Highlights
-
•
Six deformable registration algorithms (ANTs, DRAMMS, ART, NiftyReg, SSD-FFD, and NMI-FFD) were compared for the registration of longitudinal breast DCE-MRI scans.
-
•
The effect of these registration methods was evaluated on the predictive value of radiomic features to model RFS in women undergoing NAC for locally advanced breast cancer.
-
•
The accuracies of the different registration methods were also evaluated with expert-defined anatomical landmarks as a gold standard.
Keywords: Breast cancer, Deformable image registration, Longitudinal breast MRI, Parametric response maps, Radiomic features, Tumor heterogeneity, Recurrence-free survival analysis
Abstract
Purpose
Image registration plays a vital role in spatially aligning multiple MRI scans for better longitudinal assessment of tumor morphological features. The objective was to evaluate the effect of registration accuracy of six established deformable registration methods(ANTs, DRAMMS, ART, NiftyReg, SSD-FFD, and NMI-FFD) on the predictive value of extracted radiomic features when modeling recurrence-free-survival(RFS) for women after neoadjuvant chemotherapy(NAC) for locally advanced breast cancer.
Methods
130 women had DCE-MRI scans available from the first two visits in the ISPY1/ACRIN-6657 cohort. We calculated the transformation field from each of the different deformable registration methods, and used it to compute voxel-wise parametric-response-maps(PRM) for established four kinetic features.104-radiomic features were computed from each PRM map to characterize intra-tumor heterogeneity. We evaluated performance for RFS using Cox-regression, C-statistic, and Kaplan-Meier(KM) plots.
Results
A baseline model(F1:Age, Race, and Hormone-receptor-status) had a 0.54 C-statistic, and model F2(baseline + functional-tumor-volume at early treatment visit(FTV2)) had 0.63. The F2+ANTs had the highest C-statistic(0.72) with the smallest landmark differences(5.40±4.40mm) as compared to other models. The KM curve for model F2 gave p=0.004 for separation between women above and below the median hazard compared to the model F1(p=0.31). A models augmented with radiomic features, also achieved significant KM curve separation(p<0.001) except the F2+ART model.
Conclusion
Incorporating image registration in quantifying changes in tumor heterogeneity during NAC can improve prediction of RFS. Radiomic features of PRM maps derived from warping the DCE-MRI kinetic maps using ANTs registration method further improved the early prediction of RFS as compared to other methods.
Introduction
Breast cancer is the second leading cancer in the United States and continues to increase in incidence [1]. An estimated 281,550 new cases of invasive breast cancer will be diagnosed among women in 2021 that will result in approximately 43,600 breast cancer deaths in women in the same year [1]. This emphasizes the need for early tumor diagnosis and accurate assessment of treatment response. In the budding precision medicine era, it is accepted that a predefined therapy treatment may not provide effective treatment for all patients or yield identical responses. Identifying the non-responders at an early stage may direct these patients to different therapies, leading to potentially better outcomes [2,3]. This highlights the importance of early predictions of patient response to cancer treatments or therapies [4,5]. The longitudinal patterns of tumor response may be a significant predictor in evaluating treatment response and measuring the likelihood of overall survival during neoadjuvant chemotherapy (NAC). At present, NAC has become a standard preoperational treatment paradigm for locally advanced breast cancer patients, which enables monitoring the longitudinal changes [6,7].
Breast cancer is a heterogeneous disease, with intra-tumor heterogeneity manifesting in imaging modalities both spatially and temporally. Recent studies have shown that analysis of intra-tumor heterogeneity manifests in raw image data [8,9] or parametric maps of kinetic features [10,11], which can help in the evaluation of breast cancer therapy response. Regional analysis like voxel-based analysis can capture heterogeneity better than average-based whole-tumor analysis [12,13]. Conventional biomarkers have fallen short of fully capturing intra-tumor heterogeneity as reported in previous studies [14], [15], [16]. Several studies have shown that voxel-level heterogeneity measures derived from dynamic-contrast-enhanced MRI (DCE-MRI) can be associated with NAC outcomes [11,17].
During treatment of locally advanced breast cancer, monitoring tumor changes during NAC effectively and predicting the long‐term pathologic response and recurrence-free survival (RFS) are important for patient management [18]. For this purpose, clinicians refer to longitudinal breast MR images acquired in multiple visits, days or weeks apart [7]. In contrast, to quantify heterogeneous tumor changes, the challenge is to evaluate patients over multiple longitudinal MRI acquisitions to assess the morphological and functional tumor changes during treatment. For a better assessment of tissue morphological features, inter-visit image registration can play a vital role in bringing multiple MRI scans in the same space [19]. Deformable image registration techniques can also evaluate the spatial heterogeneity of voxel-level changes [18,20,21]. There are several image registration methods that exist and can be used for this task. Moreover, a comprehensive evaluation of these image registration methods remains an unexplored topic, motivating our work in this article.
The purpose of our study is to evaluate the effect of the registration accuracy of several deformable image registration methods on the predictive value of extracted radiomic features to model RFS in women undergoing NAC for locally advanced breast cancer. We also evaluate and compare six deformable image registration methods based on 2,380 landmarks individually marked by two radiologists in an independent dataset of longitudinal breast MR scans.
Methods
Study population and MRI data acquisitions
There was no consent required for this retrospective study as de-identified data was obtained from “The Cancer Imaging Archive” (TCIA) [22]. This study has been approved by the Institutional Review Board of the University of Pennsylvania. The patient population analyzed in our study was a subset of patients with longitudinal breast DCE-MRI scans, publicly available at TCIA, from the ISPY1/ACRIN 66577 trial. The women enrolled in the study had T3 breast tumors measuring 3cm and received anthracycline-cyclophosphamide neoadjuvant chemotherapy. The imaging protocols were previously described according to the ISPY1/ACRIN 66577.
All MRI experiments were performed at 1.5 Tesla using a dedicated breast coil. Four longitudinal MRI examinations were performed at different time points during the therapy. In this study, we used MRIs from the first two visits, focusing on the ability to predict likelihood of survival early in the course of the therapy. The first MRI (T1, MRI1 (pre-treatment)) was performed at four weeks before anthracycline-cyclophosphamide (AC) chemotherapy. The second MRI (T2, MRI2 (early-treatment)) was performed at least two weeks after the first cycle of AC and before the second cycle of AC.
