Abstract
‘Candidatus Phytoplasma’ is an uncultivated, intracellular bacterial plant pathogen transmitted by phloem-feeding insect vectors. Among the group of phytoplasmas, the Peanut Witches' Broom or 16SrII group of phytoplasmas associated with various diseases cause severe crop losses every year in India. The ‘Ca. Phytoplasma sp.’ strain SS02 was associated with phyllody disease of sesame plants collected from New Delhi. The genome sequence of strain SS02 was obtained using its genomic DNA enrichment and hybrid assembly of sequences generated on Illumina and Oxford Nanopore Technologies MinION platforms. The hybrid assembly strategy generated a draft genome with 60 contigs totaling 553,228 bp of length with more than 400 × depth coverage and 95.21% of the estimated completeness. The SS02 genome draft sequence contains 465 protein-coding genes, 17 tRNA genes, and 3 rRNA genes. The availability of this draft genome also provided a foundation for genome-scale genotypic analyses.
Introduction
'Ca. Phytoplasma’ is an obligate plant pathogenic phloem-limited bacteria that lack the cell wall, the characteristic feature of the class Mollicutes (Phylum, Mycoplasmatota; formerly, Tenericutes). Many strains of 'Ca. Phytoplasma' were characterized at the genomic level; the prefix 'Candidatus Phytoplasma' is still retained due to the difficulty in growing them on the artificial medium. The phytoplasmas are associated with diseases in many crops like cereals, vegetables, fruits, oilseeds, legumes, plantations, ornamentals, and tree species in India (Rao et al. 2017). The Peanut Witches'Broom (PnWB) (16SrII-C and D) phytoplasma strains are widespread in India; reported in crops like soybean (Glycine max), papaya (Carica papaya), bamboo (Dendrocalamus strictus), rattle pods (Crotalaria juncea), mango (Mangifera indica), tomato (Lycopersicon esculentum), fodder crops like alfalfa (Medicago sativa), Napier Grass (Pennisetum purpureum), sesame (Sesamum indicum), fire cracker flower (Crossandra infundibuliformis), cowpea (Vigna unguiculata), and weeds like wild indigo (Tephrosia purpurea), tick weed (Cleome viscosa), silver cock's comb (Celosia argentea), cattle bush (Trichodesma zeylanicum), croton (Croton bonplandianum), Parthenium hysterophorus and many others with disease incidence ranging from 2 to 10% (Yadav et al. 2015a, b, 2016, 2014; Mahadevakumar et al. 2016; Thorat et al. 2016a, b, 2017a; Madhupriya et al. 2017; Rao et al. 2017; Kirdat et al. 2019, 2020c; Bhat et al. 2021; IRPCM 2004). Among these, sesame phyllody is responsible for a yield loss of up to 100% in severe cases (Rao et al. 2015). There are four phytoplasma groups (the Aster Yellows, 16SrI; PWB, 16SrII; Clover proliferation, 16SrVI and Apple proliferation,16SrIX) belonging to 'Ca. P. asteris', 'Ca. P. aurantifolia', 'Ca. P. sp.', 'Ca. P. trifolii' and 'Ca. P. phoenicium' are associated with sesame phyllody (Asghari Tazehkand et al. 2017; Catal et al. 2013; Dubey et al. 2015; Saheli et al. 2021; Venkataravanappa V et al. 2017). In India, phytoplasma groups, viz. 16SrI-B, 16SrII-C,16SrII-D, and 16SrVI-D were associated with sesame phyllody (Ikten et al. 2011; Rao et al. 2015; Thorat et al. 2017a; Phookan et al. 2019). The 16SrII-D group ('Ca. Phytoplasma sp.') is most abundantly associated with sesame phyllody and other pulse crops (Rao et al. 2017; Thorat et al. 2017b). So far, more than 40 phytoplasma strains have been genome sequenced, including four strains of 'Ca. P. aurantifolia' of PnWB group. Given the widespread occurrence of the 16SrII-D group of phytoplasmas in India, the genome sequence information of strain SS02 was obtained and discussed in the study.
