Abstract
Background
Atopic dermatitis (AD) is a chronic relapsing inflammatory skin disease triggered by diverse factors. Microbes are one of the crucial risk factors for AD development or exacerbation. However, the effect of a fungal burden on AD has been overlooked compared to bacteria.
Objective
This study aimed to comparatively analyze cutaneous fungal distribution between AD patients and healthy individuals by polymerase chain reaction (PCR)-based analysis.
Methods
Skin samples of AD outpatients and healthy individuals collected at the Chung-Ang University were analyzed. Representative AD-associated fungal genera, Candida, dermatophytes, and Malassezia , were analyzed using specific primer and amplification methods. Amplicons were sequenced, and the fungal distribution of both groups were compared.
Results
Totally, 211 patients and 23 healthy individuals were studied. Of the 211 patients, 10.90% (23/211) had Candida species, whereas 0% (0/23) healthy individuals showed its presence. The most frequently detected species in patients was Candida albicans (5.21%) followed by Candida parapsilosis (3.79%). For dermatophytes, 1.42% (3/211) of patients showed positive results, whereas 0% (0/23) healthy individuals showed positive results. Malassezia species were identified in 20.85% (44/211) and 8.70% (2/23) in patients and healthy individuals, respectively. Malassezia restricta was the most frequently identified species in the AD patient group, and the only species found in the healthy control group.
Conclusion
The distribution of Candida spp., dermatophytes, and Malassezia spp. are altered with AD development.
Keywords: Arthrodermataceae, Atopic dermatitis, Candida, Fungi, Malassezia
INTRODUCTION
Atopic dermatitis (AD) is a chronic skin disease, which affects up to 20% of children and 2% of adults and the prevalence has increased 2 to 3 folds during the last 30 years1,2. AD is a representative complex disease because diverse factors can lead to AD, including genetic, environmental factors, and microorganisms3,4. Recent research on the effects of microbes on the skin and the underlying mechanisms has revealed that while the skin is a beneficial niche for a variety of microorganisms that exhibit mutualism or commensalism, the microbes could become pathogenic depending on the host immune condition. In the case of AD, the weakened skin barrier of patients is more vulnerable to microbial penetration. For example, it is known that Staphylococcus aureus, a representative AD-associated bacterium known to exacerbate the disease by producing superantigens, colonizes 200 folds more in the skin of patients with AD than healthy individuals5,6. Concerning AD, comprehensive research on cutaneous fungal microbiota is few compared to that of bacterial microbiota because of lower interest and limited methodological approach. Although sufficient research has not been constructed yet, many suggestions are emphasizing the fungal burden on disease development. For example, AD patients who responded poorly to conventional treatment showed improvement when treated with antifungal drugs4,7. Moreover, it is known that the cutaneous fungal diversity and richness is altered in AD patients compared with healthy individuals8,9. Although the fungus is not a direct causative agent, dysbiosis of the fungal community appears to contribute to AD onset or exacerbation. This makes it important to study the association between the fungal community and AD, which can provide a novel perspective on understanding microbes as contributor to AD and identifying appropriate solution to a disease.
In this study, we aimed to comparatively analyze the cutaneous distribution of Candida, dermatophytes, and Malassezia between the lesional skin of AD patients and healthy individuals. Since culture-based analysis does not accurately reflect fungal community due to cultural limitations, we conducted a polymerase chain reaction (PCR)-based analysis.