Cohort for evaluating registration accuracy
Prior to analyzing the cohort for RFS, an independent “test-retest” sample of 14 subjects with retrospectively collected breast MR images, also publicly available by the ISPY/ACRIN 6657, were used to evaluate the registration accuracy in this study [23,24]. This dataset was previously used and described in the literature to evaluate the registration accuracy for different registration methods (DRAMMS, NMI-FFD and SSD-FFD) [19]. Briefly, two experienced breast imaging radiologists annotated landmarks individually in the images. A total of 2,380 landmarks (median 78 (IQR:63.25-107.25) per patient) were marked and labeled at breast boundaries, nipples, internal milk ducts, chest walls, glandular structures, vessels, and tumors. We registered the follow-up image to the baseline image for each patient (median 17.5 (IQR:14-55.75) days apart). The expert-defined landmarks were used as the gold standard. In the current study, we included and compared the registration accuracy of ANTs, ART and NiftyReg methods with above mentioned methods.
Cohort for the prediction of NAC RFS
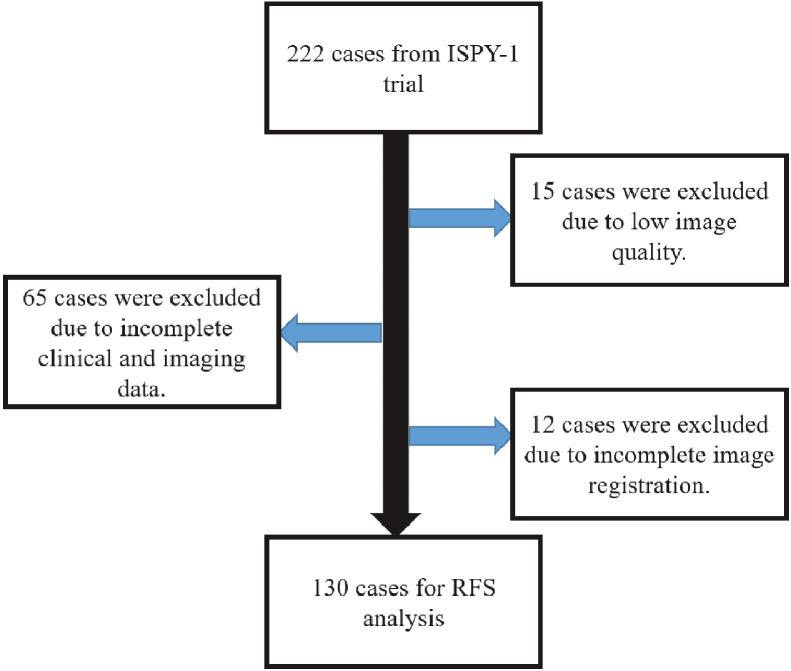
The functional tumor volume (FTV) calculated at pre-treatment (FTV1) and early-treatment visits (FTV2) was available for each DCE-MRI scan. According to STEEP criteria [25], RFS outcomes were calculated for each woman, defined as the time from the first chemotherapy cycle to the event (i.e., recurrence or death). The inclusion and exclusion criteria used to attain the final study cohort are summarized in Figure 1. After excluding 65 scans due to incomplete clinical and/or imaging data at the first two visits, 15 scans due to the low image quality, and 12 scans for which image registration failed, our final analysis included 130 women with 38 events from the ISPY1/ACRIN 66577 cohort for RFS analysis. Pathologic Complete Response (pCR) information was missing for 5 participants, leaving 125 patients for pCR analysis, having an overall tumor response rate for our study sample of about 28.5%.
Fig. 1.
Inclusion and exclusion criteria for our study.
Image pre-processing
All images were pre-processed using N3 bias-field normalization to minimize intensity variations caused by the imaging sequences or artifacts [26] . Histogram matching was applied between pre- and early-treatment images for more accurate registration [10]. These image pre-processing steps were applied before registration and quantitative analysis.
Registration methods and parameter setting
After the pre-processing steps, we used six different deformable image registration methods outlined in Supplementary Table S1 (ANTs, DRAMMS, ART, NiftyReg, SSD-FFD, and NMI-FFD) for spatial and anatomical alignment of the early-treatment MRI scans to their corresponding pre-treatment MRIs. Each deformable image registration method is usually described by three important components: 1) deformation model, 2) similarity metric, and 3) regularization, as shown in Table S1. We chose these methods because they are publicly available and previously reported in the literature to register MR images in general [19,[27], [28], [29], [30], [31]]. Below we summarize the registration methods evaluated in our study. The parameters stated below were not set arbitrarily. Generally, the authors of the below-mentioned studies evaluated a wide variety of parameter settings and reported the ones that gave the most accurate alignment of longitudinal breast MR images.
Advanced normalization tools (ANTs)
ANTs [32] splits the diffeomorphic deformation into two symmetric components. Instead of deforming the first image into the space of the second image, which is usually not symmetric to the input images, ANTs work by deforming two input images, each towards the “midpoint” image. As a result, the formulation is symmetric to the input images. We used cross-correlation with symmetric image normalization method (SyN) transformation in this study. The ANTs registration tool can be found at http://stnava.github.io/ANTs/.
Deformable registration via attribute matching and mutual-saliency weighting (DRAMMS)
This deformable image registration method [33] is based on attribute-matching and mutual-saliency-weighting, which uses high-dimensional Gabor texture attributes obtained from the multi-scale and multi-orientation neighborhoods of voxels [31]. The DRAMMS registration tool is publicly available at http://www.cbica.upenn.edu/sbia/software/dramms/. The regularization weight (g) is an important parameter that may need to be tuned in the DRAMMS registration method. It is commonly set between 0 and 1. We used g=0.3, which led to slightly smoother deformations than the default setting (g=0.2).