DNA Preparation for Genome Sequencing Sesame (Sesamum indicum L.) plant samples exhibiting typical phyllody and witches' broom symptoms were collected from New Delhi, India. The genomic DNA was extracted from 100 mg of symptomatic leaf tissues by a CTAB method (Doyle 1990). The presence of phytoplasma was confirmed by amplifying the 16S rRNA gene using primers P1 (Deng and Hiruki 1991) and P7 (Schneider 1995) followed by a nested PCR with primers R16F2n and R16R2 (Gundersen and Lee 1996). The 16S rRNA gene sequences were obtained directly by sequencing using bacterial universal primers 343R, 704F, 907R, 1028F and 1492R (Baker et al. 2003) on ABI® 3730xl DNA Analyser. The assembled sequences were analyzed using the EzBioCloud database (Yoon et al. 2017) to search phylogenetically closest relative. Further, the strain SS02 was genome sequenced by the procedure described earlier (Kirdat et al. 2020a, b). Briefly, total nucleic acids were extracted from the strain SS02 plant sample and enriched for prokaryotic DNA selection using the NEBNext microbiome enrichment kit (Cat. No. E2612, New England BioLabs, USA). The enriched DNA of strain SS02 was amplified using illustra Ready-To-Go GenomiPhi V3 DNA amplification kits (Cat. No. 25–6601, GE Healthcare, USA). Following the manufacturer's instructions, it was sequenced on the Illumina NovaSeq 6000 platform. Simultaneously, the enriched and amplified DNA of SS02 strain was sequenced on the Oxford Nanopore Technology (ONT) MinION platform by following the manufacturer's instructions.
All Illumina reads were quality checked with FastQC v0.11.8 (Brown et al. 2017). The ONT sequencing data were base-called with quality filtering (> Q7) using GUPPY v3.5.4. All QC-passed Illumina reads (> Q30) were subjected to metagenomic assembly using MEGAHIT v1.1.3 (Li et al. 2016). This assembly was subjected to binning, using MetaBAT2 v2.12.1 (Kang et al. 2015). Raw reads were mapped on bins corresponding to phytoplasma. Additionally, Illumina reads were mapped on PnWB phytoplasma NTU2011 (AMWZ00000000) (Chung et al. 2013), 'Ca. P. aurantifolia' WBDL (MWKN00000000) and 'Ca. P. aurantifolia' NCHU2014 (CP040925) (Chang et al. 2015) genome sequences, using Bowtie2 v2.2.6 (Langmead and Salzberg 2012). Finally, all mapped reads and QC-passed ONT reads were used to generate the hybrid assembly in Unicycler v0.5.0 (Wick et al. 2017), followed by polishing using Pilon (Walker et al. 2014). Bin quality check, genome completeness, and taxonomic assignments were performed using CheckM v1.0.14 (Parks et al. 2015). Assembly quality was checked using QUAST v5.0.2 (Gurevich et al. 2013). The genome coverage was calculated using BBMap (Bushnell 2014). All scaffolds were used as the queries to run a BLASTX search against the GenBank non-redundant protein database to identify contaminating scaffolds. The genome was annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Zhao et al. 2012). All contigs and raw sequencing data were deposited in the DDBJ/ENA/GenBank database with accession number JAHBAJ020000000 under the BioSample ID SAMN19066185 BioProject ID PRJNA727971. Further, the comparison of orthologous gene clusters among SS02, WBDL, NTU2011 and NCHU2014 genomes using OrthoVenn2 (Xu et al. 2019). The eggNOG-mapper v2 was used for functional annotation (Clusters of Orthologous Groups, COG) of the PGAP annotated proteins of SS02 and closely related strains viz. WBDL, NTU2011 and NCHU2014 genomes (Huerta-Cepas et al. 2019; Cantalapiedra et al. 2021).
