Abstract
Human salivary histatin-5 (Hsn-5) is a 24-residue peptide that possesses potent antifungal activity in vitro. The MUC7 gene encodes human salivary low-molecular-weight mucin (MG2). The candidacidal activity of MUC7 domain 1 (MUC7 D1, the N-terminal 51 amino acid residues of MUC7) in vitro has also been demonstrated. In this study, we have investigated the antifungal therapeutic potential of Hsn-5, its two variants, R12I/K17N and R12I/H21L, and MUC7 D1. First, these peptides were tested for activities against different clinically important fungi. We found them to possess broad-spectrum antifungal activities; specifically, most exhibited excellent in vitro activity against eight clinically important fungal strains tested, including Candida albicans and Candida glabrata and their azole-resistant counterparts and Cryptococcus neoformans and its amphotericin B-resistant counterpart. These findings also suggest that the mechanism of action of both Hsn-5 and MUC7 D1 for these fungi is different from that of amphotericin B or azole antifungal agents. Second, we examined the stability of these peptides in whole human saliva and human serum. In saliva, the Hsn-5 variants R12I/K17N and R12I/H21L and MUC7 D1 degraded at a lower rate than Hsn-5. In human serum, MUC7 D1 was also more stable than Hsn-5; both peptides were more stable in serum than in saliva. Third, we examined the cytotoxicity of these peptides using human erythrocytes and two human cell lines (KB and HSG). No (or very low) hemolytic activity was observed with any of the four peptides, even at the highest protein concentration tested (200 μM), while amphotericin B caused 100% hemolysis at only 12.5 μM. The toxic effects of Hsn-5 and MUC7 D1 toward KB and HSG cells were also much lower than that of amphotericin B as measured by trypan blue exclusion. Together, these findings indicate that the investigated peptides possess high antifungal therapeutic potential, in particular for the treatment of drug-resistant fungal strains associated with immunocompromised (particularly human immunodeficiency virus-infected) patients. The same peptides could also be used as components of artificial saliva for patients with salivary dysfunction.
Medical advances have led to increased numbers of immunocompromised patients living longer. At the same time, however, there have been dramatic increases in the incidence of fungal infections. Unfortunately, most of the currently available antifungal drugs have undesirable toxic and other side effects. Also, widespread use of the limited numbers of antifungal agents to treat infections has led to the rapid development of drug-resistant strains, which are the main cause for fungal infection treatment failures (1). Of all the strategies which have been identified to overcome resistance to antifungal drugs, the development of new antifungal drugs is likely to have the most significant future impact on the treatment as well as the management of drug-resistant fungal infections.
Histatin-5 (Hsn-5), a 24-amino-acid (aa) polypeptide secreted by human salivary glands, has been demonstrated to possess potent properties against a number of medically important fungi in vitro (7, 23). However, Hsn-5 has been shown to be very vulnerable to degradation by human saliva. This degradation was found to be mediated mainly by a trypsin-like enzyme activity, and the degraded Hsn-5 was found to be less effective in killing Candida albicans (24). In our effort to produce Hsn-5 variants with enhanced protective function and/or increased resistance to proteolysis, we produced two Hsn-5 variants fulfilling these criteria (19, 20). In the first variant, the targets of the trypsin-like enzyme, lysine-12 and arginine-17, were changed to isoleucine and asparagine, respectively (designated R12I/K17N). This variant, which possesses high candidacidal activity but not increased activity (relative to Hsn-5), appeared to be more resistant to degradation based on the fact that we were able to purify from bacterial cultures significantly higher quantities of it than of other variants or of wild-type Hsn-5. In the second variant, R12I/H21L, arginine was substituted with isoleucine and histidine was substituted with leucine. This variant was found to have a slightly lower but statistically significant effective dose than Hsn-5 for killing C. albicans.
Human submandibular-sublingual saliva also contains low- and high-molecular-weight mucins, MG2 and MG1, respectively. Previous studies implied that these mucins are structurally and functionally distinct and function in lubrication, tissue coating, digestion, and microbial interactions (13, 14). MG2 isolated from saliva is a glycoprotein that has an apparent molecular mass of ∼125 kDa and that contains about 30% protein. The cDNA that encodes the apopeptide moiety of this mucin (designated MUC7) was previously isolated and characterized by our group (3). The secreted MUC7 apopeptide is composed of 357 aa residues and has a molecular mass of 37 kDa. Domain 1 of MUC7 (MUC7 D1) constitutes the N-terminal 51 aa residues of MUC7. A sequence near the N terminus, spanning 15 aa residues (R3 to Q17), showed 53% sequence similarity with Hsn-5 and was therefore designated the histatin-like domain (5). This 15-aa histatin-like domain was chemically synthesized and tested for its candidacidal activity. The results showed that it was about sixfold less potent than Hsn-5; intact MUC7 did not show any candidacidal activity (5). Later on, the entire 51-aa MUC7 D1, which includes the 15-aa histatin-like domain, was synthesized; this domain was found to be as potent as Hsn-5 in the killing of C. albicans and its azole-resistant counterpart (J. Satyanarayana, S. Narasimhamurthy, N. Bhayani, H. Situ, L. A. Bobek, and M. J. Levine, submitted for publication). These findings suggested that MUC7 D1 could also act as an antimicrobial agent in the oral cavity and could be used as an antifungal therapeutic agent.
Therefore, the aims of this study were to determine if Hsn-5, its two variants, and MUC7 D1 are suitable antifungal therapeutic agents and/or components of artificial saliva for patients with salivary dysfunction. These goals were accomplished by investigating their (i) activities against different clinically important fungi, (ii) stability in whole human saliva and human serum, and (iii) toxicity to human red blood cells and human cell lines.
MATERIALS AND METHODS
General materials.
Trypsin, aprotinin, and phenylmethylsulfonyl fluoride (PMSF) were from Sigma Chemical Co. (St. Louis, Mo.). Azocoll was from Calbiochem-Behring Co. (La Jolla, Calif.). Sabouraud dextrose agar (SAB) was from Difco Laboratories (Detroit, Mich.). Amphotericin B (Sigma) was dissolved in dimethyl sulfoxide and diluted in phosphate-buffered saline (PBS) for experimental use. FBS, DMEM, and DMEM/F-12 were from Grand Island Biological Company (GIBCO, Grand Island, N.Y.).
Peptides.