Automatic registration toolbox (ART)
In the ART [28,34] method, the intensity values of all the neighborhood voxels are stacked to generate a high dimensional feature vector that characterizes a voxel. The inner-product of two feature vectors and an idempotent and symmetric matrix that removes the mean of the vector it pre-multiplies defines the similarity of two voxels. ART only searches correspondences at voxels whose gradient norms are in a certain upper percentile of the gradient magnitude histogram when analyzing the highest image resolution. For ART, sd is defined as specifying the degree of smoothing applied to the displacement vector field, which typically ranges from 5.0 to 12.0 mm. we set sd=8.0 in our evaluation. This ART registration tool is publicly available at http://www.nitrc.org/projects/art/.
Free form deformation (FFD)
FFD is a geometric transformation model and combined with normalized mutual information (NMI) similarity metric in the original work [30]. It can be used along with other similarity metrics. Two important parameters are control point spacing δ with a larger spacing that capture more global deformations; and regularization weight λ with larger weights that provide smoother deformations. We used δ = 10 mm, larger spacing means capturing more global deformations; and λ =0.01 in our evaluation. NMI-FFD and SSD-FFD were included in this study. This software tool is publicly available at http://www.doc.ic.ac.uk/∼dr/software/.
Nifty registration (NiftyReg)
This method is used for global and local image registration [27,30]. The global registration uses the block-matching technique whereas cubic B-splines to capture the local registration. This method is based on free form deformation [30], further extended as fast free form deformation [27] to speed-up registration. Normalized Mutual Information (NMI) and the Bending-Energy are used in this study. This software tool is publicly available at https://www.nitrc.org/projects/niftyreg/.
Evaluation criterion for registration accuracy
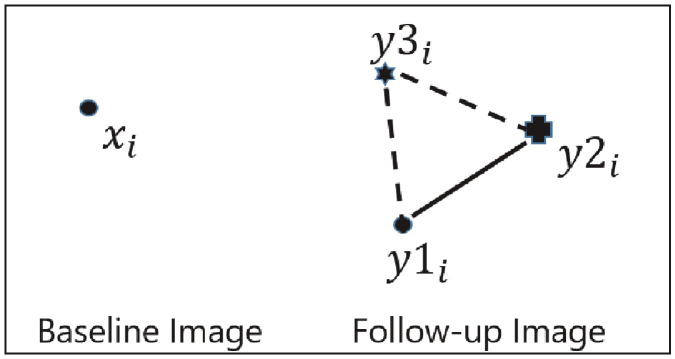
The inter-expert landmark differences and algorithm‐to‐expert landmark differences were first computed to evaluate the registration accuracy using the previously described method [19]. Two radiologists independently found their corresponding points in the follow-up image, denoted as y1i, and y2i. The inter-expert landmark difference (the length of the solid line as shown in Figure 2), for each landmark, was computed as the Euclidean distance between the landmark points designated by each of the two experts.
| d (y1i, y2i), for i=1,2,3,……………….……,k | (1) |
Fig. 2.
Depiction of inter-expert and algorithm versus expert landmark differences.
The algorithm‐to‐expert landmark difference (the average length of the dashed lines in Figure 2) was computed as:
| [d (y3i, y1i) + d (y3i, y2i)]/2, for i=1,2,3,…………,k | (2) |
where d (•,•) is again the Euclidean distance and i varies over the k landmarks.
Computation of parametric response maps (PRM) and their radiomic features for the prediction of RFS
DCE-MRI images consisted of a pre-contrast (t0) image and images at two times, t1 and t2, following injection of the contrast agent. The signal intensity of each voxel was computed at each time point S(t). Four voxel-wise kinetic features were calculated from the t0, t1, and t2 intensity values at each voxel, for both pre- and early-treatment images: peak enhancement (PE), signal enhancement ratio (SER), wash-in slope (WIS), and wash-out slope (WOS) [18]. We also computed the difference between the kinetic feature maps from pre- and early-treatment MR images without registration. Resampling value of 1 mm was used for higher accuracy. Rotation variant LBP was used and a total of 27 neighbors were considered for computation of LBP. Linear Interpolation was used for the MRI images. We extracted 104 radiomic texture features (detailed list of radiomic features in Supplementary Table S2) from each without registered kinetic features map, using the publicly available software, Cancer Phenomics Toolkit (CaPTk) [35], applied principal component (PC) analysis to the 104-dimensional feature vector after calculating the z-score, and retained the first four PCs for modeling (i.e., one covariate for every 10 events in our cohort). To quantify voxel scale changes between visits, corresponding voxels between pre- and early-treatment visits maps must be determined. The transformation field derived from the image registration allowed us to construct the voxel-wise maps of change in a given kinetic feature (also called the PRM) between corresponding points at the pre- and early treatment visits. For each of the six different deformable registration methods (ANTs, DRAMMS, ART, NiftyReg, NMI-FFD, and SSD-FFD), we calculated the transformation field and used it to warp the kinetic maps obtained from early-treatment MR images. For a voxel x in the pre-treatment image, let T12(x) indicate the voxels in the early-treatment image, which was warped to x. For a given Kinetic Feature (KF), one of PE, SER, WIS, or WOS, we computed at the pre- or early-treatment visit(KF1 or KF2), the PRM value at each voxel x18 is calculated as:
| PRM(x)=Jacobian (KF2(T12(x)) - KF1(x)) | (3) |
Jacobian is the proportional volume change at x between pre- and early-treatment visits, which scales the value when a voxel in one image is the larger or smaller volume in the corresponding image. The kinetic features (and their PRMs and radiomic features) were only measured within tumor. A signal enhancement ratio method was used to analyze DCE-MR images and segment functional tumor volumes (FTV1 and FTV2). A total of 104 radiomic features were extracted from each PRM kinetic map, using CaPTk [35], applied principal component (PC) analysis to the 104-dimensional feature vector after calculating the z-score, and retained the first four PCs for modeling (i.e., one covariate for every 10 events in our cohort). Figure 3 shows the flow chart of the analysis performed in this study.
Fig. 3.
