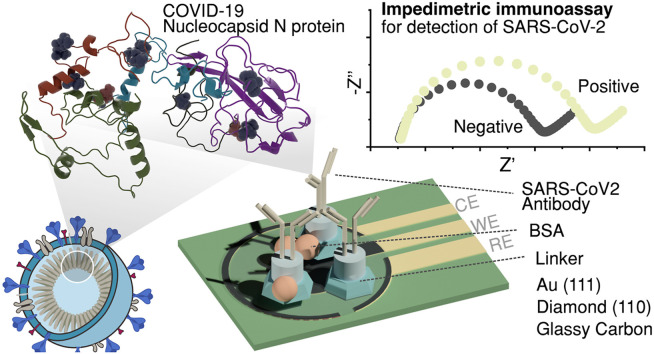
Abstract
The 21st century has already brought us a plethora of new threats related to viruses that emerge in humans after zoonotic transmission or drastically change their geographic distribution or prevalence. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first spotted at the end of 2019 to rapidly spread in southwest Asia and later cause a global pandemic, which paralyzes the world since then. We have designed novel immunosensors targeting conserved protein sequences of the N protein of SARS-CoV-2 based on lab-produced and purified anti-SARS-CoV-2 nucleocapsid antibodies that are densely grafted onto various surfaces (diamond/gold/glassy carbon). Titration of antibodies shows very strong reactions up to 1:72 900 dilution. Next, we showed the mechanism of interactions of our immunoassay with nucleocapsid N protein revealing molecular recognition by impedimetric measurements supported by hybrid modeling results with both density functional theory and molecular dynamics methods. Biosensors allowed for a fast (in less than 10 min) detection of SARS-CoV-2 virus with a limit of detection from 0.227 ng/ml through 0.334 ng/ml to 0.362 ng/ml for glassy carbon, boron-doped diamond, and gold surfaces, respectively. For all tested surfaces, we obtained a wide linear range of concentrations from 4.4 ng/ml to 4.4 pg/ml. Furthermore, our sensor leads to a highly specific response to SARS-CoV-2 clinical samples versus other upper respiratory tract viruses such as influenza, respiratory syncytial virus, or Epstein-Barr virus. All clinical samples were tested simultaneously on biosensors and real-time polymerase chain reactions.
Graphical abstract
1. Introduction
Towards the end of 2019, a new infectious disease appeared in Wuhan, Hubei Province, China. This disease caused by the new coronavirus is called the COVID-19 disease, while the virus was denoted as SARS-CoV-2. Millions of tests for infection have been performed worldwide, reported by 135 countries. Cases of infection were confirmed predominantly via nucleic acid amplification tests for viral RNA. Such testing requires specialized laboratories, and although some automation of the process is possible, mass testing can be very labor-intensive, with several points along the path of performing a single test where errors may occur. Alternative diagnostic methods are biosensors which are generally faster and less labor-intensive. To improve the specificity and selectivity of such diagnostic tests, it is crucial to select targets that are predominant and not highly variable due to natural selective pressure.
Although the observed genetic diversity of SARS-CoV-2 variants was low at the beginning of the pandemic, its rapid global spread provides the virus with many opportunities for natural selection to act upon rare but favorable mutations (Forni et al., 2021). Even though SARS-CoV-2 is a new threat, the accumulation of such mutations within the spike protein was reported. Up to date, according to the Centers for Disease Control and Prevention (CDC), fourteen mutations are prevalent within SARS-CoV-2 variants (CDC, 2020). Moreover, recent studies reveal that rapid diagnostic of SARS-CoV-2 is highly desired due to the reinfection cases with the virus (Tillett et al., 2020).
The current standard in virus diagnostics is a quantitative polymerase chain reaction (qPCR), which detects the pathogen's amplified genetic material from a patient's fluid sample and requires highly specialized equipment and trained personnel. The time required for the qPCR analysis is still acceptable (45–90 min). Nevertheless, the logistics of collected samples to the centralized laboratory followed by isolation of the nucleic acids often exceeds 24–48 h. Next, a variety of different tests were developed and approved involving amplification of RNA or protein interaction detection. A couple of novel testing platforms were reported, but unfortunately, none of them were experimentally validated and capable of achieving reproducible and sensitive results (Asif et al., 2020; Pokhrel et al., 2020; Samson et al., 2020; Xu et al., 2020). These platforms include tailoring and monitoring the key symptoms (Jeong et al., 2020), enzyme-linked immunosorbent assay (ELISA) or chemiluminescence (Espejo et al., 2020), surface-enhanced Raman scattering (Liu et al., 2021) plasmonic (Qiu et al., 2020) or field-effect transistor-based biosensors (Seo et al., 2020a), loop-mediated isothermal amplification (Thi et al., 2020a), and various PCR tactics (Carter et al., 2020; Mahapatra and Chandra, 2020). Nevertheless, more sensitive and specific tests with simplified procedures and high yield fabrication approaches should become accessible (Weissleder et al., 2020a). Therefore, the design of novel and fast diagnostic tests based on conserved proteins is vitally needed to enable a test-and-trace strategy (Hussein et al., 2020).
In the recent decade, affinity ligand-based biosensing devices have received rapid development. Such nano-biosensors have wide application potential with various detection formats (Lima et al., 2021). Immuno-based biosensing devices as a fast and real-time platform approach would help to monitor the pandemic (Balkourani et al., 2021; Lu et al., 2021; Mahshid et al., 2021; Weissleder et al., 2020b). Rashed et al. (2021) demonstrated electrochemical impedance based on commercially available sensing electrodes precoated with receptor-binding domain (RBD) and tested with samples of anti-SARS-CoV-2 monoclonal antibody CR3022. Next, a nano-Pd thin film electrode was reported to detect modulators that affect the SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2) interaction (Kiew et al., 2021). It was also shown that the surface ligand plays a critical role in the efficient design of biosensors (Szunerits et al., 2021). Chaibun et al. (2021) manifested an electrochemical biosensor based on multiplex RCA for the rapid detection of the N and S genes of SARS-CoV-2 from clinical samples on carbon screen-printed electrodes by differential pulse voltammetry (DPV). Moreover, the electrode-tethered sensors using chronoamperometry were applied to detect viral particles by the kinetic response of the complexation state of the antibody (Yousefi et al., 2021). Recent approaches for immunosensor detection of SARS-CoV-2 were based on spike protein (Witt et al., 2021). Mahari et al. (2020) and Perdomo et al. (2021) developed in-house built devices using bio-modified screen-printed carbon electrodes (SPCE) with immobilized monoclonal antibodies recognizing spike protein. By the end of 2020, Torrente-Rodríguez et al. (Torrente-Rodríguez et al., 2020, p.) described a platform prepared with commercially available SARS-CoV-2 reagents such as proteins and antibodies. The proposed graphene-based multiplexed platform allowed for the detection of full-length nucleocapsid protein and anti-spike IgG and IgM as well as C-reactive protein. Despite the substantial potential of this approach, still, the cross-reactivity with other respiratory viruses was not shown.
The future perspective on immunosensor design should take into consideration the fact that new variants of the virus emerge. For that, it is crucial to choose conserved regions within structural proteins less prone to mutation. In this report, we design and critically compare real-time immunoassays (boron-doped diamond/gold/glassy carbon) targeting conserved sequences of inner components of the viral particle from a patient's sample of nasopharyngeal fluid. In contrast to other affinity biosensing devices or detection methods ( Table 1 ), our solution is based on custom-made conserved protein sequences of the N protein of SARS-CoV-2. We estimated the grafting density of the molecular recognition system at different electrodes and the detection mechanism using hybrid modelling with both density functional theory (DFT) and molecular dynamics (MD) methods. The binding of SARS-CoV-2 antibody to the nucleoprotein leads to a change in electrochemical impedance that is transduced into a highly sensitive electrical detection signal. The platform was tested on real clinical samples received from the diagnostic laboratory and verified with RT-PCR.
Table 1.
Comparison of available diagnostic techniques for SARS-CoV-2.
| Detection platform | Turnaround time | Sensitivity | Selectivity | Limit of detection | Reference |
|---|---|---|---|---|---|
| Surface plasmon resonance (LSPR) | Few-minutes | – | – | 0.22 pM | Seo et al. (2020b) |
| RT-PCR | >4 h | 95% | – | 3.6 copies/reaction | Corman et al. (2020) |
| Digital polymerase chain reaction (ddPCR) | >2h | 94% | 100% | 1.8 copies/reaction | Corman et al. (2020) |
| Reverse-transcription LAMP with turbidimetric detection | ∼1 h | 100% | 100% | 20 copies/reaction | Yan et al. (2020) |
| CRISPR-based LAMP with lateral flow assay | <45 min | 95% | 100% | 10 copies/μL | Broughton et al. (2020) |
| FET-based biosensor | ∼30 min | – | – | 242 copies/mL | Thi et al. (2020b) |
| LAMP with colorimetric readout | <1 h | – | – | 4.8 copies/μL | Zhang et al. (2020) |
| Digital PCR | <1 h | 100% | 80% | 1 copy/μL | Lu et al. (2020) |
| RCA with magnetic nanoparticles | <2 h | – | – | 2 fM | Tian et al. (2020) |
| RT-LAMP with colorimetric readout | <2 h | 97.5% | 99.7% | 100 copies/μL | Thi et al. (2020b) |
| RT-LAMP with colorimetric readout | <1 h | – | – | 80 copies/mL | Huang et al. (2020) |
| Electrochemical Impedance Spectroscopy | Few minutes | – | – | – | Rashed et al. (2021) |
| Electrochemical Impedance Spectroscopy | Few minutes | – | – | 200 copies/ml | (Zhao et al., 2021), (Chaibun et al., 2021) |
| Reagent-free sensing of viral particles using an electrochemical approach monitoring the kinetics of transport for a DNA−antibody complex | 10 min | > ng/mL | – | 4 × 103 viral particles per mL | Yousefi et al. (2021) |
| Magnetic beads combined with carbon black-based screen screen-printed electrodes | ∼30 min | 6.5 × 103 PFU/mL | – | untreated saliva: equal to 19 ng/mL In saliva clinical samples and cultured SARS-CoV-2: 8 ng/mL |
Fabiani et al. (2021) |
| Ultrasensitive supersandwich-type electrochemical sensor | ∼2h | – | Confirmed patients 85.5% Recovery patients 46.2% |
200 copies/ml | Zhao et al. (2021) |
| Electrochemical Impedance Spectroscopy | Few minutes | – | – | Can detect clinically relevant antibody concentrations | Rashed et al. (2021) |
| Ligand on the surface of electrochemical biosensors | Few minutes | Depending on the ligand, above 95 | – | Depending on the ligand, for example, 80 fM, 120 fM for the S1 protein | Szunerits et al. (2021) |
| RT-PCR with viral RNA | ∼2–4 h | High sensitivity | – | – | Mahshid et al. (2021) |
| Lateral flow immunoassay | Few minutes | 95% | – | – | Liu et al. (2021) |
| MIP-based electrochemical sensor | ∼15 min | – | – | 15fM-50fM | Raziq et al. (2021) |
| Enzymatic test | 13 min | – | – | 125 copies/ml | Singh et al. (2021) |
| Electrochemical impedance spectroscopy-based biosensing platform | 15–20 min | High sensitivity | High selectivity | – | Kiew et al. (2021) |
2. Materials and methods
2.1. Reagents
Potassium hexacyanoferrate (III), methanol, sodium nitrite, and hydrochloric acid were purchased from Chempur (Poland). Phosphate-buffered saline (PBS), 4-aminobenzoic acid (4-ABA), N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC), Tris-buffered saline (TBS), IPTG, HIS-Select® Nickel Affinity Gel, Tris-HCl, Glycerol, 4-aminothiphenol (4-ATP), glutaraldehyde (GA), bovine serum albumin (BSA), ethanol, demineralized/ultrapure water (ddH2O), hydrochloric acid (37%), sodium nitrite and ethylenediaminetetraacetic acid (EDTA) were purchased from Sigma Aldrich (Germany). Codon optimization for E. coli, ALICator Ligation Independent Cloning, B-PER Complete Bacterial Protein Extraction Reagent, Gene Art Gene Synthesis, Slide-A-Lyzer Dialysis Cassettes, Wedge Well Gel, Super Signal West Pico Plus Chemiluminescent Substrate, and 1-Step Turbo TMB-ELISA were purchased from Thermo Fisher Scientific (USA). pFastBac1 was purchased from Invitrogen (USA). 15 cm Chromatography column, Trans-Blot Turbo RTA Transfer Kit were purchased from Biorad (USA). Protein A-agarose affinity matrix was purchased from Roche (Swiss). Instant Blue was purchased from Expedeon (United Kingdom). Secondary goat anti-rabbit HRP antibodies were purchased from Jackson ImmunoResearch (United Kingdom). 96-well ELISA plate was purchased from Greiner Microlon (Germany).