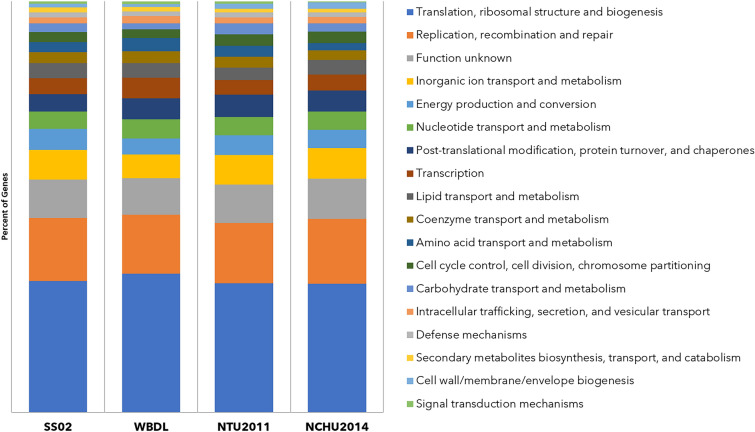
The obtained 16S rRNA gene sequences of SS02 showed 98.76% similarity with the reference sequence of 'Ca. P. aurantifolia' strain WBDL (U15442) when analyzed on EzBioCloud database (Yoon et al. 2017). The genome sequencing of the strain SS02 resulted in 42,296,107 reads (150 × 2 chemistry) using the Illumina NovaSeq 6000 platform, while the ONT MinION sequencing data generated 33,607 reads. The final SS02 genome assembly contained 60 scaffolds corresponding to 553,228 bp in length. The estimated coverage for Illumina was 407x, while that for ONT was 4.3x. The completeness of assembly was 95.21% of the estimated genome size, and the GC content was 23.55%. The SS02 genome was predicted to have 465 protein-coding genes, 17 transfer RNA (tRNA), and one rRNA operon (Table 1). The COG functional category distribution of genes in the strain SS02 revealed approximately 106 (22.79%) genes for translation, ribosomal structure, and biogenesis, 51 (10.96%) genes for replication, recombination, and repair, and 31 (6.6%) genes were assigned for putative functions as the abundant assigned categories. Approximately 83 (17.84%) genes were categorized with transport mechanisms. The distribution of gene components between SS02 and its closely related strain genomes describe the similar composition of COG functional categories as shown in Fig. 1. Further, the Orthovenn analysis assigned 465 proteins into 412 clusters shared by the strain SS02 and closely related strains (WBDL, NTU2011and NCHU2014) and 53 singletons. Figure 2 shows overlapping cluster numbers shared between SS02 and closely related species.
Table 1.
Genome statistics of 'Ca. Phytoplasma sp.' strain SS02 (JAHBAJ020000000)
| Genome statistics | SS02 | Tools used |
|---|---|---|
| Assembly | Unicycler | |
| Genome size in contigs (bp) | 553,228 | Quast |
| GC content (%) | 23.55 | Quast |
| Number of contigs | 60 | Quast |
| Contig N50 length (bp) | 30,531 | Quast |
| L50 | 7 | Quast |
| Genome Coverage | 407.962x | BBMap |
| Annotation | Tools used | |
| Protein-coding genes | 465 | PGAP |
| Genome completeness (%) | 95.21 | CheckM |
| No. of tRNA genes | 17 | tRNAscan-SE |
| No. of rRNA genes | 3 | PGAP |
Fig. 1.
The COG functional category distribution of genes in SS02 and closely related strains, viz. WBDL, NTU201, and NCHU2014 using the eggNOG-mapper v2
Fig. 2.