Outline of the process using different deformable image registration methods. (a) Acquiring dynamic contrast-enhanced magnetic resonance imaging before and during neoadjuvant chemotherapy. (b) Computing different kinetic maps from both pre-treatment and early treatment MR images. (c) Applying different deformable image registration method to obtain the transformation field, followed by warping the kinetic maps obtained from early treatment MR images. (d) Parametric response maps (PRM) showing kinetic feature variation. (e) Computing radiomic features extraction followed by principal component analysis. (f) Building multi-variable models using the meta feature and comparing their performance in predicting recurrence-free survival (RFS).
Statistical analysis
Cox proportional hazards regression was used to model 5-year RFS for each set of covariates, comparing eight models: 1) First, a model with the baseline covariates of age, race, and hormone receptor status (model F1), 2) model F1 plus functional tumor volume at the early-treatment visit (FTV2) (model F2), 3) model F2 plus radiomic feature without registration(model F3) and 4-9) the covariates in F2 with the addition of the radiomic feature PCs derived from each registration method (ANTs, DRAMMS, ART, NiftyReg, SSD-FFD, and NMI-FFD). The C-statistic [36] was calculated both on the full models as well as 3-fold cross-validation averaged over 100 replicates. The full models were used to generate corresponding KM plots, and the log-rank p-value was used for estimating significance of KM separation and hazard ratios (HR) in Cox modeling. Each subject's risk signature was dichotomized at the median into high- and low-risk groups. For a given model, the risk signature of each subject was defined as that subject's values of the covariates in the model (age, race, hormone receptor status, FTV2, and selected features) weighted by the corresponding coefficients of those covariates in the model, to arrive at a predicted risk score. For every model, we also used a randomization test to calculate a p-value versus the null hypothesis that the improvement in the C statistic, obtained by adding the radiomic data, was due only to chance. This randomization test was performed for both full-model and 3-cross-validated with 100 repetitions C-statistic. The p-value of 0.05 cutoff was used to indicate statistical significance throughout this article. The radiomic features were extracted using Cancer Imaging Phenomics Toolkit (CaPTK) [35] version 1.7.2. The statistical analysis was computed using R version 3.3.2.
Results
Landmark-based registration accuracy
The mean and SD of landmark difference(mm) are listed for different registration methods (Table 1). The mean inter-expert landmark differences was 3.12 ± 2.84 mm. The landmark differences of all the automated registration methods evaluated were larger than the inter-expert differences. Among the automated registration methods included in our study, ANTs had the smallest landmark differences (5.40 ± 4.40 mm) and was closest to the inter-expert differences, followed by the DRAMMS (6.05 ± 4.86 mm), NiftyReg (7.02±4.23), NMI-FFD (8.21 ± 3.81 mm), and SSD-FFD (9.46 ± 4.55 mm). ART had the largest landmark differences (10.33 ± 2.79 mm) from the inter-expert differences.
Table 1.
Mean and Standard Deviation (SD) of Landmark differences(mm) for different registration methods and inter-expert.
| Inter-expert | ANTs | DRAMMS | ART | NiftyReg | NMI-FFD | SSD-FFD | |
|---|---|---|---|---|---|---|---|
| Mean | 3.12 | 5.40 | 6.05 | 10.33 | 7.02 | 8.21 | 9.46 |
| SD | 2.84 | 4.40 | 4.86 | 2.79 | 4.23 | 3.81 | 4.55 |
Recurrence-free survival
130 women were used in our study for RFS analysis. The clinical, demographic, and histopathologic information were available for each patient, as shown in Table 2. In this study cohort, 38 women (29.2%) had recurrence/death events, and 92 women (70.8%) were event-free until their last available follow-up (Table 2).
Table 2.
Patient characteristics for our study from the ISPY-1/ACRIN 6657 cohort.
| Non-recurrent cases N=92(70.8%) |
Recurrent/death cases N=38(29.2%) |
P-Value | |
|---|---|---|---|
| Age | |||
| Median age (IQR), years | 47.73(41.31-53.30) | 48.77(39.16-53.69) | |
| Laterality | p = 0.78 | ||
| Left | 46(50%) | 18(47.4%) | |
| Right | 46(50%) | 20(52.6%) | |
| Race | p = 0.63 | ||
| White or Hispanic | 69(75%) | 30(78.9%) | |
| Others | 23(25%) | 8(21.1%) | |
| Progesterone Receptor Status | p = 0.89 | ||
| Negative | 52(56.5%) | 21(55.3%) | |
| Positive | 40(43.5%) | 17(44.7%) | |
| Hormone Response Status | p = 0.38 | ||
| HER2+ | 30(32.6%) | 17(44.7%) | |
| HR+ and HER2- | 39(42.4%) | 12(31.6%) | |
| Triple-negative | 23(25%) | 9(23.7%) | |
The baseline model (F1: age, race, and hormone receptor status) provided a C-statistic of 0.54 with a full-model and mean 3-fold cross-validated with 100 repetitions C-statistic of 0.44 (SD:0.07) (Table 3). Fitting this baseline (F1) model to the full dataset, none of the covariates (age, race, and hormone receptor status) showed a statistically significant association with RFS (Table 4). When adding FTV2 to the baseline model (F2), this F2 model provided a full-model C-statistic of 0.63 and a mean 3-fold cross-validated with 100 repetitions C-statistic of 0.53 (SD:0.08) (Table 3). Fitting this model to the full dataset, only FTV2 showed a statistically significant association with RFS (hazard ratio: 1.62, p < 0.001), as shown in Table 5.
Table 3.
C-statistics of the F1: baseline and F2: baseline + FTV2 for full and 3-fold cross-validation (with 100 rep) Cox models. SD: Standard Deviation; rep: repetitions.
| Model | Covariates | C-statistic (Full model) | Mean C-statistic (SD) (3-fold cross-validation with 100 rep) |
|---|---|---|---|
|
|
|
|
| F2: F1+FTV2 | Age, Race, Hormone receptor status, FTV2 |
|
|
| F3: F2+Without Registration | Age, Race, Hormone receptor status, FTV2, radiomics features without registration. |
|
|
Table 4.
