Abstract
Enterobacteria in fecal flora are often reported to be highly resistant. Escherichia coli is the main species; resistance data on other species are rare. To assess the effect of the host's environment, antimicrobial resistance was determined in fecal species of the family Enterobacteriaceae from three populations: healthy people (HP)(n = 125) with no exposure to antimicrobials for 3 months preceding sampling, university hospital patients (UP) (n = 159) from wards where the antibiotic use was 112 defined daily doses (DDD)/bed/month, and geriatric long-term patients (LTP) (n = 74) who used 1.8 DDD/bed/month. The mean length of hospital stay was 5 days for the UP and 22 months for the LTP. The isolates were identified to at least genus level, and MICs of 16 antimicrobials were determined. From the university hospital, resistance data on clinical Enterobacteriaceae isolates were also collected. Resistance data for on average two different isolates per sample (range, 1 to 5) were analyzed: 471 E. coli isolates and 261 other Enterobacteriaceae spp. Resistance was mainly found among E. coli; even in HP, 18% of E. coli isolates were resistant to two or more antimicrobial groups, with MIC patterns indicative of transferable resistance. Other fecal enterobacteria were generally susceptible, with little typically transferable multiresistance. Clinical Klebsiella and Enterobacter isolates were significantly more resistant than fecal isolates. The resistance patterns at both hospitals mirrored the patterns of antibiotic use, but LTP E. coli isolates were significantly more resistant than those from UP. Conditions permitting an efficient spread may have been more important in sustaining high resistance levels in the LTP. E. coli was the main carrier of antimicrobial resistance in fecal flora; resistance in other species was rare in the absence of antimicrobial selection.
High frequencies of antimicrobial resistance have been found in enterobacteria, in fecal flora as well as in clinical isolates. Escherichia coli isolates from numerous environments have been studied. Data on other enterobacterial species are usually found only for clinical strains, which tend to be relatively resistant. Very patchy data on other enterobacteria in the fecal flora have been published, and thus we know surprisingly little about how their resistance is acquired and maintained.
Part of the problem is the lack of specificity of the data in studies screening for resistance in fecal flora or environmental strains. They usually focus on coliforms, or aerobic gram-negative rods isolated on, e.g., MacConkey agar (8, 10, 15, 16, 27) or concentrate on E. coli (1, 14, 22). There could be differences in the spread of resistance genes in different species, which are not detected when studying coliforms. Since endogenous (chromosomal) resistance is common among enterobacteria, it is imperative to know which species are tested. For example, a high level of ampicillin resistance is very significant in E. coli, while it would be natural in most other enterobacteria. E. coli and Shigella, Salmonella, and Klebsiella spp. are the only ones generally susceptible to narrow-spectrum cephalosporins (4, 17, 18). Similarly, Proteus spp. and Providencia rettgeri are naturally resistant to tetracycline (3) and Morganella morganii is naturally resistant to sulfamethoxazole (36), while these resistance factors are a sign of transferable resistance in other species.
When studying enterobacteria isolated from food, we found that Enterobacter, Klebsiella, and Citrobacter spp. and Hafnia alvei, which were the members of the Enterobacteriaceae most commonly encountered, were susceptible to most antibiotics tested (26, 29). The lack of multidrug-resistance patterns, indicative of transferable resistance and typically seen in clinical strains, and fecal E. coli strains (27), was particularly noted. There is no evidence to show that environmental strains of a species are completely different from human strains; on the contrary, human, animal, and environmental clones have been shown to be indistinguishable in E. coli and Pseudomonas aeruginosa (25, 31, 33). Thus, the question arose as to what species were responsible for the high levels of resistance in fecal flora. We decided to do a between-species comparison of antimicrobial resistance in enterobacteria in human fecal flora. Three different human populations were included to study how resistance varies depending on the host's environment.
MATERIALS AND METHODS
Collection of isolates.
Human fecal samples were collected from three populations: (i) healthy persons (HP) that had not been treated with antibiotics for at least 3 months preceding sampling, in 1993 and 1994, as previously described (29); (ii) long-term patients (LTP) (mean duration of hospitalization, 22 months) at two medicine wards at Turku City Hospital in 1994 (13); and (iii) university hospital patients (UP) (mean duration of hospitalization, 5 days) at the Turku University Hospital in 1997. The UP, who were treated in the Departments of Medical Intensive Care, Cardiology, Gastroenterology, Infectious Diseases, Hematology, and Nephrology at the Turku University Hospital, were taking part in a screening program for carriage of fecal vancomycin-resistant enterococci; at the same time, gram-negative isolates were collected. Briefly, the samples were diluted in saline and plated onto MacConkey agar. Plates with discrete colonies were chosen, and at least five colonies per sample, or all different-looking colonies, were selected. Enterococci were ignored.
From the university hospital, data on antimicrobial consumption were obtained from the hospital pharmacy, and data on hospitalization times were taken from the hospital records. Data on overall antimicrobial use at the long-term hospital were calculated from an earlier work by our group (13); data on individual use was extracted directly from patient records.
