Stand-first:
Ferroptosis is a regulated form of non-apoptotic cell death implicated in pathological settings. To be exploited clinically, ferroptosis requires reagents that unequivocally detect ferroptosis in human and animal tissues. Such tools may enable development of ferroptosis-based medicines for diverse diseases.
Cell death is a critical physiological process that can occur through unregulated necrosis or in regulated manner. The past two decades have uncovered a variety of regulated cell death modalities that are distinct from apoptosis1. Ferroptosis, coined as a term in 2012, is a regulated form of cell death that is initiated by iron-dependent peroxidation of phospholipids with polyunsaturated fatty acyl tails2. Over the past decade, ferroptosis has become a critical and rapidly growing field of research. Many investigators have revealed key regulators of this process, including ferroptosis-inhibitory processes (i.e., the system-xc-/glutathione/GPX4 axis, the FSP1/ubiquinol axis, the GCH1/DHFR/tetrahydrobiopterin axis and the DHODH/ubiquinol axis), cellular processes inducing lipid peroxidation, iron dependency, and metabolic pathways that modulate ferroptosis3–9. These findings have shaped our understanding of the molecular actions driving and suppressing ferroptosis and have highlighted its potential importance in diverse pathological settings, including neurodegeneration, stroke, traumatic brain injury, ischemia-reperfusion injury, cardiomyopathy, and kidney degeneration10,11. In addition, recent studies have demonstrated that induction of ferroptosis is potentially useful as a novel strategy to attack responsive cancers12–14.
Hence, studies in the past decade have largely concentrated on mechanistic discoveries of ferroptosis-regulating pathways, and generating small molecule modulators to interrogate ferroptosis in cell models (Fig. 1).
Figure 1:
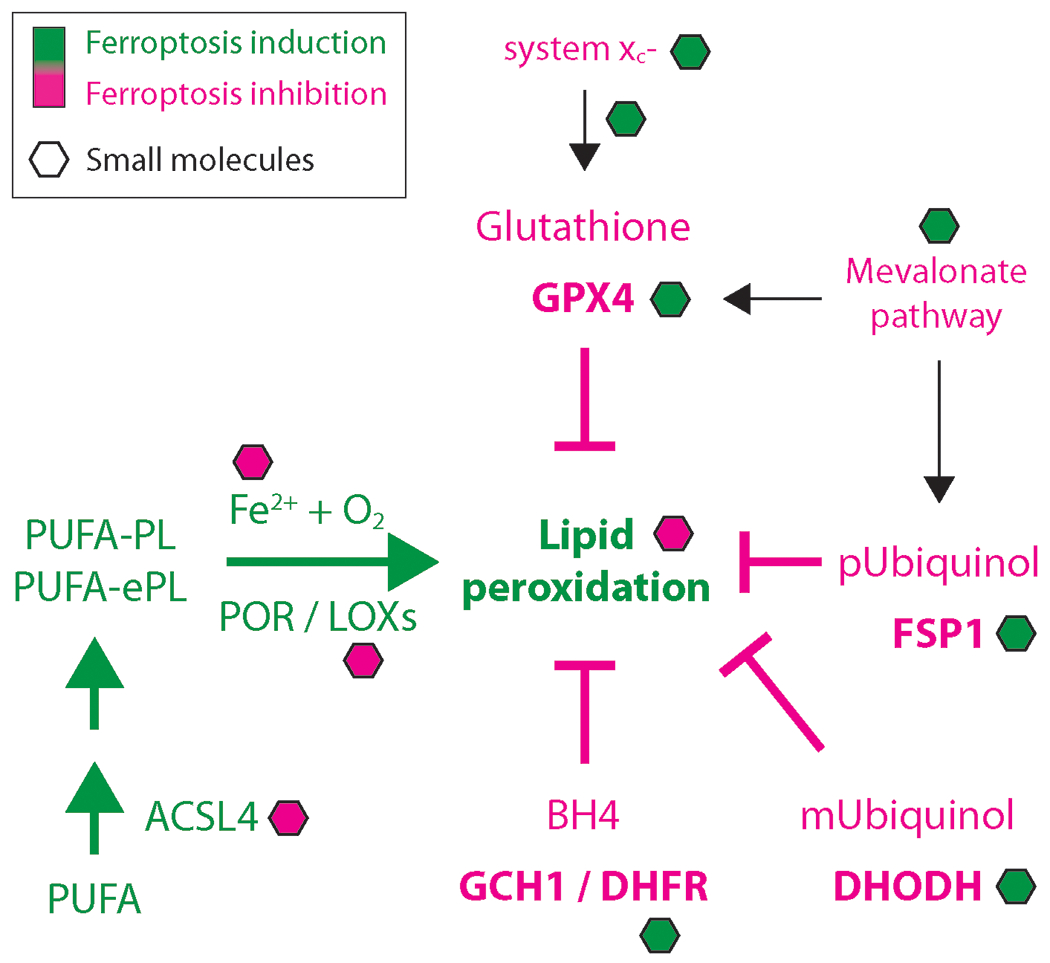
Major ferroptosis regulators and small molecule modulators. Components inducing ferroptosis are depicted in green, and components inhibiting ferroptosis are shown in pink. Small molecules are shown as hexagons. pUbiquinol: ubiquinol in plasma membrane; mUbiquinol: mitochondrial ubiquinol.