Multi-variable analyses of the F1: baseline model using the full dataset for hazard ratio assessment.
| Covariate | Hazard ratio | 95% CI | p-value |
|---|---|---|---|
| Age | 0.99 | 0.95-1.03 | 0.72 |
| Race | 0.91 | 0.41-1.99 | 0.82 |
| Hormone receptor status | 1.11 | 0.74-1.67 | 0.59 |
Table 5.
Multi-variable analyses of the F2: baseline and FTV2 model using the full dataset for hazard ratio assessment. * p <0.05
| Covariate | Hazard ratio | 95% CI | p-value |
|---|---|---|---|
| Age | 0.98 | 0.95-1.02 | 0.59 |
| Race | 0.85 | 0.39-1.88 | 0.69 |
| Hormone receptor status | 0.99 | 0.64-1.52 | 0.97 |
| FTV2 | 1.62 | 1.19-2.20 | <0.001* |
For both full and 3-fold cross-validated with 100 repetitions models, the C-statistic of Cox models including F2 + PCs of PRM radiomic features, as derived from different registration methods were higher than the C-statistic of the baseline (F1) model, F2 model and the model incorporating radiomic features without registration. The F2 + radiomics without registration model (F3 model) provided a C-statistic of 0.66 and a mean 3-fold cross-validated with 100 repetitions C-statistic of 0.55 (SD:0.07). Among the automated registration methods (Table 6), F4(F2+ANTs) model had the highest performance for the full model (C-statistic of 0.72 with p = 0.01) as well as a mean 3-fold cross-validated with 100 repetitions (C-statistic of 0.63(SD: 0.07) with p = 0.01). F5(F2+DRAMMS) model provided a C-statistic of 0.70 with a full-model and mean 3-fold cross-validated with 100 repetitions C-statistic of 0.58(SD:0.07), followed by F2+NiftyReg, F2+NMI-FFD, and F2+SSD-FFD. F6 (F2 + ART) had the lowest performance for both full (C-statistic of 0.66) and 3-fold cross-validation with 100 repetitions (mean C-statistic of 0.54(SD: 0.08)).
Table 6.
Mean C-statistics of full and 3-fold cross validation (with 100 rep) Cox models including F2 + PCs of PRM radiomic features, as derived from each registration method. P value computed using randomization test. SD:Standard Deviation.* p <0.05
| Model | C-statistic (Full model) | p-value | Mean C-statistic (SD) (3-fold cross-validation with 100 rep) | p-value |
|---|---|---|---|---|
| F4: F2+ANTs | 0.72 | 0.01* | 0.63(0.07) | 0.01* |
| F5: F2+DRAMMS | 0.70 | 0.05 | 0.58(0.07) | 0.07 |
| F6: F2+ART | 0.66 | 0.20 | 0.54(0.08) | 0.16 |
| F7: F2+NiftyReg | 0.68 | 0.06 | 0.56(0.08) | 0.20 |
| F8: F2+NMI-FFD | 0.67 | 0.11 | 0.55(0.07) | 0.07 |
| F9: F2+SSD-FFD | 0.67 | 0.14 | 0.56(0.08) | 0.17 |
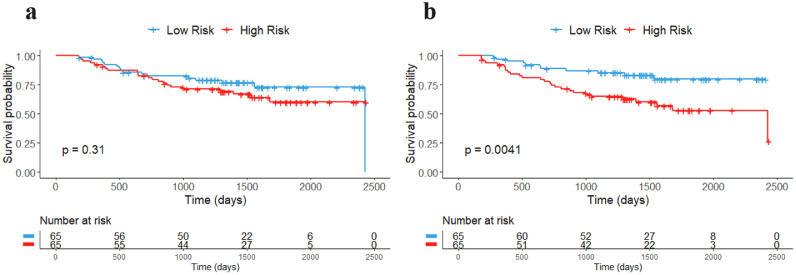
When combining the FTV2 with the baseline predictors, the KM plots (Figure 4) showed a greater separation between high-risk and low-risk patients based on their median risk score with log-rank p = 0.004 as compared to the baseline predictors (log-rank p = 0.31).
Fig. 4.
Kaplan-Meier plots of RFS for full models. a) F1: Baseline, and b) F2: F1+FTV2.
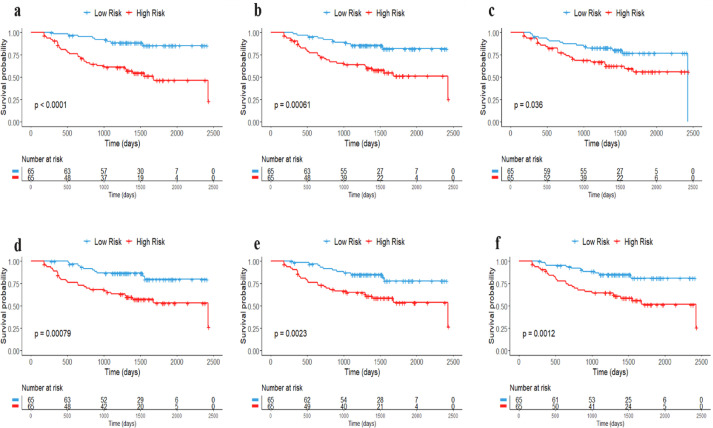
The F4 model gave a greater separation between high-risk and low-risk patients with log-rank p<0.0001 as compared to other models (F5: log-rank p = 0.00061, F6: log-rank p = 0.036, F7: log-rank p=0.00079, F8: log-rank p = 0.0023, and F9: log-rank p = 0.0012) as shown in Figure 5.
Fig. 5.
Kaplan-Meier plots of RFS (full models) for different registration methods. a) F2+ANTs, b) F2+DRAMMS, c) F2+ART, d) F2+Niftyreg, e) F2+NMI-FFD, and f) F2+SSD-FFD.
Discussion and conclusion
It has been shown that early-treatment response assessment helps in optimizing patient care and treatment alteration [37,38]. Voxel-wise 3-D volumetric changes can be automatically captured by deformable image registration. This makes it a potentially suitable choice for quantifying the spatially heterogeneous changes within tumors over the course of therapy. This study evaluated six different deformable registration algorithms (ANTs, DRAMMS, ART, NiftyReg, SSD-FFD, and NMI-FFD) for registration of longitudinal breast DCE-MRI scans. We evaluated the effect of the registration accuracy of these registration methods on the predictive value of radiomic features to model RFS in women undergoing NAC for locally advanced breast cancer. We also compared the accuracies of different registration methods using expert-defined landmarks as gold standard.
