Abstract
Asthma is associated with chronic changes in the airway epithelium, a key target of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Many epithelial changes, including goblet cell metaplasia, are driven by the type 2 cytokine IL-13, but the effects of IL-13 on SARS-CoV-2 infection are unknown. We found that IL-13 stimulation of differentiated human bronchial epithelial cells (HBECs) cultured at air–liquid interface reduced viral RNA recovered from SARS-CoV-2–infected cells and decreased double-stranded RNA, a marker of viral replication, to below the limit of detection in our assay. An intact mucus gel reduced SARS-CoV-2 infection of unstimulated cells, but neither a mucus gel nor SPDEF, which is required for goblet cell metaplasia, were required for the antiviral effects of IL-13. Bulk RNA sequencing revealed that IL-13 regulated 41 of 332 (12%) mRNAs encoding SARS-CoV-2–associated proteins that were detected in HBECs (>1.5-fold change; false discovery rate < 0.05). Although both IL-13 and IFN-α each inhibit SARS-CoV-2 infection, their transcriptional effects differed markedly. Single-cell RNA sequencing revealed cell type–specific differences in SARS-CoV-2–associated gene expression and IL-13 responses. Many IL-13–induced gene expression changes were seen in airway epithelium from individuals with type 2 asthma and chronic obstructive pulmonary disease. IL-13 effects on airway epithelial cells may protect individuals with type 2 asthma from COVID-19 and could lead to identification of novel strategies for reducing SARS-CoV-2 infection.
Keywords: COVID-19, SARS-CoV-2, asthma, IL-13, airway epithelium
One remarkable feature of the coronavirus disease (COVID-19) pandemic is the wide range of disease severity seen after severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Host factors, including age and male sex (1), inborn or acquired disorders of type I interferon-mediated antiviral immunity (2–4), and various preexisting medical conditions (5) influence the risk of severe disease. There have been concerns that COVID-19 risks would also be increased in persons with asthma, which affects ∼339,000,000 individuals worldwide (6). These concerns arose from experience with other respiratory viruses that trigger asthma exacerbations and can be associated with worse outcomes in individuals with preexisting asthma (7). However, asthma was underrepresented in early studies of patients with COVID-19 as well as prior studies of SARS (8). Subsequent studies have failed to find consistent evidence of increased risk of COVID-19 diagnosis, hospitalization, or mortality due to asthma (1, 9, 10), and some studies concluded that the risks of acquiring and being hospitalized for COVID-19 are lower in those with asthma (10, 11). Factors that might provide protection against COVID-19 in individuals with asthma (12–15) include increased attention to limiting viral exposure, younger age, absence of comorbidities, use of inhaled corticosteroids, and a variety of biological features of asthma, including chronic airway inflammation, mucus hypersecretion, and altered expression of SARS-CoV-2 receptor (12). However, direct evidence that these features alter SARS-CoV-2 infection has been lacking.
Asthma is associated with changes in the structure and function of the airway epithelium, a critical site for SARS-CoV-2 infection (16, 17). Airway epithelial gene expression changes attributable to the type 2 cytokine IL-13 are seen in approximately half of individuals with asthma (18, 19). One prominent effect of IL-13 is the induction of goblet cells that produce and secrete the airway mucin MUC5AC, a process that depends on the transcription factor SPDEF. IL-13 stimulation of airway epithelial cells decreases expression of ACE2, which encodes the SARS-CoV-2 receptor, and increases expression of TMPRSS2, which encodes a transmembrane protease that primes the viral spike protein (20–22). Similar changes are seen in airways of individuals with type 2–high asthma (21, 22). IL-13 has also been reported to protect against other RNA viruses, including respiratory syncytial virus (23) and rhinovirus (24), that do not rely on ACE2 and TMPRSS2 for entry, indicating that other IL-13–regulated genes can also protect against viral infection. We therefore hypothesized that direct effects of IL-13 on the airway epithelium, which are seen in a large subset of individuals with asthma, reduce susceptibility of these cells to SARS-CoV-2 infection.
Some of the results of these studies have been previously reported in the form of a preprint (bioRxiv, [2021 Feb 25] http://biorxiv.org/lookup/doi/10.1101/2021.02.25.432762).
Methods
Additional details are provided in the data supplement.
Human Bronchial Epithelial Cell Culture
Primary human bronchial epithelial cells (HBECs) from 14 individuals listed in Table E1 (see data supplement) were cultured at air–liquid interface as previously described (25, 26). For cytokine stimulation, cultures were stimulated by addition of cytokines (10 ng/ml) to the basolateral medium (IFN-α: 24 h; IFN-γ: 24 h; IL-13: 7 d; IL-17: 7 d). The University of California, San Francisco Committee on Human Research approved these studies.
SARS-CoV-2 Infection
SARS-CoV-2 virus (USA-WA1/2020 strain) was provided by Dr. Melanie Ott and propagated in Vero E6 cells. HBECs were cultured in the absence or presence of IL-13 (10 ng/ml). Mucus was allowed to accumulate for 3 days or was removed immediately before viral infection by washing the apical surface with a prewarmed solution of 10 mM DTT (Thermo) in PBS for 10 minutes (26) and then washing twice with PBS without DTT. Cells were inoculated by adding virus to the apical surface. After 2 hours, the apical surface was washed twice with PBS, and cells were returned to the incubator. Cells were harvested 48 hours after infection for analysis of viral RNA (by qRT-PCR) and double-stranded RNA (dsRNA) staining (by immunofluorescence).
SPDEF Targeting of HBECs
HBECs were subjected to two successive rounds of electroporation with complexes comprising guide RNAs (gRNAs) targeting SPDEF or a nontargeting control sequence and recombinant Cas9 as previously described (27). Cells were subsequently cultured as described above.
RNA Sequencing
RNA was isolated from cytokine-stimulated HBECs derived from six individuals, and bulk RNA sequencing (RNA-seq) was performed as previously described (28, 29). We previously reported other analyses based on data from unstimulated cells and cells stimulated with individual cytokines (see data supplement for data availability statement [28, 29]); data from cells stimulated with a combination of IL-13 and IFN-α have not been previously reported. For single-cell RNA sequencing (scRNA-seq), single-cell suspensions were generated from HBECs from four of the donors used for bulk RNA-seq and analyzed using the 10X Genomics platform.