2.2. Biomaterials preparation and identification
Expression and purification of RNA binding domain of nucleocapsid protein of Wuhan strain in E. coli: The DNA sequence coding for SARS-CoV-2 protein N (amino acid sequence: MGGSGGGSGYPYDVPDYAGSSSGGSSSGGSSSGVDENLYFQSSWFTALTQHGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAGLPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSGHHHHHHG) was synthesized with codon optimization for E. coli by Thermo Fisher Scientific. For recombinant 6xHis-protein isolation, synthetic fragment of N plasmid was cloned into the pLATE31 expression vector using ALICator Ligation Independent Cloning [Thermo Fisher Scientific] with primers CoV–31F: AGAAGGAGATATAACTATGGGTGGTAGCGGTGGTGGTAGT; CoV-31R: GTGGTGGTGATGGTGATGGCCGCTACCTTCGGCATAAAAACC. The recombinant protein was expressed in E. coli BL21(DE3) by induction with 0.3 mM IPTG (Sigma-Aldrich) at 37 °C for 3 h. The cells were harvested by centrifugation (12.000×g, 30 min, 4 °C). Whole-cell lysates were prepared with B-PER Complete Bacterial Protein Extraction Reagent (Thermo Scientific). Recombinant Protein (Wuhan-N) was purified under native conditions by Ni-NTA affinity chromatography using HIS-Select® Nickel Affinity Gel (Sigma Aldrich). The purified recombinant protein was dialyzed to TBS buffer (150 mM NaCl (Sigma Aldrich), 50 mM Tris-HCl (Sigma Aldrich), pH 7.5) containing 5% glycerol (Sigma-Aldrich) and 10 mM EDTA (Sigma-Aldrich).
Expression of full-length nucleocapsid protein in insect cells: DNA coding sequence of nucleocapsid protein of SARS-CoV-2 with insect enhancer sequence added on the N-end was synthesized by Gene Art Gene Synthesis (Thermo Fisher Scientific). The synthetic sequence was cloned under the polyhedrin promotor into pFastBac1 (Invitrogen, Carlsbad, CA) using NheI and KpnI restriction sites. Recombinant baculoviruses were generated according to the manufacturer's protocol. Briefly: DH10Bac™ competent E. coli cells were transfected with pFB1-N plasmid to obtain recombinant bacmid DNA, which was used to generate recombinant baculovirus (rBV-N) in Spodoptera frugiperda (Sf9) insect cells. For the production of nucleocapsid protein, Sf9 cells in suspension culture were infected with recombinant baculovirus at a multiplicity of infection (MOI) of 3 and harvested 60 h post-infection. Cell lysates containing full-N protein were used as a positive control.
Production of polyclonal antibodies: To obtain specific polyclonal antibodies against the nucleoprotein of SARS-CoV-2, a 9 months old rabbit was immunized twice with 100–200 μg purified Wuhan-N protein mixed with Addavax adjuvant, and boosted 3 weeks later with the same dose of purified Wuhan-N recombinant protein in Freund's incomplete adjuvant. The animal blood was collected 2 weeks after the second injection.
Purification of IgG rabbit antibodies using Protein A affinity chromatography: A 15 cm column (BioRad) was packed with 3 ml of Protein A-agarose affinity matrix (Roche). The column was pre-equilibrated with 5 bed volumes of Starting buffer (100 mMTris-HCl, pH 8.0, degassed and filtered). 2 mL of serum sample was filtered through a 0.2-μm syringe filter to remove insoluble debris. An aliquot of filtered serum (1 mL) was mixed with 100 μl of 1.0 M Tris, pH 8.0 to adjust the pH of the crude antibody sample to pH 7.5–8.0. Next, the serum sample was loaded on a protein A-agarose gravity column. The column was washed with 10 bed volumes of Washing buffer 1 (100 mM Tris-HCl, pH 8.0 degassed and filtered) followed by washing with 10 bed volumes of Washing buffer 2 (10 mM Tris-HCl, pH 8.0, degassed and filtered). The rabbit IgG was eluted using 2 bed volumes of Elution buffer (100 mM glycine, pH 3.0, degassed and filtered). 6 × 1 ml fractions were collected into collection tubes containing 200 μl of Neutralization buffer (1.0 M Tris, pH 8.0). 6 ml of purified rabbit IgG was dialyzed against PBS buffer using Slide-A-Lyzer Dialysis Cassettes (Thermo Fisher Scientific). Purified rabbit IgG was analyzed on SDS-PAGE gel and stored at 4 °C for further use. The concentration of purified antibodies was measured by BCA assay reaching 0.5 mg/ml (Thermo Fisher Scientific).
SDS-PAGE and western blotting: Samples were loaded on 10–20% precast Wedge Well Gel (Thermo Fisher Scientific) and run at the constant voltage of 165V. After electrophoresis, proteins were stained with Instant Blue (Expedeon) or transferred onto PVDF membranes using the semi-dry immune transfer (Trans-Blot Turbo RTA Transfer Kit, Biorad). PVDF membranes were blocked for 1 h in a 5% semi-skimmed milk solution (5%milk/TBS/0.01%Tween20) and incubated for 1 h with purified rabbit anti-N antibodies (dilution 1:1000 from 0.5 mg/ml stock). Next, the membranes were washed (TBS/0.01%Tween20) and incubated for 1 h with secondary goat anti-rabbit HRP antibodies (1:4000, Jackson Immuno Research). A reaction was developed with Super Signal West Pico Plus Chemiluminescent Substrate (Thermo Fisher Scientific).
Tittering of the Wuhan-N-ab by Enzyme-linked immunosorbent assay (ELISA): A 96-well ELISA plate (Greiner Microlon High-Binding, clear) was coated with 100 ng/well of purified Wuhan-N protein in PBS for 16 h at 4 °C. Then, the plate was washed 4 times for 5 min with 200 μl/well of washing buffer (PBS/0,05%Tween20) and blocked for 1 h at 37 °C with 250 μl/well of blocking buffer (3%BSA/PBS/0,05%Tween20). After the incubation had ended, the blocking buffer was discarded and the plate was washed as above. Then 100 μl/well of serial dilutions of rabbit Wuhan-N-ab were added and the plates were incubated for 1 h at 37 °C. The antibody solution was then discarded, and the plate was washed as previously. 100 μl/well of secondary goat anti-rabbit HRP antibodies (1:4000, Jackson Immuno Research) were added and incubated for 1 h at room temperature. Finally, following the last plate-washing step (6 times for 5 min with 200 μl/well) 100 μl/well of HRP-substrate solution was added (1-Step Turbo TMB-ELISA, Thermo Fisher Scientific). The reaction was stopped by adding 50 μL of 0.5M sulfuric acid to each well. Signal intensity was measured at 450 nm using a plate reader (Epoch, Biotek).
Cross-reactivity testing by Enzyme-linked immunosorbent assay (ELISA): A 96-well ELISA plate (Greiner Microlon High-Binding, clear) was coated with 100 μl/well of inactivated viruses: Influenza A Virus (IVA), Influenza B virus (IVB), Respiratory Syncytial Virus (RSV) and Epstein-Barr Virus (EBV) (SkyWayBiolab, Poland) in PBS for 16 h at 4 °C. Then, the plate was washed 4 times for 5 min with 200 μl/well of washing buffer (PBS/0,05%Tween20) and blocked for 1 h at 37 °C with 250 μl/well of blocking buffer (3%BSA/PBS/0,05%Tween20). After the incubation had ended, the blocking buffer was discarded and the plate was washed as above. Then 100 μl/well of rabbit Wuhan-N-ab (dilution 1:1000 from 0.5 mg/ml stock) was added and the plates were incubated for 1 h at 37 °C. The antibody solution was then discarded, and the plate was washed as previously. 100 μl/well of secondary goat anti-rabbit HRP antibodies (1:4000, Jackson Immuno Research) were added and incubated for 1 h at room temperature. Finally, following the last plate-washing step (6 times for 5 min with 200 μl/well) 100 μl/well of HRP-substrate solution was added (1-Step Turbo TMB-ELISA, Thermo Fisher Scientific). The reaction was stopped by adding 50 μL of 0.5M sulfuric acid to each well. Signal intensity was measured at 450 nm using a plate reader (Epoch, Biotek).