MATERIALS AND METHODS
Subjects and skin sample collection
Skin samples were collected from 211 outpatients with AD and 23 healthy controls between January 2011 and February 2013 at Chung-Ang University Hospital (Seoul, Republic of Korea). A total of 234 skin swab samples were used in this study. The inclusion criteria for AD patients were as follows: 1) patients diagnosed with AD by a dermatologist in Chung-Ang University Hospital; 2) patients who had AD for more than 10 years; 3) patients who had typical AD skin lesion on the antecubital fossa. The inclusion criteria for healthy control was subjects without AD or any other active skin diseases confirmed by thorough history taking and physical examination by a dermatologist in Chung-Ang University Hospital. For both groups, exclusion criteria were as follows: 1) participants who had any active skin diseases, exclusive of AD; 2) participants with a history of receiving oral or topical antifungal agents (used within 4 weeks of study enrolment); 3) participants a history of serious cardiovascular, respiratory, endocrine system, and central nervous system diseases, or who have been diagnosed with a mental illness that can significantly affect this clinical trial; 4) participants who are judged unsuitable for participation in clinical trials by other investigators. The study protocol was approved by the Institutional Review Board (IRB) of Chung-Ang University Hospital (C2010003, 298), and informed consent was obtained according to the IRB’s policy. Because of the lack of previous data, the sample size was determined based on feasibility considerations rather than power analysis. A total of 234 skin swab samples were collected from the antecubital area from both groups using the skin swab method. The detailed swab method was as follows: The cotton swabs premoistened in sterilized saline were repeated rubbed at least 10 times against the lesional sites of the antecubital fossae of AD patients and healthy individuals. The cotton swabs used in skin swab were stored with 500 µl of lysis buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0], 1% sodium dodecyl sulfate, 2% Triton X-100). Subsequently, the DNAs are extracted and used for amplification.
Real-time qPCR screening using pan-fungal primers
Real-time qPCR screening was performed for comparative analysis of fungal distribution between AD patients and healthy individuals. To perform amplification, we used pan-fungal primers (Table 1)10,11,12, which are universally selected for targeting the fungal ITS1 region. Real-time qPCR was conducted with iTaq Universal SYBR Green PCR master mix (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions on a CFX96 real-time PCR detection system. The amplicons were purified using a PCR purification kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The purified amplicons were sequenced using BigDye® Terminator V3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) with forward and reverse primers initially used in PCR amplification. To ascertain species identity, the characterized sequence was analyzed using BLASTn to align sequence similarity with intended target genes in NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table 1. Primers used in the real-time qPCR assay.
| Name | Sequence (5’-3’) | Characterization |
|---|---|---|
| ITS1 F | TGC GTT CTT CAT CGA TGC GA | Pan-fungal primer (Forward) |
| ITS4 R | TAA GCG CAA GTC ATC AGC TTG CGT | Pan-fungal primer (Reverse) |
| ITS1 F | TAA GCG CAA GTC ATC AGC TTG CGT T | Pan-Candida primer (Forward) |
| 5.8S1R | TGC GTT CTT CAT CGA TGC GA | Pan-Candida primer (Reverse) |
| ITS1 F2 | SSC CCC ATT CTT GTC TAC MTY AC | Trichophyton detection (Forward) |
| ITS R2 | AAC GCT CAG ACT GAC AGC TCT TC | Trichophyton detection (Reverse) |
| MC F | CCT AAG CGG TGG GTG GTT ACT | Microsporum detection (Forward) |
| MC R | TGA AAG AAC ATA CCG TCT GAG CG | Microsporum detection (Reverse) |
Real-time qPCR screening using genus-specific primers
We carried out further analysis with genus-specific primers as compensation for the possibly poor sensitivity. Pan-candida primer pair, designed for amplifying a variable segment of 28S ribosomal gene of Candida spp. was used for Candida spp. detection. For dermatophytes detection, a genus-specific primer for Trichophyton and Microsporum was designed, which are listed in Table 1. The amplicons were analyzed in the same way as in pan-fungal primer amplification.
