Highlights
-
•
Notch3 promotes proliferation and enhances the osteoclastogenic potential of multiple myeloma cells.
-
•
Notch3 mediates the communication between myeloma cells and osteocytes that leads to tumor proliferation.
-
•
Newly diagnosed MM patients with high Notch3 expression exhibit upregulation of gene signatures associated with cell proliferation and osteoclast formation.
-
•
Genetic inhibition of Notch3 in MM cells markedly decreases tumor growth and bone destruction in an immunocompetent mouse model of MM disease.
Keywords: Notch, Osteocytes, Myeloma, Cancer, Bone, Tumor microenvironment
Abbreviations: MM, Multiple myeloma; BM, Bone marrow; MMRF, Multiple Myeloma Research Foundation; GO, Gene Ontology; CTX, C-telopeptide of type 1 collagen; miCT, MicroComputed tomography; NICD, Notch intracellular domain; KD, Knock down
Abstract
In multiple myeloma (MM), communication via Notch signaling in the tumor niche stimulates tumor progression and bone destruction. We previously showed that osteocytes activate Notch, increase Notch3 expression, and stimulate proliferation in MM cells. We show here that Notch3 inhibition in MM cells reduced MM proliferation, decreased Rankl expression, and abrogated the ability of MM cells to promote osteoclastogenesis. Further, Notch3 inhibition in MM cells partially prevented the Notch activation and increased proliferation induced by osteocytes, demonstrating that Notch3 mediates MM-osteocyte communication. Consistently, pro-proliferative and pro-osteoclastogenic pathways were upregulated in CD138+ cells from newly diagnosed MM patients with high vs. low NOTCH3 expression. These results show that NOTCH3 signaling in MM cells stimulates proliferation and increases their osteoclastogenic potential. In contrast, Notch2 inhibition did not alter MM cell proliferation or communication with osteocytes. Lastly, mice injected with Notch3 knock-down MM cells had a 50% decrease in tumor burden and a 50% reduction in osteolytic lesions than mice bearing control MM cells. Together, these findings identify Notch3 as a mediator of cell communication among MM cells and between MM cells and osteocytes in the MM tumor niche and warrant future studies to exploit Notch3 as a therapeutic target to treat MM.
Introduction
Multiple myeloma (MM) is a hematological cancer characterized by the expansion of malignant monoclonal plasma cells in the bone marrow (BM) and the development of osteolytic lesions that increase fracture risk and cause bone pain [1,2]. MM cells locate in specialized niches in the bone microenvironment where they interact with multiple cell types. MM cells are highly dependent on these interactions, which transform the bone/BM niche into an ideal microenvironment for the migration, proliferation, and survival of MM cells [3]. Thus, current research efforts focus on finding aberrant signaling pathways in both MM cells and cells of the host niche to identify novel molecular targets that can be exploited as new therapeutic targets.
Notch signaling is a highly conserved mechanism for communication between adjacent cells and regulates many aspects of cancer biology [4]. MM cells use Notch to communicate with adjacent MM cells and other cells in the tumor niche [5]. Activation of Notch stimulates MM growth, increases osteoclastogenesis and bone destruction, and contributes to the development of drug resistance [6], [7], [8], [9], [10], [11]. Thus, Notch signaling is an important target in the MM tumor niche for the treatment of MM [5]. Pharmacologic inhibition of Notch signaling with gamma-secretase inhibitors (GSIs) blocks signaling downstream the four Notch receptors, decreases MM growth, and reduces bone destruction in vitro and in vivo [12], [13], [14]. However, the use of GSI in the clinic is limited due to acute gut toxicity and immunosuppression [15], [16], [17]. Understanding the specific role of each Notch component (receptors and ligands) is paramount to effectively and safely target this pathway in the MM-niche.
In earlier studies, we demonstrated that osteocytes, the most abundant and long-lived cells in bone [18], interact with MM cancer cells and are part of the vicious cycle in the tumor niche that fuels the growth of MM cells in bone [6]. Notch signaling is activated in MM cells upon contact with osteocytes, resulting in rapid upregulation of Notch3 expression and increased proliferation [6]. While Notch1 signals are known to regulate MM cell growth [8,19], the specific role of Notch3 in MM cell biology is practically unknown. In the current study, we employed well-established in vitro, ex vivo, and in vivo approaches, and in silico tools to examine the role of Notch3 in MM. We report that homotypic Notch3 signaling among MM cells stimulates MM cell proliferation and osteoclastogenic potential; and that heterotypic Notch3 signaling in MM cells is required for the full stimulation of MM proliferation induced by osteocytes. Moreover, we found that newly diagnosed patients with high NOTCH3 expression have enrichment in gene sets involved in pathways controlling cell proliferation and osteoclast differentiation. Consistent with these findings, we show that genetic inhibition of Notch3 in MM cells results in lower tumor burden and bone lytic disease in a mouse model of MM,
Materials and Methods
Reagents
RPMI 1640 media, Minimum Essential Media (MEM) α, fetal bovine serum, bovine calf serum, normocin, antibiotics (penicillin/streptomycin), Trizol, MTT, and DiI cell tracker were purchased from Invitrogen Life Technologies (Grand Island, NY, USA). Trypan blue was purchased from Sigma Aldrich (St. Louis, MO, USA). γ-secretase inhibitor XX (GSI-XX) was purchased from Calbiochem (San Diego, CA, USA). Anti-GAPDH (Cat#2118S, RRID:AB_561053), NOTCH1 (Cat #4380S, RRID:AB_10691684), and NOTCH2 (Cat #5732S, RRID:AB_10693319) antibodies were purchased from Cell Signaling Technologies (Danvers, MA, USA); anti-NOTCH3 (ab23426, RRID:AB_776841) and NOTCH4 (ab166605) antibodies were from Abcam (Cambridge, MA, USA).
Study population
The expression of Notch receptors and transcriptome analysis was performed in MM cells from newly diagnosed patients to avoid the potential influence of therapy on gene expression. We obtained gene expression data on newly diagnosed MM patients from the Multiple Myeloma Research Foundation (MMRF) CoMMpass registry (NCT01454297, version IA13). The CoMMpass study was initiated in 2011 as a large-scale prospective observational study in MM that has collected tissue samples, genetic information, quality of life, and clinical outcomes from over 1100 patients with newly diagnosed MM at 90 different sites worldwide. From an initial 992 patients with accessible gene expression data in the CoMMpass registry, 768 samples at baseline were selected. Salmon gene count data were imported into and normalized using the R package DESeq2 [20,21]. We segregated high vs. low NOTCH3 patients using quartile measures, which spread gene expression values above and below the mean by dividing the distribution into four groups. The top 25% (i.e., the upper quartile) was considered high NOTCH3 expression, the bottom 25% (i.e., the lower quartile) was considered low NOTCH3 expression [22], [23], [24], [25]. Comparative gene expression analysis was performed using the top vs. bottom quartiles (Figure 1a). DESeq2 was used for differential expression analysis with an adjusted p-value cutoff of 0.05 (see Supplementary Table 1 for a list of differentially expressed genes).
Fig. 1.
NOTCH receptor expression in CD138+ cells from newly diagnosed MM patients and human MM cell lines. (a) NOTCH1, 2, 3, and 4 expression distribution in a population of 768 newly diagnosed MM patients. Dashed lines indicate the top and bottom quartiles used to separate low vs. high NOTCH receptor expression. (b) Comparison of NOTCH1, 2, 3, and 4 expression levels in newly diagnosed MM patients. *p<0.05 vs. NOTCH1 expression. (c) Protein levels of the activated form of NOTCH3 (NICD3) in human MM cell lines. Two independent samples per cell line are shown.
Bioinformatic analysis of gene expression data
The PIANO package was used to perform the gene-set enrichment analysis of Gene Ontology (GO) using adjusted p-value and log2 fold changes as the input to calculate the enrichment p-value of the GO terms [26]. The most significant twenty biological processes GO terms are provided in Supplementary Table 2. A subset of differential GO terms associated with cell proliferation or osteoclasts was plotted as a network plot using the PIANO package to show the number of overlap genes among the differential GO terms [26].