Identification.
Isolates that were found to be gram-negative rods, oxidase negative, and able to ferment glucose, and that thus belonged to the Enterobacteriaceae, were selected for further identification. All isolates were first screened for β-glucuronidase activity and indole production; isolates positive in both tests were presumed to be E. coli and were not further tested. All others were tested for the following: β-galactosidase (with o-nitrophenyl-β-d-galactopyranoside [ONPG] discs), production of gas from glucose, citrate and urea utilization; Voges-Proskauer and methyl red; ornithine, arginine, and lysine decarboxylase; production of dihydrogen sulfide; motility; DNase activity at +25°C; pigmentation; and fermentation of the following sugars: mannitol, maltose, lactose, arabinose, sorbitol, inositol, raffinose, and trehalose. Media and reagents were from BBL (Becton Dickinson Microbiology Systems, Cockeysville, Md.), Rosco (Taastrup, Denmark), Sigma Chemical Co. (St. Louis, Mo.), and Oxoid (Unipath Ltd., Basingstoke, United Kingdom). The tests were essentially performed as described in the manufacturer manuals, and in the Clinical Microbiology Procedures Handbook (7).
The results were compared with a database, generated by combining the lists of identification of Enterobacteriaceae in Bergey's Manual of Determinative Bacteriology (6) and the Manual of Clinical Microbiology (3), using the Excel 97 spreadsheet program (Microsoft). Unclear cases, with the genus determined at a probability lower than 65%, were retested and in some instances were tested with the API-E test (bioMérieux, Lyon, France).
Determination of susceptibility.
MICs were determined by a standard agar dilution method (23) on Mueller-Hinton II medium (BBL). NCCLS breakpoints were applied. The following antimicrobials were tested (range): ampicillin (0.25–256 mg/liter), trimethoprim (0.12–1024 mg/liter), sulfamethoxazole (0.5–1024 mg/liter), chloramphenicol (2–128 mg/liter), cephalothin (0.25–64 mg/liter), cefuroxime (0.06–64 mg/liter), cefotaxime (0.06–32 mg/liter), gentamicin (0.25–64 mg/liter), tetracycline (0.12–64 mg/liter), and nalidixic acid (0.5–128 mg/liter), all from Sigma; amoxicillin-clavulanic acid (0.5–64 and 0.25–32 mg/liter, respectively; SmithKline Beecham Pharmaceuticals, Rixensart, Belgium), aztreonam (0.25–64 mg/liter, Bristol-Myers Squibb, Italy), imipenem (0.25–64 mg/liter; Merck, Sharp & Dohme, Westpoint, Pa.), ciprofloxacin (0.06–8 mg/liter; Bayer, Leverkusen, Germany).
Resistance data on clinical enterobacterial isolates.
Resistance data were collected from the same wards as those from which the patients were sampled at the Turku University Hospital, and from the same time period when the fecal samples were collected. All types of isolates were included, except fecal cultures. The data were obtained with the WHONET program (available from J. Stelling, World Health Organization/EMC, Geneva, Switzerland) from the records of the Microbiological Laboratory. These isolates had been tested with the disk diffusion method, using Oxoid disks with Iso-Sensitest medium (Oxoid). The NCCLS interpretive criteria were used (23).
REA.
Restriction enzyme analysis (REA) was performed on all trimethoprim-resistant E. coli isolates from the LTP. Bacterial DNA was isolated from overnight cultures in Luria broth by the method of Wilson (37). The DNA was cut with restriction enzymes (HindIII, EcoRI, BamHI, SacI, SalI, and PstI; Promega, Madison, Wis.) overnight with an oil overlay and run into 0.6% agarose gels at 15 to 20 V for 20 h. The patterns were visually compared.
Statistics.
The sample groups were compared in a pairwise fashion, using the chi-square analysis-of-contingency table test or Fisher's exact test as applicable. A P value of <0.05 was considered statistically significant.
RESULTS
Samples and isolates.
We studied 125 HP, 74 LTP, and 159 UP fecal samples. Of the five isolates picked per sample, isolates that appeared to be of the same strain based on biochemical and MIC profiles were excluded from the analysis. On average two isolates per sample were included in the resistance analysis. Escherichia, Klebsiella, Enterobacter, and Citrobacter were the most common genera (Table 1). A total of 471 E. coli isolates and 261 other enterobacterial isolates were studied. Data on clinical isolates from the university hospital were obtained for E. coli and Klebsiella and Enterobacter spp.
TABLE 1.