Nested PCR and direct sequencing for Malassezia spp. detection
Cutaneous Malassezia communities were comparatively analyzed between AD patients and healthy individuals by conventional nested PCR due to the low concentration of DNA. In the first-round amplification, the fungal ITS1 region was amplified by using a universal primer pair (Table 2)13, and 1 µl of the first-round PCR product was added as template DNA in the second-round amplification. We used 7 Malassezia species-specific primers in the second-round amplification. After amplification, the PCR product was identified on 2% agarose gel, following purification using a PCR purification kit (Qiagen). Purified PCR products were sequenced using BigDye® Terminator V3.1 cycle sequencing kit (Applied Biosystems) with primers used in second-round PCR amplification. For more accurate results, the characterized sequence was analyzed using BLASTn to align sequence similarity with intended target genes in NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table 2. Primers used in the nested PCR assay.
| Name | Sequence (5’-3’) | Characterization | |
|---|---|---|---|
| First-round amplification | |||
| ITS1 | TCC GTA GGT GAA CCT GCG G | ITS target primer (Forward) | |
| ITS4 | TCC TCC GCT TAT TGA TAT GC | ITS target primer (Reverse) | |
| Second-round amplification | |||
| M. restricta F | CTT GGT TGG ACC GTC ACT GG | Malassezia restricta detection (Forward) | |
| M. restricta R | AGG CGG ATG CAA AGT GTC TC | M. restricta detection (Reverse) | |
| M. globosa F | CAA TAA GTG TGT CTC TGC GG | Malassezia globosa detection (Forward) | |
| 5.8S R | TTC GCT GCG TTC TTC ATC GA | M. globosa detection (Reverse) | |
| M. sympodialis F | CGG ACG CAA ACA CGT CTC TG | Malassezia sympodialis detection (Forward) | |
| 5.8S R | TTC GCT GCG TTC TTC ATC GA | M. sympodialis detection (Reverse) | |
| M. furfur F | CTA CTC GCG TAC AAC GTC TCT G | Malassezia furfur detection (Forward) | |
| 5.8S R | TTC GCT GCG TTC TTC ATC GA | M. furfur detection (Reverse) | |
| M. pachydermatis F | CTG CCA TAC GGA TGC GCA AG | Malassezia pachydermatis detection (Forward) | |
| 5.8S R | TTC GCT GCG TTC TTC ATC GA | M. pachydermatis detection (Reverse) | |
| M. obtusa F | ACC CGT GTG CAC ACT GTT GAG | Malassezia obtusa detection (Forward) | |
| 5.8S R | TTC GCT GCG TTC TTC ATC GA | M. obtusa detection (Reverse) | |
| M. slooffiae F | ACG CAC GCT AAC ACA ACG TG | Malassezia slooffiae detection (Forward) | |
| 5.8S R | TTC GCT GCG TTC TTC ATC GA | M. slooffiae detection (Reverse) | |
The primer sequences shown in this table were previously published13.
Statistical analysis
All samples from all participants were used in statistical analysis. The chi-square test was used for statistical analysis, and the significance levels were set at p<0.05. Datasets were entered into Microsoft Excel 2016 (Microsoft, Redmond, WA, USA) and analyzed using R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
Distribution of Candida
The abundance and diversity of Candida spp. in the skin were higher in AD patients (10.90%; 23/211) than healthy controls (0%) (Fig. 1). We also confirmed that the pan-fungal primer was much more sensitive than the pan-candida primer for Candida detection (data not shown). The most frequently detected species in the AD group was Candida albicans (5.21%; 11/211), a representative clinically isolated species. Candida parapsilosis was second-most frequent among AD patients (3.79%; 8/211). Four more samples from AD patients tested positive for Candida, but we could not characterize the exact species because of the low homology score despite further sequencing analysis including of the D1/D2 domain (Table 3).
Fig. 1. Detection frequency of Candida spp., dermatophytes, and Malassezia spp. in healthy controls and patients with atopic dermatitis (AD). Candida spp. were detected in 10.90% AD samples and 0% samples from healthy controls. Dermatophytes were detected in 1.42% AD samples and 0% samples from healthy controls. Malassezia spp. were detected most frequently in 20.85% AD samples and 8.70% samples from healthy controls.