Recombinant Hsn-5 and its two variants, R12I/K17N and R12I/H21L, were produced in Escherichia coli by use of the pET-30b(+) vector. Because the cDNAs encoding the Hsn-5 variants R12I/K17N and R12I/H21L were subcloned previously only into the pGEX-2T expression vector (20), they had to be recloned into pET-30b(+). The cloning, expression, and purification of these peptides were as described previously for the cloning of unaltered Hsn-5 into pET-30b(+) (17). Recombinant MUC7 D1 was also produced by use of the E. coli pET-30b(+) expression system. Synthetic MUC7 D1 peptide was prepared by linear solid-phase 9-fluorenylmethoxy carbonyl chemistry (Satyanarayana et al., submitted). The synthesis, purification to homogeneity (by conventional reversed-phase high-pressure liquid chromatography with two columns connected in series), and characterization of the peptide were as described elsewhere (Satyanarayana et al., submitted). The concentrations of all peptides were determined by amino acid analysis. Table 1 shows the amino acid sequences of the four peptides.
TABLE 1.
Amino acid sequences of Hsn-5, Hsn-5 variants R12I/K17N and R12I/H21L, and MUC7 D1a
| Peptide | Amino acid sequence |
|---|---|
| 1 10 20 24 | |
| Hsn-5 | D-S-H-A-K-R-H-H-G-Y-K-R-K-F-H-E-K-H-H-S-H-R-G-Y |
| R12I/K17N | D-S-H-A-K-R-H-H-G-Y-K-I-K-F-H-E-N-H-H-S-H-R-G-Y |
| R12I/H21L | D-S-H-A-K-R-H-H-G-Y-K-I-K-F-H-E-K-H-H-S-L-R-G-Y |
| 1 10 20 | |
| MUC7 D1 | E-G-R-E-R-D-H-E-L-R-H-R-R-H-H-H-Q-S-P-K- |
| 21 30 40 | |
| S-H-F-E-L-P-H-Y-P-G-L-L-A-H-Q-K-P-F-I-R- | |
| 41 50 | |
| K-S-Y-K-C-L-H-K-R-C-R |
The amino acids replaced in Hsn-5 variants are indicated in bold.
Yeast and fungal strains.
C. albicans azole-sensitive clinical isolate (2-76) and azole-resistant clinical isolate (12-99) were kindly provided by Theodore C. White (Department of Pathology, School of Public Health and Community Medicine, University of Washington, and Seattle Biomedical Research Institute, Seattle, Wash.). The azole-resistant strain was originally isolated by Spencer Redding (School of General Dentistry, University of Texas Health Sciences Center, San Antonio) from a human immunodeficiency virus-infected patient. More details regarding this isolate are described elsewhere (23). An azole-sensitive Candida glabrata strain was purchased from the American Type Culture Collection (ATCC 90030). An azole-resistant clinical isolate of C. glabrata (65C) was kindly provided by John E. Bennett (National Institute of Allergy and Infectious Diseases, Bethesda, Md.). A clinical isolate of Candida krusei was obtained from the Erie County Medical Center, Buffalo, N.Y. Amphotericin B-sensitive (CN2) and amphotericin B-resistant (CN2843) Cryptococcus neoformans strains were obtained from AIDS patients with cryptococcal meningitis and were generously provided by John H. Rex (Division of Infectious Diseases, Department of Internal Medicine, Center for the Study of Emerging and Reemerging Pathogens, University of Texas Medical School, Houston). Saccharomyces cerevisiae strain S288C was kindly provided by D. Kosman, Department of Biochemistry, State University of New York, Buffalo. All fungi were streaked and grown on SAB plates at 37°C, except for S. cerevisiae, which was grown at 30°C, until large colonies were formed. One colony was then picked and resuspended in 10 mM sodium phosphate buffer (pH 7.4), and the concentration was adjusted to 105 cells/ml for the antifungal activity assay described below.
Cell lines.
The KB cell line was obtained from ATCC (CLL-17). The HSG cell line was kindly provided by Bruce Baum at NIDCR (obtained originally as a gift from Mitsunobu Sato, Tokushima University).
Antifungal activity assay.
Twofold serial dilutions of each peptide (Hsn-5, R12I/K17N, R12I/H21L, and MUC7 D1), ranging from 100 to 1.56 μM in a 20-μl volume, were incubated with an equal volume of fungal strain (105 cells/ml) for 1.5 h at 30°C for S. cerevisiae and 37°C for the rest of the fungal strains tested in this study. At the end of the incubation, the samples were diluted 20-fold, and aliquots (∼150 cells) of each sample were plated on SAB plates as described previously (15). The loss of cell viability was plotted as a function of protein concentration.
Statistical analysis.
The molar concentrations of peptides required to kill half of the maximal number of cells (ED50) and the 95% confidence limits of the ED50 were determined by the PROBIT procedure (SPSS software package 6.1.2 for Macintosh). If the 95% confidence limits of the ED50 of the analyzed peptides do not overlap, the peptides are considered to have statistically significantly different antifungal activities.
Stability of peptides in human saliva and serum.
Stimulated whole saliva was collected from 1 male and 2 female healthy individuals during fasting. Bacterium-free whole saliva was obtained by filtration with a 0.45-μm-pore-size filter. For each time point, 4 μg of peptide in 5 μl of water was incubated with 10 μl of unfiltered or filtered stimulated whole saliva at 37°C; the samples were analyzed by native cationic polyacrylamide gel electrophoresis (PAGE) and/or sodium dodecyl sulfate (SDS)-PAGE (as described in reference 15). Human serum was obtained from a healthy individual. Experiments were performed in a fashion similar to that used for saliva. Although only 1 μl of serum was used for each reaction time point, due to a much higher concentration of total protein in serum than in saliva, the amount of peptide and the percentage of serum used in each reaction were the same as for saliva. When aprotinin and PMSF were added to the reactions (whole saliva or serum), they were added at 7.5 and 200 μg/ml, respectively.
Assay of proteolytic activity in saliva and serum with azocoll.
Standard assays for azocoll hydrolysis were carried out as previously described (4) but with some modifications. Azocoll (0.1 g) was added to 12.5 ml of 50 mM Tris-HCl (pH 7.5), and the mixture was stirred rapidly to obtain a uniform suspension. Portions of 0.5 ml (containing 4 mg of azocoll) were rapidly removed into microcentrifuge tubes, to which trypsin (25 to 200 ng) or saliva or serum (100 to 800 μl) was added. Control reaction tubes containing an equal amount of azocoll but no trypsin, saliva, or serum were assayed in parallel. Tubes were incubated at 37°C for 1 h (mixed every 15 min). Assays were stopped by immersing tubes in an ice-cold water bath. Tubes were then centrifuged at 13,000 rpm (Beckman microcentrifuge), and the absorbance at 595 nm was read against the control. The results of hydrolysis of azocoll by trypsin were plotted, and the proteolytic activities of saliva and serum were determined from the standard curve.
Preparation of red blood cells.