Identity of bacterial isolates
| Species | No. of isolates in sample population
|
||
|---|---|---|---|
| HP | UP | LTP | |
| Citrobacter braakii | 4 | ||
| Citrobacter diversus | 1 | 1 | |
| Citrobacter farmeri | 2 | ||
| Citrobacter freundii | 4 | ||
| Citrobacter sedlakii | 1 | 1 | |
| Citrobacter youngae | 4 | 9 | 4 |
| Citrobacter sp.a | 2 | 14 | 2 |
| Total | 11 | 35 | 6 |
| Enteric group 58 | 1 | 2 | 0 |
| Enterobacter aerogenes | 1 | 2 | |
| Enterobacter amnigenus | 1 | ||
| Enterobacter cloacae-Enterobacter dissolvens | 18 | 29 | 3 |
| Enterobacter sakazakii | 2 | ||
| Enterobacter taylorae | 1 | ||
| Enterobacter sp. | 4 | ||
| Total | 26 | 32 | 3 |
| Escherichia coli | 171 | 188 | 112 |
| Escherichia fergusonii-Escherichia hermanii | 0 | 0 | 4 |
| Hafnia alvei | 1 | 5 | 0 |
| Klebsiella ornithinolytica | 1 | 2 | |
| Klebsiella oxytoca | 8 | 9 | |
| Klebsiella planticola | 3 | ||
| Klebsiella pneumoniae | 23 | 33 | 3 |
| Klebsiella terrigena | 1 | ||
| Klebsiella sp. | 3 | 1 | 1 |
| Total | 35 | 47 | 4 |
| Kluyvera ascorbata | 1 | ||
| Kluyvera cryocrescens | 2 | ||
| Kluyvera sp. | 1 | ||
| Total | 3 | 1 | 0 |
| Morganella morganii | 2 | 1 | 0 |
| Proteus mirabilis | 1 | 3 | |
| Proteus penneri | 1 | ||
| Proteus vulgaris | 3 | ||
| Proteus sp. | 4 | ||
| Total | 0 | 9 | 3 |
| Salmonella sp. | 1 | ||
| Serratia fonticola | 2 | 5 | |
| Serratia liquefaciens | 1 | 1 | |
| Serratia marcescens | 1 | ||
| Serratia sp. | 1 | ||
| Total | 3 | 8 | 0 |
| Yersinia fredriksenii | 1 | 2 | |
| Yersinia intermedia | 1 | ||
| Yersinia sp. | 2 | ||
| Total | 2 | 2 | 2 |
Not determined to species level.
Antimicrobial use.
In the HP group, no antibiotics had been used for at least 3 months preceding sampling. At the University Hospital, β-lactams were the most frequently used (Table 2). Antimicrobial use at the long-term hospital was comparatively low, but individual total use could be high. Of the LTP, 21% had been treated with trimethoprim, with 15% of patients receiving this treatment for >14 days, sometimes for yearlong periods as urinary tract infection prophylaxis.
TABLE 2.
Characteristics of the wards sampled at the two hospitals
| Characteristic | University Hospital | Long-term hospital |
|---|---|---|
| Length of stay | ||
| mean | 5.1 days | 22 mo |
| max | 55.2 days | 137 mo |
| min | 1 mo | |
| median | 3 days | |
| No. of beds | 122 | 74 |
| Antibiotic group (DDDd/bed/month) | ||
| Aminoglycosides | 7.3 | 0 |
| Penicillins, penicillin with inhibitor | 11.3 | 0.1 |
| Cephalosporins | 20.9a | 0.2 |
| Meropenem, imipenem, aztreonam | 42.3 | 0 |
| Quinolonesb | 19.4 | 0.1 |
| Trimethoprim-Sulfamethoxazole | 2.9 | 1.4 |
| Othersc | 7.5 | 0 |
| Total (DDD/bed/month) | 112 | 1.8 |
55% broad spectrum.
Mainly ciprofloxacin.
Tetracyclines, rifampin, fusidic acid, and nitrofurantoin.
DDD, defined daily doses.
Antimicrobial resistance levels.
Resistance frequencies varied from 40% resistance to trimethoprim in the LTP group, to 0 to 4% resistance to broad-spectrum β-lactams, gentamicin, and ciprofloxacin in all groups (Table 3 and 4).
TABLE 3.