Immunofluorescence assay: Human airway epithelium culture was infected with SARS-CoV-2 virus. Cells were fixed with 4% formaldehyde, permeabilized with 0.3% Triton X-100 in PBS, and blocked for 1 h with 5% bovine serum albumin in PBS. Cells were stained with primary rabbit Wuhan-N-ab antibody (dilution 1:1000 from 0.5 mg/ml stock) and secondary anti-Rabbit antibody conjugated with Alexa Fluor 546 (1:400, Invitrogen, Carlsbad, CA, United States) diluted in 5% BSA with 0.5% Tween. Cell nuclei were stained with DAPI (0.1 μg/ml in PBS; Sigma-Aldrich). Fluorescent images were acquired under a Zeiss LSM 880 confocal microscope (Carl Zeiss Microscopy GmbH) at 40× magnification.
2.3. Immunosensors fabrication
2.3.1. Gold disc electrodes (Au)
Before modification, each gold disk working electrode of 1.6 mm diameter was mechanically polished on a micro cloth pad with 0.05 μm alumina slurry followed by rinsing in absolute ethanol and then dried under an argon stream. Dry electrodes were immersed in 0.1 M 4-ATP solution in absolute ethanol and left at 4 °C for 20 h. The modification process of gold electrodes involves the immobilization of antibodies based on the self-assembled monolayer of 4-ATP. It is covalently bonded to the gold surface through the amine group (Aditya Jayanthi et al., 2019), which reacts with GA cross-linker. Next, the antibody is grafted to GA by the amine groups through imine group formation. The use of cross-linker results in an enhanced immobilization of antibodies. Electrodes were thoroughly washed with ethanol again and dried under an Ar stream. Every next incubation step was followed by rinsing with ultrapure water and drying under Ar stream. Then 10 μL of 2.5% GA solution in ultrapure water was placed on each electrode surface and left at room temperature for 15 min in the dark medium. Functionalized electrodes were then exposed to direct antibody immobilization by dropping 10 μL of 0.1 mg/ml antibody solution on the electrode surface for 90 min at 4 °C. In the last step, the electrodes were incubated in 10 μL of 0.1% concentrated solution of BSA dissolved in 0.01M PBS of pH = 7.45 for 30 min at 4 °C to block possible remaining uncovered active sites of the electrode surface. The modified gold disk electrodes were then used for impedimetric measurements, being ready to selectively anchor SARS-CoV-2 antigen.
2.3.2. Boron doped diamond (BDD) and glassy carbon (GC) electrodes
BDD electrodes were synthesized using the microwave plasma-enhanced chemical vapor deposition (MWPECVD) system (SEKI Technotron AX5400S, Japan). BDD electrodes were grown on p-type (100)-oriented silicon substrates. Substrates were seeded by sonication in nanodiamond suspension (crystallite size of 5–10 nm) for 1 h. The substrate temperature was kept at 700 °C during the deposition process. Excited plasma was ignited by microwave radiation (2.45 GHz). The plasma microwave power, optimized for diamond synthesis, was kept at 1300 W (Bogdanowicz et al., 2014). The gas mixture ratio was 1% of the molar ratio of CH4–H2 at a gas volume of 300 sccm of total flow rate. The base pressure was about 10−6 Torr, and the process pressure was kept at 50 Torr. All samples were doped by using diborane (B2H6) dopant precursor; [B]/[C] ratio was 10 000 ppm in plasma (Bogdanowicz et al., 2013). The time of synthesis was 12h resulting in the film thickness of ca. 6 μm.
Before electrochemical measurements, both BDD and GC (Stanlab, Poland) were cleaned using an ultrasonic cleaner for 15-min in methanol and demineralized water. Then the electrodes were modified by polarizing the sample eleven times in a previously prepared deoxidized solution of diazonium salt (Zhang et al., 2014). 20 mg of 4-ABA was dissolved in 2 ml of 37% HCl (stirring for 15 min - average stirring speed 400 rpm). Then it was cooled to 0 °C. Next, 2 ml of demineralized water was added to the mixture. The mixture was then stirred for a further 15 min to dissolve the precipitated 4-ABA chloride. Then 25 mg NaNO2 dissolved in 3 mL ddH2O was added dropwise for 30 min. After the addition of sodium nitrite, the compounds were stirred at 0 °C for about 10 min.
Modification of GC and BDD surface was achieved by voltammetric electroreduction of the aryldiazonium reagents. The nitrosonium ion is formed formation, which subsequently activates the amino group on 4-ABA. During CV sweeping, irreversible reduction peaks occur at a potential around 0.2 V. These peaks form due to the reduction of the diazonium precursor reagents by single electron transfer. Modification by electrode polarization from 0 V vs Ag/AgCl to -1V vs Ag/AgCl eleven times at a speed of 100 mV/s was prepared with deoxidized diazonium salt solution with an Ag/AgCl (3M KCl) electrode as reference electrode (RE), and a platinum mesh as a counter electrode (CE). The geometric surface of the sample in this processing process is 0.38 cm2.
The samples were then washed with a strong stream of ddH2O, dried with a stream of argon, and 50 μl of protein A solution with EDC and NHS (protein A = 0.5 mg/ml, EDC = 4.80 mg/ml, NHS = 2.87 mg/ml) was placed on them. This process lasted an hour and occurred at 4 °C. The samples were then washed with ddH2O, and incubated with 50 μl of 0.1 μg/ml antibody solution and left for 24h at 4 °C. Then the antibody-modified electrodes were washed with ddH2O, dried with argon, and treated with 1% BSA for 15 min. After this stage, proper measurements were started consisting of adding the specified amount of the tested agent (PBS, protein, pathogen) to the measuring system.
2.4. Electrochemical experiments
The cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) were conducted using a Palmsens 4 potentiostat/galvanostat system (Methrom, Autolab, Netherlands) in the standard three-electrode configuration. BDD/Au/GC surfaces were used as a working electrode, the Pt mesh was used as a counter electrode, and Ag/AgCl/0.1M KCl as a reference electrode. All electrochemical tests were carried out in 5 mM K3[Fe(CN)6]/K4[Fe(CN)6]/in 0.01 M PBS that was previously deaerated. In the case of the EIS, the frequency ranged from 10 kHz to 1 Hz for BDD, and to 0.2 Hz for Au/GC with 40 points. The amplitude of the alternating current (AC) signal was 10 mV. Each potential was held constant for 5 s before each measurement to obtain steady-state conditions. Obtained data were subjected to analysis using EIS Spectrum Analyzer according to the proposed electric equivalent circuit (EEQC) (Fig. S2).
2.5. Clinical samples
Clinical samples were received thanks to the courtesy of GeneMe Ltd, Piotrkowska 41C, 80–180 Gdańsk. GeneMe is a diagnostic laboratory in Poland working for the service of Polish Health Ministry. Therefore, as the sole activity of GeneMe is to analyze patient samples under the cooperation with Polish Health Ministry there was no necessity to apply for a bioethical comity approval. Residual saliva swab material was anonymized and delivered to the Institute of Biotechnology and Molecular Medicine to be utilized for the study. The samples were tested according to the WHO regulations – Anatolya Gene Works qRT-PCR (Bosphore, Turkey) Kit was both genes with Ct max 38. Negative SARS-CoV-2 samples had only amplification in E gene or none.
3. Results
3.1. Production, purification, and validation of antibodies revealed a universal selective character towards the nucleocapsid protein of SARS-CoV-2
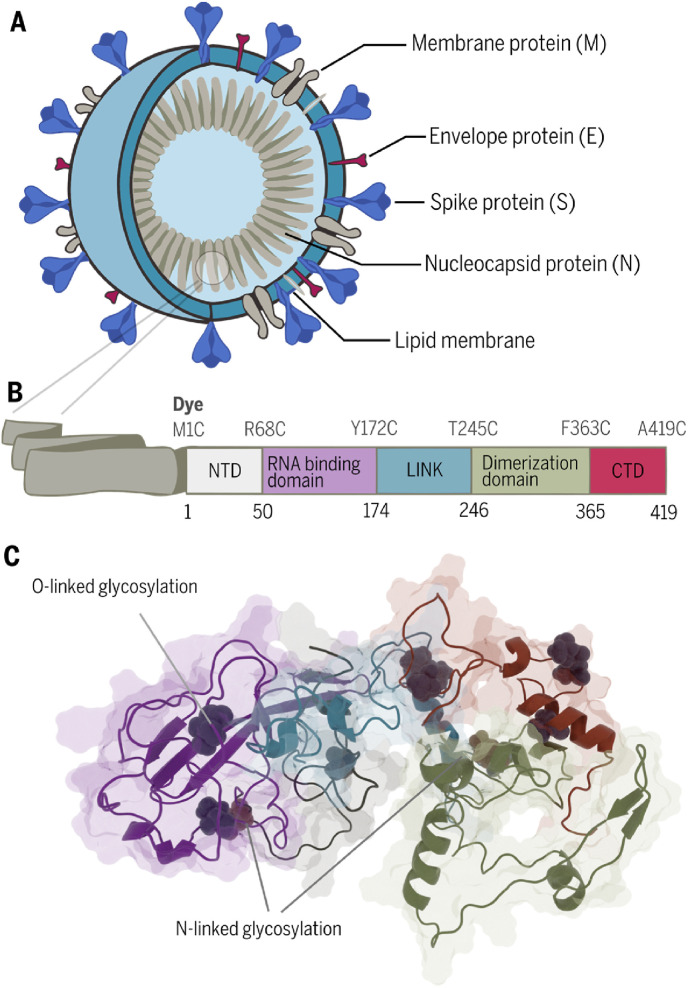
The SARS-CoV-2 is an enveloped virus with three main structural proteins: membrane protein (M), an envelope protein (E), spike protein (S), and one multifunctional structural nucleocapsid protein (N). The N protein is the main component of the virus nucleocapsid and determines the replication cycle through binding the viral RNA (Kang et al., 2020).
Structurally, the nucleocapsid protein can be divided into five distinct regions (Fig. 1 B): the N-terminal domain (NTD), the RNA binding domain, the central Ser/Arg (SR)-rich linker (LINK), the dimerization domain, and the C-terminal domain (CTD) (Dutta et al., 2020).
Fig. 1.