Whole blood (10 ml) from a healthy individual was collected into a conical tube containing heparin as an anticoagulant (blood group O+ was used). Erythrocytes were harvested by centrifugation for 10 min at 1,000 × g and room temperature and washed three times in PBS solution. The top layer (plasma) and the next, milky layer (buffy coat with a layer of platelets on top of it) were then carefully aspirated and discarded. The cell pellet was resuspended in 10 ml of PBS solution and mixed by gentle aspiration with a Pasteur pipette. This cell suspension was used immediately.
Hemolytic assay.
Hsn-5, Hsn-5 variants R12I/K17N and R12I/H21L, and MUC7 D1, at concentrations ranging from 1.56 to 200 μM, were incubated with an equal volume of 1% human red blood cells in PBS at 37°C for 1 h. Amphotericin B was tested simultaneously. Nonhemolytic and 100% hemolytic controls were the buffer alone and the buffer containing 1% Triton X-100, respectively. Cell lysis was monitored by measuring the release of hemoglobin at 540 nm. Percent hemolysis was calculated as follows: [(A540 of sample treated with peptide − A540 of sample treated with buffer)/(A540 of sample treated with Triton X-100 − A540 of sample treated with buffer)] × 100. The difference in the hemolytic activity of Hsn-5, R12I/K17N, R12I/H21L, MUC7 D1, and amphotericin B at each concentration was assessed by analysis of variance by use of the StatView SE+ Graphics software package on a Macintosh computer.
Cell culture. KB and HSG cells were propagated in Dulbecco's modified Eagle's medium (DMEM) and DMEM/F12 supplemented with 12% heated inactivated FBS. Approximately 70% confluent cells were used for the viability assay.
Viability assay using trypan blue dye exclusion.
KB and HSG cells were diluted to 1.5 × 104/ml with medium–12% FBS. An aliquot (75 μl) of the cell solution was incubated with an equal volume of peptides (1.5625 to 50 μM) at 37°C in a 5% CO2 atmosphere. After a 48-h exposure, cells were harvested with trypsin, and then aliquots of the 1:1 dilution of the cell suspension with 0.4% trypan blue were removed and scored for viability using a hemacytometer. The controls were cells incubated with media alone. Percentage cytolysis was calculated as (1 − amount of viable cells in the test group)/(amount of viable cells in the control group) × 100. Concentrations resulting in a 50% inhibition of growth relative to growth of control cells (IC50s) were determined by PROBIT (SPSS software package 6.1.2 for Macintosh).
RESULTS
Antifungal activity.
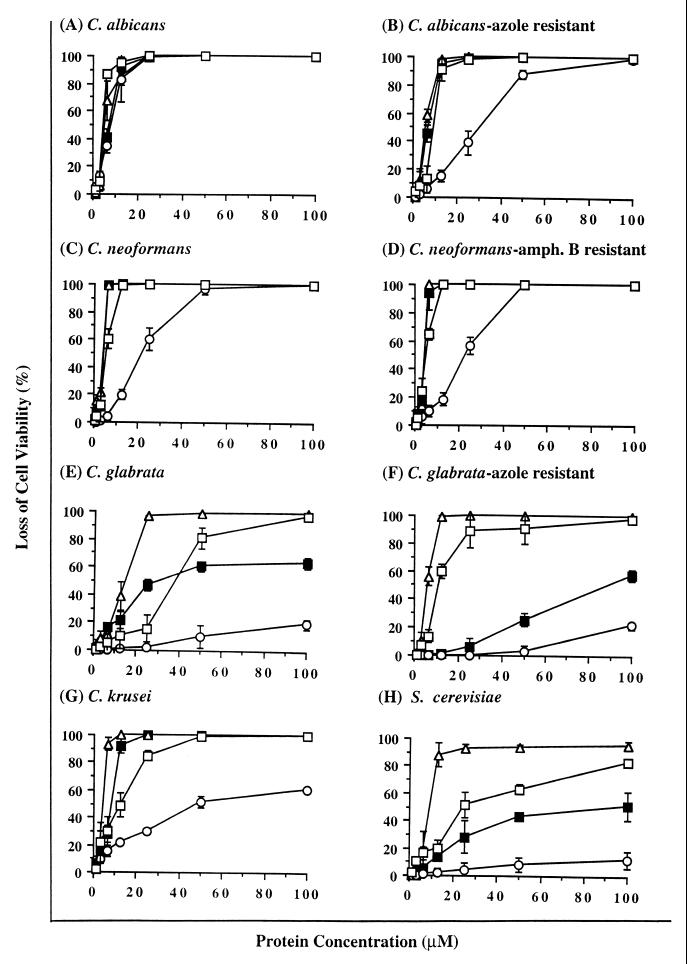
The results of antifungal activity assays of Hsn-5, Hsn-5 variants R12I/K17N and R12I/H21L, and MUC7 D1 (see Table 1 for amino acid sequences of these peptides) against eight clinically important fungal strains, including azole-resistant C. albicans and C. glabrata and amphotericin B-resistant C. neoformans, are summarized in Fig. 1 and Table 2. Figure 1 shows the loss of fungal cell viability as a function of peptide concentration, and Table 2 shows the ED50 (and the 95% confidence limits of the ED50) of the investigated peptides against various fungi. Overall, all four peptides were quite active against all fungi tested, but the C. neoformans isolates, both amphotericin B-sensitive and -resistant, were the two most susceptible isolates to killing by the four investigated peptides.
FIG. 1.
Fungicidal activities of Hsn-5 (closed squares), Hsn-5 variant R12I/H21L (open squares), Hsn-5 variant R12I/K17N (open circles), and MUC7 D1 (open triangles) against various fungi. amph., amphotericin. Results represent the mean of triplicate experiments.
TABLE 2.
ED50 of Hsn-5, Hsn-5 variants R12I/K17N and R12I/H21L, and MUC7 D1 against various fungal strains
| Fungus | ED50 [μM] (95% confidence limits)a
|
|||
|---|---|---|---|---|
| Hsn-5 | R12I/K17N | R12I/H21L | MUC7 D1 | |
| Candida albicans | 6.68 (6.05–7.37) | 6.83 (6.11–7.64) | 5.12 (4.5–6.14) | 6.02 (4.85–7.43) |
| Candida albicans (azole resistant) | 6.4 (5.59–7.32) | 24.55 (19.7–30.92) | 7.67 (5.49–10.97) | 5.24 (4.75–5.80) |
| Cryptococcus neoformans | 3.71 (1.92–5.60) | 17.2 (14.33–20.69) | 5.16 (4.25–6.32) | 3.27 (2.77–3.89) |
| Cryptococcus neoformans (amphotericin B resistant) | 3.72 (2.9–4.87) | 6.76 (5.84–7.83) | 4.56 (4.15–5.01) | 3.67 (2.8–4.52) |
| Candida glabrata | 38.70 (30.81–50.70) | >100 | 31.6 (21.38–49.85) | 11.46 (8.34–16.06) |
| Candida glabrata (azole resistant) | 85.35 (78.44–94.05) | >100 | 17.2 (14.18–20.9) | 5.5 (4.99–6.08) |
| Candida krusei | 6.47 (5.79–7.55) | 56.4 (46.87–70.10) | 9.5 (7.52–12) | 3.82 (3.49–4.19) |
| Saccharomyces cerevisiae | 74.03 (58.06–101.35) | >100 | 12.04 (9.36–15.41) | 9.73 (5.66–5.83) |
The results are based on three separate experiments, each run in triplicate.