Resistance frequencies in E. coli from fecal samples and clinical isolates from the university hospitala
| Antimicrobial (resistance breakpoint [mg/liter]) | HP
|
UP
|
LTP
|
C
|
Pd
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. tested | R% | No. tested | R% | No. tested | R% | No. tested | R% | HP-UP | HP-LTP | HP-C | UP-LTP | UP-C | LTP-C | |
| AMP (≥32) | 171 | 12 | 188 | 14 | 112 | 25f | 136 | 32f | 0.05 | <0.0001 | 0.03 | 0.0002 | ||
| AMC (≥32/16) | 165 | 0 | 188 | 3 | 112 | 1 | 19 | 5 | ||||||
| CEX (≥32) | 165 | 1 | 188 | 5 | 112 | 8e | 154 | 18f | 0.008 | <0.0001 | <0.0001 | 0.01 | ||
| CXM (≥32) | 170 | 0 | 188 | 1 | 112 | 3 | 146 | 1 | ||||||
| CTX (≥64) | 165 | 0 | 188 | 0 | 112 | 0 | ND | ND | ||||||
| IPM (≥16) | 165 | 0 | 188 | 0 | 112 | 0 | 139 | 0 | ||||||
| ATM (≥32) | 165 | 0 | 188 | 0 | 112 | 0 | ND | ND | ||||||
| GEN (≥16) | 165 | 0 | 188 | 3 | 112 | 0 | 151 | 1 | ||||||
| STR (≥32)b | 156 | 18 | 188 | 14 | ND | ND | ND | ND | ||||||
| CHL (≥32) | 171 | 4 | 188 | 7 | 112 | 3 | ND | ND | ||||||
| TET (≥16) | 171 | 14 | 188 | 13 | 112 | 25e | ND | ND | 0.05 | 0.2 | ||||
| SUL (≥512) | 171 | 16 | 188 | 13 | 112 | 28e | ND | ND | 0.02 | 0.002 | ||||
| TMP (≥16) | 171 | 9 | 188 | 12 | 112 | 40f | 127 | 26e | <0.0001 | 0.0001 | <0.0001 | 0.002 | 0.03 | |
| SXT (≥4/76)c | 171 | 8 | 188 | 8 | 112 | 15 | 150 | 24f | 0.0001 | <0.0001 | ||||
| NAL (≥32) | 171 | 1 | 188 | 4e | 112 | 13f | ND | ND | 0.04 | <0.0001 | 0.008 | |||
| CIP (≥4) | 171 | 0 | 188 | 0 | 112 | 0 | 143 | 4e | 0.008 | 0.006 | 0.04 | |||
Abbreviations for antimicrobial agents: AMP, ampicillin; AMC, amoxicillin-clavulanic acid; CEX, cephalothin; CXM, cefuroxime; CTX, cefotaxime; IPM, imipenem; ATM, aztreonam; GEN, gentamicin; STR, streptomycin; CHL, chloramphenicol; TET, tetracycline; SUL, sulfamethoxazole; TMP, trimethoprim; SXT, co-trimoxazole; NAL, nalidixic acid; CIP, ciprofloxacin. Other abbreviations: R%, resistance frequency; C, clinical isolates (university hospital); ND, not determined.
Set by authors, on the basis of histogram distribution.
Tested in combination only for UP and C; for HP and LTP the numbers are for strains resistant to SUL and TMP, tested separately. The UP were tested for both SUL and TMP as well as SXT resistance; the result of combining SUL and TMP resistance was the same as that produced by SXT.
Comparison between subject groups (significant values).
Significantly more resistant.
Very significantly more resistant.
TABLE 4.
Resistance frequencies in Enterobacteriaceae other than E. coli from fecal samples and clinical isolates from the university hospitala
| Antimicrobial (resistance breakpoint [mg/liter]) | HP
|
UP
|
C
|
LTP
|
P
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. tested | R% | No. tested | R% | No. tested | R% | No. tested | R% | HP-UP | HP-C | HP-LTP | UP-C | UP-LTP | C-LTP | |
| CMX (≥32)a | 83 | 2 | 143 | 26b | 62 | 26b | 22 | 0 | <0.0001 | 0.0007 | 0.002 | 0.02 | ||
| CTX (≥64) | 64 | 0 | 143 | 0 | ND | ND | 22 | 0 | ||||||
| IPM (≥16) | 83 | 1 | 143 | 0 | 61 | 0 | 22 | 0 | ||||||
| ATM (≥32) | 84 | 0 | 143 | 3 | ND | ND | 22 | 0 | ||||||
| GEN (≥16) | 84 | 0 | 143 | 0 | 53 | 0 | 22 | 0 | ||||||
| STR (≥32) | 83 | 5 | 143 | 10 | ND | ND | ND | ND | ||||||
| CHL (≥32) | 83 | 2 | 143 | 20b | ND | ND | 22 | 5 | 0.0003 | |||||
| TET (≥16) | 83 | 2 | 134 | 11b | ND | ND | 22 | 5 | 0.02 | |||||
| SUL (≥512) | 83 | 5 | 143 | 5 | ND | ND | 22 | 14 | ||||||
| TMP (≥16) | 82 | 0 | 143 | 5 | 33 | 24b | 22 | 36b | <0.0001 | <0.0001 | 0.006 | <0.0001 | ||
| SXT (≥4/76) | 82 | 0 | 143 | 3 | 67 | 11b | 22 | 14b | 0.005 | 0.008 | ||||
| NAL (≥32) | 83 | 0 | 143 | 10b | ND | ND | 22 | 0 | 0.008 | |||||
| CIP (≥4) | 58 | 0 | 143 | 1 | 62 | 2 | 22 | 0 | ||||||
See footnotes to Table 3.
Significantly more resistant.
Resistance in E. coli compared to other enterobacteria.