The universal character of nucleocapsid protein. (A)Structure of the virus. (B) Domain architecture of SARS-CoV-2 N protein. (C) Predicted structure of nucleocapsid protein with post-translational modification prepared using the Robetta comparative modelling tools and two templates, the NMR structure of the RNA-binding domain (PDB:1SSK) and the X-ray structure of the dimerization domain (PDB:6YUN) (details in Fig. S8.). The identity of nucleocapsid and spike proteins of different human coronaviruses has been listed in Tables S1 and S2.
The biosensing device presented in this report is based on the anti-RNA binding domain of nucleocapsid protein antibodies of SARS-CoV-2. Our research for the first time compared different electrode surfaces and their modification procedures which allow for the adjustment and development of a wide range of biological recognition systems for target pathogens with great freedom for customization. Thus, for our study, we chose the nucleocapsid protein, which is an inner virus component that, similarly to spike protein, shows little sequence variation (for both amino acids and nucleotides sequences) between isolates from around the world as listed in Tables S1 and S2 in Supplementary materials.
The nucleocapsid protein is present in multiple copies in the viral particle and is one of the most abundant proteins in the virus particle (Fig. 1A). Because the nucleoprotein is translated during the early stages of virus replication, multiple copies of N-protein are present within cells at the beginning of the virus infection. Moreover, the N protein has few post-translational modifications such as phosphorylation and N and O glycosylation (Supekar et al., 2020). Most modifications are present in the linker region, the dimerization domain, and the C-terminal domain.
In contrast, the spike protein is heavily glycosylated (22 glycosylation sites) (Watanabe et al., 2020) which requires long and complex maturation processes in ER-Golgi compartments (endoplasmic reticulum). These can have a negative impact on the binding affinity to antibodies dedicated to detecting the virus. Even a single change in one amino acid can cause changes in the glycosylation pattern. For our study, we chose the RNA-binding domain of N protein, which lacks N-glycosylation sites and has only two predicted O-glycosylation sites (Fig. 1C). This makes the truncated N protein (RNA binding domain) an exceptionally good target for detecting SARS-CoV-2 infection. All the above features make the N protein the perfect candidate for a diagnostic target.
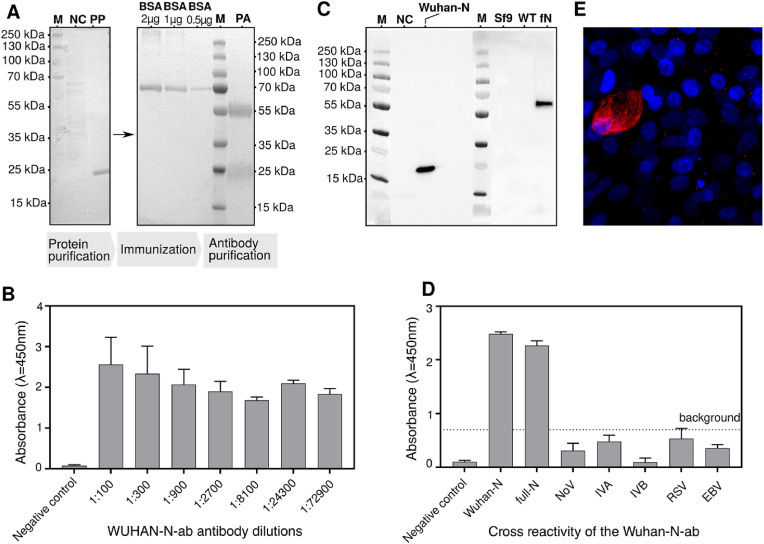
For sensor preparation, the RNA-binding domain of N protein was produced in bacteria, purified using a two-step purification process, and used for rabbit vaccination (Fig. 2 A). Antibodies obtained from rabbits were validated, showing great reactivity with the RNA-binding domain of N protein produced in bacteria (Fig. 2B) and with the full-length N protein produced in insect cells (Fig. 2C). Both recombinant proteins show reactivity with the obtained antibodies, which may suggest that the glycosylation status does not play a role in the protein-antibody interaction (Fig. 2C). Wuhan-N-ab antibodies show no cross-reactivity with other upper respiratory tract viruses (Influenza A Virus (IVA), Influenza B virus (IVB), Respiratory Syncytial Virus (RSV), and Epstein-Barr Virus (EBV) as well as with saliva proteins as was shown in ELISA assay (Fig. 2D). Antibodies were reactive with the whole virus, which was confirmed on cells infected with SARS-CoV-2 using an immunofluorescence assay (Fig. 2E).
Fig. 2.
Production, purification, and validation of antibodies. (A) A pipeline of antibody production: expression and purification of the recombinant protein – Wuhan-N (truncated form of N protein); immunization of the rabbit; purification of rabbit antibodies. Right panel: SDS PAGE gel showing purified Wuhan-N protein, Left panel: SDS PAGE gel showing BSA standard titration and purified Wuhan-N-ab antibodies (NC – Negative control, PP – Purified Wuhan-N, PA – Purified Wuhan-N-ab). (B) Titration of the Wuhan-N-ab antibodies in ELISA tests, ELISA plates were coated with Wuhan-N protein, bacterial cell lysate from BL21 E.coli served as a negative control. (C) Cross-reactivity of the Wuhan-N-ab antibody with different forms of nucleoprotein antigens produced in heterologous expression systems. Purified Wuhan-N protein was produced in a bacterial system, full-length N protein - full-N was produced in insect cells. Bacterial cell lysate from BL21 E.coli, Sf9 insect cell lysate, and wild-type baculovirus lysate served as a negative control (NC - BL21 E.coli cell lysate, SF9 - Sf9 cell lysate, WT - WT baculovirus lysate and fN - full-N) (D) Cross-reactivity of the Wuhan-N-ab antibody with other respiratory tract viruses. Influenza A Virus (IVA), Influenza B virus (IVB), Respiratory Syncytial Virus (RSV), and Epstein-Barr Virus (EBV) inactivated viruses were used to coat the ELISA plate. Norovirus (NoV) served as a non-respiratory tract virus control, Wuhan-N and full-N served as a positive control, swab from the healthy individual served as a negative control. (E) Detection of SARS-CoV-2 virus using immunofluorescence assays in infected human airway epithelial cells using the Wuhan-N-ab antibody. Red: SARS-CoV-2 virus, blue: nucleus. Images were obtained at 40x magnification.
3.2. Design of the COVID 19 immunosensors for real-time multiplatform: GC, BDD, and Au electrode surfaces
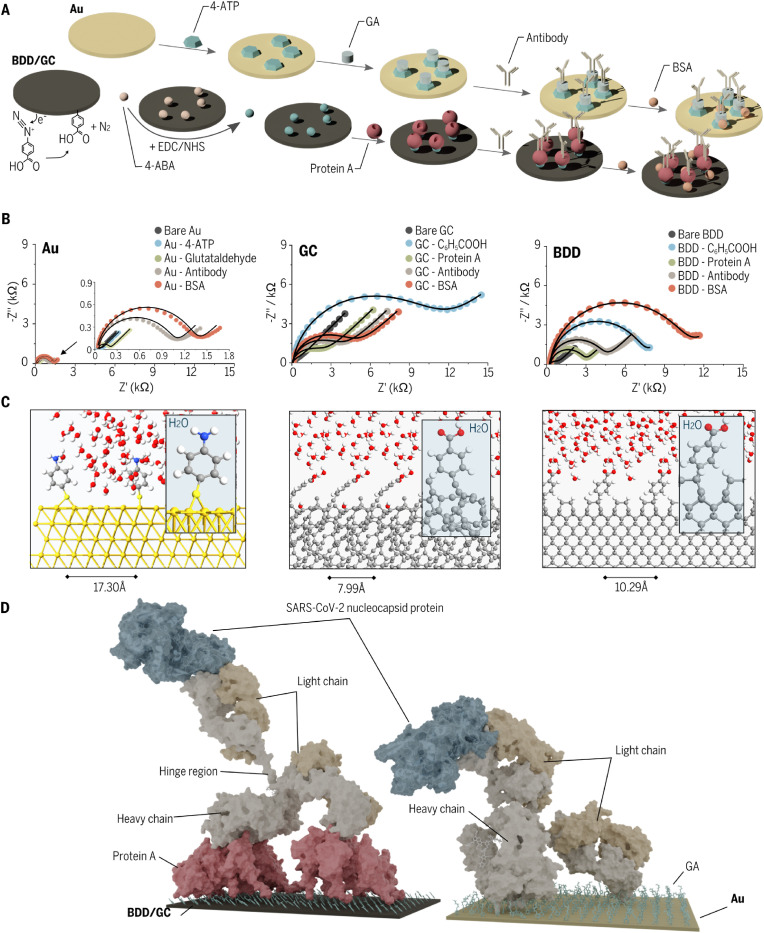
We chose three-electrode materials for the various procedures to form antibody-modified immunoassays. We present here the optimized procedures revealing favored sensing performance toward virus proteins (Fig. 3 A). The pretreatment approach for gold differed significantly from that for carbon-based (GC, BDD) materials. The Au electrode was modified mainly by chemisorption using a two-part linker (4-ATP and GA). Next, antibodies were attached to the surface by incubation of the initially functionalized electrode material in a solution of antibodies. Finally, the unsaturated sites of electrode material have been blocked by the solution of BSA. This procedure lasted around 13 h.
Fig. 3.
Functionalization process of immunosensors. (A) Schematic representation of the modification process for gold and carbon electrodes. (B) EIS spectra restored at the formal potential of K3 [Fe(CN)6] 3-/4- in the Nyquist projection recorded at each stage of the procedure (dots) with fitting data (lines). (C) Atomic structures of ab-initio simulated electrode surfaces covered by linkers. (D) Model of binding of N proteins to antibodies grafted onto various electrode surfaces based on molecular dynamics investigations.
The carbon-based surfaces (BDD and GC) undergoing an analogous procedure turned out to be slightly parametrically different after advanced optimization. Laboratory equipment was required for diazonium salt synthesis and potentiostat for its electro reduction in the first step. Before anchoring antibodies, protein A was linked via EDC/NHS to the 4-ABA acid electro-grafted BDD/GC-C6H5COOH. This step is presented in Fig. 3A where lighter spheres represent the 4-ABA molecules at the surface of BDD, while darker spheres represent the products of the reaction between 4-ABA and EDC/NHS, later replaced by antibody amino groups. The next step was blocking the unsaturated binding sites with BSA as conducted for Au surface. The total time required is 26 h, which is 2-times longer than for Au. The main difference in BDD and GC procedures was for diazonium salt concentration (stock for BDD, 1000-times diluted for GC). This was due to the high surface area of the developed polycrystalline diamond structure and the relatively smaller one for GC (Niedzialkowski et al., 2020a).