Several more conclusions can be made on the basis of statistical analysis of the ED50 of the different peptides against the different fungi. First, the four investigated peptides showed comparable activities only against C. albicans (indicated by overlapping 95% confidence limits of their ED50). Second, three out of four peptides, Hsn-5, R12I/H21L, and MUC7 D1, possessed comparable activities against azole-resistant C. albicans and amphotericin B-sensitive and -resistant C. neoformans (indicated by overlapping 95% confidence limits of their ED50). The fourth peptide, R12I/K17N, showed lower activity against these fungal strains (the 95% confidence limits of its ED50 did not overlap the values for the other three peptides). Third, MUC7 D1 was more active than Hsn-5 and its two variants against azole-sensitive and -resistant C. glabrata, C. krusei, and S. cerevisiae (the 95% confidence limits of the MUC7 D1 ED50 against these strains did not overlap those of Hsn-5 and its two variants). Fourth, Hsn-5 variant R12I/H21L was also more active than Hsn-5 and variant R12I/K17N against azole-resistant C. glabrata and S. cerevisiae. Fifth, R12I/K17N was comparable in activity to the other three peptides against C. albicans, slightly less active against amphotericin B-resistant C. neoformans, and much less active against the other strains tested in this study.
Stability of peptides in human saliva.
The following experiments were performed using whole saliva from three donors. The results were monitored by gel electrophoresis. Because very similar results were obtained with saliva samples from all three donors, the gel patterns are shown only for saliva from one donor.
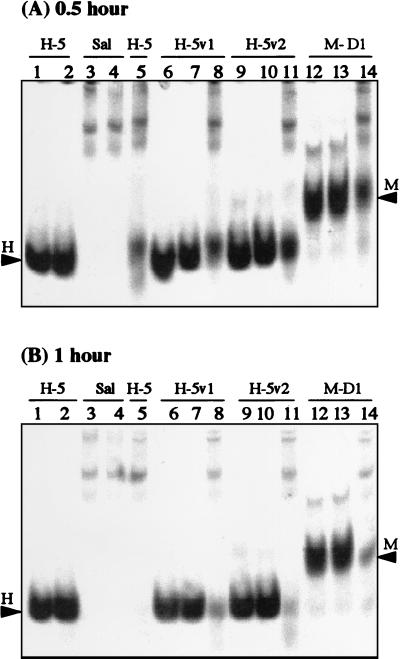
Figure 2 shows the stability of peptides in unfiltered whole saliva at the 0.5- and 1-h incubation time points, as monitored by gel electrophoresis on native gels (cationic PAGE). After incubation with saliva for 0.5 h, only portions of the added Hsn-5, R12I/K17N, R12I/H21L, and MUC7 D1 were recovered intact (Fig. 2A, lanes 5, 8, 11, and 14, respectively), with the band intensities of R12I/H21L (lane 11) and MUC7 D1 (lane 14) being slightly stronger than those of Hsn-5 (lane 5) and R12I/K17N (lane 8). After incubation with whole saliva for 1 h, no Hsn-5 was recovered, while portions of R12I/K17N, R12I/H21L, and MUC7 D1 were still intact (Fig. 2B, lanes 5, 8, 11, and 14, respectively). These results showed that in whole saliva, Hsn-5 variants and MUC7 D1 were more stable than Hsn-5.
FIG. 2.
Stability of Hsn-5 (H-5), Hsn-5 variant R12I/K17N (H-5v1), Hsn-5 variant R12I/H21L (H-5v2), and MUC7 D1 (M-D1) in whole saliva at 37°C, as monitored by 15% cationic PAGE, after incubation for 0.5 h (A) and 1 h (B). Lane 1, Hsn-5 (unincubated); lane 2, Hsn-5 (37°C); lane 3, whole saliva (Sal) (unincubated); lane 4, whole saliva (37°C); lane 5, Hsn-5 plus whole saliva (37°C); lane 6, R12I/K17N (unincubated); lane 7, R12I/K17N (37°C); lane 8, R12I/K17N plus whole saliva (37°C); lane 9, R12I/H21L (unincubated); lane 10, R12I/H21L (37°C); lane 11, R12I/H21L plus whole saliva (37°C); lane 12, MUC7 D1 (unincubated); lane 13, MUC7 D1 (37°C); lane 14, MUC7 D1 plus whole saliva (37°C). Arrowheads labeled H and M indicate the migration positions of Hsn-5 (or Hsn-5 variants) and MUC7 D1, respectively.
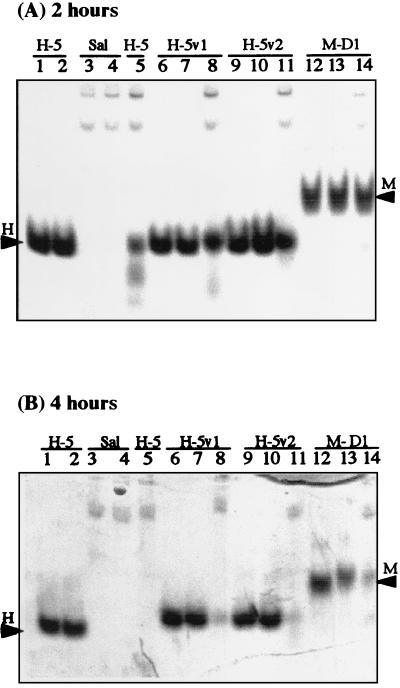
In order to determine if the “disappearance” of the investigated peptides is due at least partially to the presence of bacteria and their proteolytic enzymes in whole saliva, we also investigated their stability in filtered and thus bacterium-free saliva. Figure 3 shows the resulting patterns on native gels (cationic PAGE) with the same donor saliva as that used in Fig. 2. After incubation with filtered whole saliva for 2 h, portions of Hsn-5, R12I/K17N, R12I/H21L, and MUC7 D1 were recovered intact (Fig. 3A, lanes 5, 8, 11, and 14, respectively); in this case, however, the band intensities of both Hsn-5 variants (R12I/K17N and R12I/H21L, lanes 8 and 11) and MUC7 D1 (lane 14) were slightly stronger than that of Hsn-5 (lane 5). After incubation with filtered whole saliva for 4 h, Hsn-5 was totally gone, while portions of R12I/K17N, R12I/H21L, and MUC7 D1 were still intact (Fig. 3B, lanes 5, 8, 11, and 14, respectively). These results showed that, as in whole saliva, in bacterium-depleted saliva, Hsn-5 variants and MUC7 D1 were more stable than Hsn-5. These results also showed that all peptides were more stable in filtered saliva than in unfiltered saliva, indicating that their disappearance was due at least partially to the presence of bacteria and their proteolytic enzymes in unfiltered saliva. More specifically, no Hsn-5 was recovered after incubation with unfiltered whole saliva in about 1 h, but after 2 h of incubation with filtered whole saliva, there still was some intact Hsn-5. The band intensities of Hsn-5 variants and MUC7 D1 incubated with filtered whole saliva for 2 h were stronger than those of these peptides incubated with unfiltered whole saliva for 1 h.