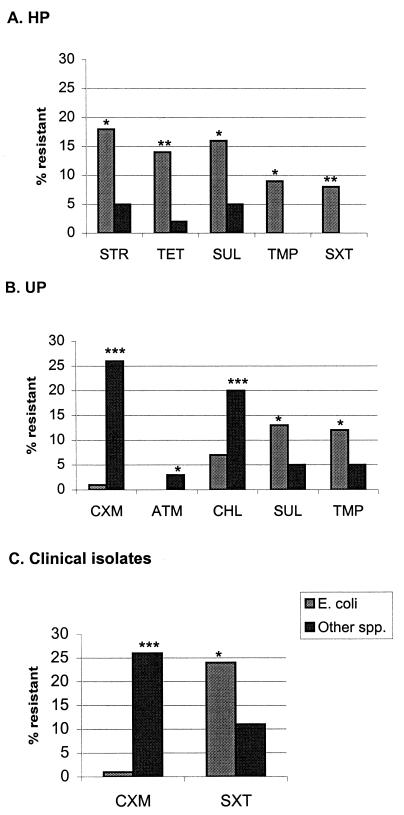
Among HP, E. coli (Table 3) was responsible for most of the resistance found. For all other enterobacterial species together (Table 4), resistance did not exceed 5% for any of the antibiotics tested (antibiotics to which these species have intrinsic resistance were not included). The difference was statistically significant for resistance to streptomycin, tetracycline, sulfamethoxazole, trimethoprim, and trimethoprim and sulfamethoxazole resistance combined (Fig. 1A).
FIG. 1.
Statistically significant differences in resistance between E. coli and other Enterobacteriaceae. Abbreviations are explained in footnote a to Table 3. ∗, P < 0.05; ∗∗, P < 0.005; ∗∗∗ P < 0.0005.
In the UP, E. coli was still slightly more resistant than the other enterobacterial species to sulfamethoxazole and trimethoprim, but the other species were more resistant than E. coli to chloramphenicol, cefuroxime, and aztreonam (Fig. 1B). Among the clinical isolates it is noteworthy that E. coli and the other species were equally resistant to trimethoprim. The only significant differences were for cefuroxime and trimethoprim-sulfamethoxazole (Fig. 1C), the latter evidently as a consequence of sulfamethoxazole resistance being rarer in the other species, since trimethoprim resistance levels were equal.
Comparison of E. coli isolates from different sources.
E. coli isolates from the HP samples were the most susceptible, while LTPs yielded the most resistant isolates (Table 3): trimethoprim resistance in particular was unusually high (40%, compared to 9% in the HP). The UP fecal samples were less resistant than the clinical isolates. The HP and the UP populations differed only in nalidixic acid resistance. In the LTP samples, nalidixic acid resistance was even higher.
In the LTP, a group (n = 10) of nalidixic acid-resistant strains with trimethoprim MICs in the range of 16 to 128 mg/liter were prominently featured. These were shown by REA to represent a single clone, which had spread to eight patients in five rooms; the samples yielding these strains were collected over 2 weeks. Thus, 22% of the LTP's trimethoprim resistance was caused by this clone.
Comparison of other enterobacteria from different sources.
A similar comparison of other Enterobacteriaceae showed a greater difference between UP and HP; a more prevalent cefuroxime, chloramphenicol, and tetracycline resistance and also nalidixic acid resistance among the UP was noticeable (Table 4). On the other hand, sulfamethoxazole resistance remained rare at 5%. In the clinical data, more trimethoprim, and trimethoprim-sulfamethoxazole resistance was seen. Although there were only 22 LTP isolates, two things are noteworthy: the resistance frequency of trimethoprim was as high as 36%, paralleling the high frequency in LTP E. coli, and cefuroxime resistance was missing, similar to the HP's other enterobacteria.
Multidrug resistance and indications of transferable resistance.
Multidrug resistance patterns among the HP, UP, and LTP populations were compared to give a picture of the prevalence of transferable resistance in the different populations. Strains resistant (not intermediately resistant) to at least two of the following antimicrobials—ampicillin (only in E. coli), tetracycline, sulfamethoxazole, trimethoprim, streptomycin, and chloramphenicol—were designated multidrug resistant. This selection was based on the fact that these resistance traits are very often found together on transferable elements, e.g., Tn21 and related transposons (21).
(i) E. coli.
The numbers of multidrug resistant E. coli strains were the same in the HP and UP populations (18 and 19%, respectively, of all isolates), and sulfamethoxazole resistance occurred in 83 and 71%, respectively, of these. In the LTP strains multidrug resistance had increased to 33% (of which 81% were sulfamethoxazole resistant). The MICs were in all cases within the highest part of the tested range for at least one of the antibiotics.
(ii) Other enterobacteria.
In the HP group only 3 isolates (4%) were multidrug resistant. In the UP group, 18 isolates (13%) were multidrug resistant, but only 9 had MIC patterns that were similar to that of the multidrug resistant E. coli. The others had low-level chloramphenicol (32 to 64 mg/liter) and tetracycline (16 mg/liter) resistance, together with a borderline nalidixic acid MIC (16 to 32 mg/liter).