Analysis of Gene Expression in Asthma and Chronic Obstructive Pulmonary Disease
We correlated measures of type 2 activity from studies of asthma (three-gene mean [28]) and chronic obstructive pulmonary disease (COPD) (type 2 score [30]) with IL-13–induced SARS-CoV-2–associated genes in HBECs using Pearson’s correlation coefficient and a linear regression model that adjusted for age and sex.
Results
IL-13 Protects HBECs from SARS-CoV-2 Infection
We tested whether IL-13 restricted SARS-CoV-2 infection by pretreating HBECs with IL-13 before inoculation with SARS-CoV-2 and quantifying viral RNA 48 hours after inoculation. Mucus produced by HBECs was left in place or removed by washing the apical surface immediately before inoculation. In the first experiment using donor 10-75, we found that the presence of mucus decreased the amount of SARS-CoV-2 RNA detected after infection of unstimulated cells by 74% compared with cells infected after removal of mucus (Figure 1A). Prestimulation with IL-13 markedly lowered levels of SARS-CoV-2 RNA detected when HBECs were infected after removal of mucus (95% reduction) or when mucus was present (97% reduction). In a second experiment with a different HBEC donor (14-30; Figure 1B), mucus was more effective in inhibiting infection (90–97% reduction for three different viral inocula). In cells infected after removal of mucus, IL-13 prestimulation reduced viral RNA by 82–92%.
Figure 1.

IL-13 stimulation and mucus reduce severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus RNA levels in infected human bronchial epithelial cells (HBECs). (A) HBECs from donor 10-75 were left unstimulated (−) or stimulated with IL-13 (+), washed with a DTT-containing solution (Mucus Removed) or left unwashed (Mucus Intact), and inoculated with SARS-CoV-2 (0.3 plaque-forming units [pfu] based on titration in Vero E6 cells). SARS-CoV-2 mRNA was measured 48 hours after infection. (B) In a second experiment, cells from donor 14-30 were studied using the same protocol, except that three different inocula (0.3, 1.0, and 1.3 pfu) from another virus preparation were used. Each point represents a separate Transwell culture (n = 3 per condition except as shown). **P < 0.01 and ***P < 0.0001 for the effects of IL-13 by ANOVA with Tukey-Kramer post hoc tests. For cells not stimulated with IL-13, viral RNA load was lower in infections performed with mucus intact compared with infections performed with mucus removed (P < 0.0001 for all viral inocula in both experiments, except for P = 0.01 for the 0.3 pfu inoculum in the second experiment, by ANOVA with Tukey-Kramer post hoc tests). For viral RNA, 1 unit represents the amount of viral RNA present in 1 pfu from the viral stock, based on titration in Vero E6 cells.
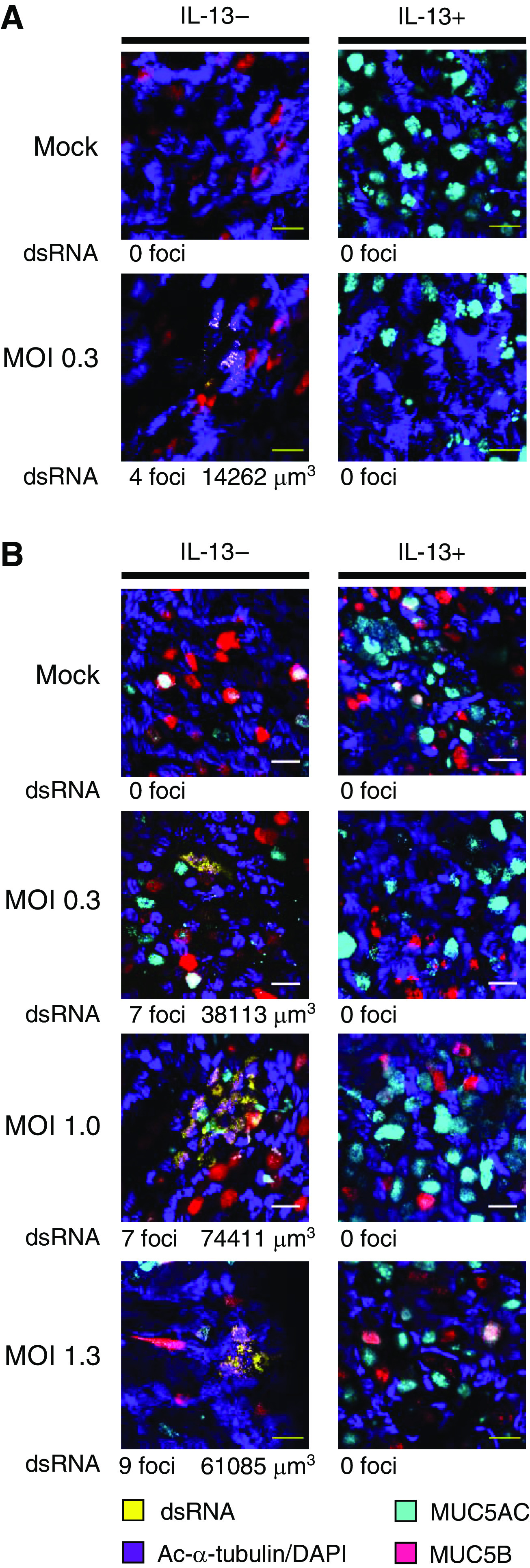
We also assessed the effects of mucus and IL-13 on dsRNA, which is produced during SARS-CoV-2 viral replication. dsRNA was not detected in uninfected cells but was detected within isolated cells or clusters of cells in unstimulated SARS-CoV-2–infected cultures. Analysis of cultures inoculated with varying amounts of virus after removal of mucus revealed a total of 27 dsRNA-stained foci with a mean volume of 6,958 μm3 (Figure 2). In contrast, only three foci (mean, 1,246 μm3) were seen in paired cultures inoculated without removing mucus (Figure E1). In cultures prestimulated with IL-13, no foci were observed whether or not mucus was removed. The observation that relatively small amounts of viral RNA were detectable in IL-13–stimulated cultures (Figure 1), but dsRNA staining was not evident under these conditions (Figures 2 and E1), might indicate that dsRNA staining is less sensitive than qRT-PCR for viral RNA. Alternatively, it is possible that IL-13 completely prevented viral replication and that viral RNA detected in IL-13–stimulated cells was residual RNA from the viral inoculum. Most dsRNA-containing infected cells costained for the ciliated cell marker acetylated-α-tubulin, although dsRNA was occasionally seen in nonciliated cells that stained for mucins. Because PCR analysis of viral RNA indicated that the protective effects of mucus were greater in donor 14-30, it is noteworthy that mucin expression differed between the two donors. In the absence of IL-13, cultures from donor 10-75 had MUC5B-containing cells but no detectable MUC5AC-containing cells (Figures 2A and E1B), whereas cultures from donor 14-30 had both MUC5AC- and MUC5B-containing cells (Figures 2B and E1B). IL-13 stimulation increased MUC5AC in cells from both donors and caused an obvious decrease in MUC5B in donor 10-75; these effects of IL-13 are consistent with those observed in our previous studies (25) and people with type 2 asthma (19). Based on viral RNA measurements and dsRNA staining, we conclude that both the presence of a mucus gel and IL-13 stimulation reduced viral infection and that the effects of IL-13 were seen even after removal of the mucus layer prior to infection.