The characterization of all electrodes during modification was conducted utilizing simultaneously CV and EIS. Fig. 3B presents the impedance spectra for bare and modified Au, GC, and BDD. For the EIS data analysis, an EEQC was elaborated, as displayed in Fig. S2. EEQC was used to estimate the electrochemical parameters of electrode surface such as resistance charge transfer (Rct). Except for Rct, the constant phase element (CPE, Q) parameter was used to define material heterogeneity and the n exponent was used to measure the degree of homogeneity (tables in Fig. S3, formula in SI) (Wang et al., 2004). Also to better compare the different samples of immunosensor the relative change in Rct parameter, also called sensor response, has been calculated according to the formula (1):
| (1) |
where RBasic ct is Rct before the test (blank, positive sample, negative sample) and RTest ct is Rct after the test. The sensor response can be interpreted as the ratio of the modified surface to the total surface of the sample (Muñoz et al., 2017) and can nihilate differences between results that are caused by slight differences of samples surface initial state.
The Rct resistance for gold increased by a factor of two in the presence of glutaraldehyde. For GC and BDD, the diazonium salt linkage was more efficient, delivering 94.20% and 88.60% of impedance changes. These high values indicate a dense-packed layer of linkers to enable further protein linkage. One difference in resistance was observed for BDD and GC after GC/BDD-protein A immobilization: Rct decreased 2.7-times (BDD) and 5-times (GC), compared to measurements on the electro-grafted electrode. This may be caused by differences in the molecular charge of this protein and its impact on electron transfer (Furst and Francis, 2019; Niedzialkowski et al., 2020b; Wang et al., 2011) as proved also by impaired potentials in CV curves and decreased reduction/oxidation couple currents (Fig. S3).
The Rct resistance increased significantly after protein immobilization, indicating effective anchoring of the antibodies and BSA adsorption – for more detailed data, please, see Tables S3–S5 in Supplementary Information. The degree of electrode coverage with antibodies was calculated to be comparable to the prior coverage by linkers. We conclude that a thick biolayer was formed successively, so we investigated it further as a sensing platform.
3.3. Ab-initio and molecular dynamics of RNA binding domain of nucleocapsid protein of Wuhan strain of SAR-CoV-2 antibody (WUHAN–N-ab) system grafted onto various electrode surfaces
To get a deeper insight into the antibody surface coverage, we performed ReaxFF reactive force field (Schneider et al., 2017) simulations of the Au(111), Diamond(110), and Glassy Carbon molecular surfaces followed by manual placement of the antibody model on such surfaces with very short MD simulations (the time required to fully equilibrate such systems multiple starting positions of the antibody is beyond the current computational capabilities). MD simulation at the atomic scale for a periodic system consisting of the antibody and model surfaces. The Au(111), Diamond(110), and Glassy Carbon molecular surface models were prepared and then activated with 4-ATP (Gold) and 4-ABA (BDD, GC) molecules. Each model was then relaxed in a water box using the ReaxFF reactive force field with the Limited-memory Broyden–Fletcher–Goldfarb–Shanno algorithm (Liu and Nocedal, 1989) using the Quantum ATK software (“Atomistic Simulation Software | QuantumATK - Synopsys,” n.d.) (Fig. 3C).
The optimized cells were recalculated with different surface areas for single linker molecules. Those calculations showed the optimal coverage of linkers on each studied surface (minimal energetic configuration in Fig. S1) of the linker (Fig. 3C). The optimized coverage was taken and the work function of the surfaces was computed using DFT approach to determine the change in surfaces’ work function due to linker adsorption (Smidstrup et al., 2017). The calculated results show that 4-ATP increases work function (WF) by 1.46 eV at Gold surface increasing the energy needed for electron removal from the surface. In the case of GC and BDD, WF is decreased by 1.92 eV and 0.18 eV respectively (Fig. S1B). The minimal surface area per molecule was calculated to 129.68 Å (Gold), 105.75 Å (BDD), and 64.32 Å (GC) per linker molecule.
The surface coverage of linkers did not influence ligand coverage (Fig. S1B). We then prepared a covalently linked antibody (PDB code: 1IGT (Harris et al., 1997) to surfaces either via the K384/K445/K446 lysine cluster of the antibody heavy chain connected to three 4-aminothiophenol moieties on the Au surface via imine bonds to glutaral aldehyde or via K446 connected to the protein A (via amide bonds) immobilized on the Diamond surface via amide bonds to benzoic acid moieties. The lysine cluster was chosen based on the analysis of all lysine moieties found on the antibody surface, which led to the assumption that the K384/K445/K446 triad is the most likely candidate to bind three different linker molecules during grafting. Short canonical ensemble NVT MD runs of 10 ns of the entire antibody on surfaces followed by 1 μs NPT MD runs were conducted (Figs. S7–S10.). Fig. 3D displays the final snapshot of the MD for both studied surfaces. Additionally, the nucleocapsid protein was docked to the antibody, subjected to a 5 ns MD run, minimized, and superimposed onto the final MD snapshot. The nucleocapsid–antibody pose was obtained via protein-protein docking of the nucleocapsid protein model (described earlier) to the crystal structure of the antibody.
Our calculations predict a relatively flat orientation of the antibody on the surface, with both the heavy chains and one of the light chains close to the surface while the second light chain points away from the surface (Fig. 3D). Such an orientation should favor the interaction of the second light chain with the nucleocapsid protein, allowing for an unperturbed recognition process (Fig. S10). A 5 ns of isothermal–isobaric ensemble NPT MD of the antibody light chain complex with the N protein, followed by minimization and molecular mechanics/generalized Born surface area (MM/GBSA) calculations allowed to estimate the Gibbs free binding energy for this system at −129.2 kcal/mol, suggesting a very strong binding.
3.4. Electrochemical validation of immunosensors (GC, BDD, and Au) by impedimetric detection of the truncated form of nucleocapsid N protein of Wuhan strain of SARS-Cov-2
The validation of immunosensors included the optimization of time needed for protein N to be detected, defining the limit of the detection, and cross-reactivity test with pathogens other than the target pathogen.
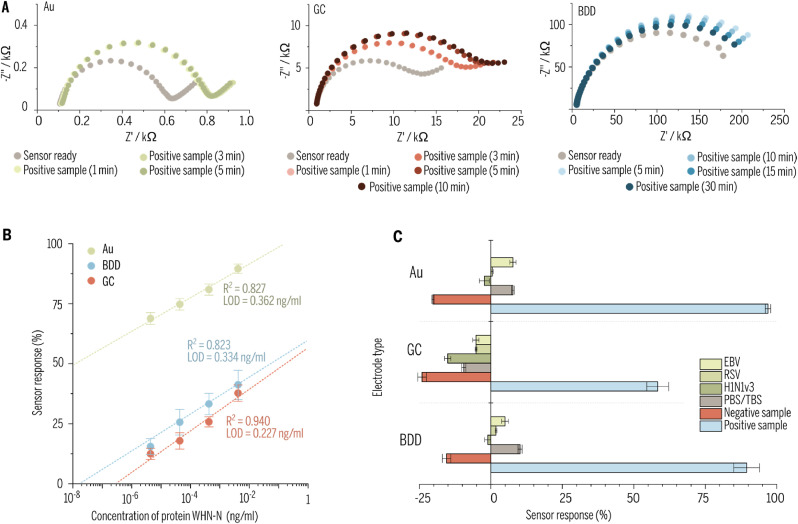
To estimate the optimal time for detection of target pathogen the immunosensor based on Au, BDD and GC were prepared according to the presented procedure then the solution of protein N with a concentration of 266 μg/mL (positive sample) was diluted in PBS was spotted on the surface of each electrode and incubated at a temperature of 5 °C for different periods. Next, their impedance spectra were recorded to estimate changes in electrode surface properties. Incubation of the sample at the temperature of 5 °C suppresses potential structural changes in proteins. The measurements were performed at room temperature until no change in the impedance spectra of each sensor was observed. The results are presented in Fig. 4 A. It can be seen that Au and GC impedance spectra recorded after 3 and 5 min of incubation in a solution of protein N haven't changed which would lead to the suggestion that incubation in protein solution for 5 min is enough for those immunosensors. On the other hand, the BDD has shown other behavior where after a short time of incubation (10 min) the change in impedance spectra was the most significant and decrease with increasing time. Such behavior may be caused by a change of the conformation of adsorbed molecules of protein N on the surface of the electrode or even occurring the desorption process. Such observations lead authors to statement that 10 min is the most optimal time needed for detection by immunosensor deigned with BDD.
Fig. 4.
Data from tests of sensitivity, selectivity, and linearity range of the prepared biological sensors. (A) Study of the optimal response - incubation of the electrode in a protein N solution for different periods. (B) Dependence of the responses of various sensors (Au, BDD, GC) versus concentrations of Wuhan-N protein (RNA binding domain of nucleocapsid protein of Wuhan strain of SARS-CoV-2). (C) Cross-reactivity with other human upper respiratory tract viruses. Selectivity of different types of electrodes was tested for inactivated viruses. Epstein-Barr virus (EBV), respiratory syncytial virus (RSV), influenza A virus H1N1 Virginia 3 subtype (H1N1v3), PBS/TBS buffer, recombinant N protein from a different human coronavirus (HCoV-NL63) served as a negative control, and swabs from COVID-19 positive patients served as a positive sample.
After optimization of time detection, the EIS was used to investigate the metrological performance of the biosensor detecting the SARS-CoV-2 virus protein N by spotting the solutions with a different concentration (4.4 pg/mL, 44.4 pg/mL, 444 pg/mL, and 4.44 ng/mL – diluted in PBS) on the surface of electrodes and incubating them for the optimal time. Next, based on the data obtained from EEQC fitting to impedance spectrum (more specifically - from Rct) the sensor response has been calculated by the formula (1). The relationship between protein N concentrations and sensor response is presented in Fig. 4B for all three electrode materials.
The linear regression equations, presented in Fig. 4B, for Au, GC, BDD electrodes with the corresponding values of the correlation coefficient (R2) and the limit of detection (LOD), can be expressed as:
| ΔRct[%]Au = 22.427·CWHN–N [ng/ml] + 71.394 R2 = 0.827 LOD = 0.362 ng/ml |
| ΔRct[%]GC = 27.791·CWHN–N [ng/ml] + 13.290 R2 = 0.940 LOD = 0.227 ng/ml |
| ΔRct[%]BDD = 36.438·CWHN–N [ng/ml] + 17.281 R2 = 0.823 LOD = 0.334 ng/ml |
The limit of detection was calculated from the relation LOD = 3 x SD/slope, where SD is the standard deviation in the low concentration range. For all tested surfaces, a wide linear range of concentrations from 4.4 ng/ml to 4.4 pg/ml was obtained. Error bars were calculated as relative standard deviation (RSD) (For detailed data, please see Table S6 in Supplementary information), while their low levels indicate high repeatability of conducted measurements.