DISCUSSION
We found that E. coli was the main carrier of the resistance combinations that are typically transferable in the fecal flora of all three human populations in this study. The lack of resistance in other enterobacteria was particularly noticeable in the samples from the HP. The high levels of resistance reported in fecal gram-negative flora in HP (16, 19, 27) are thus at least in Finland specific for the E. coli part of the flora. It should be noted that the high levels were maintained even in the absence of direct antimicrobial selection. Similar species differences have previously been reported in detail only by Platt et al. (30), in fecal isolates from hospital patients.
Patterns of multidrug resistance, with combinations of ampicillin, sulfamethoxazole, trimethoprim, tetracycline, streptomycin, and chloramphenicol, were seen in one-fifth of the HP and UP E. coli isolates, but they were nearly completely missing from other enterobacteria. Approximately half of all sulfamethoxazole resistance is mediated by the sulI gene (32), which has so far only been found as an integrated part of the class 1 integron structure. This is a recombinatorial hot spot for transferable resistance, often found on transposons (5). A 13 to 16% sulfamethoxazole resistance frequency in E. coli would indicate a presence of integrons in about 7% of isolates. In contrast, although other enterobacteria from the UP were more resistant than those from the HP, sulfamethoxazole resistance remained at 5%, indicating that integrons were rare. In the clinical Klebsiella and Enterobacter isolates on the other hand, trimethoprim-sulfamethoxazole resistance was much higher (11%), suggesting a fundamental difference between the way resistance is acquired in clinical and commensal isolates of species other than E. coli. Enterobacteria other than E. coli are in fact often perceived as very resistant, as a result of the spread of multidrug-resistant epidemic clones through hospitals (24, 34). In reports of resistance levels in clinical isolates, there are generally no large differences in resistance between E. coli and other enterobacteria (e.g., see reference 20). Either pathogenic strains of other enterobacteria are better at acquiring transferable resistance than their commensal counterparts or the laboratory data are somehow biased. One source of bias might be that samples yielding bacteria are commonly obtained from patients with treatment failures.
Since the isolates in this study were selected in the same way as in our previous work on resistance in bacteria from food (26, 29), it is possible to compare the data directly. (We found practically no E. coli in the food, therefore it remains unknown if resistance in strains from nature would be as common in the absence of antimicrobial selection as it is in the HP.) The resistance levels found in this study in other Enterobacteriaceae from the feces of the HP were in fact as low as those found on vegetables and in hamburger meat. There is thus no major input of resistance from these foods in Finland, but occasional transfer of resistance genes might still occur. The low levels of resistance in feces indicate that at least in the absence of antimicrobial selection, there is no enrichment of such resistance in Enterobacter, Klebsiella, and Citrobacter in fecal flora. This is no reason for complacency, however; under the right circumstances, even minority strains can be dangerous.
In the hospital environment, antimicrobial use plays an essential role in the emergence of resistant bacterial strains and, subsequently, in producing a selection pressure causing the spread of resistant clones. The effects of antibiotic use were seen in both of the study hospitals. In the university hospital, species other than E. coli had high levels of cefuroxime resistance, probably as a consequence of the extended use of β-lactams. These species generally have inducible class C β-lactamases, which frequently mutate to a derepressed state, conveying resistance to, e.g., cefuroxime (18). In this hospital environment, mutant strains would have a selective advantage. From our data, however, it is not possible to determine which factor is the most important: spread, caused by selection pressure, or de novo emergence of mutant strains. In the long-term hospital, a prolonged use of trimethoprim by 15% of the patients was probably the cause of the unusually high trimethoprim resistance level (40%) in E. coli. The trimethoprim use also caused a rise in multidrug resistance by the coselection of other resistance traits, thus causing the difference between the HP and LTP populations.
Besides antimicrobial use, other factors are evidently important for patient colonization in the hospital. A person's fecal flora is not an isolated entity but is part of the immediate environment, and all factors affecting the total bacterial flora in this environment are gradually also affecting the individual flora. This was clearly exemplified in the present study. Although the university hospital used over 60 times more systemic antimicrobials than the long-term hospital, resistance was higher at the latter. Nevertheless, the impact of the magnitude of antimicrobial therapy as compared to other factors cannot be assessed here, since there were major differences between the two inpatient groups: the UP had a shorter hospital stay (mean, 5 days), were in general younger, and had acute diseases, while the LTP (mean hospital stay, 22 months) were geriatric and had chronic diseases. Patient-to-patient transmission via staff hands could have been one major factor contributing to the spread of resistant strains on the geriatric wards. The finding of the multidrug-resistant E. coli clone which had spread through five rooms at one ward at the long-term hospital supports such an assumption.