FIG. 3.
Stability of Hsn-5 (H-5), Hsn-5 variant R12I/K17N (H-5v1), Hsn-5 variant R12I/H21L (H-5v2), and MUC7 D1 (M-D1) in filtered whole saliva at 37°C, as monitored by 15% cationic PAGE, after incubation for 2 h (A) and 4 h (B). Sample loading order is the same as in Fig. 2, as are arrowheads.
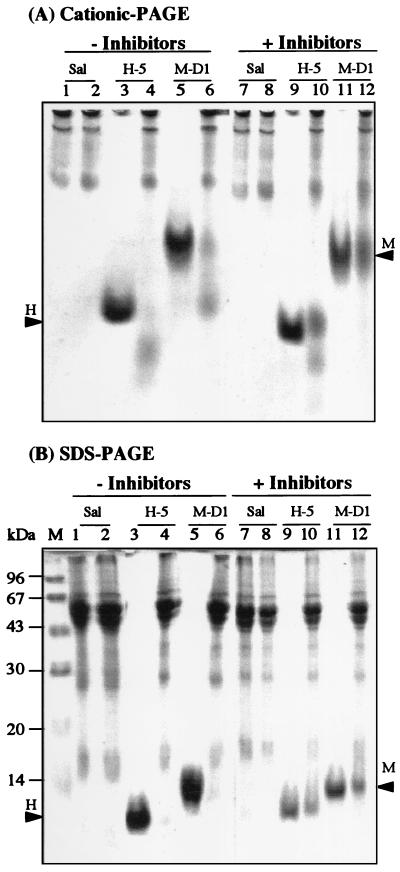
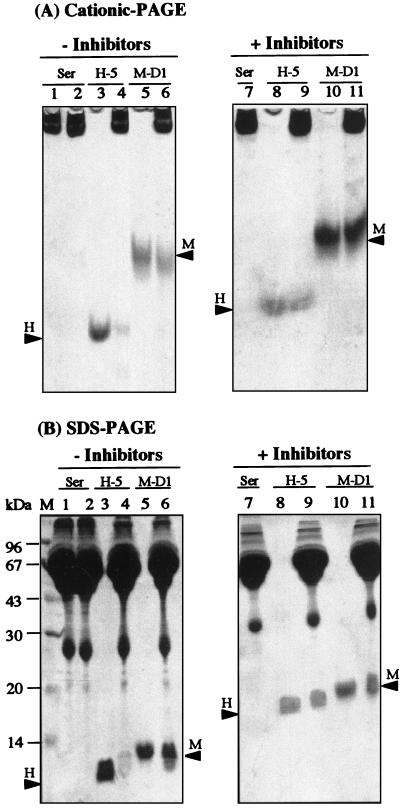
Several earlier studies indicated that salivary molecules are likely to form homotypic and heterotypic complexes. It was postulated that these complexes, mediated primarily by noncovalent ionic forces (dissociated by SDS-PAGE), may potentiate the biological properties of the individual molecules and may provide protection against proteolysis in the oral environment (2, 8, 12, 13); see the Discussion for more details. To address the question of whether the disappearance of the peptides in saliva on native gels (cationic PAGE) is due only to degradation by proteases or to complexing with other salivary molecules, we performed the following experiment. The peptides (in this case, only Hsn-5 and MUC7 D1) were incubated with unfiltered whole saliva for 0.5 h without or with the serine protease inhibitors aprotinin and PMSF. Degradation of Hsn-5 by human saliva was previously shown to be mediated mainly by a trypsin-like enzyme activity (24). After incubation, the samples were resolved by both cationic PAGE (Fig. 4A) and SDS-PAGE (Fig. 4B). Without the protease inhibitors, only small portions of the added Hsn-5 and MUC7 D1 were recovered intact, and no increased amounts of peptides were recovered by SDS-PAGE (compare lanes 4 and 6 in Fig. 4), indicating that no SDS-dissociable complexes were formed within the time of incubation. In the presence of protease inhibitors, much larger amounts of intact peptides were recovered on both types of gels (as shown in lanes 10 and 12 of Fig. 4), indicating that the peptides were at least partially protected from proteolysis. Again, no increased amounts of peptides were recovered by SDS-PAGE compared to cationic PAGE, indicating no formation of SDS-dissociable complexes. In summary, our data indicate that the disappearance of the investigated peptides in whole saliva is due primarily to their degradation by proteolytic enzymes present in saliva rather than to complexing with other salivary molecules. However, we cannot rule out the possible formation of complexes that are not dissociated by SDS-PAGE.
FIG. 4.
Stability of Hsn-5 (H-5) and MUC7 D1 (M-D1) in whole saliva at 37°C, without or with protease inhibitors, as monitored by 15% cationic PAGE (A) and SDS-PAGE (B), after incubation for 0.5 h. Lanes 1 and 7, whole saliva (Sal) (unincubated); lanes 2 and 8, whole saliva (37°C); lanes 3 and 9, Hsn-5 (37°C); lanes 4 and 10, Hsn-5 plus whole saliva (37°C); lanes 5 and 11, MUC7 D1 (37°C); lanes 6 and 12, MUC7 D1 plus whole saliva (37°C). Protease inhibitors were added to the whole saliva used in lanes 7 to 12. Arrowheads labeled H and M indicate the migration positions of Hsn-5 and MUC7 D1, respectively. Lane M, molecular mass markers.