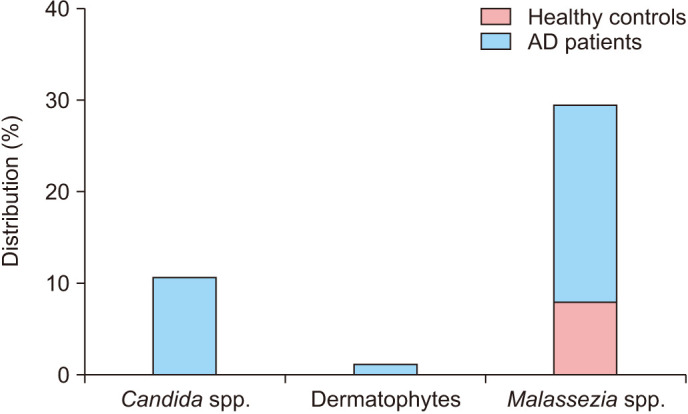
Table 3. Distribution of Candida, dermatophytes, and Malassezia in atopic dermatitis patients and healthy controls.
| Distribution | Species | Atopic dermatitis (n=211) | Healthy control (n=23) |
|---|---|---|---|
| Candida spp. | Total | 23 (10.90) | - |
| Candida albicans | 11 (5.21) | - | |
| Candida parapsilosis | 8 (3.79) | - | |
| Candida spp. | 4 (1.90) | - | |
| Dermatophytes | Total | 3 (1.42) | - |
| Trichophyton rubrum | 2 (0.95) | - | |
| Microsporum canis | 1 (0.47) | - | |
| Malassezia spp. | Total | 44 (20.85) | 2 (8.70) |
| Malassezia restricta | 32 (15.17) | 2 (8.70) | |
| Malassezia globosa | 8 (3.79) | - | |
| Malassezia furfur | 3 (1.42) | - | |
| Malassezia sympodialis | 1 (0.47) | - | |
| Malassezia pachydermatis | - | - | |
| Malassezia obtusa | - | - | |
| Malassezia slooffiae | - | - |
Values are presented as number (%). Cutaneous distribution of Candida, dematophytes, Malassezia from atopic dermatitis patients and healthy individuals is shown in this table. Cutaneous fungal distribution between two groups were not statistically significant (Candida spp.: not applicable, dermatophyte: not applicable, Malassezia spp.: p=0.163575).
Distribution of dermatophytes
Three samples from AD patients tested positive for dermatophytes, whereas none in the group of healthy individuals tested positive (Table 3). Unlike for Candida, the genus-specific primer was more effective to detect dermatophytes than a pan-fungal primer. In the AD patient group, Trichophyton rubrum was detected in two samples, and Microsporum canis was detected in one (Table 3).
Distribution of Malassezia
We comparatively analyzed 7 Malassezia species between AD patients and healthy controls. Overall, 20.85% (44/211) AD patients were identified to have Malassezia spp. In the case of healthy controls, only 8.70% (2/23) had Malassezia (Fig. 1). Both the species abundance and diversity of Malassezia were higher in AD patients than in healthy individuals, but this was not statistically significant (p=0.163575). Malassezia restricta was the most frequently detected species in AD patients (15.17%; 32/211) and simultaneously the only found species in healthy controls (Table 3). In AD patients, Malassezia globosa was detected second-most frequently (3.79%; 8/211), followed by Malassezia furfur and Malassezia sympodialis (1.42% and 0.47%, respectively) (Table 3). Malassezia pachydermatis, Malassezia obtusa, or Malassezia slooffiae was not identified in both groups.
DISCUSSION
Recent research has focused on the role of microbes in AD pathogenesis. While bacteria have been studied extensively, the effects of fungi have been overlooked4. Fungi are not a direct causative agent of AD because they are present in the normal skin of healthy individuals, but they appear to contribute to the onset and exacerbation of AD14,15,16. A change in the composition of the fungal community in lesional or non-lesional skin of AD patients has been demonstrated in local and global trials8,9,17. This change could be because of the altered physiological skin environment as AD progresses. For example, AD patients showed epidermal barrier dysfunction resulting from filaggrin mutation18,19. The weakened skin barrier of AD patients is more sensitive to microbial infections and allergenic reactions. The intact healthy skin produces antimicrobial peptides such as defensins and maintains pH weakly acidic thereby protecting it from external environments3,20. However, the skin of AD patients is deficient in this respect, resulting in alteration of skin microbial flora21,22.