Cell culture
5TGM1 murine MM cells (RRID:CVCL_VI66) were obtained from Dr. B. Oyajobi (University of Texas at San Antonio, TX, USA) and MLO-A5 murine osteocyte-like cells (RRID:CVCL_0P24) were provided by Dr. L. Bonewald (Indiana University, IN, USA). JJN3 cells (RRID:CVCL_2078) were obtained from Dr. N. Giuliani (University of Parma, Italy). OPM2 (RRID:CVCL_1625), MM1.S (RRID:CVCL_8792), and U266 (RRID:CVCL_0566) human MM cells were obtained from Dr. G.D. Roodman (Indiana University, IN, USA). MM cell lines were cultured in RPMI with 10% FCS and 1% P/S [6]. Osteocyte-like cells were cultured on calf skin collagen type I-coated plates in αMEM media with 2.5% FBS, 2.5% BCS, and 1% P/S [6]. Cell line authentication was routinely examined for proper morphology, population doubling, and paraprotein production. MM cells were treated with GSI (5uM) [6]. Treatments were refreshed every 24h. Direct MM:osteocyte cell-to-cell co-cultures were established by adding MM cells on top of osteocyte-like cells in a 1:5 (osteocyte:MM) ratio [6]. In these co-cultures, MM cells were stained with the fluorescent cell-tracker DiI following the manufacturer's recommendations. DiI fluorescence was read at 520 to 565 nm. EDTA incubations were used to separate MM cells from the osteocytes, as previously described [6]. All cells were cultured under 37⁰C and 5% CO2 conditions.
Lentiviral-mediated gene knock-down in MM cells
Murine 5TGMI MM cells were transduced with lentiviral particles (MOI=10; GenTarget Inc, San Diego, CA, USA) containing sequences for Red Fluorescent Protein (RFP) and validated shRNA against Notch3 or Notch2. 5TGM1 MM cells transduced with shRNA-scramble were used as control. 72h after transduction, 5TGM1 MM cells were sorted for RFP expression and cultured for 7 days in RPMI complete media supplemented with 3uL/mL of puromycin for additional selection. Transduced MM cells were expanded and maintained in complete media supplemented with 3uL/mL of puromycin.
Cell Viability and Apoptosis Assays
MM cell viability was determined using the MTT assay or Trypan Blue exclusion following protocols established by the manufacturer. Apoptosis in MM cells was assayed by flow cytometry, using the Annexin V apoptosis Detection kit (BD Biosciences) following the manufacturer's recommendations. Samples were analyzed in a BD FACSCalibur (UAMS Core Facility for Flow cytometry) within 1h. At least 10,000 cells were used for each group, and the data was analyzed by FlowJo software to detect different cell populations.
Gene Expression
Total RNA was isolated from MM cells using Trizol and converted to cDNA (Invitrogen Life Technologies), following the manufacturer's directions. Gene expression was quantified by quantitative real-time PCR (qPCR) using Taqman assays from Applied Biosystems (Foster City, CA, USA), following the manufacturer's directions. Gene expression levels were calculated using the comparative threshold (CT) method and were normalized to the housekeeping gene GAPDH.
Western Blot
Cell lysates (50μg) were boiled in the presence of SDS sample buffer (NuPAGE LDS sample buffer; Invitrogen) for 10 minutes and subjected to electrophoresis on 10% SDS-PAGE (Bio-Rad Laboratories). Proteins were transferred to PVDF membranes using a semidry blotter (Bio-Rad) and incubated in blocking solution (5% nonfat dry milk in TBS containing 0.1% Tween-20) for 1 hour to reduce nonspecific binding. Immunoblots were performed using anti-GAPDH (1:1000), NOTCH1-3 (1:1000), and NOTCH4 (1:500), antibodies followed by goat anti-rabbit secondary antibodies, conjugated to horseradish peroxidase (1:2000) in 5% milk (Santa Cruz Biotechnology). Western blots were developed using an enhanced chemiluminescence detection assay following the manufacturer's directions (Bio-Rad). Protein bands were quantified using Image Lab 6.0.1 (Bio-Rad).
Osteoclast Formation
Bone marrow cells were flushed out from long bones of 8 week-old C57BL/6 mice and cultured in α-MEM containing 10% FBS overnight. Non-adherent cells were harvested and enriched for CD11b+ mononuclear cells using the Miltenyi Biotec MACS magnetic cell-sorting system, following the manufacturer's recommendations. Conditioned media was collected from 5TGM1 MM cells cultured at a density of 1 × 106 cells/mL in 10 mL of RPMI complete media after 72h of incubation and concentrated to 2 mL using a Centriprep Centrifugal filter (YM-10) by centrifuging for 30min at 1500g. 2 × 105 CD11b cells/well were plated and cultured in αMEM media with M-CSF (10 ng/ml) for 3 days. Then conditioned media from the MM cells was added to the cultures for 4 additional days. No recombinant RANKL was used in these experiments. Osteoclasts with 3 or more nuclei were enumerated in TRAP-stained cell cultures using a leukocyte acid phosphatase kit (Sigma-Aldrich).
Ex vivo organ cultures
Ex vivo intact bone organ culture systems recapitulate the spatial dimension, cellular diversity, and molecular networks of the tumor niche, and contain authentic osteocytes in their 3D microenvironment. A step-by-step guide on establishing the ex vivo MM-bone organ cultures used in this study is provided here [27]. Briefly, calvarial bones from 9 day-old C57BL/6 pups were cut into 5mm disks with a biopsy punch and placed into 96-well plates. 5 × 104 5TGM1 MM cells were added on top of the bone slices (concave side up) in RPMI complete media and allowed to invade the bone for 24h. Then bones slices bearing MM cells were transferred to a new plate and cultured at 37⁰C and 5% CO2. MM cells not attached to the bones were discarded [6,27,28]. Fifty percent of the media was refreshed every 3 days. Conditioned media was collected after 11 days of culture. We have shown that at this time the bone slices are macroscopically intact, with no visible lytic lesions. However, bone resorption is increased by MM cells, as demonstrated by increased CTX levels in conditioned media from these cultures [28]. Further, MM cells continue to grow, as shown by longitudinal increases in the tumor biomarker IgG2b [28].
Enzyme-linked immunoassays (ELISA)
Myeloma cells make abnormal quantities of monoclonal proteins (or paraproteins), which accumulate in the serum and are used to diagnose and follow tumor burden in the clinic [1,29]. The levels of the tumor biomarker IgG2b, a paraprotein produced by 5TGM1 MM cells used to determine tumor growth/burden in vitro, ex vivo, or in animal models [30], [31], [32], [33], and the bone resorption biomarker C-telopeptide of type 1 collagen (CTX) were analyzed in serum (in vivo) or conditioned media (ex vivo) using commercially available specific ELISA and following manufacturer's recommendations.
In vivo mouse model of MM
Six-week-old immune-competent C57BL/KaLwRij mice were randomized by weight and injected intratibially with 105 transduced 5TGM1 MM cells or saline. Both female and male mice were used for these studies. All the groups contained an equal number of female/male mice. Mice were considered engrafted if the IgG2b levels were higher than the sum of the IgG2b average of the saline group plus two times the SD [11,34,35]. Using this threshold, we determined that all our mice were engrafted, regardless of sex or whether they were inoculated with control or shRNA-Notch3 MM cells. No differences in body weight between groups were observed. Mice were fed with a regular diet (Harlan, Indianapolis, IN), received water, and maintained on a 12-hour light/dark cycle. Studies were approved by the Institutional Animal Care and Use Committee of the Indiana University School of Medicine (protocol #11254). Institutional and national guides for the care and use of laboratory animals were followed for these studies.
Analysis of MM-induced osteolytic disease
X-Ray radiographs were taken with an UltraFocus Faxitron X-Ray System (Hologics, Marlborough, MA, USA). MicroCT analyses were performed at the cancellous bone of the proximal tibia, in an area 20 um below the growth plate, using 15 um resolution and a Scanco vivaCT 40 (Scanco Medical AG, Switzerland) [6,34]. Data analysis was performed in a blinded fashion.
Statistical Analysis
Data were analyzed using SigmaPlot 12.0 (Systat Software, Inc., San Jose, CA, USA). Values were reported as means ± SD. Differences in means were analyzed by a combination of t-test and ANOVA (Dunn's method). p≤0.05 was considered statistically significant.
Results
Notch3 is detected in newly diagnosed MM patients and MM cell lines
First, we investigated the expression pattern of Notch receptors in newly diagnosed MM patients (Figure 1a). Notch receptors were detectable in CD138+ cells from newly diagnosed MM patients and no differences in the expression distribution were found between Notch receptors. MM cells from newly diagnosed patients expressed higher levels of NOTCH1 and 2 compared to NOTCH3 or 4 (Figure 1b). Consistent with the detection of NOTCH3 in CD138+ cells from MM patients, we found expression of the Notch intracellular domain of NOTCH3 (NICD3), the activated form of the receptor, in several human MM cell lines and MM murine cells (Figures 1c and 2a-b). Together, these results demonstrate that Notch3 is expressed and activated in MM cells.