Figure 2.
IL-13 stimulation reduces SARS-CoV-2 replication in HBECs. (A and B) Additional HBEC cultures derived from cells from donor 10-75 (A) and donor 14-30 (B) were inoculated with virus after removal of mucus as part of the same experiments shown in Figure 4. After 48 hours, cells were stained with antibodies against double-stranded RNA (dsRNA) (yellow), the ciliated cell marker acetylated α tubulin (Ac-α-tubulin) and DAPI (both imaged in the same channel, purple), MUC5B (red), and MUC5AC (cyan). We surveyed the entire sample (16.6 μm) for dsRNA staining and acquired stacks encompassing each dsRNA-stained focus. Numbers of dsRNA-stained foci and total volumes of dsRNA staining are shown below representative images for each condition. Scale bars, 20 μM. MOI = multiplicity of infection.
IL-13–mediated Antiviral Effects Are Independent of SPDEF
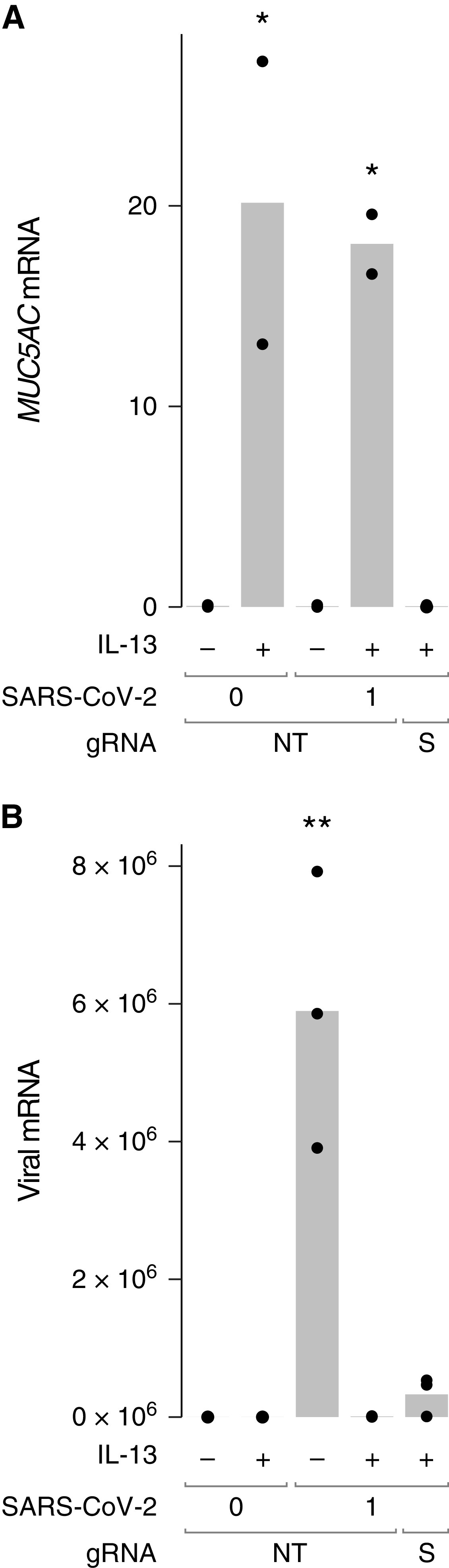
SPDEF is induced by IL-13 and encodes a transcription factor that regulates the expression of a program of genes found in mucus-producing goblet cells, including MUC5AC and MUC5B. We have previously shown that targeting of SPDEF using CRISPR-Cas9 abolished IL-13–induced MUC5AC expression and goblet cell differentiation (27). To test whether the antiviral effects of IL-13 were mediated by goblet cell differentiation, we delivered recombinant Cas9 and gRNAs targeting either a nontargeting control sequence (NT) or SPDEF to HBECs before differentiation at air–liquid interface. As expected, targeting SPDEF prevented IL-13–induced increases in MUC5AC mRNA (Figure 3A), indicating that targeting was highly effective. In cells that received the NT control gRNA, IL-13 stimulation reduced levels of viral RNA recovered after infection with SARS-CoV-2 by 99% (Figure 3B), similar to our previous experiments without CRISPR-Cas9 performed with cells from two other donors (Figure 1). Viral RNA levels were also low in IL-13–stimulated cells after SPDEF targeting (94% reduction compared with NT, no IL-13 controls), indicating that SPDEF and MUC5AC induction are not required for antiviral effects of IL-13.
Figure 3.