In the last step of sensors validation, the cross-reactivity tests were performed with different pathogens and patients' saliva swabs sick with COVID-19 as positive samples. To prepare saliva swabs for the electrochemical measurement 1 mL of it was diluted in the 1 mL of electrolyte utilized in the whole experiment, but with twice time higher concentration of electrochemical species to obtain the same concentration of [Fe(CN)6]3-/4- after dilution. The 30 μL of that mixture was then spotted on the surface of each electrode and left for the incubation time estimated in previous measurements for each electrode material. In cross-reactivity tests the same sample of saliva swab was used for multiple samples of prepared sensors, ensuring better reproducibility and comparability of results. All electrochemical measurements were conducted in the same manner as previous tests and results are shown in Fig. 4C and are elaborated in caption 3.5.
3.5. Comparison of immunosensors (GC, BDD, and Au) selectivity, repeatability, and stability studies
RSV, EBV, and H1N1v3 were used as potentially interfering viruses to investigate the selectivity of the immunosensor. The selection was based on the causatives of respiratory tract infections, especially Influenzae A which is known as a common coinfection species with SARS-CoV-2. The Wuhan-N protein concentration was kept in the same order of magnitude to receive comparable results (Fig. 4B). The repeatability and reproducibility studies involved three independent sensors. According to Fig. 4C, all negative controls did not give a substantial impedance increase. The Rct changes calculated for the PBS/TBS buffer as a negative response did not exceed the absolute threshold value of 10.2% for BDD electrode, while for glassy carbon and gold electrodes were −9.5% and 7.7%, respectively. The strongest interaction with protein was observed using the gold electrode (96.6%), next was the BDD electrode (89.4%), and the weakest was the glassy carbon electrode (58.4%). The sensor exposed to pathogenic negative control gave changes ranging from −27.85% (H1N1 on GC electrode) to 7.7% (EBV on the gold surface). The negative value of sensor response means that the decrease in Rct occurred after incubation in the studied sample – in the case of GC for any negative sample and at BDD/Au for HCov-NL63. The explanation of that phenomenon could be attributed Vroman effect (Vroman et al., 1980), meaning that some of the antibodies present on the surface of the electrode have been substituted by proteins present in the studied samples. This effect eventually could lead to the opening of pathways for electrochemical species and so to decrease in Rct values.
According to the results obtained, the immunoassays exhibited high specificity for the detection of the N protein of the Wuhan strain of SARS-Cov-2 produced in E. coli (WUHAN - N protein) without giving false-positive results in PBS/TBS buffer, and other viruses. Additionally, we found that the antibodies were not cross-reactive with other upper respiratory tract viruses (Fig. 5 D).
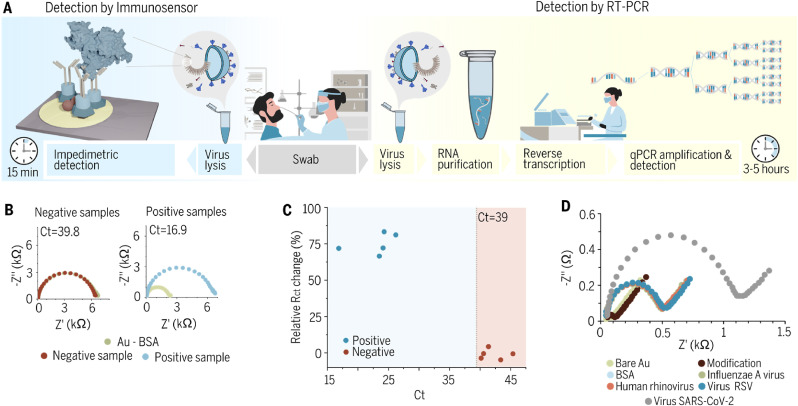
Fig. 5.
Clinical samples diagnostics employing gold immunoassays. (A) Schematic illustration of the comparison between the point-of-care ultrafast immunoassay and the standard - qRT-PCR detection methods. (B) Impedimetric spectra registered for Au sensor after incubation in a solution of positive samples (saliva swabs containing SARS-CoV-2) and negative samples (saliva swabs lacking SARS-CoV-2 but containing other pathogens (three samples) or from healthy patients (two samples)) (C) Comparison between Ct and ΔRct for the tested samples. (D) Cross-reactivity testing of the antibody with different upper respiratory tract viruses such as Influenza A, Human Rhinovirus, and RSV.
3.6. Clinical samples diagnostics of SARS-CoV-2 using the real-time Au-based immunosensor platform
In this report, we aimed to use a relatively most effective gold electrode platform to determine whether the virus can be detected in a ‘one-step’ process from aqueous samples (Malinowska et al., 2020). Clinical diagnostic of gold immunoassay specificity was determined using 10 human saliva swabs (5 positive samples, 5 negative samples) previously identified using a real-time RT-PCR test. This test is routinely used for the screening and diagnosis of SARS-CoV-2 (Fig. 5A). The immunosensor has a significantly faster analysis time (15 min) than the standard qRT-PCR (3–5 h). Saliva swabs were collected and suspended in a 0.9% NaCl solution.
According to the WHO standard, if the cycle threshold (Ct) value of the internal reference channel is less than or equal to 40, the sample is judged as positive. It follows that the threshold of the impedance immunosensor must be Ct = 39 to minimize the false positive to a false-negative ratio in determining the presence of COVID-19.
As described previously, to prepare swabs samples for electrochemical measurements they were diluted in electrolyte and then 30 μL of that solution has been spotted on the electrode. Negative samples were taken from human saliva swabs lacking SARS-CoV-2. These swabs were from both healthy and nonhealthy patients. As pre-diagnosed infected swabs contained Streptococcus pyogenes (S.py) (Ct = 41), Streptococcus pneumonia (S.pn) (Ct = 43), and Hemophilus influenzae (Hi) (Ct = 45). Two swabs were from healthy patients (Ct = 39.8 and Ct = 40.2). All samples have been tested with RT-PCR, please refer to Figs. S6–S7. Fig. 5B illustrates the impedimetric curves for the ready biosensor (gold electrode modified with antibodies) and for sensors incubated with samples for 10 min for two representative samples, for all datasets please refer to Fig. S4. Particularly, no significant rise in a semicircle was observed in the shape of the EIS spectra upon incubation of the electrode with negative samples, meaning that no specific adsorption of the biomolecules present in the swab occurred. For positive samples, the semicircles rise considerably, which is related to blockage of the electrochemically active surface by biomolecules, namely virus particles with protein N on their surface, for the electrochemically active species.
The correlation between Ct and Rct values based on the analysis applied to the impedance spectra for each sample is shown in Fig. 5C. Negative samples (Ct > 39) gave a small impedance response (up to 10% relative Rct change), which is insignificant compared to the significant (65–85%) relative Rct changes for positive samples. After electrode exposition to positive samples for just 10 min, we observed a substantial ΔRct ascent (up to 85% in Fig. 5C). The sensitivity of the applied detection methodology depends on the viral life cycle and the correlation between the number of viral copies and the type of swab used to collect the sample. It is worth noting that the quantity of RNA copies in the nasopharynx swab is 10 6–10 9, while the throat swab reaches 10 4-108. Those differences would impact the sensitivity of the assay. Moreover, the viral RNA is encapsulated by N protein. It was also confirmed that N protein is an early viral marker present in high amounts at the first hours of infection. It was reported that a single SARS-CoV-2 virion contains around 2200 copies of N protein, while only 100 copies of Spike protein (Bar-On et al., 2020). Corresponding to other studies, the Ct value may be correlated with the number of RNA copies, copies of viral particles, and PFU per ml (Brandolini et al., 2021; Pickering et al., 2021). According to Brandolini et al. (2021), a Ct value over 35 corresponds to no viral particles present in the sample. Furthermore, samples with a Ct less than 25, corresponding to approximately 1.5 × 10 6 copies/ml or 400 PFU/ml, while Ct 28 refers to approximately 1.65 × 10 5 copies/ml or 50 PFU/ml. It was recently shown that the limit of detection is approximately 250 pfu/ml for most available diagnostic tests (Stanley et al., 2022). The correlation between the concentration of N-protein is critical to assess assay applicability in clinic samples. Thus, we have estimated the concentration of N protein within the viral particle. Single virions contain up to 2200 copies of N protein which equals 168.1 ag. For Ct value around 25, the PFU reaches 400 corresponding to 67.2 fg of N protein.
4. Discussion and conclusions
In summary, the developed impedimetric immunoassays offer a competitive and time-effective alternative for SARS-CoV-2 detection approaches. Surface modification methods proposed in this work are simple and do not require specialized equipment. Thus, the biosensor fabrication process has a great potential for automation. Accessibility, simplicity, and repeatability are crucial for effective pandemic counteraction, allowing for the number of people who will be tested in the shortest time to increase. Fast and reliable diagnostic is of great importance to reduce virus transmission. The fundamental interactions between the ligand and the receptor indicating the antibody and the protein of interest are constantly analogous. Next to the golden standard nucleic acid amplification tests (NAATs), there are ELISA tests and additional immunological methods like lateral flow assay (LFA). From the last decade, rapid electrochemical assays were of the most interest due to their sensitivity and short detection time. The advantage of the electrochemical assays is fast response time, lower uses of reagents, and high specificity relating to conventional methods. The potential of the proposed approach is not only the sensing device but also custom-made biological components. Manufactured antibodies show strong and highly specific affinity to RNA binding domain of nucleocapsid protein of SARS-CoV-2. The antibodies anchored to the electrode surface result in improved selectivity and sensitivity towards SARS-CoV-2 virus. Since the conserved regions of RNA binding domain of the inner protein were chosen, probably, the antigenic drift may not influence the detection pattern. Established technological processes for all components put our biosensor one step closer to mass production.