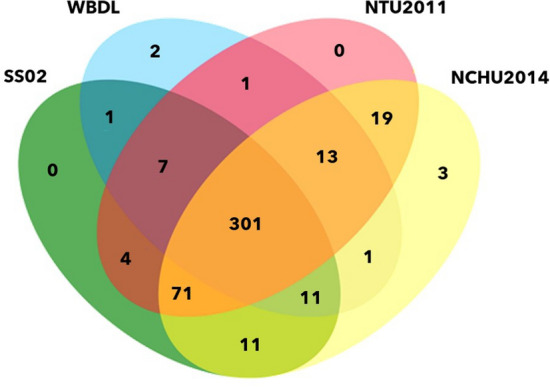
Comparison of orthologous gene clusters among SS02, WBDL, NTU2011, and NCHU2014 genomes using OrthoVenn2
Acknowledgements
The authors are thankful to the Director, ICAR-Indian Agricultural Research Institute (IARI) and Director, ICAR-National Bureau of Plant Genetic Resources (NBPGR), New Delhi, for providing facilities.
Author contributions
HR and KR collected the samples and did the primitive identification of phytoplasma in sesame samples. KK prepared the DNA samples for Illumina sequencing and performed ONT sequencing. BT assembled and analyzed the SS02 genome. HR, GR, BT, and KK wrote the first manuscript draft. AY and AS edited the manuscript; CC, KS, GR, and AY provided the funding. AY supervised the sequencing bioinformatics analysis and finalized the manuscript draft. All authors read the final draft and approved it.
Funding
The authors acknowledge the funding support by the Department of Biotechnology (DBT), Govt. of India, New Delhi, under the project BT/Ag/Network/Sesame/2019-20.
Data availability
The Whole Genome Shotgun sequence for strain SS02 have been deposited in the DDBJ/ENA/GenBank database under accession numbers JAHBAJ000000000. The version described in this paper is JAHBAJ020000000 under the BioSample ID SAMN19066185 and BioProject ID PRJNA727971.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
The first three authors Hemavati Ranebennur, Kiran Kirdat and Bhavesh Tiwarekar have contributed equally.
Contributor Information
Amit Yadav, Email: amityadav@nccs.res.in.
G. P. Rao, Email: gprao_gor@rediffmail.com
References
- Asghari Tazehkand S, Hosseinipour A, Heydarnejad J, Rahimian H, Massumi H. Identification of phytoplasmas associated with sesame phyllody disease in southeastern Iran. Arch Phytopathol Plant Prot. 2017;50:761–775. doi: 10.1080/03235408.2017.1379757. [DOI] [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Bhat S, Kepu S, Kirdat K, et al. Molecular identification of 16SrII-D phytoplasmas infecting Crossandra infundibuliformis in India. Phytopathog Mollicutes. 2021;11:23–30. doi: 10.5958/2249-4677.2021.00003.7. [DOI] [Google Scholar]
- Brown J, Pirrung M, McCue LA. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics. 2017;33:3137–3139. doi: 10.1093/bioinformatics/btx373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. BBMap: a fast, accurate, splice-aware aligner. Berkeley: Lawrence Berkeley National Lab; 2014. [Google Scholar]
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, et al (2021) eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. bioRxiv [DOI] [PMC free article] [PubMed]
- Catal M, Ikten C, Yol E, Üstün R, Uzun B. First report of a 16SrIX group (pigeon pea witches'-broom) phytoplasma associated with sesame phyllody in Turkey. Plant Dis. 2013;97:835–835. doi: 10.1094/PDIS-11-12-1100-PDN. [DOI] [PubMed] [Google Scholar]
- Chang SH, Cho ST, Chen CL, et al. Draft genome sequence of a 16SrII-A subgroup phytoplasma associated with purple coneflower (Echinacea purpurea) witches' broom disease in Taiwan. Genome Announc. 2015;3:6–7. doi: 10.1128/genomeA.01398-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W, Chen L, Lo W, Lin C, Kuo C. Comparative analysis of the peanut witches'-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLoS One. 2013;8:e62770. doi: 10.1371/journal.pone.0062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Hiruki C. Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. J Microbiol Methods. 1991;14:53–61. doi: 10.1016/0167-7012(91)90007-D. [DOI] [Google Scholar]