SPDEF targeting has little effect on the amount of viral RNA observed in HBECs pretreated with IL-13. HBEC cultures from donor 19-07 were subjected to two successive electroporations with guide RNAs (gRNAs) targeting a nonhuman, nontargeting control sequence (NT) or SPDEF (S) and fully differentiated. HBECs were left unstimulated (−) or stimulated with IL-13 (+) and washed with a DTT-containing solution before inoculation with 1 pfu SARS-CoV-2. RNA was harvested at 72 hours after infection. (A) MUC5AC mRNA was measured to assess SPDEF targeting. *P < 0.05 compared with unstimulated or IL-13–stimulated SPDEF-targeted cells by ANOVA with Tukey-Kramer post hoc test. (B) SARS-CoV-2 RNA. **P < 0.01 compared with all other conditions. Each point represents a separate Transwell culture (n = 2–3 per condition as shown). For viral RNA, 1 unit represents the amount of viral RNA present in 1 pfu from the viral stock, based on titration in Vero E6 cells.
IL-13 Effects on SARS-CoV-2– associated Gene Expression Are Distinct from IFN-α Effects
We compiled a list of 342 SARS-CoV-2– associated host genes comprising 11 genes encoding proteins implicated in viral entry: ACE2 (31), TMPRSS2 (31), the cathepsins CTSB and CTSL, FURIN(PCSK3), and the furin-like proteases PCSK1, PCSK2, and PCSK4–7 (32), plus 332 genes encoding host cell proteins shown to interact with high confidence with 26 of the 29 SARS-CoV-2 proteins in HEK293T cells (33). We evaluated the expression of these 342 SARS-CoV-2– associated genes in a previously reported bulk RNA-seq dataset that we generated using HBECs from six individuals stimulated with cytokines. In addition to IL-13, we stimulated cells with IFN-α, which plays a central role in defense against SARS-CoV-2 infection (3, 4) and has been shown to inhibit SARS-CoV-2 infection of human lung epithelial cells (34), and with IFN-γ (28) and IL-17 (35), each of which have been implicated in subsets of individuals with asthma.
We detected 332 of the 342 SARS-CoV-2– associated genes (97%; ⩾1 read per million mapped reads in ⩾50% of samples) in differentiated HBECs cultured without cytokine (Table E2). SARS-CoV-2– associated genes were substantially overrepresented among genes with high read counts (Figure 4A). Each cytokine had the expected effects on expression of known cytokine-responsive genes (Figure 4B). Of the 332 SARS-CoV-2– associated genes detected in HBECs, IL-13 regulated 41 genes, IFN-α regulated 19 genes, the combination of IL-13 and IFN-α regulated 63 genes, IFN-γ regulated 14 genes, and IL-17 regulated 21 genes (false discovery rate [FDR] q ⩽ 0.05 and absolute fold change ⩾ 1.5; Figure 4C and Table E2).
Figure 4.
SARS-CoV-2–associated genes are highly expressed in HBECs, and many are regulated by cytokines. HBECs from six donors were cultured without cytokine (−), or with IL-13, IFN-α, a combination of IL-13 and IFN-α, IFN-γ, or IL-17 and analyzed by RNA sequencing (RNA-seq). (A) Comparison of read counts between SARS-CoV-2–associated genes, including ACE2 and TMPRSS2, and all detected genes (⩾1 read per million mapped reads in ⩾50% of samples) in unstimulated HBECs. (B and C) Heatmap illustrating canonical cytokine-regulated genes (B), and cytokine regulated SARS-CoV-2–associated genes (C) (false discovery rate [FDR] q ⩽ 0.05; absolute fold change ⩾ 1.5 for any cytokine).
Previous SARS-CoV-2–related studies have focused on the viral entry genes ACE2 and TMPRSS2. In our dataset, ACE2 was reduced by IL-13 (29% decrease, q = 0.003), consistent with a prior report (24), was the most highly upregulated interferon-stimulated gene (IFN-α [451% increase, q = 3 × 10−74] or IFN-γ [185% increase, q = 9 × 10−29]), and was less strongly upregulated following IL-17 stimulation (31% increase, q = 0.02). Analysis of ACE2 splicing confirmed prior reports that interferon stimulation increased expression of the decoy isoform (36), but we found no significant effect of IL-13 on levels of this isoform (mean decoy ACE2 normalized reads: unstimulated, 62; IFN-α–stimulated: 588; IL-13–stimulated, 61). TMPRSS2 was increased by IL-13 (61% increase, q = 7 × 10−15) and IL-17 (22% increase, q = 0.005) but modestly decreased by IFN-α (17% decrease, q = 0.01) and IFN-γ (16% decrease, q = 0.03).
No significant correlation between the effects of IL-13 and IFN-α were observed in total (Figure E2A). We also examined the effects of costimulation with IL-13 and IFN-α. Both IL-13 (R = 0.81, P < 0.001) and, to a lesser extent, IFN-α (R = 0.49, P < 0.001) contributed to the gene expression changes seen in cells stimulated with both cytokines (Figures E2B and E2C). A strong correlation (R = 0.95, P < 0.001) was observed between the effect of the costimulation with both cytokines and the sum of the effects of stimulation with IL-13 alone and IFN-α alone, consistent with additive effects (Figure E2D). IL-13 and IFN-α therefore affect SARS-CoV-2–associated genes by different and independent mechanisms.
IL-13 Affects the Expression of Some SARS-CoV-2–associated Genes in a Cell Type–Specific Manner
We used scRNA-seq to assess cell type–specific expression of SARS-CoV-2– associated genes in unstimulated HBEC cultures from four individuals (Figure E3). Of the 332 SARS-CoV-2–associated genes detected in HBECs by bulk RNA-seq, 322 (97%) were detected in at least 10 cells. We detected ACE2 in 1.7% of basal cells, 3.1% of secretory cells, and 1.6% of ciliated cells from unstimulated HBEC cultures. A total of 113 of the 322 SARS-CoV-2 genes detected in our scRNA-seq dataset were differentially expressed between cell types (FDR q < 0.05; Table E3; selected genes shown in Figure 5A). We found similar patterns of epithelial cell subset-specific expression of these SARS-CoV-2–associated genes in a scRNA-seq dataset from human bronchial tissue (37) (Figure E4), confirming that our cell culture model recapitulated cell type–specific gene expression seen in vivo and that the expression of host cell proteins that interact with SARS-CoV-2 proteins differs between cell types.
Figure 5.