The described biosensor can be deployed as broad and repeated testing routines for saliva swabbed specimens for SARS-CoV-2 and shows no cross-reactivity with other respiratory tract viruses. Negative samples and measurement conditions reveal a satisfactorily low impact on the sensor response which leads to the high stability of the system and thus the results can be obtained with good reproducibility. However, it is recommended to incubate the sensor with the sample for at least 5 min to let the system equilibrate before a test. This may be particularly important regarding the desorption process of non-specifically bonded interfering species naturally present in the sample. The limits of detection for the biosensor detecting the SARS-CoV-2 virus protein were from 0.227 ng/ml through 0.334 ng/ml to 0.362 ng/ml for GC, BDD, and gold electrodes, respectively. Such low detection limits, combined with a linear response in a broad range of target protein concentrations and reduced influence of undesirable entities make developed sensing platforms good candidates for common use. Electrodes exhibited various selectivity behavior depending on the antibody density and orientation tuning binding of N protein, thus influencing immunoassay sensitivity.
The most critical impact on the immunoassay sensitivity has the electro-immobilization procedure of linker and architecture of antibody complexes. Their density depends on the specific surface as shown in the paper but it could be also strongly affected by electrochemical and structural heterogeneities of electrode surfaces (A. Armstrong et al., 1997; Hirst, 2006). The multi-faceted and sharply edged BDD surfaces induce a negative surficial charge attracting positively charged proteins. This allows for numerous immobilization alternatives and conformations of proteins at different regions of BDD electrode (Yates et al., 2018) followed by protein-protein accidental interactions, which could be responsible for decreased specificity and sensitivity. The orientational alterations may be also accountable for variation of the electrochemical electrode performance observed as various electron-transfer kinetic rate constants tuned by the distance between the electrode surface and the electron entrance/exit spot at the specific protein group (Morris et al., 2015). Although BDD shows the low capacity and background currents extending sensory efficiency, unfortunately, it is characterized by high heterogeneity of electrostatic interactions, often causing non-specific protein adsorption. The work of (Trouillon and O'Hare, 2010) ascribed partial blocking of BDD surface to inhomogeneous boron dopant concentration and sp 2 induced heterogeneities. On the other side, the interwoven graphite ribbons in the GC complex structure result in much homogenous adsorption of proteins even at the bare surface (Léger and Bertrand, 2008).
Next, it was observed that Gold electrode showed the most intensive interaction with N protein, while the glassy carbon revealed the weakest character. MD simulations revealed that Au results in the lowest density of coverage expressed as a large surface area per linker molecule. Area per linker for Au was almost doubled versus GC suggesting that large inter-linker area provides multi-point binding of antibody along with its unhampered motion. Although, the high sensitivity of gold is mainly attributed to the spatially expanded active antibodies immobilized at relatively lower surface coverage degree (Sharafeldin et al., 2019). The presented atomistic simulations prove details of the entire immunosensor platform architecture. The DFT and MD modelling showed a probable mode of binding of SARS-CoV-2 nucleocapsid protein to the specific antibodies functionalized on different electrode surfaces. The mechanism of interactions revealed the grafting conformation and surficial density of the molecular system, which leads to a change in electrochemical impedance that is transduced into a highly sensitive electrical detection signal. To win the race against the XXI century pandemic, test-and-trace strategy is pivotal. We believe that the proposed real-time multiplatform immunoassay may be easily used in containing current and future pandemics.
Funding
The authors gratefully acknowledge financial support from the National Science Centre (NCN) under Grant “Express Call – Research on COVID-19” No. 2020/01/0/ST7/00104. The DS funds of the Faculty of Electronics, Telecommunications and Informatics of the Gdańsk University of Technology are also acknowledged. Molecular Dynamics simulations have been performed in part at the Interdisciplinary Centre for Mathematical and Computational Modelling of the University of Warsaw (grants g86-1058 and g87-1079).
Author contributions
W.B., R.B., B.G., D.N. and K.P. conceived and designed the work. T.Ł, Y.D., B.G. and M.P. produced the recombinant protein and polyclonal antibody. M.F., M. R., Z.C., E.B. and W.B. prepared electrodes. M.B., M.K., W.B., D.B., and K.D. modified electrodes. M.B., M.K., N.M., D.F. and W.B. optimized the assays and performed analysis. W.B.; B.G., M. Pi., and R.B. wrote the manuscript with corrections from all authors. Theoretical and computational validation models were conceived and prepared by B.D, W.A.G., A.J.B., B.T and S.O. R.B , D.N. and S.Ż. were responsible for project administration and funding acquisition. All the authors analyzed the data, discussed the results, and reviewed the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank SkyWayBiolab for providing upper respiratory tract inactivated viruses that served as negative controls in our experiments and for the technological input needed for antibody production.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114222.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Armstrong F.A., Heering H.A., Hirst J. Reaction of complex metalloproteins studied by protein-film voltammetry. Chem. Soc. Rev. 1997;26:169–179. doi: 10.1039/CS9972600169. [DOI] [Google Scholar]
- Aditya Jayanthi V.S.P.K.S., Bikas Das A., Saxena U. Fabrication of an immunosensor for quantitative detection of breast cancer biomarker UBE2C. RSC Adv. 2019;9:16738–16745. doi: 10.1039/C8RA10245G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Ajmal M., Ashraf G., Muhammad N., Aziz A., Iftikhar T., Wang J., Liu H. The role of biosensors in coronavirus disease-2019 outbreak. Curr. Opinion Electrochem. 2020;23:174–184. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atomistic Simulation Software QuantumATK - Synopsys [WWW Document], n.d. URL https://www.synopsys.com/silicon/quantumatk.html (accessed 12.4.20).
- Balkourani G., Brouzgou A., Archonti M., Papandrianos N., Song S., Tsiakaras P. Emerging materials for the electrochemical detection of COVID-19. J. Electroanal. Chem. 2021;893:115289. doi: 10.1016/j.jelechem.2021.115289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanowicz R., Fabianska A., Golunski L., Sobaszek M., Gnyba M., Ryl J., Darowicki K., Ossowski T., Janssens S.D., Haenen K., Siedlecka E.M. Influence of the boron doping level on the electrochemical oxidation of the azo dyes at Si/BDD thin film electrodes. Diam. Relat. Mater. 2013;39:82–88. doi: 10.1016/j.diamond.2013.08.004. [DOI] [Google Scholar]
- Bogdanowicz R., Sawczak M., Niedziakowski P., Zieba P., Finke B., Ryl J., Karczewski J., Ossowski T. Novel functionalization of boron-doped diamond by microwave pulsed-plasma polymerized allylamine film. J. Phys. Chem. C. 2014;118:8014–8025. doi: 10.1021/jp5003947. [DOI] [Google Scholar]
- Brandolini M., Taddei F., Marino M.M., Grumiro L., Scalcione A., Turba M.E., Gentilini F., Fantini M., Zannoli S., Dirani G., Sambri V. Correlating qRT-PCR, dPCR and viral titration for the identification and quantification of SARS-CoV-2: a new approach for infection management. Viruses. 2021;13:1022. doi: 10.3390/v13061022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Coronavirus disease 2019 (COVID-19) [WWW Document]. Centers for disease control and prevention. 2020. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html URL. 2.23.22.
- Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12:802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J. Virol. 2020;94 doi: 10.1128/JVI.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo A.P., Akgun Y., Al Mana A.F., Tjendra Y., Millan N.C., Gomez-Fernandez C., Cray C. Review of current advances in serologic testing for COVID-19. Am. J. Clin. Pathol. 2020;154:293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiani L., Saroglia M., Galatà G., De Santis R., Fillo S., Luca V., Faggioni G., D'Amore N., Regalbuto E., Salvatori P., Terova G., Moscone D., Lista F., Arduini F. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens. Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Pontremoli C., Mozzi A., Pozzoli U., Clerici M., Sironi M. Antigenic variation of SARS-CoV-2 in response to immune pressure. Mol. Ecol. 2021;30:3548–3559. doi: 10.1111/mec.15730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst A.L., Francis M.B. Impedance-based detection of bacteria. Chem. Rev. 2019;119:700–726. doi: 10.1021/acs.chemrev.8b00381. [DOI] [PubMed] [Google Scholar]
- Harris L.J., Larson S.B., Hasel K.W., McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997;36:1581–1597. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
- Hirst J. Elucidating the mechanisms of coupled electron transfer and catalytic reactions by protein film voltammetry. Biochim. Biophys. Acta Bioenerg. 2006;1757:225–239. doi: 10.1016/j.bbabio.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Huang W., Lim B., Hsu C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT‐LAMP for rapid diagnosis of coronavirus SARS‐CoV‐2. Microb. Biotechnol. 2020;13 doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein H.A., Hassan R.Y.A., Chino M., Febbraio F. Point-of-Care diagnostics of COVID-19: from current work to future perspectives. Sensors. 2020;20 doi: 10.3390/s20154289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Rogers J.A., Xu S. Continuous on-body sensing for the COVID-19 pandemic: gaps and opportunities. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Ziliang, Zhou Zhechong, Chen Q., Yan Y., Zhang C., Shan H., Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B, Special Column: Res. Emerg. COVID-19 (Target, Mech. Therapeut.) 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiew L.-V., Chang C.-Y., Huang S.-Y., Wang P.-W., Heh C.-H., Liu C.-T., Cheng C.-H., Lu Y.-X., Chen Y.-C., Huang Y.-X., Chang S.-Y., Tsai H.-Y., Kung Y.-A., Huang P.-N., Hsu M.-H., Leo B.-F., Foo Y.-Y., Su C.-H., Hsu K.-C., Huang P.-H., Ng C.-J., Kamarulzaman A., Yuan C.-J., Shieh D.-B., Shih S.-R., Chung L.-Y., Chang C.-C. Development of flexible electrochemical impedance spectroscopy-based biosensing platform for rapid screening of SARS-CoV-2 inhibitors. Biosens. Bioelectron. 2021;183:113213. doi: 10.1016/j.bios.2021.113213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léger C., Bertrand P. Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem. Rev. 2008;108:2379–2438. doi: 10.1021/cr0680742. [DOI] [PubMed] [Google Scholar]