- Doyle JJ. Isolation of plant DNA from fresh tissue. Focus (madison) 1990;12:13–15. [Google Scholar]
- Dubey D, Rao G, Baranwal V, Sharma P. Molecular characterization of 'Candidatus Phytoplasma asteris' subgroup IB associated with sesame phyllody disease and identification of its natural vector and weed reservoir in India. Aust Plant Pathol. 2015;44:289–297. doi: 10.1007/s13313-015-0345-8. [DOI] [Google Scholar]
- Gundersen DE, Lee IM. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol Mediterr. 1996;35:144–151. [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: a quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Szklarczyk D, Heller D, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikten C, Yol E, Çatal M, Uzun B (2011) Frequency distribution of sesame phyllody infected by phytoplasmas in Antalya, Turkey. Emerg phytoplasma Dis stone fruits other Crop their possible impact EU Ctries 25
- IRPCM Phytoplasma/Spiroplasma Working Team-Phytoplasma Taxonomy Group 'Candidatus Phytoplasma', a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int J Syst Evol Microbiol. 2004;54:1243–1255. doi: 10.1099/ijs.0.02854-0. [DOI] [PubMed] [Google Scholar]
- Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirdat K, Thorat V, Ghole T, et al. First report of an association of multi-species phytoplasmas with Mango Phyllody and Little Leaf disease from western India. Plant Dis. 2019;103:2666. doi: 10.1094/PDIS-03-19-0599-PDN. [DOI] [Google Scholar]
- Kirdat K, Tiwarekar B, Thorat V, et al. Draft genome sequences of two phytoplasma strains associated with sugarcane grassy shoot (SCGS) and Bermuda grass white leaf (BGWL) diseases. Mol Plant-Microbe Interact. 2020;33:715–717. doi: 10.1094/MPMI-01-20-0005-A. [DOI] [PubMed] [Google Scholar]
- Kirdat K, Tiwarekar B, Thorat V, et al. 'Candidatus Phytoplasma sacchari', a novel taxon-associated with Sugarcane Grassy Shoot (SCGS) disease. Int J Syst Evol Microbiol. 2020;20:ijsem004591. doi: 10.1099/ijsem.0.004591. [DOI] [PubMed] [Google Scholar]
- Kirdat K, Tiwarekar B, Thorat V, et al. First report of association of a 16SrII group phytoplasma with a witches' broom disease of Croton bonplandianum. Phytopathog Mollicutes. 2020;10:100–103. doi: 10.5958/2249-4677.2020.00013.4. [DOI] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Luo R, Liu C-M, et al. MEGAHIT v1. 0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Madhupriya YA, Thorat V, Rao GP. Molecular detection of 16SrI-B and 16SrII-D subgroups of phytoplasma associated with flat stem and witches' broom disease of Celosia argentea L. 3 Biotech. 2017;7:3–7. doi: 10.1007/s13205-017-0962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevakumar S, Thorat V, Yadav V, et al. First report on the occurrence of 'Candidatus Phytoplasma aurantifolia' (16SrII-D) associated with virescence and phyllody disease of china aster in India. Plant Dis. 2016;101:241. doi: 10.1094/PDIS-06-16-0881-PDN. [DOI] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, et al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phookan J, Kalita MK, Rahman S, et al. Identification of sesame phyllody transmitting insect vectors in Assam, India. Phytopathog Mollicutes. 2019;9:107–108. doi: 10.5958/2249-4677.2019.00054.9. [DOI] [Google Scholar]