Single-cell RNA sequencing (scRNA-seq) reveals cell type–specific expression of many SARS-CoV-2–associated genes and cell type–specific effects of IL-13. (A) Cell type–specific expression in unstimulated HBECs. Genes were selected from a set of 113 differentially expressed SARS-CoV-2–associated genes listed in Table E3. (B) Cell type–specific differences in IL-13 responses. For 11 SARS-CoV-2– associated genes, IL-13 increased expression in at least one cell type and decreased expression in at least one other cell type (FDR q < 0.1 for both). Gene expression was determined by aggregating data from all cells from experiments with four donors. (A and B) Coloring of each dot indicates expression level relative to other cell types (A) or in IL-13–stimulated cells compared with unstimulated cells (B). The size of each dot is proportional to the percentage of cells with at least one read mapped to the gene, and black circles at the perimeter of each dot indicate that expression levels are significantly different (q < 0.1) compared with other cell types (A) or in IL-13–stimulated compared with unstimulated cells of the same type (B).
We explored the effect of IL-13 stimulation on SARS-CoV-2–associated gene expression in each cell type. Although many IL-13–regulated SARS-CoV-2–associated genes were affected similarly in each cell type, some IL-13 effects were cell type–specific, with 11 cases in which IL-13 had opposite effects (increased in one cell type and decreased in another cell type, FDR q < 0.1 for both; Figure 5B and Table E3). Notably, IL-13 upregulated TMPRSS2 expression in secretory cells but decreased expression in ciliated cells. Cell type–specific effects of IL-13 could have implications for the outcome of infection in different airway epithelial subsets.
Type 2 Signatures Are Associated with Expression of Many IL-13–Responsive SARS-CoV-2– associated Genes in Individuals with Asthma and COPD
The data presented above suggest that preexisting inflammation induced by cytokines in asthma alters the expression of SARS-CoV-2–associated host genes in the airway epithelium. We therefore assessed whether IL-13–induced SARS-CoV-2– associated genes identified in HBECs in culture were altered in asthma using a transcriptomic profiling dataset derived from endobronchial brush biopsies from individuals with mild to moderate asthma and healthy individuals (MAST [Mechanisms of Asthma Study]) (28, 38). We used the three-gene mean (TGM), an established measure of IL-13–induced airway inflammation in individuals with asthma (18, 19, 39), for this analysis. Twenty-four of 27 SARS-CoV-2–associated genes induced by IL-13 in HBECs were positively correlated with the TGM (Pearson’s R > 0); in 13 cases this correlation was statistically significant after adjustment for multiple comparisons (adjusted P < 0.05; Figure 6 and Table E4). Sixteen of 27 IL-13–induced genes were significantly associated with the TGM using a linear model that included age and sex (adjusted P < 0.05; Table E4).
Figure 6.
Expression of many IL-13–regulated SARS-CoV-2–associated genes correlates with an IL-13 signature in asthma. Correlation of IL-13–induced, SARS-CoV-2–associated genes with a type 2/IL-13 signature (the three-gene mean [TGM]) in endobronchial brushing samples from participants with asthma (red) and healthy control subjects (cyan). Values for gene expression represent log2 of normalized read counts from bulk RNA-seq. The eight SARS-CoV-2 genes with the highest Pearson’s correlations (R) are shown, and associated P values are adjusted for multiple comparisons. Correlations for the full set of IL-13–induced SARS-CoV-2–associated genes are shown in Table E4.
A study of former smokers with and without COPD found that subsets of individuals with COPD also have airway epithelial gene expression changes indicative of type 2 inflammation (40). We used data from that study, together with the type 2 score developed for use with that dataset, to analyze expression of IL-13–induced SARS-CoV-2–associated genes. Twenty-one of the 26 IL-13–inducible SARS-CoV-2– associated genes induced by IL-13 in HBECs were positively correlated with the type 2 score (Pearson’s R > 0). In 16 cases, this association was statistically significant (adjusted P < 0.05) and remained so in a model that included age and sex (Figure E5 and Table E4). Taken together, our data indicate that many IL-13–induced SARS-CoV-2–associated gene changes seen in the HBEC culture model recapitulate alterations seen in epithelial cells from individuals with asthma or COPD and preexisting type 2 inflammation.
Discussion
Our studies reveal that IL-13 stimulation of HBECs affects expression of many SARS-CoV-2–associated genes and substantially inhibits SARS-CoV-2 infection of these cells. Genes encoding most SARS-CoV-2– interacting proteins identified in a previous study of HEK293T cells were expressed in HBECs. Expression of many SARS-CoV-2–associated genes differed between basal, ciliated, and secretory cells, potentially affecting how these cell types respond to SARS-CoV-2 infection. Many IL-13–induced SARS-CoV-2–associated gene expression changes we detected in culture were also seen in bronchial epithelium obtained directly from individuals with type 2–high asthma. This provides a plausible mechanism for protection against COVID-19, although the impact of asthma on COVID-19 risk is still incompletely understood, and other factors may also influence COVID-19 risk in individuals with asthma (13–16). We also found significant associations of many IL-13–induced SARS-CoV-2–associated genes with type 2 inflammation in a large group of smokers with and without COPD, suggesting that the effects of IL-13 on SARS-CoV-2 risk may also be relevant in some individuals without asthma. The effects of IL-13 on SARS-CoV-2–associated genes were clearly different than the effects of IFN-α, suggesting that these two cytokines induce different antiviral mechanisms. Although the antiviral effects of IL-13 are independent of goblet cell differentiation, we found evidence that another barrier component, the mucus gel, also provides protection against infection. Taken together, these studies provide insights into airway epithelial responses that can protect against SARS-CoV-2 and might influence COVID-19 susceptibility and severity in individuals with asthma or other airway diseases.