- Lima L.F. de, Ferreira A.L., Torres M.D.T., Araujo W.R., Fuente-Nunez de, de la C. Minute-scale detection of SARS-CoV-2 using a low-cost biosensor composed of pencil graphite electrodes. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118 doi: 10.1073/pnas.2106724118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.C., Nocedal J. On the limited memory BFGS method for large scale optimization. Math. Program. 1989;45:503–528. [Google Scholar]
- Liu H., Dai E., Xiao R., Zhou Z., Zhang M., Bai Z., Shao Y., Qi K., Tu J., Wang C., Wang S. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sensor. Actuator. B Chem. 2021;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Zhu D.Z., Gan H., Yao Z., Fu Q., Zhang X., Jackie) Prospects and challenges of using electrochemical immunosensors as an alternative detection method for SARS-CoV-2 wastewater-based epidemiology. Sci. Total Environ. 2021;777:146239. doi: 10.1016/j.scitotenv.2021.146239. [DOI] [Google Scholar]
- Lu R., Wang J., Li M., Wang Y., Dong J., Cai W. SARS-CoV-2 detection using digital PCR for COVID-19 diagnosis, treatment monitoring and criteria for discharge. medRxiv. 2020;24:20042689. doi: 10.1101/2020.03.24.20042689. 03, 2020. [DOI] [Google Scholar]
- Mahapatra S., Chandra P. Clinically practiced and commercially viable nanobio engineered analytical methods for COVID-19 diagnosis. Biosens. Bioelectron. 2020;165:112361. doi: 10.1016/j.bios.2020.112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahari S., Roberts A., Shahdeo D., Gandhi S. Vol. 24. 2020. (eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 Antigen, a Spike Protein Domain 1 of SARS-CoV-2). bioRxiv 2020.04. 059204. [DOI] [Google Scholar]
- Mahshid S.S., Flynn S.E., Mahshid S. The potential application of electrochemical biosensors in the COVID-19 pandemic: a perspective on the rapid diagnostics of SARS-CoV-2. Biosens. Bioelectron. 2021;176:112905. doi: 10.1016/j.bios.2020.112905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowska N., Białobrzeska W., Łęga T., Pałka K., Dziąbowska K., Żołędowska S., Czaczyk E., Pala K., Nidzworski D. Antibody modified gold electrode as an impedimetric biosensor for the detection of Streptococcus pyogenes. Sensors. 2020;20:5324. doi: 10.3390/s20185324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.P., Baker R.E., Gillow K., Davis J.J., Gavaghan D.J., Bond A.M. Theoretical analysis of the relative significance of thermodynamic and kinetic dispersion in the dc and ac voltammetry of surface-confined molecules. Langmuir. 2015;31:4996–5004. doi: 10.1021/la5042635. [DOI] [PubMed] [Google Scholar]
- Muñoz J., Montes Martínez R., Baeza M. Trends in Electrochemical Impedance Spectroscopy involving nanocomposite transducers: characterization, architecture surface and bio-sensing. TrAC Trends Anal. Chem. 2017;97:201–215. doi: 10.1016/j.trac.2017.08.012. [DOI] [Google Scholar]
- Niedzialkowski P., Slepski P., Wysocka J., Chamier-Cieminska J., Burczyk L., Sobaszek M., Wcislo A., Ossowski T., Bogdanowicz R., Ryl J. Multisine impedimetric probing of biocatalytic reactions for label-free detection of DEFB1 gene: how to verify that your dog is not human? Sensor. Actuator. B Chem. 2020;323:128664. doi: 10.1016/j.snb.2020.128664. [DOI] [Google Scholar]
- Niedzialkowski P., Slepski P., Wysocka J., Chamier-Cieminska J., Burczyk L., Sobaszek M., Wcislo A., Ossowski T., Bogdanowicz R., Ryl J. Multisine impedimetric probing of biocatalytic reactions for label-free detection of DEFB1 gene: how to verify that your dog is not human? Sensor. Actuator. B Chem. 2020;323:128664. doi: 10.1016/j.snb.2020.128664. [DOI] [Google Scholar]
- Perdomo S.A., Ortega V., Jaramillo-Botero A., Mancilla N., Mosquera-DeLaCruz J.H., Valencia D.P., Quimbaya M., Contreras J.D., Velez G.E., Loaiza O.A., Gómez A., de la Roche J. SenSARS: a low-cost portable electrochemical system for ultra-sensitive, near real-time, diagnostics of SARS-CoV-2 infections. IEEE Trans. Instrum. Meas. 2021;70:1–10. doi: 10.1109/TIM.2021.3119147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering S., Batra R., Merrick B., Snell L.B., Nebbia G., Douthwaite S., Reid F., Patel A., Ik M.T.K., Patel B., Charalampous T., Alcolea-Medina A., Lista M.J., Cliff P.R., Cunningham E., Mullen J., Doores K.J., Edgeworth J.D., Malim M.H., Neil S.J.D., Galão R.P. Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: a single-centre laboratory evaluation study. The Lancet Microbe. 2021;2:e461–e471. doi: 10.1016/S2666-5247(21)00143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhrel P., Hu C., Mao H. Detecting the coronavirus (COVID-19) ACS Sens. 2020;5:2283–2296. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Rashed M.Z., Kopechek J.A., Priddy M.C., Hamorsky K.T., Palmer K.E., Mittal N., Valdez J., Flynn J., Williams S.J. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raziq A., Kidakova A., Boroznjak R., Reut J., Öpik A., Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021;178:113029. doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R., Navale G.R., Dharne M.S. Biosensors: frontiers in rapid detection of COVID-19. 3 Biotech. 2020;10:385. doi: 10.1007/s13205-020-02369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J., Hamaekers J., Chill S.T., Smidstrup S., Bulin J., Thesen R., Blom A., Stokbro K. ATK-ForceField: a new generation molecular dynamics software package. Model. Simulat. Mater. Sci. Eng. 2017;25 doi: 10.1088/1361-651X/aa8ff0. [DOI] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Sharafeldin M., McCaffrey K., Rusling J.F. Influence of antibody immobilization strategy on carbon electrode immunoarrays. Analyst. 2019;144:5108–5116. doi: 10.1039/C9AN01093A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.K., Ray P., Carlin A.F., Magallanes C., Morgan S.C., Laurent L.C., Aronoff-Spencer E.S., Hall D.A. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens. Bioelectron. 2021;180:113111. doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidstrup S., Stradi D., Wellendorff J., Khomyakov P.A., Vej-Hansen U.G., Lee M.-E., Ghosh T., Jónsson E., Jónsson H., Stokbro K. First-principles Green’s-function method for surface calculations: a pseudopotential localized basis set approach. Phys. Rev. B. 2017;96:195309. doi: 10.1103/PhysRevB.96.195309. [DOI] [Google Scholar]
- Stanley S., Hamel D.J., Wolf I.D., Riedel S., Dutta S., Cheng A., Kirby J.E., Kanki P.J. 2022. Limit of Detection for Rapid Antigen Testing of the SARS-CoV-2 Omicron Variant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar N.T., Shajahan A., Gleinich A.S., Rouhani D., Heiss C., Azadi P. SARS-CoV-2 Nucleocapsid protein is decorated with multiple N- and O-glycans. bioRxiv. 2020;26:269043. doi: 10.1101/2020.08.26.269043. 08, 2020. [DOI] [Google Scholar]
- Szunerits S., Pagneux Q., Swaidan A., Mishyn V., Roussel A., Cambillau C., Devos D., Engelmann I., Alidjinou E.K., Happy H., Boukherroub R. The role of the surface ligand on the performance of electrochemical SARS-CoV-2 antigen biosensors. Anal. Bioanal. Chem. 2021 doi: 10.1007/s00216-020-03137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi V.L.D., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Kräusslich H.-G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi V.L.D., Herbst K., Boerner K., Meurer M., Kremer L.P., Kirrmaier D., Freistaedter A., Papagiannidis D., Galmozzi C., Stanifer M.L., Boulant S., Klein S., Chlanda P., Khalid D., Miranda I.B., Schnitzler P., Kräusslich H.-G., Knop M., Anders S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Fock J., Minero G.A.S., Hansen M.F. Nicking-assisted on-loop and off-loop enzymatic cascade amplification for optomagnetic detection of a highly conserved dengue virus sequence. Biosens. Bioelectron. 2020;160:112219. doi: 10.1016/j.bios.2020.112219. [DOI] [PubMed] [Google Scholar]
- Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D., Farrell M.J., Hooser S.V., Pandori M. Genomic evidence for reinfection with SARS-CoV-2: a case study. The Lancet Infectious Diseases 0. 2020 doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020 doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillon R., O'Hare D. Comparison of glassy carbon and boron doped diamond electrodes: resistance to biofouling. Electrochim. Acta. 2010;55:6586–6595. doi: 10.1016/j.electacta.2010.06.016. [DOI] [Google Scholar]
- Vroman L., Adams A., Fischer G., Munoz P. Interaction of high molecular weight kininogen, factor XII, and fibrinogen in plasma at interfaces. Blood. 1980;55:156–159. doi: 10.1182/blood.V55.1.156.156. [DOI] [PubMed] [Google Scholar]
- Wang W., Foley K., Shan X., Wang S., Eaton S., Nagaraj V.J., Wiktor P., Patel U., Tao N. Single cells and intracellular processes studied by a plasmonic-based electrochemical impedance microscopy. Nat. Chem. 2011;3:249–255. doi: 10.1038/nchem.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wang L., Wang G., Ji X., Bai Y., Li T., Gong S., Li J. Application of impedance spectroscopy for monitoring colloid Au-enhanced antibody immobilization and antibody–antigen reactions. Biosens. Bioelectron. 2004;19:575–582. doi: 10.1016/S0956-5663(03)00252-5. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- Witt S., Rogien A., Werner D., Siegenthaler J., Lesiyon R., Kurien N., Rechenberg R., Baule N., Hardy A., Becker M. Boron doped diamond thin films for the electrochemical detection of SARS-CoV-2 S1 protein. Diam. Relat. Mater. 2021;118:108542. doi: 10.1016/j.diamond.2021.108542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Li D., Ramadan S., Li Y., Klein N. Facile biosensors for rapid detection of COVID-19. Biosens. Bioelectron. 2020;170:112673. doi: 10.1016/j.bios.2020.112673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26 doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates N.D.J., Fascione M.A., Parkin A. Methodologies for “Wiring” redox proteins/enzymes to electrode surfaces. Chem. Eur J. 2018;24:12164–12182. doi: 10.1002/chem.201800750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J.B., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., McGeer A., Christie N., Gray-Owen S., Cochrane A., Rini J.M., Sargent E.H., Kelley S.O. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- Zhang X., Rösicke F., Syritski V., Sun G., Reut J., Hinrichs K., Janietz S., Rappich J. Influence of the para-substituents of benzene diazonium salts and the solvent on the film growth during electrochemical reduction. Z. Phys. Chem. 2014;228:557–573. doi: 10.1515/zpch-2014-0450. [DOI] [Google Scholar]
- Zhang Y., Odiwuor N., Xiong J., Sun L., Nyaruaba R.O., Wei H., Tanner N.A. Vol. 26. 2020. p. 20028373. (Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP). medRxiv 2020.02. [DOI] [Google Scholar]
- Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensor. Actuator. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.