The increase in resistance in enterobacteria in the fecal flora of hospital patients has been described by several authors (2, 11, 12, 35). Yet in those previous studies, the nature of the changes in the flora remained unclear, since isolates were not classified further than the level coliforms or aerobic gram-negative bacilli. The main reason for the increase in resistance could have been a shift in species distribution, towards species naturally more resistant; a decrease of the proportion of E. coli has in fact been observed in hospital patient flora (9). This study shows that resistance increased very markedly in the E. coli population; consequently any species shift would be only partly responsible.
In conclusion, E. coli was the main carrier of resistance in fecal flora. The apparent absence of transferable multidrug resistance in other Enterobacteriaceae species in fecal flora is intriguing, and the differences between commensal and resistant pathogenic strains should be investigated on a population level. Resistance increased as a consequence of antimicrobial use, but conditions permitting an efficient spread may have been more important in sustaining high resistance levels.
ACKNOWLEDGMENTS
We thank Minna Lamppu, Anna-Liisa Lumiaho, Tarja Laustola, and Anne Nurmi for technical assistance and Jukka Heiskanen for supplying the data from the Turku University Pharmacy. Many thanks go to Heikki Arvilommi for helpful comments on the manuscript.
This study was supported by grants from the Maud Kuistila Memorial Foundation to M.Ö. and A.H. and from the Uulo Arhio Foundation to T.L.
REFERENCES
- 1.Bonten M, Stobberingh E, Philips J, Houben A. Antibiotic resistance of Escherichia coli in fecal samples of healthy people in two different areas in an industrialized country. Infection. 1992;20:258–262. doi: 10.1007/BF01710790. [DOI] [PubMed] [Google Scholar]
- 2.Datta N. Drug resistance and R factors in the bowel bacteria of London patients before and after admission to hospital. Br Med J. 1969;2:407–411. doi: 10.1136/bmj.2.5654.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer J J. Enterobacteriaceae: introduction and identification. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of Clinical Microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 438–449. [Google Scholar]
- 4.Hakanen A, Siitonen A, Kotilainen P, Huovinen P. Increasing fluoroquinolone resistance in salmonella serotypes in Finland during 1995–1997. J Antimicrob Chemother. 1999;43:145–148. doi: 10.1093/jac/43.1.145. [DOI] [PubMed] [Google Scholar]
- 5.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 6.Holt J G, Krieg N R, Smeath P H A, Staley J T, Williams S T, editors. Bergey's manual of determinative bacteriology. 9th ed. Baltimore, Md: Williams & Wilkins; 1994. [Google Scholar]
- 7.Isenberg H D. Clinical microbiology procedures handbook. Washington, D.C.: ASM Press; 1992. [Google Scholar]
- 8.Kelch W J, Lee J S. Antibiotic resistance patterns of gram-negative bacteria isolated from environmental sources. Appl Environ Microbiol. 1978;36:450–456. doi: 10.1128/aem.36.3.450-456.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LeFrock J, Ellis C A, Weinstein L. The impact of hospitalization on the aerobic fecal microflora. Am J Med Sci. 1979;277:269–274. doi: 10.1097/00000441-197905000-00004. [DOI] [PubMed] [Google Scholar]
- 10.Leistevuo T, Leistevuo J, Österblad M, Arvola T, Toivonen P, Klaukka T, Lehtonen A, Huovinen P. Antimicrobial resistance of fecal aerobic gram-negative bacilli in different age groups in a community. Antimicrob Agents Chemother. 1996;40:1931–1934. doi: 10.1128/aac.40.8.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leistevuo T, Österblad M, Toivonen P, Kahra A, Lehtonen A, Huovinen P. Colonization of resistant faecal aerobic gram-negative bacilli among geriatric patients in hospital and the community. J Antimicrob Chemother. 1996;37:169–173. doi: 10.1093/jac/37.1.169. [DOI] [PubMed] [Google Scholar]
- 12.Leistevuo T, Österblad M, Toivonen P, Kuistila M, Huovinen S, Heikkilä E, Kahra A, Lehtonen A, Huovinen P. Increase of antimicrobial resistance of faecal aerobic gram-negative bacteria in a geriatric hospital. Age Ageing. 1996;25:197–200. doi: 10.1093/ageing/25.3.197. [DOI] [PubMed] [Google Scholar]
- 13.Leistevuo T, Toivonen P, Österblad M, Kuistila M, Kahra A, Lehtonen A, Huovinen P. Problem of antimicrobial resistance of fecal aerobic gram-negative bacilli in the elderly. Antimicrob Agents Chemother. 1996;40:2399–2403. doi: 10.1128/aac.40.10.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lester S C, del Pilar Pla M, Wang F, Perez Schael I, Jiang H, O'Brien T F. The carriage of Escherichia coli resistant to antimicrobial agents by healthy children in Boston, in Caracas, Venezuela, and in Qin Pu, China. N Engl J Med. 1990;323:285–289. doi: 10.1056/NEJM199008023230501. [DOI] [PubMed] [Google Scholar]