Our work adds several important findings to the literature. We showed preliminary results of using the ANTs registration method, which had the best accuracy in registering the longitudinal breast MR images as evidenced by the smallest landmark differences and being closest to the inter-expert differences. DRAMMS had earlier demonstrated smaller landmark difference reported in previous studies [19], which were almost 25–40% smaller than the difference from other registration methods (CC-FFD, NMI-FFD, SSD-FFD, Demons, and Diffeomorphic Demons). Our results suggest that ANTs registration outperforms other methods (DRAMMS, ART, NiftyReg, SSD-FFD, and NMI-FFD) in terms of registration accuracy. The registration process improves the alignment of the input images, hence ideally resulting in improved performance of extracted PRM features. Several studies reported for prediction of RFS without integration of image registration [39], [40], [41]. Our results suggest that indeed registration methods can improve early prediction of RFS, regardless of which method is used. Furthermore, improvement in registration accuracy also results in further improvement of the extracted PRM radiomic features. Our results suggest that radiomic features of PRM maps using the ANTs registration method significantly improved the early prediction of survival during NAC compared to other methods.
In our study, we extracted PRMs derived from warping the DCE-MRI kinetic maps using registration methods. The radiomic features based on the PRMs of kinetic features are also important in capturing tumor heterogeneity to augment models of RFS. The framework of the current study consists of robust registration, voxel-wise changes of PRMs of kinetic features, radiomic feature and principal component analysis provide statistically significant improvements over previous similar analyses in predicting RFS [7,14]. By adding registration, we improved the PRM feature performance, with further improvement for registration methods that are most accurate.
Important limitations of our study must be noted. We analyzed a relatively small sample size (n=130) of the women with small number of events for RFS analysis. We plan to perform another round of evaluation when the imaging data from a larger independent validation data set such as ISPY-2 becomes available. An extension of this study can be done by applying these analyses to longitudinal images at mid- and late- treatment time points to better characterize heterogeneous tumor responses and the effects of treatment over time. The accuracy of these registration methods will also be tested for large anatomical variabilities or imaging device differences (e.g., 2-D ultrasound to 3-D MRI) in future studies. Other registration methods such as DROP, HAMMER, Plastimatch, and MIND will also be evaluated in future studies. Registration accuracy can be further improved by to utilizing multiple registration methods in a meta-analysis framework, where an underperforming or failed method in a task can be overcome by other methods. Preliminary results for the evaluation of meta-registration pipelines have been published in recent articles. For example, the combination of ANTs and DRAMMS in a multi-atlas labeling framework, results in a high-performance method, as shown in a recent MICCAI segmentation challenge [42]. Another study focused on the combination of ANTs, NiftyReg, and DROP and showed significantly reduced registration errors in pulmonary images [43]. Such a meta-registration framework can also be explored for longitudinal breast MRI in the future studies and may provide even more predictive signatures in treatment response assessment.
In conclusion, we evaluated the effect of the registration accuracy of several deformable registration methods on the predictive value of radiomic features to model RFS in women undergoing NAC for locally advanced breast cancer. Our study also performed a comprehensive evaluation of the deformable image registration methods (ANTs, DRAMMS, ART, NiftyReg, SSD-FFD, and NMI-FFD) for longitudinal breast MR images to evaluate registration accuracy, using expert-annotated landmarks as gold standard. Our results showed a two-fold conclusion that analyzing registration-quantified tumor changes improves the prediction accuracy for RFS analysis and improvement in the registration process results in further improvements in the performance of PRM radiomic features when predicting RFS. In our study, the radiomic features of PRM maps derived from warping the DCE-MRI kinetic maps using the ANTs registration method significantly improved early prediction of survival during NAC.
CRediT authorship contribution statement
Snekha Thakran: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing – original draft. Eric Cohen: Formal analysis, Writing – review & editing. Nariman Jahani: Software, Writing – review & editing. Susan P. Weinstein: Methodology, Writing – review & editing. Lauren Pantalone: Investigation, Project administration, Writing – review & editing. Nola Hylton: Investigation, Writing – review & editing. David Newitt: Investigation, Writing – review & editing. Angela DeMichele: Validation, Writing – review & editing. Christos Davatzikos: Methodology, Investigation, Writing – review & editing. Despina Kontos: Conceptualization, Methodology, Visualization, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the NIH grant: 5R01CA197000-05.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101411.