IL-13 had a substantial effect on SARS-CoV-2 infection of HBECs as demonstrated by measurements of viral RNA and dsRNA following viral inoculation. Prior studies report a variety of effects of asthma and IL-13 on development of illnesses caused by other viruses. IL-13 can increase susceptibility of HBECs to rhinovirus infection by suppressing induction of interferons (41–43), although another study reported that prolonged pretreatment with IL-13 of HBECs reduced rhinovirus infection (24). Mice with acute allergic airway inflammation (44) and people with preexisting asthma (45) are reportedly protected from H1N1 influenza. Studies in IL-13–overexpressing transgenic mice and IL-13–deficient mice showed that IL-13 reduced respiratory syncytial virus replication and severity of illness (23). Although effects of IL-13 on the airway epithelium are an important contributor to asthma pathogenesis, it is intriguing to speculate that IL-13 responses may have evolved at least in part to provide protection against viral infections. The finding that levels of IL-13 and the related type 2 cytokine IL-4 were higher in patients with moderate COVID-19 compared with severe COVID-19 or healthy control subjects is also consistent with an antiviral role for these cytokines (46).
Many mechanisms might account for IL-13–driven inhibition of SARS-CoV-2 infection. A recent study identified 65 IFN-α–stimulated genes that mediate restriction of SARS-CoV-2 infection (47), illustrating how a single cytokine can activate a large set of antiviral pathways. We found that gene expression changes induced by IL-13 were quite distinct from those induced by IFN-α, suggesting that these cytokines activate different antiviral pathways. We initially hypothesized that the effects of IL-13 were mediated by its effects on mucus, which are driven by the transcription factor SPDEF; however, minimal SARS-CoV-2 RNA or dsRNA staining was observed in IL-13–stimulated HBECs in which SPDEF was targeted using CRISPR-Cas9, indicating a SPDEF-independent mechanism. We confirmed prior studies (20, 21) showing that IL-13 induced a decrease in expression of the SARS-CoV-2 receptor ACE2, which could contribute to decreased infection. However, the reduction in ACE2 expression was modest compared with the effects of IL-13 on infection, suggesting that other IL-13 effects should also be considered. As in the previous reports, we found that IL-13 increased expression of TMPRSS2, a protease that is important for viral entry. However, we found that the IL-13 effects were cell type–dependent: TMPRSS2 expression was increased in secretory cells but decreased in ciliated cells. Because ciliated cells were the primary cell type infected in our experiments, it is possible that decreased TMPRSS2 in ciliated cells contributed to an overall reduction in infection. We recently reported a trend toward a decrease in the proportion of ciliated cells after IL-13 stimulation (26) that would compound the ciliated cell–specific decrease in TMPRSS2 and potentially restrict SARS-CoV-2 infection. Many other host cell factors influence viral entry, RNA synthesis and translation, and egress, and further studies will be required to determine which of these contribute to the antiviral effects of IL-13.
Our studies provided clear evidence that the mucus barrier produced by HBECs in cell culture inhibits SARS-CoV-2 infection. Airway mucus is a complex hydrogel that derives its characteristic viscoelastic properties from the mucin glycoproteins MUC5B and MUC5AC (48). Prior studies establish that mucins play important roles as restriction factors for other viruses, including influenza (49, 50). We found that SARS-CoV-2 infection was decreased when mucus gels were left in place at the time of viral inoculation. Differences in mucin staining and levels of protection were observed between donors, suggesting that differences in mucus gels may be important in SARS-CoV-2 infection. Changes in airway mucus volume, composition, and organization are prominent features of many airway diseases, including asthma (48). Our finding that IL-13 restricts infection in the absence of SPDEF suggests that MUC5AC induction and goblet cell differentiation does not substantially impact IL-13–induced inhibition of SARS-CoV-2 infection, however. Further studies are required to investigate what features of airway mucins and airway mucus gels confer protection to SARS-CoV-2 infection.
Although we focused principally on the effects of IL-13, the other asthma-associated cytokines that we studied, IFN-α, IFN-γ, and IL-17, also affected the expression of various SARS-CoV-2–associated genes in HBECs. ACE2 was the most highly upregulated interferon-stimulated gene in our study. In another recent study, ACE2 expression was associated with a set of interferon-inducible genes in a subset of patients with type 2–low asthma (51); these patients exhibited characteristics resembling known risk factors for severe COVID-19, supporting further relationships between asthma endotypes and SARS-CoV-2 infection. Apart from ACE2, the interferon-stimulated SARS-CoV-2– associated genes we identified were distinct from a set of IFN-α–stimulated genes recently shown to mediate restriction of SARS-CoV-2 infection (47). This illustrates that the effects of cytokines on genes other than the large set of SARS-CoV-2–associated genes we studied will also be important to study. The most IL-17–inducible gene was PLAT, which encodes tPA (tissue plasminogen activator). Elevated levels of tPA have been reported in hospitalized patients with COVID-19 and are associated with worse respiratory status and mortality, suggesting preexisting IL-17–induced inflammation could be associated with more severe disease (52). Half of the genes modulated by interferon and IL-17 encoded proteins predicted to interact with Orf8 and NSP13, viral proteins predicted to subvert host vesicle trafficking pathways. NSP13 is also predicted to interfere with type I interferon production via interactions with TBK1 and TBKBP1. TBK1 is upregulated by IFN-α stimulation in our bulk RNA-seq dataset, and autosomal-dominant deficiencies of TBK1 are associated with severe COVID-19 (21). Hence, like IL-13, IFN-α, IFN-γ, and IL-17 alter the balance of host cell proteins in the airway epithelium, potentially modifying the outcome of COVID-19 in different subsets of people with asthma. How changes in the expression of SARS-CoV-2–associated genes in response to interferons and IL-17 affect SARS-CoV-2 infection, however, requires further investigation.
Our study has some important limitations. Although we focused on a set of SARS-CoV-2–associated genes that have been defined in previous studies, other IL-13–regulated genes are also likely to be important for antiviral effects. Some IL-13–regulated genes we identified in cell culture were not associated with a type 2 signature in cells from individuals with asthma or COPD, reflecting the influence of other factors, including other asthma mediators, or differences in IL-13 responses in cell culture versus in vivo. As individual genes that contribute to inhibition of viral infection in HBECs are identified, it will be important to specifically examine the expression of those genes in asthma and COPD. Our HBEC infection studies used only one strain of SARS-CoV-2 and cells from only three donors, and further experiments with additional strains and more donors (including donors with asthma) will be required to better understand the interactions between virus, epithelial cells, and IL-13. Finally, our infection model focuses solely on the role of epithelial cells, but the effects of IL-13 on other cell types found in the lung are also deserving of further study.