To substantiate the above interpretations of our results, we assayed saliva for the presence of proteolytic enzymes using azocoll (4). Azocoll is hydrolyzed readily by a variety of proteases and yields soluble, colored peptides in proportion to enzyme concentrations at a fixed incubation time. The assay was performed as described in Materials and Methods, and trypsin was used as a standard protease. We found that assays of both trypsin and saliva gave linear rates of hydrolysis of azocoll as a function of protease concentration. From the standard curve of trypsin, we determined that 1 ml of unfiltered whole saliva contains proteolytic activity that would correspond to the activity obtained with 165 ng of trypsin. On the other hand, using the same method, we detected no measurable proteolytic enzyme activity in human serum.
Stability of peptides in human serum.
No complexing of the investigated peptides was expected with serum proteins. Nevertheless, stability was monitored with both types of gels because serum contains different spectra of proteins and a much higher concentration of total protein than saliva (mainly due to serum albumin). After incubation of peptides (Hsn-5 and MUC7 D1) in serum for 0.5 and 1 h, no degradation of peptides was evident by either cationic PAGE or SDS-PAGE (results are not shown but are consistent with the fact that no measurable proteolytic enzyme activity was detected in serum by the azocoll assay). After 2 h of incubation without the protease inhibitors, a large portion of the added Hsn-5 and a smaller portion of MUC7 D1 were degraded, as evident by both cationic PAGE (Fig. 5A; lane 4 for Hsn-5 and lane 6 for MUC7 D1) and SDS-PAGE (Fig. 5B, lane 4 for Hsn-5 and lane 6 for MUC7 D1). This finding indicated that MUC7 D1 is also more stable than Hsn-5 in serum and that there were proteases in serum, even though no measurable proteolytic enzyme activity was detected in serum. With the serine protease inhibitors (PMSF and aprotinin), large portions of both peptides were recovered intact by both cationic PAGE (Fig. 5A, lane 9 for Hsn-5 and lane 11 for MUC7 D1) and SDS-PAGE (Fig. 5B, lane 9 for Hsn-5 and lane 11 for MUC7 D1), showing the protection of peptides from degradation.
FIG. 5.
Stability of Hsn-5 (H-5) and MUC7 D1 (M-D1) in human serum at 37°C, without or with protease inhibitors, as monitored by 15% cationic PAGE (A) and SDS-PAGE (B), after incubation for 2 h. Lane 1, serum (Ser) (unincubated); lanes 2 and 7, serum (37°C); lanes 3 and 8, Hsn-5 (37°C); lanes 4 and 9, Hsn-5 plus serum (37°C); lanes 5 and 10, MUC7 D1 (37°C); lanes 6 and 11, MUC7 D1 plus serum (37°C). Protease inhibitors were added to the serum used in lanes 7 to 11. Arrowheads are as in Fig. 4. Lane M is as in Fig. 4.
Peptide toxicity studies.
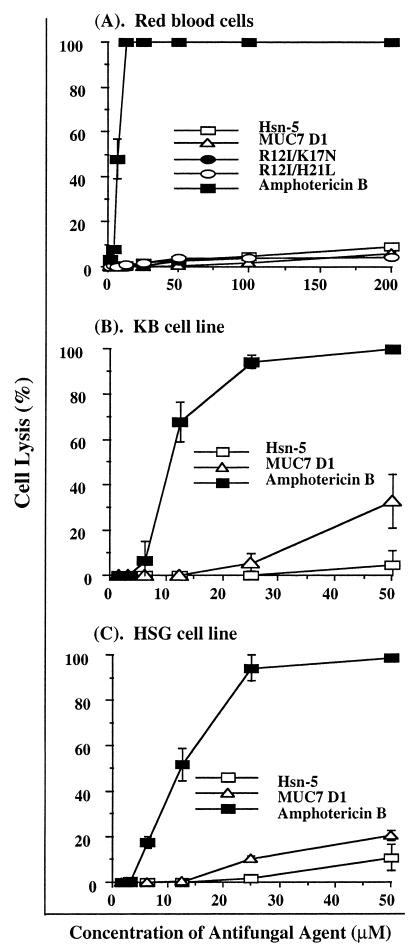
Human red blood cells provide a handy tool for toxicity studies of peptides or other compounds, because they are readily available, their membrane properties are well known, and their lysis is easy to monitor by measuring the release of hemoglobin. The hemolytic activities of Hsn-5, R12I/K17N, R12I/H21L, MUC7 D1, and amphotericin B on human red blood cells (as a function of peptide concentration) are shown in Fig. 6. At the highest concentration tested in this study (200 μM), none of the four salivary peptides caused even 10% hemolysis, while amphotericin B caused 100% hemolysis at only 12.5 μM. Amphotericin B consistently displayed significantly higher levels of hemolytic activity than Hsn-5, its two variants, and MUC7 D1 at all the concentrations tested (P < 0.05). In addition to red blood cells, KB cells (human oral epithelial carcinoma cell line) and HSG cells (a neoplastic epithelial duct cell line established from an irradiated human submandibular gland) were used for the toxicity studies of Hsn-5 and MUC7 D1. After a 48-h exposure, at the highest concentration tested in this study (50 μM), neither peptide caused 50% cytotoxic effect, while the IC50s of amphotericin B for KB and HSG were 9.8 and 11.9 μM, respectively. Together, our results indicate that all four peptides were significantly less cytotoxic than amphotericin B, one of the most widely used antifungal therapeutic agents.
FIG. 6.
Cytotoxicities of antifungal agents against red blood cells (A), KB cells (B), and HSG cells. The data are means of duplicate experiments.
DISCUSSION
In one of our early studies, we showed that both azole-resistant and azole-sensitive isolates of C. albicans and C. glabrata were susceptible to killing by Hsn-5 (17). Later, we demonstrated for the first time that Hsn-5 is also effective in killing another medically important opportunistic pathogen, C. neoformans (three different clinical isolates) (18). In a recent study, we demonstrated for the first time that the N-terminal 51-residue peptide of MUC7, MUC7 D1, has fungicidal activity against C. albicans comparable to that of Hsn-5 (Satyanarayana et al., submitted). In this study, we demonstrated effective killing of the aforementioned fungi with the two Hsn-5 variants (with the exception of R12I/K17N for C. glabrata) and, very importantly, with the MUC7 D1 peptide. The four investigated peptides were also effective in killing C. krusei and S. cerevisiae (except for R12I/K17N) and very effective in killing amphotericin B-resistant C. neoformans. Another recent study (7) showed that Hsn-5 and an amphipathic analog of the C-terminal 14 aa of Hsn-5 (designated dhvar-4) were effective against amphotericin B-resistant C. albicans, C. krusei, and Aspergillus fumigatus and against fluconazole-resistant C. glabrata. Collectively, these studies showed that Hsn-5 has broad-spectrum antifungal activity and is effective against amphotericin B-resistant and azole-resistant fungi. In addition, this study showed that MUC7 D1 exhibits the same broad-spectrum antifungal activity as Hsn-5 but, interestingly, is somewhat more potent than Hsn-5 in killing C. krusei and much more potent in killing C. glabrata (both azole-sensitive and azole-resistant isolates). In fact, the activities of MUC7 D1 against C. albicans and C. glabrata are comparable. MUC7 D1 is also much more potent in killing S. cerevisiae (see ED50 in Table 2). Therefore, it is of interest to determine the molecular mechanism of MUC7 D1 antifungal action and whether the mechanism is different from that of Hsn-5.