- 15.Levy S B. Antibiotic-resistant bacteria in food of man and animals. In: Woodbine M, editor. Antimicrobials and agriculture. London, United Kingdom: Butterworth; 1984. pp. 525–531. [Google Scholar]
- 16.Levy S B, Marshall B, Schluederberg S, Rowse D, Davis J. High frequency of antimicrobial resistance in human fecal flora. Antimicrob Agents Chemother. 1988;32:1801–1806. doi: 10.1128/aac.32.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livermore D M. Are all β-lactams created equal? Scand J Infect Dis. 1996;101:33–43. [PubMed] [Google Scholar]
- 18.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.London N, Nijsten R, van de Bogaard A, Stobberingh E. Antibiotic resistance of faecal Enterobacteriaceae isolated from healthy volunteers, a 15-week follow-up study. J Antimicrob Chemother. 1993;32:83–91. doi: 10.1093/jac/32.1.83. [DOI] [PubMed] [Google Scholar]
- 20.Manninen R, Auvinen H, Huovinen P The Finnish Study group for Antimicrobial Resistance (FiRe) Resistance to second- and third-generation cephalosporins among Escherichia coli and Klebsiella species is rare in Finland. Clin Microbiol Infect. 1997;3:408–413. doi: 10.1111/j.1469-0691.1997.tb00276.x. [DOI] [PubMed] [Google Scholar]
- 21.Martinez E, de la Cruz F. Genetic elements involved in Tn21 site-specific integration, a novel mechanism for the dissemination of antibiotic resistance. EMBO J. 1990;9:1275–1281. doi: 10.1002/j.1460-2075.1990.tb08236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray B E, Mathewson J J, DuPont H L, Ericsson C D, Reves R R. Emergence of resistant fecal Escherichia coli in travellers not taking prophylactic antimicrobial agents. Antimicrob Agents Chemother. 1990;34:515–518. doi: 10.1128/aac.34.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards (ed.). 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Neuwirth C, Siebor E, Lopez J, Pechinot A, Kazmierczak A. Outbreak of TEM-24-producing Enterobacter aerogenes in an intensive care unit and dissemination of the extended-spectrum β-lactamase to other members of the family Enterobacteriaceae. J Clin Microbiol. 1996;34:76–79. doi: 10.1128/jcm.34.1.76-79.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Österblad M, Kilpi E, Hakanen A, Palmu L, Huovinen P. Antimicrobial resistance of enterobacteria isolated from minced meat. J Antimicrob Chemother. 1999;44:298–299. doi: 10.1093/jac/44.2.298. [DOI] [PubMed] [Google Scholar]
- 27.Österblad M, Leistevuo J, Leistevuo T, Järvinen H, Pyy L, Tenovuo J, Huovinen P. Antimicrobial and mercury resistance in aerobic gram-negative bacilli in fecal flora among persons with and without dental amalgam fillings. Antimicrob Agents Chemother. 1995;39:2499–2502. doi: 10.1128/aac.39.11.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Österblad M, Leistevuo T, Huovinen P. Screening for antimicrobial resistance in fecal samples by the replica plating method. J Clin Microbiol. 1995;33:3146–3149. doi: 10.1128/jcm.33.12.3146-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Österblad M, Pensala O, Peterzéns M, Helenius H, Huovinen P. Antimicrobial susceptibility of Enterobacteriaceae isolated from vegetables. J Antimicrob Chemother. 1999;43:503–509. doi: 10.1093/jac/43.4.503. [DOI] [PubMed] [Google Scholar]
- 30.Platt D J, Chesham J S, Kristinsson K G. R-plasmid transfer in vivo: a prospective study. J Med Microbiol. 1986;21:325–330. doi: 10.1099/00222615-21-4-325. [DOI] [PubMed] [Google Scholar]
- 31.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rådström P, Swedberg G, Sköld O. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob Agents Chemother. 1991;35:1840–1848. doi: 10.1128/aac.35.9.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Römling U, Wingender J, Müller H, Tümmler B. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl Environ Microbiol. 1994;60:1734–1738. doi: 10.1128/aem.60.6.1734-1738.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon K, Stapleton P, Xiang X, Johnson A, Beattie H, El Bakri F, Cookson B, French G. Extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains causing nosocomial outbreaks of infection in the United Kingdom. J Clin Microbiol. 1998;36:3105–3110. doi: 10.1128/jcm.36.10.3105-3110.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw E J, Datta N. Effect of stay in hospital and oral chemotherapy on the antibiotic sensitivity of bowel coliforms. J Hyg. 1973;71:529–534. doi: 10.1017/s0022172400046519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock I, Wiedemann B. Identification and natural antibiotic susceptibility of Morganella morganii. Diagn Microbiol Infect Dis. 1998;30:153–165. doi: 10.1016/s0732-8893(97)00243-5. [DOI] [PubMed] [Google Scholar]
- 37.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular microbiology. New York, N.Y: Greene Publishing and Wiley Interscience; 1988. pp. 2.4.1–2.4.5. [Google Scholar]