Fig. 2.
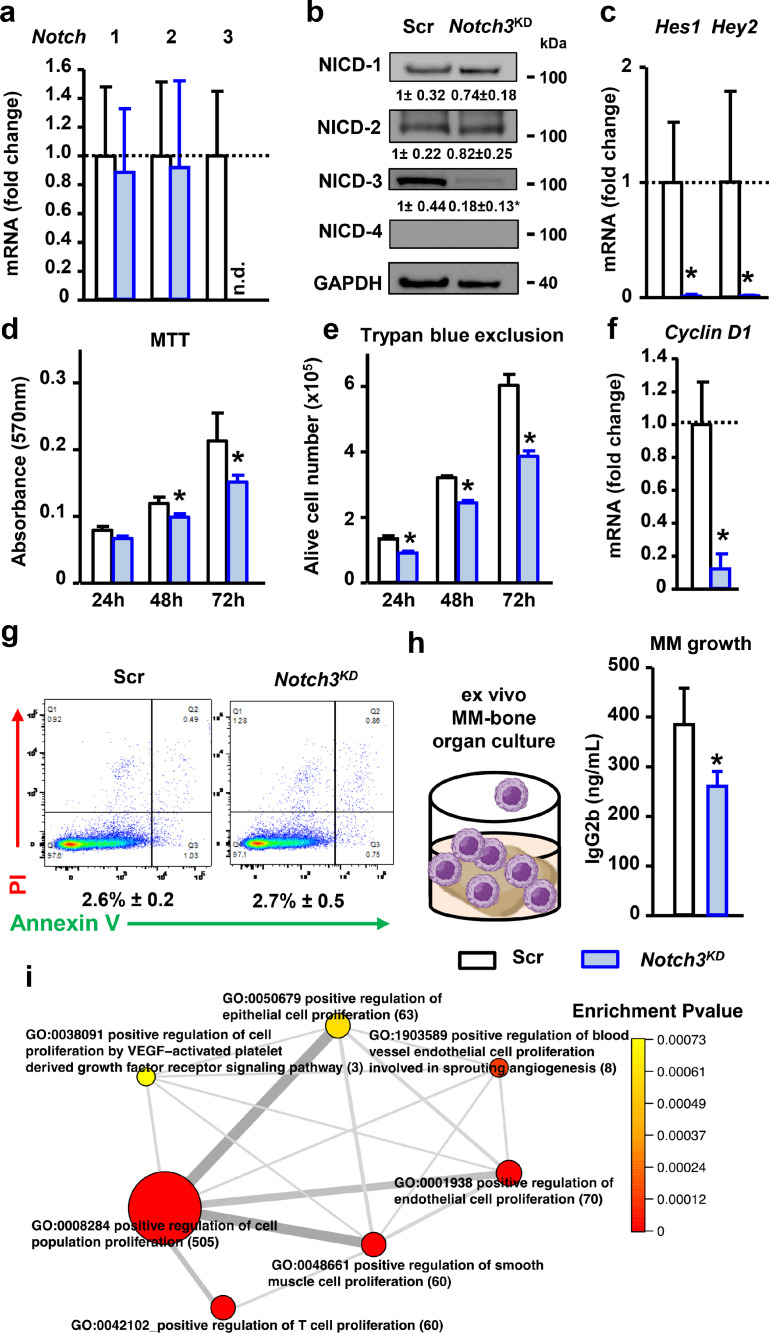
Genetic inhibition of Notch3 decreases Cyclin D1 expression and proliferation in MM cells. (a)Notch1-4 mRNA gene expression in shRNA-scramble (Scr) or shRNA-Notch3 (Notch3KD) MM cells. (b) Western blot images for NICD1-4 in 5TGM1 MM cells. Bands were quantified and normalized by GAPDH. (c) mRNA gene expression of Notch target genes Hes1 and Hey2 in control or shRNA-Notch3-transduced MM cells. (d) Cell viability by MTT assay, (e) number of alive MM cells by Trypan Blue exclusion, and (f)Cyclin D1 mRNA gene expression in Scr and Notch3KD MM cells. For mRNA gene expression, fold change calculations vs. Scr are shown. (g) Apoptosis assay using flow cytometry after staining with annexin V/propidium iodide (PI). Representative scatter plots of PI (y-axis) vs. annexin V (x-axis) and percentage of apoptotic cells are shown. Representative experiments out of 3. n=4/group. n.d. = not detected.*p<0.05 vs Scr vehicle. (h) Levels of the tumor biomarker IgG2b in conditioned media collected from ex vivo MM-bone organ cultures show less MM growth in bones bearing Notch3KD MM cells after 11 days of culture. Representative experiment out of 2. *p<0.05 vs. Scr, n=4-5/group. (i) GO terms network plot of selected upregulated functional enrichment analysis of GO terms related to proliferation in MM patients with high vs. low NOTCH3 expression. The size of the circles represents the number of genes in the individual GO terms. The thickness of the lines represents the number of overlapped genes between the individual GO terms.
Homotypic Notch3 signaling activates Notch in MM cells
To assess the impact of Notch3 inhibition in MM cells, we knocked down Notch3 expression using shRNA-mediated gene inhibition in murine 5TGM1 cells, a MM cell line that allows the study of the pathobiology of MM and disease progression in immunocompetent mouse models [32,36]. MM cells stably transduced with shRNA-Notch3 (Notch3KD cells) had undetectable mRNA levels of Notch3 mRNA and an 80% decrease in the levels of NICD3 compared to control cells expression (Figure 2a-c). Notch3 knock-down did not affect the mRNA expression of NICD protein levels of Notch1 or Notch2, which remained similar to those found in control MM cells (Figure 2a-c). As previously reported [6], we did not detect expression of Notch4 in this MM cell line. Notch3KD cells exhibited a marked decrease in the expression of the Notch target genes Hes1 and Hey2 (Figure 2c). Collectively, these findings show that Notch3 is essential to maintain endogenous levels of Notch signaling in MM cells.
Homotypic Notch3 signaling regulates cell proliferation in MM cells
Notch3KD cells had a 30-35%, time-dependent, decrease in MM cell number compared to control MM cells (Figure 2d-e). Further, Notch3KD cells displayed a 90% reduction in Ccnd1 (Cyclin D1) mRNA expression (Figure 2f). In addition, no changes in apoptosis were found between Notch3KD cells and control cells (Figure 2g). To further analyze the effects of silencing Notch3 signaling in MM on tumor growth, we used ex vivo MM-bone organ cultures that reproduce the complexity of the MM bone microenvironment in a controlled setting [6,28] (Figure 2h). After 11 days of culture, we found that MM growth was lower in bones bearing Notch3KD MM cells than those bearing control MM cells, as determined by the lower levels of the tumor biomarker IgG2b in conditioned media (CM). Next, we compared the transcriptome of CD138+ cells isolated from newly diagnosed MM patients with high vs. low NOTCH3 expression (Figure 1a) in CD138+ MM cells (see Supplementary Table 1 for differentially expressed genes) and investigated the impact of NOTCH3 expression on biological processes based on gene ontology annotation (see Supplementary Table 2 for top 20 upregulated processes). In line with our in vitro observations, MM patients with high NOTCH3 expression exhibited upregulation and enrichment in genes associated with processes related to positive regulation of cell proliferation (Figure 2i). The list of genes and corresponding fold changes included in the positive regulation of cell population biological process are shown in Supplementary Table 3. These data indicate Notch3 signals between MM cells regulate MM proliferation.