Appendix. Supplementary materials
References
- 1.American Cancer Society. Cancer Society . American Cancer Society; Atlanta: 2021. Cancer Facts & Figures 2021. [Google Scholar]
- 2.Wistuba I.I., Gelovani J.G., Jacoby J.J., Davis S.E., Herbst R.S. Methodological and practical challenges for personalized cancer therapies. Nat. Rev. Clin. Oncol. 2011;8:135–141. doi: 10.1038/nrclinonc.2011.2. [DOI] [PubMed] [Google Scholar]
- 3.Huang E., McNeese MD, Strom EA, Perkins GH, Katz A, Hortobagyi GN, Valero V, Kuerer HM, Singletary SE, Hunt KK, Buzdar AU, Buchholz TA. Locoregional treatment outcomes for inoperable anthracycline-resistant breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2002;53:1225–1233. doi: 10.1016/s0360-3016(02)02878-x. [DOI] [PubMed] [Google Scholar]
- 4.Dowsett M., Dunbier A.K. Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clin. Cancer Res. 2008;14:8019–8026. doi: 10.1158/1078-0432.CCR-08-0974. [DOI] [PubMed] [Google Scholar]
- 5.McDermott U., Settleman J. Personalized cancer therapy with selective kinase inhibitors: an emerging paradigm in medical oncology. J. Clin. Oncol. 2009;27:5650–5659. doi: 10.1200/JCO.2009.22.9054. [DOI] [PubMed] [Google Scholar]
- 6.Fangberget A., Nilsen L.B., Hole K.H., Holmen M.M, Engebraaten O., Naume B., Smith H.J., Olsen D.R., Seierstad T. Neoadjuvant chemotherapy in breast cancer-response evaluation and prediction of response to treatment using dynamic contrast-enhanced and diffusion-weighted MR imaging. Eur. Radiol. 2011;21:1188–1199. doi: 10.1007/s00330-010-2020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hylton N.M., Blume J.D., Bernreuter W.K., Pisano E.D., Rosen M.A., Morris E.A., Weatherall P.T., Lehman C.D., Newstead G.M., Polin S., Marques H.S., Esserman L.J., Schnall M.D. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy -- Results from ACRIN 6657/I-SPY TRIAL. Radiology. 2012;263:663–672. doi: 10.1148/radiol.12110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teruel J.R., Heldahl M.G., Goa P.E., Pickles M., Lundgren S., Bathen T.F., Gibbs P. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed. 2014;27:887–896. doi: 10.1002/nbm.3132. [DOI] [PubMed] [Google Scholar]
- 9.Parikh J., Selmi M., Charles-Edwards G., Glendenning J., Ganeshan B., Verma H., Mansi J., Harries M., Tutt A., Goh V. Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology. 2014;272:100–112. doi: 10.1148/radiol.14130569. [DOI] [PubMed] [Google Scholar]
- 10.Golden D.I., Lipson J.A., Telli M.L., Ford J.M., Rubin D.L. Dynamic contrast-enhanced MRI-based biomarkers of therapeutic response in triple-negative breast cancer. J. Am. Med. Informatics Assoc. 2013;20:1059–1066. doi: 10.1136/amiajnl-2012-001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashraf A., Gaonkar B., Mies C., DeMichele A., Rosen M., Davatzikos C., Kontos D. Breast DCE-MRI kinetic heterogeneity tumor markers: Preliminary associations with neoadjuvant chemotherapy response. Transl. Oncol. 2015;8:154–162. doi: 10.1016/j.tranon.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahrooghy M., Ashraf A.B., Daye D., McDonald E.S., Rosen M., Mies C., Feldman M., Kontos D. Pharmacokinetic tumor heterogeneity as a prognostic biomarker for classifying breast cancer recurrence risk. IEEE Trans. Biomed. Eng. 2015;62:1585–1594. doi: 10.1109/TBME.2015.2395812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Arlinghaus L.R., Ayers G.D., Chakravarthy A.B., Abramson R.G., Abramson V.G., Atuegwu N., Farley J., Mayer I.A., Kelley M.C., Meszoely I.M., Means-Powell J., Grau A.M, Sanders M., Bhave S.R., Yankeelov T.E. DCE-MRI analysis methods for predicting the response of breast cancer to neoadjuvant chemotherapy: pilot study findings. Magn. Reson. Med. 2014;71:1592–1602. doi: 10.1002/mrm.24782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hylton N.M., Gatsonis C.A, Rosen M.A., Lehman C.D., Newitt D.C., Partridge S.C., Bernreuter W.K., Pisano E.D., Morris E.A., Weatherall P.T., Polin S.M., Newstead G.M., Marques H.S., Esserman L.J., Schnall M.D., ACRIN 6657 Trial Team and I-SPY 1 TRIAL Investigators Neoadjuvant chemotherapy for breast cancer : functional tumor volume by mr imaging predicts recurrence-free survival — results from the ACRIN 6657 /CALGB 150007 I-SPY 1 TRIAL. Radiology. 2015;279:44–55. doi: 10.1148/radiol.2015150013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J.H., Ko E.S., Lim Y., Lee K.S., Han B.K., Ko E.Y., Hahn S.Y., Nam S.J. Breast cancer heterogeneity : MR imaging texture analysis and survival outcomes. Radiology. 2016;282:665–675. doi: 10.1148/radiol.2016160261. [DOI] [PubMed] [Google Scholar]
- 16.Wu J., Cao G., Sun X., Lee J., Rubin D.L., Napel S., Kurian A.W., Daniel B.L., Li R. Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology. 2018;288(1):26–35. doi: 10.1148/radiol.2018172462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhooshan N, Giger ML, Jansen SA, Lan Li. Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology. 2010 Mar;254(3):680–690. doi: 10.1148/radiol.09090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahani N., Cohen E., Hsieh M.K., Weinstein S.P., Pantalone L., Hylton N., Newitt D., Davatzikos C., Kontos D. Prediction of treatment response to neoadjuvant chemotherapy for breast cancer via early changes in tumor heterogeneity captured by DCE-MRI registration. Sci. Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-48465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou Y., Weinstein S.P., Conant E.F., Englander S., Da X., Gaonkar B., Hsieh M.K., Rosen M., DeMichele A., Davatzikos C., Kontos D. Deformable registration for quantifying longitudinal tumor changes during neoadjuvant chemotherapy. Magn. Reson. Med. 2017;73(6):2343–2356. doi: 10.1002/mrm.25368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurtado D.E., Villarroel N., Andrade C., Retamal J., Bugedo G., Bruhn A. Spatial patterns and frequency distributions of regional deformation in the healthy human lung. Biomech. Model. Mechanobiol. 2017;16(4):1413–1423. doi: 10.1007/s10237-017-0895-5. [DOI] [PubMed] [Google Scholar]