We comparatively analyzed three representative AD-associated fungi between AD patients and healthy individuals by conducting a PCR-based analysis. Candida, a commensal yeast that mainly colonizes mucosal surfaces, is thought to worsen AD via the secretion of allergens and antigens23. Candida has been cultured more frequently from the gastrointestinal tract of AD patients than healthy individuals, but there is little information about its skin colonization23,34. In the present study, we confirmed that the skin distribution of Candida spp. is higher in AD patients than in healthy individuals. As a representative resident flora, Candida does not directly trigger AD progression, but impaired immune systems of AD patients might be susceptible to Candida infections. Dermatophytes are also known to contribute to AD; however, the association is still largely unknown. Jones et al.25 found that chronic dermatophyte infections are more common in AD patients, and cases of chronic dermatophyte infection are more severe and difficult to treat. Although AD patients with dermatophyte infections may show improvement when treated with antifungal drugs, a more detailed link between dermatophytes infection and AD progression remains to be established26. Malassezia spp. are the main eukaryotes composing the microbial flora in normal human skin27. In healthy individuals, colonization of Malassezia spp. accounts for 53% to 80% of all fungi at different locations28,29. Malassezia is also associated with several human skin diseases and disorders especially, Malassezia restricta is a predominant species in patients with seborrheic dermatitis and AD30,31. M. restricta and M. globosa are thought to play a major role in AD development because these two Malassezia species are abundantly present in almost all AD patients, thus supporting our findings. The prevalence of Malassezia is commonly studied in sebum-rich areas such as the scalp, chest, and back because of their lipophilic nature15,32. However, in this study, all swab samples were collected from the antecubital area because antecubital fossa is the most common site of AD and an easy area to collect samples. In addition, skin microbiome shows phylogenic diversity according to body parts. Therefore, we conducted the study by limiting the sample collection area to the antecubital area in both the patient group and the control group.
A hallmark of human skin microbiota communities is high diversity and high interpersonal variation. However, the skin microbiota of a healthy adult remains stable over time, despite environmental perturbations. Adults stably maintain the composition of their skin microbial communities as assessed for at least 2 years33. So, considering that the participants in this study were all adults, it is believed that the intra-individual fungal microbiota was not significantly affected by the environment. Costello et al.34 found that skin sites including the palms, fingers, and forearm had greater phylogenetic diversity than communities in the gut, external auditory canal, or oral cavity. Therefore, we conducted the study by limiting the sample collection area to the antecubital area in both the patient group and the control group.
There are some limitations to be considered when interpreting our findings. The number of specimens from healthy controls was relatively smaller than that from AD patients. In addition, the lack of clinical information including gender, age, and disease severity made it impossible to analyze the results reflecting it. Despite these limitations, our study has two strengths. First, our analysis included 234 many clinical samples. Second, we designed diverse primers and amplification methods according to analysis purposes.
In conclusion, we compared the distribution of Candida, dermatophytes, and Malassezia in the skin of AD patients and healthy individuals. The distribution of fungi in all three genera was altered in AD patients. In our study, both the species abundance and diversity of Malassezia were higher in AD patients than in healthy individuals. It is speculated that the impaired skin barrier in AD allows colonization with more different Malassezia than healthy skin. Vice versa, the altered mycobiome may cause activation of the skin immune system leading to inflammation and eczema. Considering the composition of normal skin microflora, fungi are not a direct causative agent of AD, but an imbalance in their composition appears to be associated with AD development or exacerbation. Further studies focusing on fungal immune response or allergenic mechanisms will be required to better understand the role of fungi in AD progression.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING SOURCE: This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT); No.2020R1A5A1018052 and No. 2019R1A2C1090226.