Homotypic Notch3 signaling confers osteoclastogenic potential to MM cells
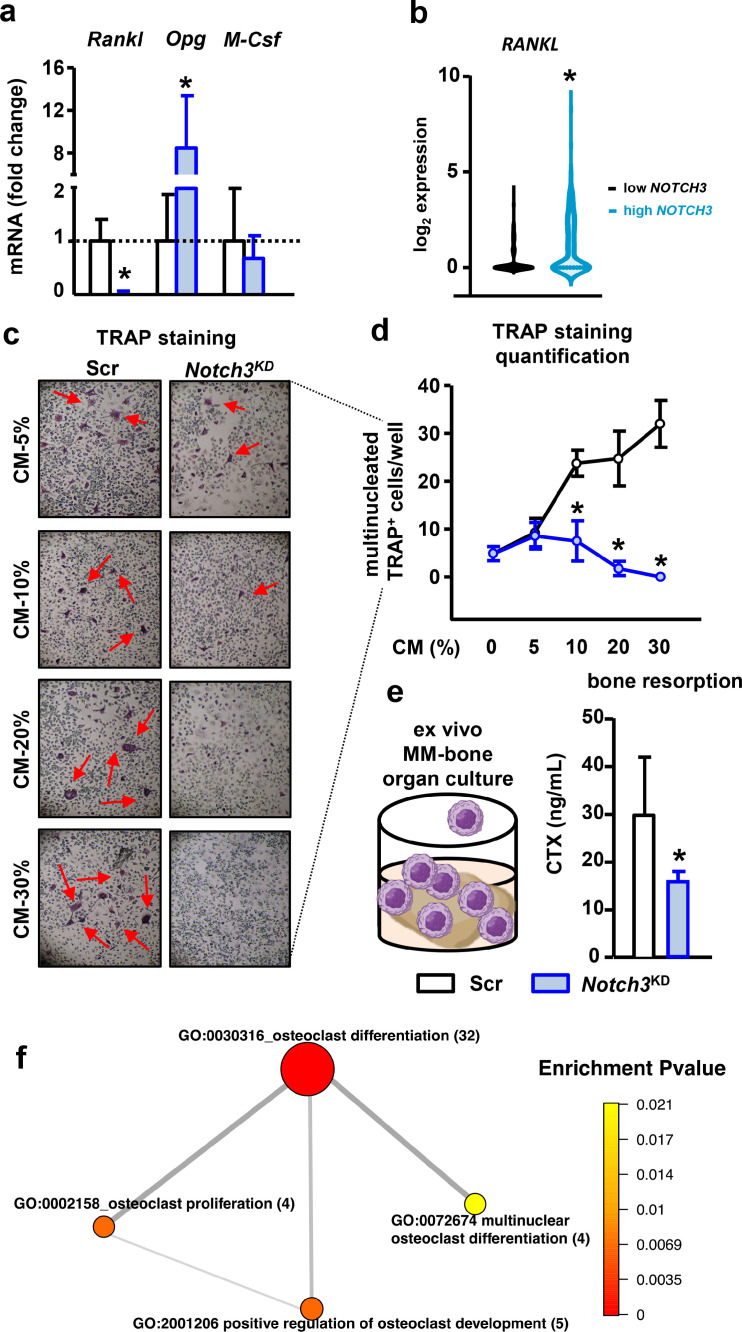
Next, we assessed the consequences of Notch3 knock-down on the expression of pro- and anti-osteoclastogenic cytokines in MM cells. Compared to control cells, Notch3KD cells exhibited a 95% decrease in the expression of the pro-osteoclastogenic cytokine, Tnfsf11 (Rankl), which was accompanied by a 6-fold increase in the mRNA levels of the anti-osteoclastogenic cytokine, Tnfrsf11b (Opg). Csf1 (M-Csf) expression was unchanged in Notch3KD cells compared to control cells (Figure 3a). Consistent with this observation, newly diagnosed MM patients with high NOTCH3 expression exhibited increased RANKL expression than patients with low NOTCH3 expression (Figure 3b). To determine if the Rankl/Opg ratio reduction affects the ability of Notch3KD MM cells to stimulate osteoclast formation, we treated osteoclast precursors with CM collected from control and Notch3KD MM cells. CM from control MM cells increased osteoclast formation in a dose-dependent manner, while the number of osteoclasts did not increase in cultures treated with CM from Notch3KD MM cells (Figure 3c-d). These studies were performed without exogenous recombinant RANKL, making MM-derived RANKL the only source to induce osteoclast differentiation, suggesting that reduced RANKL production by Notch3KD cells is responsible for the decreased osteoclast differentiation. Further, the bone resorption marker CTX levels were also reduced in ex vivo MM-bone organ cultures bearing Notch3KD MM cells compared to control MM cells (Figure 3e). Supporting the role of Notch3 signaling as a regulator of the osteoclastogenic potential of MM cells, in silico analysis of the transcriptome of MM patients revealed that MM patients with high NOTCH3 expression have upregulation and enrichment of biological processes related to osteoclast differentiation and proliferation (Figure 3f). The list of genes and corresponding fold changes (high vs. low NOTCH3) included in the osteoclast differentiation biological process are shown in Supplementary Table 3. Results from this set of experiments support that Notch3 signaling in MM cells regulates the osteoclastogenic potential of MM cells.
Fig. 3.
Notch3 inhibition decreases the osteoclastogenic potential of MM cells. (a)Rankl, Opg, and M-Csf mRNA gene expression in shRNA-scramble (Scr) or shRNA-Notch3 (Notch3KD) MM cells. (b)RANKL expression in newly diagnosed MM patients with high vs. low NOTCH3 expression. *p<0.05 vs. low NOTCH3 MM patients. For mRNA gene expression, fold change calculations vs. Scr are shown. (c) Representative images of TRAP+ cells in pre-osteoclast cultures treated with conditioned media (CM) from control or Notch3KD MM cells without exogenous recombinant RANKL. (d) Quantification of the number of multinucleated TRAP+ cells in pre-osteoclast cultures treated with increasing concentrations of CM from control or Notch3KD MM cells. Representative experiment out of 3. *p<0.05 vs. Scr. n=4-6/group. (e) Levels of the bone resorption marker CTX in CM collected from ex vivo MM-bone organ cultures show less bone resorption in bones bearing Notch3KD MM cells after 11 days of culture. Representative experiment out of 2. *p<0.05 vs. Scr, n=4-5/group. (f) GO terms network plot of selected upregulated functional enrichment analysis of GO terms related to osteoclast differentiation and proliferation in MM patients with high vs. low NOTCH3 expression. The size of the circles represents the number of genes in the individual GO terms. The thickness of the lines represents the number of overlapped genes between the individual GO terms.
Notch3 signaling mediates heterotypic communication between MM cells and osteocytes
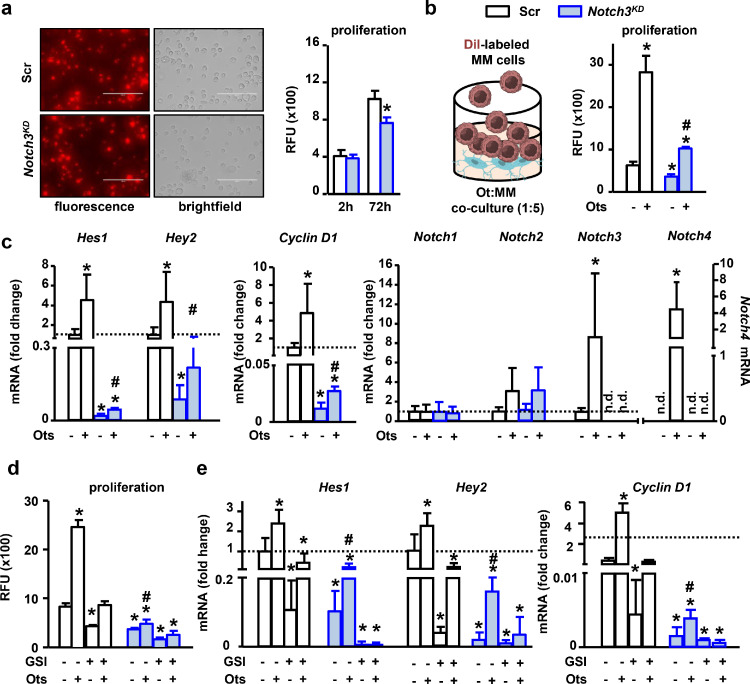
Because we previously showed that osteocytes promote the cell proliferation of MM cells by activating Notch signaling [6], we next studied if Notch3 contributes to this communication. First, we examined MM cell growth using the red fluorescent cell tracker DiI. After 72h of culture, relative fluorescence units (RFU) increased in both control and Notch3KD MM cells. However, consistent with the results in Figure 2, the increase in RFU in Notch3KD cells was 20% lower than control cells (Figure 4a), thus validating the use of DiI-RFU as an indirect measurement of MM proliferation. As shown before [6], contact with osteocytes induced a 3-fold increase in proliferation in control MM cells compared to control cells cultured alone. In contrast, Notch3KD cells in co-culture with osteocytes only exhibited a 1.5-fold increase in proliferation compared to cells cultured alone (Figure 4b). Compared with MM cells cultured alone, co-culture with osteocytes increased Hes1 and Hey2 expression by 4.5 fold in control MM cells (Figure 4c), whereas osteocytes only increased the expression of these genes by 2.5-fold in Notch3KD MM cells. Similarly, contact with osteocytes increased Cyclin D1 expression by a 4.8-fold in control MM cells versus a 2.2-fold in Notch3KD cells, compared to their respective counterparts cultured alone (Figure 4c). Similar to our previously published findings [6], osteocytes induced a 9-fold increase in Notch3 mRNA expression in control cells. In contrast, Notch3 expression was not detected in Notch3KD cells cultured either alone or with osteocytes. Notch2 expression increased in both control and Notch3KD cells in contact with osteocytes, although it did not reach statistical significance. Notch4 expression was detected only in control MM cells in co-culture with osteocytes, but its expression remained undetectable in all other culture conditions (Figure 4c). Treatment with the pan Notch inhibitor GSI fully prevented the increase in MM cell proliferation induced by osteocytes in both control and Notch3KD MM cells (Figure 4d) and decreased the osteocyte-induced upregulation of Notch target genes and Cyclin D1 expression in Notch3KD cells (Figure 4e). Together, these results indicate that Notch3 mediates the Notch communication between MM cells and osteocytes, leading to MM cell proliferation.