- 21.Li X., Arlinghaus L.R., Chakravarthy A.B., Farley J., Mayer I.A., Abramson V.G., Kelley M.C., Meszoely I.M., Means-Powell J., Yankeelov T.E. Early DCE-MRI changes after longitudinal registration may predict breast cancer response to neoadjuvant chemotherapy. Biomed. Image Regist. 2012;229–230 [Google Scholar]
- 22.Clark K., Vendt B., Smith K., Freymann J., Kirby J., Koppel P., Moore S., Phillips S., Maffitt D., Pringle M., Tarbox L., Prior F. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esserman L.J., Perou C., Cheang M., DeMichele A., Carey L., Van ’t Veer L.J., Gray J., Petricoin E., Conway K., Hylton N., Berry D. Breast cancer molecular profiles and tumor response of neoadjuvant doxorubicin and paclitaxel: The I-SPY TRIAL (CALGB 150007/150012, ACRIN 6657) J. Clin. Oncol. 2009;27(18 Suppl):515. [Google Scholar]
- 24.Barker A.D., Sigman C.C, Kelloff G.J., Hylton N.M., Berry D.A., Esserman L.J. I-SPY 2 : an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Nature. 2009;86:97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 25.Hudis C.A., Barlow W.E., Costantino J.P., Gray R.J., Pritchard K.I., Chapman J.A., Sparano J.A., Hunsberger S., Enos R.A., Gelber R.D., Zujewski J.A. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials:The STEEP System. J. Clin. Oncol. 2007;25:2127–2132. doi: 10.1200/JCO.2006.10.3523. 252127-2132. [DOI] [PubMed] [Google Scholar]
- 26.Sled J.G., Zijdenbos A.P., Evans A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 27.Modat M., Ridgway G.R., Taylor Z.A., Lehmann M., Barnes J., Hawkes D.J., Fox N.C., Ourselin S. Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed. 2010;98:278–284. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Ardekani B.A., Braun M., Hutton B.F., Kanno I., Iida H. A fully automatic multimodality image registration algorithm. J. Comput. Assisted Tomography. 1995;19:615–623. doi: 10.1097/00004728-199507000-00022. [DOI] [PubMed] [Google Scholar]
- 29.Diez Y., Gubern-Merida A., Wang L., Diekmann S., Marti J., Platel B., Kramme J., Marti R. Comparison of methods for current-to-prior registration of breast DCE-MRI. Lecture Notes in Computer Sci. 2014;8539:689–695. [Google Scholar]
- 30.Rueckert D., Sonoda L.I., Hayes C., Hill D.L.G., Leach M.O., Hawkes D.J. Nonrigid registration using free-form deformations : application to breast MR Images. IEEE Trans. Med. Imaging. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 31.J. Wu, Y. Ou, S.P. Weinstein, E.F. Conant, N. Yu, V. Hoshmand, B. Keller, A.B. Ashraf, M. Rosen, A. DeMichele, C. Davatzikos, Quantification of tumor changes during neoadjuvant chemotherapy with longitudinal breast DCE-MRI registration. Medical Imaging 2015: Computer-Aided Diagnosis 9414(2015), 502-508.
- 32.Avants B.B., Epstein C.L., Grossman M., Gee J.C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ou Y., Sotiras A., Paragios N., Davatzikos C. DRAMMS: Deformable registration via attribute matching and mutual-saliency weighting. Med. Image Anal. 2011;15:622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ardekani B.A., Guckemus S., Bachman A., Hoptman M.J., Wojtaszek M., Nierenberg J. Quantitative comparison of algorithms for inter-subject registration of 3D volumetric brain MRI scans. J. Neurosci. Methods. 2005;142:67–76. doi: 10.1016/j.jneumeth.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Davatzikos C., Rathore S., Bakas S., Pati S., Bergman M., Kalarot R., Sridharan P., Gastounioti A., Jahani N., Cohen E., Akbari H., Tunc B., Doshi J., Parker D., Hsieh M., Sotiras A., Li H., Ou Y., Doot R.K., Bilello M., Fan Y., Shinohara R.T., Yushkevich P., Verma R., Kontos D. Cancer imaging phenomics toolkit : quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imag. 2018;5(1) doi: 10.1117/1.JMI.5.1.011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uno H., Cai T., Pencina M.J., Agostino R.B.D., Wei L.J. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat. Med. 2011;30:1105–1117. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma U., Danishad K.K.A., Seenu V., Jagannathan N.R. Longitudinal study of the assessment by MRI and diffusion-weighted imaging of tumor response in patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy. NMR Biomed. 2008;22:104–113. doi: 10.1002/nbm.1245. [DOI] [PubMed] [Google Scholar]
- 38.Rousseau C., Devillers A., Sagan C., Ferrer L., Bridji B., Campion L., Ricaud M., Bourbouloux E., Doutriaux I., Clouet M., Berton-Rigaud D., Bouriel C., Delecroix V., Garin E., Rouquette S., Resche I., Kerbrat P., Chatal J.F., Campone M. Monitoring of early response to neoadjuvant chemotherapy in stage II and III breast cancer by [18F]fluorodeoxyglucose positron emission tomography. J. Clin. Oncol. 2006;24(34):5366–5372. doi: 10.1200/JCO.2006.05.7406. [DOI] [PubMed] [Google Scholar]
- 39.Bae M.S., Shin S.U., Ryu H.S., Han W., Im S.A., Park I.A., Noh D.Y., Moon W.K. Pretreatment MR imaging features of triple-negative breast cancer: Association with response to neoadjuvant chemotherapy and recurrence-free survival. Radiology. 2016;281:392–400. doi: 10.1148/radiol.2016152331. [DOI] [PubMed] [Google Scholar]
- 40.Lee J., Kim S.H., Kang B.J. Prognostic factors of disease recurrence in breast cancer using quantitative and qualitative magnetic resonance imaging (MRI) parameters. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-64564-6. 7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazurowski M.A., Grimm L.J., Zhang J., Marcom P.K., Yoon S.C., Kim C., Ghate S.V., Johnson K.S. Recurrence-free survival in breast cancer is associated with MRI tumor enhancement dynamics quantified using computer algorithms. Eur. J. Radiol. 2015;84:2117–2122. doi: 10.1016/j.ejrad.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Doshi J.J., Erus G., Ou Y., Davatzikos C. Ensemble-based medical image labeling via sampling morphological appearance manifolds. MICCAI Chall. Work. Segmentation Algorithms, Theory Appl. 2013 [Google Scholar]
- 43.Muenzing S.E.A., Ginneken B.V, Viergever M.A., Pluim J.P.W. DIRBoost-An algorithm for boosting deformable image registration: application to lung CT intra-subject registration. Med. Image Anal. 2014;18:449–459. doi: 10.1016/j.media.2013.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.