DATA SHARING STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann Nutr Metab. 2015;66 Suppl 1:8–16. doi: 10.1159/000370220. [DOI] [PubMed] [Google Scholar]
- 2.Hanifin JM. Epidemiology of atopic dermatitis. Monogr Allergy. 1987;21:116–131. [PubMed] [Google Scholar]
- 3.Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat Rev Dis Primers. 2018;4:1. doi: 10.1038/s41572-018-0001-z. [DOI] [PubMed] [Google Scholar]
- 4.Thammahong A, Kiatsurayanon C, Edwards SW, Rerknimitr P, Chiewchengchol D. The clinical significance of fungi in atopic dermatitis. Int J Dermatol. 2020;59:926–935. doi: 10.1111/ijd.14941. [DOI] [PubMed] [Google Scholar]
- 5.Geoghegan JA, Irvine AD, Foster TJ. Staphylococcus aureus and atopic dermatitis: a complex and evolving relationship. Trends Microbiol. 2018;26:484–497. doi: 10.1016/j.tim.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Lee MK, Park KY, Jin T, Kim JH, Seo SJ. Rapid detection of staphylococcus aureus and methicillin-resistant S. aureus in atopic dermatitis by using the BD Max StaphSR assay. Ann Lab Med. 2017;37:320–322. doi: 10.3343/alm.2017.37.4.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lintu P, Savolainen J, Kortekangas-Savolainen O, Kalimo K. Systemic ketoconazole is an effective treatment of atopic dermatitis with IgE-mediated hypersensitivity to yeasts. Allergy. 2001;56:512–517. doi: 10.1034/j.1398-9995.2001.056006512.x. [DOI] [PubMed] [Google Scholar]
- 8.Han SH, Cheon HI, Hur MS, Kim MJ, Jung WH, Lee YW, et al. Analysis of the skin mycobiome in adult patients with atopic dermatitis. Exp Dermatol. 2018;27:366–373. doi: 10.1111/exd.13500. [DOI] [PubMed] [Google Scholar]
- 9.Edslev SM, Andersen PS, Agner T, Saunte DML, Ingham AC, Johannesen TB, et al. Identification of cutaneous fungi and mites in adult atopic dermatitis: analysis by targeted 18S rRNA amplicon sequencing. BMC Microbiol. 2021;21:72. doi: 10.1186/s12866-021-02139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Hung GC, Nagamine K, Li B, Tsai S, Lo SC. Development of Candida-specific real-time PCR assays for the detection and identification of eight medically important Candida species. Microbiol Insights. 2016;9:21–28. doi: 10.4137/MBI.S38517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyajima Y, Satoh K, Uchida T, Yamada T, Abe M, Watanabe S, et al. Rapid real-time diagnostic PCR for Trichophyton rubrum and Trichophyton mentagrophytes in patients with tinea unguium and tinea pedis using specific fluorescent probes. J Dermatol Sci. 2013;69:229–235. doi: 10.1016/j.jdermsci.2012.11.589. [DOI] [PubMed] [Google Scholar]
- 12.Brillowska-Dabrowska A, Swierkowska A, Lindhardt Saunte DM, Arendrup MC. Diagnostic PCR tests for Microsporum audouinii, M. canis and Trichophyton infections. Med Mycol. 2010;48:486–490. doi: 10.3109/13693780903312454. [DOI] [PubMed] [Google Scholar]
- 13.Sugita T, Suto H, Unno T, Tsuboi R, Ogawa H, Shinoda T, et al. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J Clin Microbiol. 2001;39:3486–3490. doi: 10.1128/JCM.39.10.3486-3490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faergemann J. Atopic dermatitis and fungi. Clin Microbiol Rev. 2002;15:545–563. doi: 10.1128/CMR.15.4.545-563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada K, Saito M, Sugita T, Tsuboi R. Malassezia species and their associated skin diseases. J Dermatol. 2015;42:250–257. doi: 10.1111/1346-8138.12700. [DOI] [PubMed] [Google Scholar]