In conclusion, we found that the central asthma mediator IL-13 has a strong inhibitory effect on SARS-CoV-2 infection of HBECs. The mechanisms that account for this effect are independent of SPDEF, but widespread effects of IL-13 on expression of SARS-CoV-2–associated genes that are distinct from those induced by interferons suggest that some of these mechanisms may be novel. Although the use of IL-13 itself as a therapeutic may well be prevented by the proasthmatic effects of this cytokine, identification of IL-13–induced antiviral pathways could help address the urgent need for development of novel targeted treatments for COVID-19.
Acknowledgments
Acknowledgment
The authors thank Paul Wolters (UCSF) for providing HBECs from transplant recipients; Dingyuan Sun for assistance with HBEC culture; Melanie Ott (Gladstone Institute of Virology), Semil Choksi (UCSF), and Jeremy Reiter (UCSF) for providing advice; and the staff of the UCSF Center for Advanced Light Microscopy for technical assistance.
Footnotes
Supported by National Institutes of Health (NIH) awards U19 AI 077439 and R35 HL145235 (D.J.E.) and R00 HL135403 (W.L.E.) and University of California, San Francisco Program for Breakthrough Biomedical Research (PBBR; L.R.B.).
Author Contributions: L.R.B., W.L.E., L.R., and D.J.E. contributed to study conception and design. L.R.B., L.R., K.D.K., K.G., and L.T.Z. performed experiments. L.R.B., W.L.E., J.S., K.D.K., S.C., P.G.W., and D.J.E. analyzed and interpreted data. W.E.F. and D.J.E. supervised study execution. L.R.B. and D.J.E. drafted the manuscript. All authors reviewed the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2021-0364OC on January 4, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature . 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science . 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, et al. HGID Lab; NIAID-USUHS Immune Response to COVID Group; COVID Clinicians; COVID-STORM Clinicians; Imagine COVID Group; French COVID Cohort Study Group; Milieu Intérieur Consortium; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science . 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Combes AJ, Courau T, Kuhn NF, Hu KH, Ray A, Chen WS, et al. UCSF COMET Consortium Global absence and targeting of protective immune states in severe COVID-19. Nature . 2021;591:124–130. doi: 10.1038/s41586-021-03234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention 2020https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/evidence-table.html.
- 6. Vos T, Abajobir AA, Abate KH, Abbafati C, Abbas KM, Abd-Allah F, et al. GBD 2016 Disease and Injury Incidence and Prevalence Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet . 2017;390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Busse WW, Lemanske RF, Jr, Gern JE. Role of viral respiratory infections in asthma and asthma exacerbations. Lancet . 2010;376:826–834. doi: 10.1016/S0140-6736(10)61380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Halpin DMG, Faner R, Sibila O, Badia JR, Agusti A. Do chronic respiratory diseases or their treatment affect the risk of SARS-CoV-2 infection? Lancet Respir Med . 2020;8:436–438. doi: 10.1016/S2213-2600(20)30167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu Z, Hasegawa K, Ma B, Fujiogi M, Camargo CA, Jr, Liang L. Association of asthma and its genetic predisposition with the risk of severe COVID-19. J Allergy Clin Immunol . 2020;146:327–329.e4. doi: 10.1016/j.jaci.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Terry PD, Heidel RE, Dhand R. Asthma in adult patients with COVID-19: prevalence and risk of severe disease. Am J Respir Crit Care Med . 2021;203:893–905. doi: 10.1164/rccm.202008-3266OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sunjaya AP, Allida SM, Di Tanna GL, Jenkins C. Asthma and risk of infection, hospitalization, ICU admission and mortality from COVID-19: systematic review and meta-analysis. J Asthma . 2021 doi: 10.1080/02770903.2021.1888116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farne H, Singanayagam A. Why asthma might surprisingly protect against poor outcomes in COVID-19. Eur Respir J . 2020;56:2003045. doi: 10.1183/13993003.03045-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Skevaki C, Karsonova A, Karaulov A, Xie M, Renz H. Asthma-associated risk for COVID-19 development. J Allergy Clin Immunol . 2020;146:1295–1301. doi: 10.1016/j.jaci.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Beasley R, Hills T, Kearns N. Asthma and COVID-19: preconceptions about predisposition. Am J Respir Crit Care Med . 2021;203:799–801. doi: 10.1164/rccm.202102-0266ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martinez FD. Asthma in the time of COVID-19. Am J Respir Crit Care Med . 2021;203:785–786. doi: 10.1164/rccm.202102-0389ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA . 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med . 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA . 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med . 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jackson DJ, Busse WW, Bacharier LB, Kattan M, O’Connor GT, Wood RA, et al. Association of respiratory allergy, asthma, and expression of the SARS-CoV-2 receptor ACE2. J Allergy Clin Immunol . 2020;146:203–206.e3. doi: 10.1016/j.jaci.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sajuthi SP, DeFord P, Li Y, Jackson ND, Montgomery MT, Everman JL, et al. Type 2 and interferon inflammation regulate SARS-CoV-2 entry factor expression in the airway epithelium. Nat Commun . 2020;11:5139. doi: 10.1038/s41467-020-18781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kimura H, Francisco D, Conway M, Martinez FD, Vercelli D, Polverino F, et al. Type 2 inflammation modulates ACE2 and TMPRSS2 in airway epithelial cells. J Allergy Clin Immunol . 2020;146:80–88.e8. doi: 10.1016/j.jaci.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou W, Hashimoto K, Moore ML, Elias JA, Zhu Z, Durbin J, et al. IL-13 is associated with reduced illness and replication in primary respiratory syncytial virus infection in the mouse. Microbes Infect . 2006;8:2880–2889. doi: 10.1016/j.micinf.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roberts N, Al Mubarak R, Francisco D, Kraft M, Chu HW. Comparison of paired human nasal and bronchial airway epithelial cell responses to rhinovirus infection and IL-13 treatment. Clin Transl Med . 