Fig. 4.
Notch3 signaling is required for the increased MM cell proliferation induced by osteocytes. (a) Representative images of shRNA-scramble (Scr) or shRNA-Notch3 (Notch3KD) MM cells stained with DiI. Relative fluorescence units (RFU) in DiI labeled Scr or Notch3KD MM cells at basal levels and after 72h of culture. (b) DiI RFU in Scr or Notch3KD MM cells cultured with/without osteocytes. (c) mRNA gene expression of Hes1, Hey2, Cyclin D1, and Notch receptors in Scr or Notch3KD MM cells cultured with/without osteocytes. (d) DiI RFU and (e) mRNA gene expression in Scr or Notch3KD MM cells in the absence and presence of osteocytes, with/without GSI (5µM). Representative experiments out of 3. *p<0.05 vs. Scr vehicle/alone. #p<0.05 vs Notch3KD vehicle/alone. n=6/group. For mRNA gene expression, fold change calculations vs. Scr are shown.
Notch2 signaling does not contribute to heterotypic Notch communication between MM cells and osteocytes
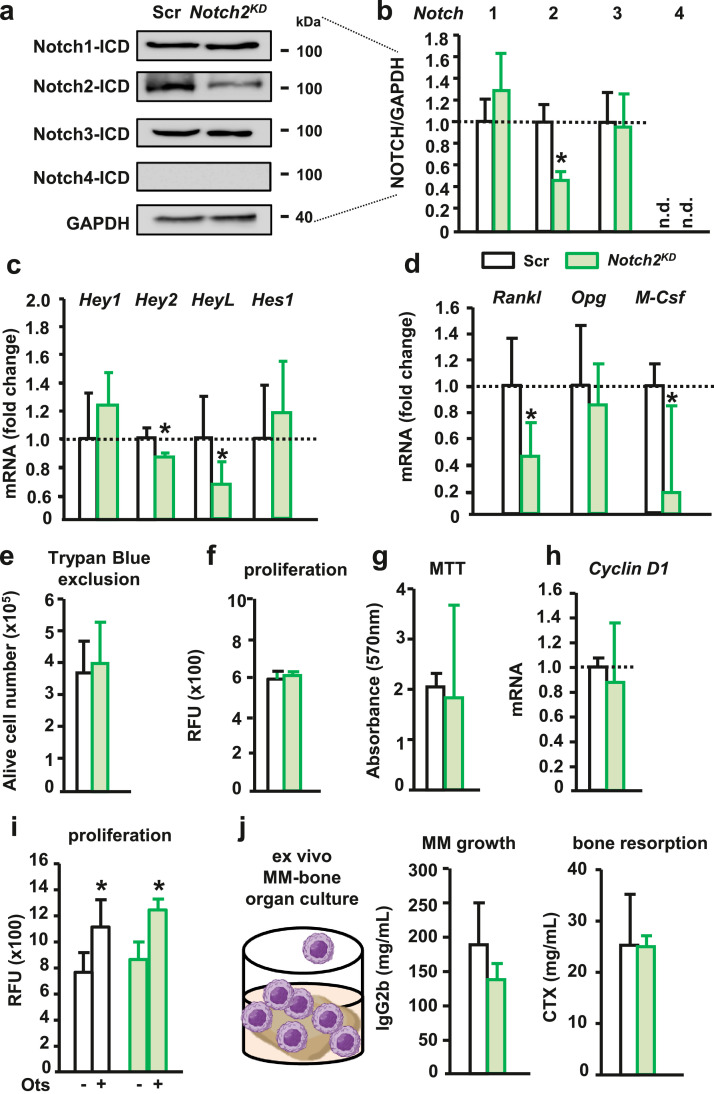
To further understand Notch receptor specificity in MM cells, we next knocked-down Notch2 signaling in MM cells. Protein expression of the activated form of NOTCH2 was decreased by 40% in MM cells silenced for Notch2 (Notch2KD cells) compared to control MM cells (Figure 5a-b), whereas no differences were detected in the activation of other Notch receptors. The expression of the Notch target genes Hey2 and HeyL was decreased in Notch2KD cells, but to a lesser extent than in Notch3KD cells (Figure 5c). Rankl expression was also reduced in Notch2KD cells compared to control MM cells (Figure 5d). Notch2 knock-down did not affect MM cell proliferation or Cyclin D1 expression (Figure 5e-h). We also examined the contribution of Notch2 signaling in MM cells to their communication with osteocytes and found that osteocytes increased MM cell proliferation to the same extent in both the control and Notch2KD cells (Figure 5i). Lastly, we used ex vivo MM-bone cultures to examine the effects of Notch2 signaling in MM cells in a more physiological microenvironment. Analysis of the tumor biomarker IgG2b in the CM revealed no differences in tumor growth between bones bearing Notch2KD cells and control MM cells (Figure 5j). Also, the levels of the bone resorption marker CTX did not differ between these two groups. Altogether, these observations indicate that Notch2 is not required for the heterotypic Notch signaling between MM cells and osteocytes.
Fig. 5.
Notch2 homotypic and heterotypic signaling in MM cells. (a-b) Protein levels of activated Notch receptors in 5TGM1 MM cells transduced with shRNA-scramble (Scr) or shRNA-Notch2 (Notch2KD) after 72h of culture. Representative western blot images for NICDs 1-4 in MM cells. (c-d) mRNA gene expression for Notch target genes, Rankl, Opg, and M-Csf mRNA in Notch2KD MM cells. (e-h) Number of alive MM cells by Tryan Blue exclusion, MM cell proliferation by DiI fluorescence (relative fluorescence units, RFU) or MTT assay, Cyclin D1 mRNA gene expression, in Scr or Notch2KD MM cells at 72h of culture. For mRNA gene expression, fold change calculations vs. Scr are shown. (i) DiI RFU of Scr or Notch2KD MM cells co-cultured with osteocytes for 72h. (j)Ex vivo MM-bone organ culture established with Scr or Notch2KD MM cells. (j) IgG2b and CTX levels in conditioned media after 11 days. *p<0.05 vs. Scr. Representative experiments out of 2-3. n=3-8/group. n.d.=not detected.
Inhibition of Notch3 signaling in MM cells decreases tumor growth and MM bone disease in vivo in an immunocompetent mouse model of MM
We next addressed in vivo the significance of our in vitro/ex vivo/in silico findings using an immunocompetent mouse model of MM. Tumor progression was slower in mice injected with Notch3KD MM cells compared to mice bearing control cells (Figure 6a). After 5 weeks, mice bearing Notch3KD MM cells had a 50% less tumor burden than mice bearing control MM cells (Figure 6b). Moreover, mice bearing Notch3KD MM cells had 50% fewer cortical osteolytic lesions than control mice injected with control MM cells (Figure 6c-d). Consistent with this observation, microCT 3D reconstruction of cortical bone revealed more cortical osteolysis in mice injected with control cells versus Notch3KD cells (Figure 6e). Further, mice injected with control MM cells exhibited a 40% decrease in proximal tibial cancellous bone volume (BV/TV), whereas mice bearing Notch3KD MM cells had 30% more cancellous bone than mice bearing control MM cells (Figure 6f). Results from this in vivo preclinical study provides in vivo evidence of Notch3's role as a mediator of signals from the MM niche supporting tumor growth and bone destruction.