- 16.Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S. Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol. 2016;55:494–504. doi: 10.1111/ijd.13116. [DOI] [PubMed] [Google Scholar]
- 17.Kim JE, Kim HS. Microbiome of the skin and gut in atopic dermatitis (AD): understanding the pathophysiology and finding novel management strategies. J Clin Med. 2019;8:444. doi: 10.3390/jcm8040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Regan GM, Sandilands A, McLean WHI, Irvine AD. Filaggrin in atopic dermatitis. J Allergy Clin Immunol. 2008;122:689–693. doi: 10.1016/j.jaci.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Rehbinder EM, Advocaat Endre KM, Lødrup Carlsen KC, Asarnoj A, Stensby Bains KE, Berents TL, et al. Predicting skin barrier dysfunction and atopic dermatitis in early infancy. J Allergy Clin Immunol Pract. 2020;8:664–673.e5. doi: 10.1016/j.jaip.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Hata TR, Gallo RL. Antimicrobial peptides, skin infections, and atopic dermatitis. Semin Cutan Med Surg. 2008;27:144–150. doi: 10.1016/j.sder.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 22.Leung DY. Infection in atopic dermatitis. Curr Opin Pediatr. 2003;15:399–404. doi: 10.1097/00008480-200308000-00008. [DOI] [PubMed] [Google Scholar]
- 23.Savolainen J, Lammintausta K, Kalimo K, Viander M. Candida albicans and atopic dermatitis. Clin Exp Allergy. 1993;23:332–339. doi: 10.1111/j.1365-2222.1993.tb00331.x. [DOI] [PubMed] [Google Scholar]
- 24.Buslau M, Menzel I, Holzmann H. Fungal flora of human faeces in psoriasis and atopic dermatitis. Mycoses. 1990;33:90–94. doi: 10.1111/myc.1990.33.2.90. [DOI] [PubMed] [Google Scholar]
- 25.Jones HE, Reinhardt JH, Rinaldi MG. A clinical, mycological, and immunological survey for dermatophytosis. Arch Dermatol. 1973;108:61–65. [PubMed] [Google Scholar]
- 26.Wilson BB, Deuell B, Mills TA. Atopic dermatitis associated with dermatophyte infection and Trichophyton hypersensitivity. Cutis. 1993;51:191–192. [PubMed] [Google Scholar]
- 27.Grice EA, Dawson TL., Jr Host-microbe interactions: Malassezia and human skin. Curr Opin Microbiol. 2017;40:81–87. doi: 10.1016/j.mib.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Gao Z, Perez-Perez GI, Chen Y, Blaser MJ. Quantitation of major human cutaneous bacterial and fungal populations. J Clin Microbiol. 2010;48:3575–3581. doi: 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang E, Tanaka T, Tajima M, Tsuboi R, Nishikawa A, Sugita T. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol. 2011;55:625–632. doi: 10.1111/j.1348-0421.2011.00364.x. [DOI] [PubMed] [Google Scholar]
- 30.Lee YW, Byun HJ, Kim BJ, Kim DH, Lim YY, Lee JW, et al. Distribution of malassezia species on the scalp in Korean seborrheic dermatitis patients. Ann Dermatol. 2011;23:156–161. doi: 10.5021/ad.2011.23.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagielski T, Rup E, Ziółkowska A, Roeske K, Macura AB, Bielecki J. Distribution of Malassezia species on the skin of patients with atopic dermatitis, psoriasis, and healthy volunteers assessed by conventional and molecular identification methods. BMC Dermatol. 2014;14:3. doi: 10.1186/1471-5945-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev. 2012;25:106–141. doi: 10.1128/CMR.00021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh J, Byrd AL, Park M, Kong HH, Segre JA NISC Comparative Sequencing Program. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


