple-institution data. This may result in improved generalizability. The ability of each participating institution to optimize and verify model performance based on their own data raises the safety profile of the AI model, hence overcoming one of the major hurdles of AI implementation in medicine (ie, implementation gap) [17]. Like many issues in health care, the implementation gap became more evident during the COVID-19 pandemic. With heightened interest in this entity, many AI models have been developed but they have had little to no impact on the pandemic [3, 4, 5, 6,18]. One of the major obstacles in this rapidly changing environment is the current inability of many practices to optimize the AI models for their local (data) environment, and external platforms such as the AI-LAB may facilitate this activity. Lastly, continuous learning has been proposed as a method to preserve AI model robustness and promote adaption to changes in the local environment [19]. Engagement of radiology departments in codeveloping and testing of AI models has been proposed as a method to develop an environment for continuous learning of AI models. Platforms such as AI-LAB and MIDRC might facilitate achieving this goal.
Limitations
There are a few limitations to our study. First, this study involves using AI-LAB at a single institution. Assessment at multiple institutions will be important for future scaling of this work. Second, because some patients had multiple frontal CXR images obtained in a single study accession (due to challenging patient positioning or body habitus), we had to manually select which CXR image to load into AI-LAB. This problem with selecting the correct series is a barrier to scaling such models and needs to be addressed in future studies. Third, we tested a CXR-based model in this study; however, models using different modalities like CT and MRI may have other challenges for deployment using AI-LAB. Fourth, the data and images for this experiment were collected during the first surge of the pandemic in the United States, which mostly affected older patients. This is almost certainly associated with a higher pretest probability of a poorer outcome from a COVID-19 infection (such as intubation and death) than would be expected in a younger population. During the second surge of pandemic in the United States, relatively younger patients with fewer comorbidities were more frequently affected [20]. The performance of the AI model may have been impacted by this demographic shift.
Take-Home Points
-
▪
This demonstration represents first time the ACR AI-LAB on-premise platform was used to expedite transfer and assessment of a COVID-19 CXR AI algorithm on local imaging data without the need for image or clinical data exchange between institutions.
-
▪
The degree of correlation of the pulmonary X-ray severity score model with radiologists’ assessment and clinical outcomes in an external institution demonstrate the generalizability of model for assessment of lung disease severity in COVID-19 patients.
-
▪
The inherent ability of AI algorithms to (repeatedly) execute inference on entire cohorts of patients within or across institutions points to its potential utility in context with managerial decision making (supply chain, human resources).
-
▪
For AI models to achieve widespread clinical use, software platforms such as the freely available AI-LAB, which facilitate local testing and inference application, will be important to close the AI implementation gap across imaging practices.
Acknowledgments
We thank Deepak Kattil Veettil, Laura Coombs, and other ACR-AI LAB staff for their help to establish and update the platform. This work was supported by departmental funds. This research was carried out in part at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies, P41EB015896, a P41 Biotechnology Resource Grant supported by the National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health.
Footnotes
Dr Kalpathy-Cramer reports grants from GE Healthcare, nonfinancial support from AWS, and grants from Genentech Foundation, outside the submitted work. The other authors state that they have no conflict of interest related to the material discussed in this article. The authors are non-partner/non-partnership track/employees.
References
- 1.Shakespeare, William. The tempest, Act V, Scene I, ll 203–206.
- 2.American College of Radiology ACR AI-LAB™. https://ailab.acr.org/Account/Home Available at.
- 3.Rajaraman S., Siegelman J., Alderson P.O., Folio L.S., Folio L.R., Antani S.K. Iteratively pruned deep learning ensembles for COVID-19 detection in chest x-rays. IEEE Access. 2020;8:115041–115050. doi: 10.1109/ACCESS.2020.3003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L., Qin L., Xu Z., et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murphy K., Smits H., Knoops A.J.G., et al. COVID-19 on chest radiographs: a multireader evaluation of an artificial intelligence system. Radiology. 2020;296:E166–E172. doi: 10.1148/radiol.2020201874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wehbe R.M., Sheng J., Dutta S., et al. DeepCOVID-XR: an artificial intelligence algorithm to detect COVID-19 on chest radiographs trained and tested on a large US clinical dataset. Radiology. 2021;299:E167–E176. doi: 10.1148/radiol.2020203511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.American College of Radiology ACR recommendations for the use of chest radiography and computed tomography (CT) for suspected COVID-19 infection. https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection Available at.
- 8.Li M.D., Arun N.T., Gidwani M., et al. Automated assessment and tracking of COVID-19 pulmonary disease severity on chest radiographs using convolutional Siamese neural networks. Radiol Artif Intell. 2020;2 doi: 10.1148/ryai.2020200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mushtaq J., Pennella R., Lavalle S., et al. Initial chest radiographs and artificial intelligence (AI) predict clinical outcomes in COVID-19 patients: analysis of 697 Italian patients. https://doi.org/10.1007/s00330-020-07269-8 Eur Radiol Published online 2020. [DOI] [PMC free article] [PubMed]
- 10.Signoroni A, Savardi M, Benini S, et al. End-to-end learning for semiquantitative rating of COVID-19 severity on chest X-rays. Published online 2020.
- 11.Li M.D., Chang K., Bearce B., et al. Siamese neural networks for continuous disease severity evaluation and change detection in medical imaging. NPJ Dig Med. 2020;3:48. doi: 10.1038/s41746-020-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M.D., Arun N.T., Aggarwal M., et al. Improvement and multi-population generalizability of a deep learning-based Chest Radiograph Severity Score for COVID-19 [preprint] medRxiv. 2020 doi: 10.1101/2020.09.15.20195453. 2020.09.15.20195453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren M.A., Zhao Z., Koyama T., et al. Severity scoring of lung oedema on the chest radiograph is associated with clinical outcomes in ARDS. Thorax. 2018;73:840–846. doi: 10.1136/thoraxjnl-2017-211280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Github.com. ACRCode/AILAB_documentation. Available at: https://github.com/ACRCode/AILAB_documentation/wiki/AILAB-Inference-Model-Standards.
- 15.Wang X., Liang G., Zhang Y., Blanton H., Bessinger Z., Jacobs N. Inconsistent performance of deep learning models on mammogram classification. J Am Coll Radiol. 2020;17:796–803. doi: 10.1016/j.jacr.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Zech J.R., Badgeley M.A., Liu M., Costa A.B., Titano J.J., Oermann E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLOS Med. 2018;15 doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seneviratne M.G., Shah N.H., Chu L. Bridging the implementation gap of machine learning in healthcare. BMJ Innov. 2020;6:45. [Google Scholar]
- 18.Hu Y., Jacob J., Parker G.J.M., Hawkes D.J., Hurst J.R., Stoyanov D. The challenges of deploying artificial intelligence models in a rapidly evolving pandemic. Nat Mach Intell. 2020;2:298–300. [Google Scholar]
- 19.Pianykh O.S., Langs G., Dewey M., et al. Continuous learning AI in radiology: implementation principles and early applications. Radiology. 2020;297:6–14. doi: 10.1148/radiol.2020200038. [DOI] [PubMed] [Google Scholar]
- 20.Boehmer T.K., DeVies J., Caruso E., et al. Changing age distribution of the COVID-19 pandemic—United States, May–August 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1404–1409. doi: 10.15585/mmwr.mm6939e1. [DOI] [PMC free article] [PubMed] [Google Scholar]