Fig. 6.
Genetic inhibition of Notch3 signaling in MM reduces tumor progression and bone destruction in a mouse model of MM disease. (a-b) Tumor progression in mice injected with saline, shRNA-scramble (Scr), or shRNA-Notch3 (Notch3KD) MM cells. Linear regression shows that the slopes of the IgG2b (tumor biomarker) progression curves are statistically different between mice bearing Scr vs. Notch3KD MM cells. (c-d) Representative X-Ray images of tibias bearing Scr or Notch3KD MM cells and quantification of the number of cortical osteolytic lesions per tibia. (e) MicroCT 3D reconstruction of tibias injected with saline, Scr, or Notch3KD MM cells. (f) Analysis of bone volume over tissue volume (BV/TV) in the cancellous bone of the proximal tibia. *p<0.05 vs. Scr. Saline: 4F/3M; MM-Scramble: 6F/6M; MM-Notch3KD: 4F/4M.
Discussion
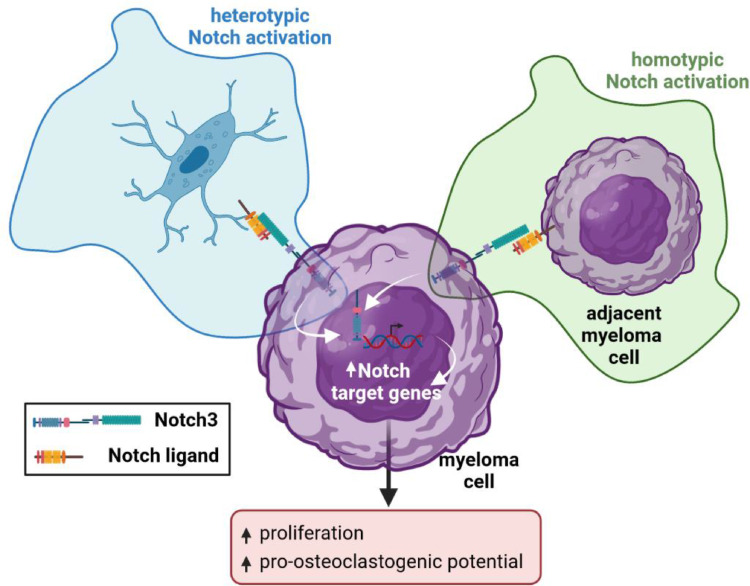
Accumulating evidence supports a central role of Notch signaling in MM onset and progression, making this pathway an attractive therapeutic target for the treatment of MM patients [5]. However, the specific contribution of each Notch receptor to MM progression and bone disease is not fully understood. In particular, limited information is available about Notch3, as most of the work studying Notch receptors in MM has focused on Notch1 [4,10,17]. Using genetic and in vitro, ex vivo, and in vivo approaches, and in silico data mining of MM patient databases, we demonstrate that Notch3 signaling contributes to tumor proliferation and bone destruction in MM. In concert, our results unravel previously unknown functions of Notch3 in the integration of homotypic (among MM cells) and heterotypic (between MM cells and osteocytes) Notch signals (Figure 7) and identify Notch3 as a mediator of proliferative and osteoclastogenic signals in the MM tumor microenvironment.
Fig. 7.
Notch3 signaling in MM cells mediates homotypic and heterotypic communication via Notch between neighboring MM cells and between MM cells and osteocytes in the tumor niche. Homotypic Notch3 signaling between MM cells is required for normal Notch signaling, proliferation, survival, and osteoclastogenic potential. Heterotypic Notch3 signaling mediates the activation of Notch and the stimulation of proliferation in MM cells induced by osteocytes.
Our results show that Notch3 signaling contributes to tumor growth by promoting cell proliferation and survival. This conclusion is consistent with the observation that MM patients with high NOTCH3 expression have upregulation of pro-proliferative biological programs. This proliferative advantage conferred by Notch3 appears to be mediated by upregulation of Cyclin D1 levels, which enables MM cells to proliferate and avoid cell cycle arrest. Notch receptors can have both overlapping and specialized functions [37,38], which are cell and location-dependent. However, our findings suggest that Notch receptors have distinct roles in MM proliferation. Previous findings showed that Notch1 signaling indirectly stimulates MM proliferation by increasing IL-6 production in both MM and stromal cells [7]. In contrast, we show here that homotypic Notch2 signaling is not required for MM growth. Together, these findings identify Notch3 signaling, together with Notch1, as an important contributor to MM growth via homotypic Notch communication.
Our findings support that Notch3 signaling in MM cells increases their ability to stimulate osteoclastogenesis and destroy the bone. These preclinical results are supported by the upregulation of osteoclast differentiation and proliferation biological processes seen in newly diagnosed MM patients with high NOTCH3 expression. Moreover, our observations align with recent findings suggesting Notch signaling regulates the expression of the pro-osteoclastogenic cytokine RANKL in MM cells and is essential for osteoclastogenesis [39]. Although inhibition of Notch2 did not affect bone resorption, it also decreased Rankl expression in MM cells, suggesting that Notch2 and Notch3 could have redundant functions in the regulation of Rankl.
MM cells utilize Notch signaling to physically communicate with other cells in the tumor niche, particularly with stromal cells in the BM [17,19]. Although embedded in the mineral matrix, it is well established that osteocytes have long cytoplasmatic extensions that form a complex network that directly connects osteocytes to other cells in the bone marrow and blood vessels [18,40]. Using acid-etching scanning electron microscopy, we demonstrated that these cytoplasmic projections reach areas of the BM infiltrated with MM cells and mediate cell-to-cell interactions with MM cells, even without overt lytic lesions [6]. More recently, direct cell contact between cancer cells and deeply embedded osteocytes in cortical bone was described by other investigators using a similar mouse model to the one employed in our study [41]. We also showed that cell-to-cell interactions lead to heterotypic, bidirectional Notch signaling between MM cells and osteocytes, contributing to MM growth and bone disease [6]. Our current results indicate that this crosstalk requires Notch3 in MM cells, but not Notch2. Interestingly, blockade of Notch signaling downstream the four Notch receptors was required to fully inhibit osteocyte-induced MM proliferation, suggesting MM cells can use other Notch receptor(s) to communicate with osteocytes. We showed earlier that osteocytes induce the expression of Notch4 in MM cells [6], and future studies are warranted to examine the role of this receptor in MM-osteocyte communication. In addition, further studies are needed to determine if MM cells also employ Notch3 signaling to communicate with other cells in the tumor niche (i.e., osteoblasts, stromal cells).
Treatment with GSI has significant safety drawbacks, including gastrointestinal toxicity, limiting its use in the clinical setting [15,16]. We recently generated a Notch inhibitor specifically targeted to the MM niche and showed it exerts potent anti-MM and anti-resorptive activities without the associated gut toxicity, thus providing a new approach to target Notch safely in MM patients [11]. Our Notch3 findings offer an additional avenue to inhibit Notch communication in the MM niche while circumventing the unwanted side-effects of systemic pan Notch inhibition. Several neutralizing antibodies against NOTCH3 are currently under development; however, none have been tested against MM. Importantly, preclinical and clinical results have shown that pharmacologic inhibition of Notch3 has anti-tumor activity, minimal intestinal toxicity, and a manageable safety profile [42]. Further, activation of Notch3 in the skeleton leads to increased Rankl expression and bone loss [43,44], suggesting that blockade of Notch3 in bone could decrease bone resorption. Altogether, these findings support the notion that targeting Notch3 in the MM microenvironment may have dual anti-MM and anti-resorptive effects. Yet, whether pharmacologic inhibition of Notch3 can become a safe and effective therapeutic approach to treat MM remains to be determined.
Osteocyte-like cells are well-established models to study the biology of osteocytes in vitro and are widely used in the bone field [45]. However, they do not reproduce all the morphological and functional features of authentic osteocytes. Moreover, the co-culture of osteocytic and MM cell lines might not entirely represent the interactions between osteocytes and MM cells in the tumor microenvironment. However, our findings using osteocyte-like cells and the cell-to-cell co-culture system have been reproduced in authentic osteocytes ex vivo, using osteocyte-enriched MM bone organ cultures, in vivo, in preclinical mouse models of MM [11,34,46], and in osteocytes in bones from MM patients [47]. Further, most of the results obtained with these approaches have been validated by other investigators [34,[46], [47], [48], [49]]. Despite the limitations, these cell lines and in vitro systems constitute valuable tools to study the crosstalk between osteocytes and MM cells. We performed all our experiments with murine 5TGM1 cells for the current study. Yet, our key observations were validated in primary CD138+ cells from newly diagnosed MM patients, adding translational value and clinical relevance to our findings.