The azole-based drugs competitively inhibit lanosterol 14 α-demethylase (a cytochrome P-450 enzyme), one of the enzymes involved in the biosynthetic pathway of ergosterol, the major sterol of the fungal plasma membrane (9, 22). The gene encoding this enzyme in C. albicans (ERG160) has been isolated and sequenced (10). The azole-resistant C. albicans strain tested in this study was characterized to have an increased level of mRNA for this enzyme; other defects, however, may also contribute to the azole resistance of this strain (23). The fungicidal activity assays showed that the azole-resistant strains of C. albicans and C. glabrata were susceptible to Hsn-5 and MUC7 D1 killing to a similar degree as the azole-sensitive strains, suggesting that the cellular target of both Hsn-5 and MUC7 D1 is different from that of azole-based antifungal drugs. The interesting finding is that MUC7 D1 kills azole-resistant C. glabrata much more efficiently than Hsn-5.
The reason for the development of amphotericin B-resistant C. neoformans is not known. In general, though, resistance to amphotericin B is associated with an alteration of membrane lipids, especially ergosterol, the target of amphotericin B in the fungal cell membrane (21). The fact that Hsn-5 and MUC7 D1 kill amphotericin B-sensitive and -resistant C. neoformans strains equally well suggests that the mechanism of interaction of either peptide with C. neoformans is distinct from that of amphotericin B; the latter, through binding to a fungal membrane sterol, particularly ergosterol, forms channels that increase membrane ionic permeability and disrupt the cell membrane, leading to leakage of intracellular components and cell death (16). In this case, MUC7 D1 kills amphotericin B-resistant C. neoformans with an efficiency similar to that of Hsn-5.
No or very low cell toxicity was observed with all four peptides. In addition, R12I/K17N, R12I/H21L, and MUC7 D1 degraded at a lower rate than Hsn-5 in whole saliva, indicating their greater proteolytic resistance. The latter property would be especially valuable if these peptides were to be used as topical oral antifungal therapeutic agents.
As mentioned in Results, several earlier studies indicated that salivary molecules are likely to form homotypic and heterotypic complexes (2, 8, 12, 13). Most of these involve complex formation between salivary mucins and other salivary proteins. Mucins may act as carrier molecules which bind and concentrate other salivary proteins at oral tissue-environmental interfaces. Specifically, a study by Biesbrock et al. (2) showed that MG2 (MUC7) isolated from the human submandibular-sublingual saliva (the full-length glycosylated mucin) complexed with secretory immunoglobulin A. In a recent study (8), in which salivary mucin MG1 was isolated under mild conditions (no dialysis, lyophilization, use of denaturing agents, or covalent modification), it was shown that native MG1 formed heterotypic complexes with histatins (and with amylase, proline-rich proteins, and statherins). On the other hand, MG2, secretory immunoglobulin A alpha chain and secretory component, and cystatins did not form complexes with MG1 that were detectable by the immunological methods used. Histatins were found in type I complexes with MG1; these were dissociated by SDS-PAGE. In addition, type III complexes were formed that were also dissociated by SDS-PAGE, but the released proteins themselves appeared to be complexes containing two or more immunoreactive components: amylase, proline-rich proteins, statherins, and histatins. These findings suggested that these could be covalent complexes or other strong complexes not dissociated by SDS-PAGE, possibly cross-linked by a transglutaminase enzyme. This possibility was further substantiated by the recent finding that some of the lysine residues present in histatins could participate in the cross-linking reaction catalyzed by oral transglutaminase (26). Based on the results of the present study, we suggest that the peptides added to saliva do not form SDS-dissociable complexes with other salivary molecules. However, we cannot rule out the possible formation of complexes that are not dissociated by SDS-PAGE. Further, we conclude that the peptide disappearance is due primarily to peptide degradation by proteolytic enzymes present in saliva. Since only partial protection from proteolysis was obtained with serine proteases, other types of proteases may contribute to degradation.
In summary, this study showed that the investigated Hsn-5, its two variants, and MUC7 D1 have broad-spectrum antifungal activities and are effective against azole- and amphotericin B-resistant fungal strains and that the two Hsn-5 variants and MUC7 D1 exhibit lower susceptibility to degradation in human saliva. In human serum, MUC7 D1 is also more stable than Hsn-5, and both peptides are more stable in serum than in saliva. None of the peptides is toxic to human red blood cells. Very low cytotoxic activities against KB or HSG cells were observed with Hsn-5 and MUC7 D1 as well. These properties strongly indicate that these four peptides have promising antifungal therapeutic potential, in particular for the treatment of drug-resistant fungal strains associated with immunocompromised (especially human immunodeficiency virus-infected) patients. The same molecules could be used as components of artificial saliva for patients with salivary dysfunction. As always, however, the translation of in vitro activity into clinical efficacy must be established.
Further investigation of these exciting new agents, in particular their mechanism of action and the development of effective delivery systems, is needed. For topical use in the oral cavity, formulation in oral rinses would be one possible type of delivery. More ideal would be some type of slow-release device in the oral cavity, so that these peptides would not have to be used several times a day. It is known that in order for Hsn-5 to kill C. albicans, it has to be internalized by C. albicans (6, 11, 25) and cannot be conjugated to other molecules (unpublished results). However, the mechanism of killing of other fungi by Hsn-5 is not known; neither is the mechanism by which the MUC7 D1 peptide kills any fungi. Problems arise when these peptides (or any peptides) are considered for systemic delivery. If administered orally, rapid presystemic elimination renders peptides ineffective. Thus, either direct injections or transdermal or transmucosal delivery, bypassing hepatogastrointestinal clearance, would be needed. Indeed, as mentioned above, if histatins and other peptides are to be used as effective antifungal therapeutic agents, effective delivery systems must be developed.
ACKNOWLEDGMENTS
We thank M. J. Levine and J. Satyanarayana for providing synthetic MUC7 D1.