- Rao GP, Kumar A, Baranwal VK. Classification of the sesame phytoplasma strains in India at the 16Sr subgroup level. J Plant Pathol. 2015;97:25. [Google Scholar]
- Rao G, Madhupriya VT, Manimekalai R, et al. A century progress of research on phytoplasma diseases in India. Phytopathog Mollicutes. 2017;7:1–38. doi: 10.5958/2249-4677.2017.00001.9. [DOI] [Google Scholar]
- Salehi M, Faghihi MM, Ebadi N, Salehi E. First report of a' Candidatus Phytoplasma phoenicium'-related strain (16SrIX-C) associated with phyllody disease of Reseda lutea. Aust Plant Dis Notes. 2021;16:1–4. doi: 10.1007/s13314-021-00445-9. [DOI] [Google Scholar]
- Schneider B. Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasma. Mol Diagn Proced Mycoplasmol. 1995;1:369–380. doi: 10.1016/B978-012583805-4/50040-6. [DOI] [Google Scholar]
- Thorat V, Bhale U, Sawant V, et al. Alternative weed hosts harbors 16SrII group phytoplasmas associated with little leaf and witches' broom diseases of various crops in India. Phytopathog Mollicutes. 2016;6:50. doi: 10.5958/2249-4677.2016.00009.8. [DOI] [Google Scholar]
- Thorat V, More V, Jadhav P, et al. First report of a 16SrII-D group phytoplasma associated with witches’-broom disease of soybean (Glycine max) in Maharashtra. India Plant Dis. 2016;100:2521. doi: 10.1094/PDIS-05-16-0741-PDN. [DOI] [Google Scholar]
- Thorat V, Kirdat K, Takawale P, Yadav A. First report of 16SrII-D phytoplasmas associated with fodder crops in India. Phytopathog Mollicutes. 2017;7:106–110. doi: 10.5958/2249-4677.2017.00015.9. [DOI] [Google Scholar]
- Venkataravanappa V, Reddy CNL, Manjunath M, et al. Detection, characterization and in-silico analysis of 'Candidatus Phytoplasma australasia' associated with phyllody disease of sesame. Adv Plants Agric Res. 2017;7(3):288–300. doi: 10.15406/apar.2017.07.00256. [DOI] [Google Scholar]
- Walker BJ, Abeel T, Shea T, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Dong Z, Fang L, et al. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A, Bhale U, Thorat V, Shouche Y. First Report of new subgroup 16SrII- M 'Candidatus Phytoplasma aurantifolia’ associated with “witches broom” disease of Tephrosia purpurea in India. Plant Dis. 2014;98:990. doi: 10.1094/PDIS-11-13-1183-PDN. [DOI] [PubMed] [Google Scholar]
- Yadav A, Thorat V, Bhale U, Shouche Y. Association of 16SrII-C and 16SrII-D subgroup phytoplasma strains with witches’ broom disease of Parthenium hysterophorus and insect vector Orosius albicinctus in India. Aust Plant Dis Notes. 2015;10:31. doi: 10.1007/s13314-015-0181-2. [DOI] [Google Scholar]
- Yadav A, Thorat V, Shouche Y. Candidatus Phytoplasma aurantifolia (16SrII group) associated with witches’ broom disease of Bamboo (Dendrocalamus strictus) in India. Plant Dis. 2015;100:209. doi: 10.1094/PDIS-05-15-0534-PDN. [DOI] [PubMed] [Google Scholar]
- Yadav V, Thorat V, Collection MC, Yadav A (2016) First report of the association of the 16SrII-D phytoplasma subgroup with little leaf disease of Crotalaria in Karnataka, India, pp 5–7. 10.1094/PDIS-06-16-0888-PDN
- Yoon S-H, Ha S-M, Kwon S, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wu J, Yang J, et al. PGAP: pan-genomes analysis pipeline. Bioinformatics. 2012;28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Whole Genome Shotgun sequence for strain SS02 have been deposited in the DDBJ/ENA/GenBank database under accession numbers JAHBAJ000000000. The version described in this paper is JAHBAJ020000000 under the BioSample ID SAMN19066185 and BioProject ID PRJNA727971.