In summary, our in vitro, ex vivo, in vivo, and in silico studies reveal a previously unknown role of Notch3 signaling in the Notch-dependent dialog between MM cells and cells of the tumor niche conducive to tumor progression and bone destruction. In addition, our findings further support the notion that osteocytes are an abundant and durable source of signals in the MM tumor microenvironment and can support MM progression. Lastly, these studies provide the framework for additional preclinical and clinical studies examining the efficacy of pharmacologic approaches inhibiting Notch3 to treat MM and its associated skeletal disease.
Author contributions
T.B., G.D.R., J.D.C. conceived and supervised the project. T.B., G.D.R., J.D.C. designed the experiments; H.M.S., T.A., C.A., D.H., J.A., M.C., M.S., I.N., and N.K performed the experiments, H.M.S., T.A., C.A., D.H., J.A., M.C., M.S., I.N., and N.K contributed to the data analysis and interpretation. J.D.C and H.M.S. wrote the manuscript. All authors reviewed the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could influence the work reported in this paper.
Acknowledgments
This work was supported by the National Institutes of Health (R37-CA251763 to J.D.C., R01-CA209882 to G.D.R. and T.B., and R01-CA241677 to G.D.R.), the Arkansas COBRE program (NIGMS P20GM125503) to J.D.C. and I.N., and a Scholar Award by the American Society of Hematology (to J.D.C). The CoMMpass data used in this study were generated as part of the Multiple Myeloma Research Foundation Personalized Medicine Initiatives (https://research.themmrf.org and www.themmrf.org).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100785.
Appendix. Supplementary materials
References
- 1.Rajkumar S.V. Myeloma today: Disease definitions and treatment advances. Am. J. Hematol. 2016;91:90–100. doi: 10.1002/ajh.24236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terpos E., Berenson J., Raje N., Roodman G.D. Management of bone disease in multiple myeloma. Expert Rev. Hematol. 2014;7:113–125. doi: 10.1586/17474086.2013.874943. [DOI] [PubMed] [Google Scholar]
- 3.Lomas O.C., Tahri S., Ghobrial I.M. The microenvironment in myeloma. Curr. Opin. Oncol. 2020;32:170–175. doi: 10.1097/CCO.0000000000000615. [DOI] [PubMed] [Google Scholar]
- 4.Meurette O., Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34:536–548. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Sabol H.M., Delgado-Calle J. The multifunctional role of Notch signaling in multiple myeloma. J. Cancer Metast. Treat. 2021;7:20. doi: 10.20517/2394-4722.2021.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delgado-Calle J., Anderson J., Cregor M.D., Hiasa M., Chirgwin J.M., Carlesso N., Yoneda T., Mohammad K.S., Plotkin L.I., Roodman G.D., Bellido T. Bidirectional notch signaling and osteocyte-derived factors in the bone marrow microenvironment promote tumor cell proliferation and bone destruction in multiple myeloma. Cancer Res. 2016;76:1089–1100. doi: 10.1158/0008-5472.CAN-15-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombo M., Galletti S., Bulfamante G., Falleni M., Tosi D., Todoerti K., Lazzari E., Crews L.A., Jamieson C.H., Ravaioli S., Baccianti F., Garavelli S., Platonova N., Neri A., Chiaramonte R. Multiple myeloma-derived Jagged ligands increases autocrine and paracrine interleukin-6 expression in bone marrow niche. Oncotarget. 2016;7:56013–56029. doi: 10.18632/oncotarget.10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo D., Li C., Teng Q., Sun Z., Li Y., Zhang C. Notch1 overexpression promotes cell growth and tumor angiogenesis in myeloma. Neoplasma. 2013;60:33–40. doi: 10.4149/neo_2013_005. [DOI] [PubMed] [Google Scholar]
- 9.Chiron D., Maiga S., Descamps G., Moreau P., Le G.S., Marionneau S., Ouiller T., Moreaux J., Klein B., Bataille R., Amiot M., Pellat-Deceunynck C. Critical role of the NOTCH ligand JAG2 in self-renewal of myeloma cells. Blood Cells Mol. Dis. 2012;48:247–253. doi: 10.1016/j.bcmd.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Xu D., Hu J., Xu S., De B.E., Menu E., Van C.B., Vanderkerken K., Van V.E. Dll1/Notch activation accelerates multiple myeloma disease development by promoting CD138+ MM-cell proliferation. Leukemia. 2012;26:1402–1405. doi: 10.1038/leu.2011.332. [DOI] [PubMed] [Google Scholar]
- 11.Sabol H.M., Ferrari A.J., Adhikari M., Amorim T., McAndrews K., Anderson J., Vigolo M., Lehal R., Cregor M., Khan S., Cuevas P.L., Helms J.A., Kurihara N., Srinivasan V., Ebetino F.H., Boeckman R.K., Jr., Roodman G.D., Bellido T., Delgado-Calle J. Targeting notch inhibitors to the myeloma bone marrow niche decreases tumor growth and bone destruction without gut toxicity. Cancer Res. 2021;81:5102–5114. doi: 10.1158/0008-5472.CAN-21-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarzer R., Nickel N., Godau J., Willie B.M., Duda G.N., Schwarzer R., Cirovic B., Leutz A., Manz R., Bogen B., Dorken B., Jundt F. Notch pathway inhibition controls myeloma bone disease in the murine MOPC315.BM model. Blood Cancer J. 2014;4:e217. doi: 10.1038/bcj.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M., Chen F., Clifton N., Sullivan D.M., Dalton W.S., Gabrilovich D.I., Nefedova Y. Combined inhibition of Notch signaling and Bcl-2/Bcl-xL results in synergistic antimyeloma effect. Mol. Cancer Ther. 2010;9:3200–3209. doi: 10.1158/1535-7163.MCT-10-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramakrishnan V., Ansell S., Haug J., Grote D., Kimlinger T., Stenson M., Timm M., Wellik L., Halling T., Rajkumar S.V., Kumar S. MRK003, a gamma-secretase inhibitor exhibits promising in vitro preclinical activity in multiple myeloma and non-Hodgkin’s lymphoma. Leukemia. 2012;26:340–348. doi: 10.1038/leu.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imbimbo B.P. Therapeutic potential of gamma-secretase inhibitors and modulators. Curr. Top. Med. Chem. 2008;8:54–61. doi: 10.2174/156802608783334015. [DOI] [PubMed] [Google Scholar]
- 16.Searfoss G.H., Jordan W.H., Calligaro D.O., Galbreath E.J., Schirtzinger L.M., Berridge B.R., Gao H., Higgins M.A., May P.C., Ryan T.P. Adipsin, a biomarker of gastrointestinal toxicity mediated by a functional gamma-secretase inhibitor. J. Biol. Chem. 2003;278:46107–46116. doi: 10.1074/jbc.M307757200. [DOI] [PubMed] [Google Scholar]
- 17.Colombo M., Galletti S., Garavelli S., Platonova N., Paoli A., Basile A., Taiana E., Neri A., Chiaramonte R. Notch signaling deregulation in multiple myeloma: A rational molecular target. Oncotarget. 2015;6:26826–26840. doi: 10.18632/oncotarget.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delgado-Calle J., Bellido T. The osteocyte as a signaling cell. Physiol. Rev. 2022;102:379–410. doi: 10.1152/physrev.00043.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nefedova Y., Cheng P., Alsina M., Dalton W.S., Gabrilovich D.I. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- 20.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taron M., Rosell R., Felip E., Mendez P., Souglakos J., Ronco M.S., Queralt C., Majo J., Sanchez J.M., Sanchez J.J., Maestre J. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum. Mol. Genet. 2004;13:2443–2449. doi: 10.1093/hmg/ddh260. [DOI] [PubMed] [Google Scholar]