This work was supported by Public Health Service grant DE09820 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Alexander B D, Perfect J R. Antifungal resistance trends towards the year 2000. Drugs. 1997;54:657–678. doi: 10.2165/00003495-199754050-00002. [DOI] [PubMed] [Google Scholar]
- 2.Biesbrock A R, Reddy M S, Levine M J. Interaction of a salivary mucin-secretory immunoglobulin A complex with mucosal pathogens. Infect Immun. 1991;59:3492–3497. doi: 10.1128/iai.59.10.3492-3497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobek L A, Tsai H, Biesbrock A R, Levine M J. Molecular cloning, sequence, and specificity of expression of the gene encoding the low molecular weight human salivary mucin (MUC7) J Biol Chem. 1993;268:20563–20569. [PubMed] [Google Scholar]
- 4.Chavira R, Jr, Burnett T J, Hageman J H. Assaying proteinases with azocoll. Anal Biochem. 1984;136:446–450. doi: 10.1016/0003-2697(84)90242-2. [DOI] [PubMed] [Google Scholar]
- 5.Gururaja T L, Levine J H, Tran D T, Naganagowda G A, Ramalingam K, Ramasubbu N, Levine M J. Candidacidal activity prompted by N-terminus histatin-like domain of human salivary mucin (MUC7) Biochim Biophys Acta. 1999;1431:107–119. doi: 10.1016/s0167-4838(99)00034-5. [DOI] [PubMed] [Google Scholar]
- 6.Helmerhorst E J, Breeuwer P, van't Hof W, Walgreen-Weterings E, Oomen L C, Veerman E C, Amerongen A V, Abee T. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J Biol Chem. 1999;274:7286–7291. doi: 10.1074/jbc.274.11.7286. [DOI] [PubMed] [Google Scholar]
- 7.Helmerhorst E J, Reijnders I M, Van't Hof W, Simmons-Smit I, Veerman E C I, Nieuw Amerongen A V. Amphotericin B- and fluconazole-resistant Candida spp., Aspergillus fumigatus, and other newly emerging pathogenic fungi are susceptible to basic antifungal peptides. Antimicrob Agents Chemother. 1999;43:703–704. doi: 10.1128/aac.43.3.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iontcheva I, Oppenheim F G, Troxler R E. Human salivary mucin MG1 selectively forms heterotypic complexes with amylase, proline-rich proteins, statherin, and histatins. J Dent Res. 1997;76:734–743. doi: 10.1177/00220345970760030501. [DOI] [PubMed] [Google Scholar]
- 9.Joly V, Bolard J, Yeni P. In vitro models for studying toxicity of antifungal agents. Antimicrob Agents Chemother. 1992;36:1799–1804. doi: 10.1128/aac.36.9.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirsch D R, Lai M H, O'Sullivan J. Isolation of the gene for cytochrome P450L1A1 (lanosterol 14 alpha-demethylase) from Candida albicans. Gene. 1988;68:229–237. doi: 10.1016/0378-1119(88)90025-x. [DOI] [PubMed] [Google Scholar]
- 11.Koshlukova S E, Lloyd T L, Araujo M W, Edgerton M. Salivary histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J Biol Chem. 1999;274:18872–18879. doi: 10.1074/jbc.274.27.18872. [DOI] [PubMed] [Google Scholar]
- 12.Levine M J. Development of artificial salivas. Crit Rev Oral Biol Med. 1993;4:270–286. doi: 10.1177/10454411930040030401. [DOI] [PubMed] [Google Scholar]
- 13.Levine M J, Reddy M S, Tabak L A, Loomis R E, Bergey E J, Jones P C, Cohen R E, Stinson M W, Al-Hashimi I. Structural aspects of salivary glycoproteins. J Dent Res. 1987;66:436–441. doi: 10.1177/00220345870660020901. [DOI] [PubMed] [Google Scholar]
- 14.Prakobphol A, Levine M J, Tabak L A, Reddy M S. Purification of a low-molecular-weight, mucin-type glycoprotein from human submandibular-sublingual saliva. Carbohydr Res. 1982;108:111–122. doi: 10.1016/s0008-6215(00)81896-0. [DOI] [PubMed] [Google Scholar]
- 15.Situ H, Tsai H, Bobek L A. Construction and characterization of human salivary histatin-5 multimers. J Dent Res. 1999;78:690–698. doi: 10.1177/00220345990780020901. [DOI] [PubMed] [Google Scholar]
- 16.Smith C M, Reynard A M. Textbook of pharmacology. Philadelphia, Pa: The W. B. Saunders Company; 1992. [Google Scholar]
- 17.Tsai H, Bobek L A. Studies of the mechanism of human salivary histatin-5 candidacidal activity with histatin-5 variants and azole-sensitive and -resistant Candida species. Antimicrob Agents Chemother. 1997;41:2224–2228. doi: 10.1128/aac.41.10.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsai H, Bobek L A. Human salivary histatin-5 exerts potent fungicidal activity against Cryptococcus neoformans. Biochim Biophys Acta. 1997;1336:367–369. doi: 10.1016/s0304-4165(97)00076-7. [DOI] [PubMed] [Google Scholar]
- 19.Tsai H, Bobek L A. Human salivary histatins: promising anti-fungal therapeutic agents. Crit Rev Oral Biol Med. 1998;9:480–497. doi: 10.1177/10454411980090040601. [DOI] [PubMed] [Google Scholar]
- 20.Tsai H, Raj P A, Bobek L A. Candidacidal activity of recombinant human salivary histatin-5 and variants. Infect Immun. 1996;64:5000–5007. doi: 10.1128/iai.64.12.5000-5007.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanden Bossche H, Dromer F, Improvisi I, Lozano-Chiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36(Suppl. 1):119–128. [PubMed] [Google Scholar]
- 22.Vanden Bossche H, Willemsens G, Marichal P. Anti-candida drugs—the biochemical basis for their activity. Crit Rev Microbiol. 1987;15:57–72. doi: 10.3109/10408418709104448. [DOI] [PubMed] [Google Scholar]
- 23.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increase in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Lal K, Santarpia III R P, Pollock J J. Salivary proteolysis of histidine-rich polypeptides and the anti-fungal activity of peptide degradation products. Arch Oral Biol. 1993;38:277–283. doi: 10.1016/0003-9969(93)90133-7. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Ambudkar I, Yamagishi H, Swaim W, Walsh T J, O'Connell B C. Histatin 3-mediated killing of Candida albicans: effect of extracellular salt concentration on binding and internalization. Antimicrob Agents Chemother. 1999;43:2256–2262. doi: 10.1128/aac.43.9.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao Y, Lamkin M S, Oppenheim F G. Pellicle precursor proteins: acidic proline-rich proteins, statherin, and histatins, and their crosslinking reaction by oral transglutaminase. J Dent Res. 1999;78:1696–1703. doi: 10.1177/00220345990780110601. [DOI] [PubMed] [Google Scholar]