2018;7:13. doi: 10.1186/s40169-018-0189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bonser LR, Zlock L, Finkbeiner W, Erle DJ. Epithelial tethering of MUC5AC-rich mucus impairs mucociliary transport in asthma. J Clin Invest . 2016;126:2367–2371. doi: 10.1172/JCI84910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonser LR, Koh KD, Johansson K, Choksi SP, Cheng D, Liu L, et al. Flow-cytometric analysis and purification of airway epithelial-cell subsets. Am J Respir Cell Mol Biol . 2021;64:308–317. doi: 10.1165/rcmb.2020-0149MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koh KD, Siddiqui S, Cheng D, Bonser LR, Sun DI, Zlock LT, et al. Efficient RNP-directed human gene targeting reveals SPDEF is required for IL-13-induced mucostasis. Am J Respir Cell Mol Biol . 2020;62:373–381. doi: 10.1165/rcmb.2019-0266OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhakta NR, Christenson SA, Nerella S, Solberg OD, Nguyen CP, Choy DF, et al. IFN-stimulated gene expression, type 2 inflammation, and endoplasmic reticulum stress in asthma. Am J Respir Crit Care Med . 2018;197:313–324. doi: 10.1164/rccm.201706-1070OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Christenson SA, van den Berge M, Faiz A, Inkamp K, Bhakta N, Bonser LR, et al. An airway epithelial IL-17A response signature identifies a steroid-unresponsive COPD patient subgroup. J Clin Invest . 2019;129:169–181. doi: 10.1172/JCI121087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Steiling K, van den Berge M, Hijazi K, Florido R, Campbell J, Liu G, et al. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med . 2013;187:933–942. doi: 10.1164/rccm.201208-1449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell . 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res . 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature . 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, et al. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem . 2020;295:13958–13964. doi: 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Östling J, van Geest M, Schofield JPR, Jevnikar Z, Wilson S, Ward J, et al. U-BIOPRED Study Group IL-17-high asthma with features of a psoriasis immunophenotype. J Allergy Clin Immunol . 2019;144:1198–1213. doi: 10.1016/j.jaci.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 36. Blume C, Jackson CL, Spalluto CM, Legebeke J, Nazlamova L, Conforti F, et al. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat Genet . 2021;53:205–214. doi: 10.1038/s41588-020-00759-x. [DOI] [PubMed] [Google Scholar]
- 37. Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med . 2019;25:1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 38. Solberg OD, Ostrin EJ, Love MI, Peng JC, Bhakta NR, Hou L, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med . 2012;186:965–974. doi: 10.1164/rccm.201201-0027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bhakta NR, Solberg OD, Nguyen CP, Nguyen CN, Arron JR, Fahy JV, et al. A qPCR-based metric of Th2 airway inflammation in asthma. Clin Transl Allergy . 2013;3:24. doi: 10.1186/2045-7022-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Christenson SA, Steiling K, van den Berge M, Hijazi K, Hiemstra PS, Postma DS, et al. Asthma-COPD overlap: clinical relevance of genomic signatures of type 2 inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med . 2015;191:758–766. doi: 10.1164/rccm.201408-1458OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Contoli M, Message SD, Laza-Stanca V, Edwards MR, Wark PAB, Bartlett NW, et al. Role of deficient type III interferon-lambda production in asthma exacerbations. Nat Med . 2006;12:1023–1026. doi: 10.1038/nm1462. [DOI] [PubMed] [Google Scholar]
- 42. Lachowicz-Scroggins ME, Boushey HA, Finkbeiner WE, Widdicombe JH. Interleukin-13-induced mucous metaplasia increases susceptibility of human airway epithelium to rhinovirus infection. Am J Respir Cell Mol Biol . 2010;43:652–661. doi: 10.1165/rcmb.2009-0244OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Contoli M, Ito K, Padovani A, Poletti D, Marku B, Edwards MR, et al. Th2 cytokines impair innate immune responses to rhinovirus in respiratory epithelial cells. Allergy . 2015;70:910–920. doi: 10.1111/all.12627. [DOI] [PubMed] [Google Scholar]
- 44. Samarasinghe AE, Woolard SN, Boyd KL, Hoselton SA, Schuh JM, McCullers JA. The immune profile associated with acute allergic asthma accelerates clearance of influenza virus. Immunol Cell Biol . 2014;92:449–459. doi: 10.1038/icb.2013.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Myles PR, Semple MG, Lim WS, Openshaw PJM, Gadd EM, Read RC, et al. Influenza Clinical Information Network (FLU-CIN) Predictors of clinical outcome in a national hospitalised cohort across both waves of the influenza A/H1N1 pandemic 2009-2010 in the UK. Thorax . 2012;67:709–717. doi: 10.1136/thoraxjnl-2011-200266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Su Y, Chen D, Yuan D, Lausted C, Choi J, Dai CL, et al. ISB-Swedish COVID19 Biobanking Unit Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell . 2020;183:1479–1495.e20. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martin-Sancho L, Lewinski MK, Pache L, Stoneham CA, Yin X, Becker ME, et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol Cell . 2021;81:2656–2668.e8. doi: 10.1016/j.molcel.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bonser LR, Erle DJ. Airway mucus and asthma: the role of MUC5AC and MUC5B. J Clin Med . 2017;6:112. doi: 10.3390/jcm6120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zanin M, Baviskar P, Webster R, Webby R. The interaction between respiratory pathogens and mucus. Cell Host Microbe . 2016;19:159–168. doi: 10.1016/j.chom.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ehre C, Worthington EN, Liesman RM, Grubb BR, Barbier D, O’Neal WK, et al. Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc Natl Acad Sci USA . 2012;109:16528–16533. doi: 10.1073/pnas.1206552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Camiolo M, Gauthier M, Kaminski N, Ray A, Wenzel SE. Expression of SARS-CoV-2 receptor ACE2 and coincident host response signature varies by asthma inflammatory phenotype. J Allergy Clin Immunol . 2020;146:315–324.e7. doi: 10.1016/j.jaci.2020.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zuo Y, Warnock M, Harbaugh A, Yalavarthi S, Gockman K, Zuo M, et al. Plasma tissue plasminogen activator and plasminogen activator inhibitor-1 in hospitalized COVID-19 patients. Sci Rep . 2021;11:1580. doi: 10.1038/s41598-020-80010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]