- 23.Shalapour S., Hof J., Kirschner-Schwabe R., Bastian L., Eckert C., Prada J., Henze G., von Stackelberg A., Seeger K. High VLA-4 expression is associated with adverse outcome and distinct gene expression changes in childhood B-cell precursor acute lymphoblastic leukemia at first relapse. Haematologica. 2011;96:1627–1635. doi: 10.3324/haematol.2011.047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuster S.R., Kortuem K.M., Zhu Y.X., Braggio E., Shi C.X., Bruins L.A., Schmidt J.E., Ahmann G., Kumar S., Rajkumar S.V., Mikhael J., Laplant B., Champion M.D., Laumann K., Barlogie B., Fonseca R., Bergsagel P.L., Lacy M., Stewart A.K. The clinical significance of cereblon expression in multiple myeloma. Leuk. Res. 2014;38:23–28. doi: 10.1016/j.leukres.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooper L.A., Gutman D.A., Chisolm C., Appin C., Kong J., Rong Y., Kurc T., Van Meir E.G., Saltz J.H., Moreno C.S., Brat D.J. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am. J. Pathol. 2012;180:2108–2119. doi: 10.1016/j.ajpath.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varemo L., Nielsen J., Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic. Acids. Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellido T., Delgado-Calle J. Ex vivo organ cultures as models to study bone biology. JBMR Plus. 2020;4 doi: 10.1002/jbm4.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delgado-Calle J., Kurihara N., Atkinson E.G., Nelson J., Miyagawa K., Galmarini C.M., Roodman G.D., Bellido T. Aplidin (plitidepsin) is a novel anti-myeloma agent with potent anti-resorptive activity mediated by direct effects on osteoclasts. Oncotarget. 2019;10:2709–2721. doi: 10.18632/oncotarget.26831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kyle R.A., Rajkumar S.V. Criteria for diagnosis, staging, risk stratification and response assessment of multiple myeloma. Leukemia. 2009;23:3–9. doi: 10.1038/leu.2008.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dallas S.L., Garrett I.R., Oyajobi B.O., Dallas M.R., Boyce B.F., Bauss F., Radl J., Mundy G.R. Ibandronate reduces osteolytic lesions but not tumor burden in a murine model of myeloma bone disease. Blood. 1999;93:1697–1706. [PubMed] [Google Scholar]
- 31.Garrett I.R., Dallas S., Radl J., Mundy G.R. A murine model of human myeloma bone disease. Bone. 1997;20:515–520. doi: 10.1016/s8756-3282(97)00056-2. [DOI] [PubMed] [Google Scholar]
- 32.Radl J., Croese J.W., Zurcher C., Van den Enden-Vieveen M.H., de Leeuw A.M. Animal model of human disease. Multiple myeloma. Am. J. Pathol. 1988;132:593–597. [PMC free article] [PubMed] [Google Scholar]
- 33.Tsunenari T., Koishihara Y., Nakamura A., Moriya M., Ohkawa H., Goto H., Shimazaki C., Nakagawa M., Ohsugi Y., Kishimoto T., Akamatsu K. New xenograft model of multiple myeloma and efficacy of a humanized antibody against human interleukin-6 receptor. Blood. 1997;90:2437–2444. [PubMed] [Google Scholar]
- 34.Delgado-Calle J., Anderson J., Cregor M.D., Condon K.W., Kuhstoss S.A., Plotkin L.I., Bellido T., Roodman G.D. Genetic deletion of Sost or pharmacological inhibition of sclerostin prevent multiple myeloma-induced bone disease without affecting tumor growth. Leukemia. 2017;31:2686–2694. doi: 10.1038/leu.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrusca D.N., Toscani D., Wang F.M., Park C., Crean C.D., Anderson J.L., Marino S., Mohammad K.S., Zhou D., Silbermann R., Sun Q., Kurihara N., Galson D.L., Giuliani N., Roodman G.D. Growth factor independence 1 expression in myeloma cells enhances their growth, survival, and osteoclastogenesis. J. Hematol. Oncol. 2018;11:123. doi: 10.1186/s13045-018-0666-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asosingh K., Radl J., Van Riet I., Van Camp B., Vanderkerken K. The 5TMM series: a useful in vivo mouse model of human multiple myeloma. Hematol. J. 2000;1:351–356. doi: 10.1038/sj.thj.6200052. [DOI] [PubMed] [Google Scholar]
- 37.Hosseini-Alghaderi S., Baron M. Notch3 in development, health and disease. Biomolecules. 2020;10 doi: 10.3390/biom10030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aster J.C., Pear W.S., Blacklow S.C. The varied roles of notch in cancer. Annu. Rev. Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colombo M., Thummler K., Mirandola L., Garavelli S., Todoerti K., Apicella L., Lazzari E., Lancellotti M., Platonova N., Akbar M., Chiriva-Internati M., Soutar R., Neri A., Goodyear C.S., Chiaramonte R. Notch signaling drives multiple myeloma induced osteoclastogenesis. Oncotarget. 2014 doi: 10.18632/oncotarget.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dallas S.L., Prideaux M., Bonewald L.F. The osteocyte: an endocrine cell ... and more. Endocr. Rev. 2013;34:658–690. doi: 10.1210/er.2012-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemmatian H., Conrad S., Furesi G., Mletzko K., Krug J., Faila A.V., Kuhlmann J.D., Rauner M., Busse B., Jahn-Rickert K. Reorganization of the osteocyte lacuno-canalicular network characteristics in tumor sites of an immunocompetent murine model of osteotropic cancers. Bone. 2021;152 doi: 10.1016/j.bone.2021.116074. [DOI] [PubMed] [Google Scholar]
- 42.Rosen L.S., Wesolowski R., Baffa R., Liao K.H., Hua S.Y., Gibson B.L., Pirie-Shepherd S., Tolcher A.W. A phase I, dose-escalation study of PF-06650808, an anti-Notch3 antibody-drug conjugate, in patients with breast cancer and other advanced solid tumors. Invest. New Drugs. 2020;38:120–130. doi: 10.1007/s10637-019-00754-y. [DOI] [PubMed] [Google Scholar]
- 43.Yu J., Siebel C.W., Schilling L., Canalis E. An antibody to Notch3 reverses the skeletal phenotype of lateral meningocele syndrome in male mice. J. Cell. Physiol. 2020;235:210–220. doi: 10.1002/jcp.28960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Canalis E., Yu J., Schilling L., Yee S.P., Zanotti S. The lateral meningocele syndrome mutation causes marked osteopenia in mice. J. Biol. Chem. 2018;293:14165–14177. doi: 10.1074/jbc.RA118.004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pajevic P.Divieti. New and old osteocytic cell lines and 3D models. Curr. Osteoporos. Rep. 2020;18:551–558. doi: 10.1007/s11914-020-00613-3. [DOI] [PubMed] [Google Scholar]
- 46.Delgado-Calle J., Anderson J., Cregor M.D., Hiasa M., Chirgwin J.M., Carlesso N., Yoneda T., Mohammad K.S., Plotkin L.I., Roodman G.D., Bellido T. Bidirectional notch signaling and osteocyte-derived factors in the bone marrow microenvironment promote tumor cell proliferation and bone destruction in multiple myeloma. Cancer Res. 2016;76:1089–1100. doi: 10.1158/0008-5472.CAN-15-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giuliani N., Ferretti M., Bolzoni M., Storti P., Lazzaretti M., Dalla P.B., Bonomini S., Martella E., Agnelli L., Neri A., Ceccarelli F., Palumbo C. Increased osteocyte death in multiple myeloma patients: role in myeloma-induced osteoclast formation. Leukemia. 2012;26:1391–1401. doi: 10.1038/leu.2011.381. [DOI] [PubMed] [Google Scholar]
- 48.McDonald M.M., Reagan M.R., Youlten S.E., Mohanty S.T., Seckinger A., Terry R.L., Pettitt J.A., Simic M.K., Cheng T.L., Morse A., Le L.M.T., Abi-Hanna D., Kramer I., Falank C., Fairfield H., Ghobrial I.M., Baldock P.A., Little D.G., Kneissel M., Vanderkerken K., Bassett J.H.D., Williams G.R., Oyajobi B.O., Hose D., Phan T.G., Croucher P.I. Inhibiting the osteocyte-specific protein sclerostin increases bone mass and fracture resistance in multiple myeloma. Blood. 2017;129:3452–3464. doi: 10.1182/blood-2017-03-773341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toscani D., Palumbo C., Dalla P.B., Ferretti M., Bolzoni M., Marchica V., Sena P., Martella E., Mancini C., Ferri V., Costa F., Accardi F., Craviotto L., Aversa F., Giuliani N. The proteasome inhibitor bortezomib maintains osteocyte viability in multiple myeloma patients by reducing both apoptosis and autophagy: a new function for proteasome inhibitors. J. Bone Miner. Res. 2016;31:815–827. doi: 10.1002/jbmr.2741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.