Yet, in order to rigorously evaluate the therapeutic potential of ferroptosis modulation, a major challenge is to reliably detect and classify ferroptosis in animal and human tissues under pathological conditions. Currently available detection reagents9 such as probes (e.g., C11-BODIPY, TBARS and LiperFluo) and assay systems (e.g., FENIX and metabolomics/lipidomics) work well in cell culture models, but have limitations for rapid and widespread use for the analysis of tissues. Thus, biomarkers and associated detection technologies are needed to unambiguously identify ferroptotic cell death in disease tissues. In line with this, a recent study determined that antibodies against transferrin receptor 1 (TfR1) detect ferroptosis in cells and in vivo15. However, application of this reagent to tissue contexts has not yet been fully explored, and specificity versus other contexts needs to be defined. Hence, additional tools for specifically detecting ferroptosis are needed. Alongside reagent and technology development, small molecule and large molecule regulators of distinct ferroptosis targets are required to clinically modulate ferroptosis disease-relevant settings. A few demonstrated examples with efficacy in animal models12,16,17 represent foundational approaches for ultimately creating drug-like ferroptosis-regulating medicines. To translate ferroptosis biology into new therapeutics, the following two strategies have the potential to help fully leverage the past decade of ferroptosis research:
Generating a molecular compendium (biomarkers, reagents, and analysis methods) that enables specific detection of ferroptosis in animal models and human patient samples
Creating in vivo compatible drug candidates that selectively modulate ferroptosis-relevant targets
Ferroptosis biomarker and detection modalities
The most important considerations for reagent development in order to classify ferroptosis in vivo are specificity and selectivity. For instance, neurodegenerative diseases such as Alzheimer’s and Parkinson’s disease can be caused by apoptosis, ferroptosis, or necroptosis. Hence, it is critical to selectively discriminate between cell death modalities in the same disease setting in order to be able to develop specific disease-modifying medicines.
Two hallmarks of ferroptosis are oxidation of polyunsaturated-fatty-acylphospholipids (PUFA-PLs) and redox-active iron18. These cellular entities have been central elements of reagent development to detect ferroptosis in cell culture models9. However, their application for detecting ferroptosis in tissues of patient or animal origin is of limited impact to date due to several key issues: (i) these tools cannot be used on formalin-fixed paraffin-embedded (FFPE) tissue samples, which represents the major type of available tissue sources; (ii) most existing assays are technically complex and difficult for routine application in clinical practice; (iii) products of lipid peroxidation, namely malondialdehyde (MDA) and 4-hydroxynonenal (4-HNE), are good proxies for assessing lipid peroxidation, but not exclusive to ferroptosis, as they can also be produced upon general oxidative stress; (iv) in tissue settings, it is technically challenging to specifically connect iron redox state to ferroptotic cell death; and (v) oxidized lipids and redox-active iron are difficult to detect in liquid biopsies or blood samples, because of stability issues and the limited ability to connect them to their tissue of origin. Together, current biomarkers and detection technologies are not sufficient for interrogating the existence of ferroptosis in animals or patients.
Consequently, reagents that can be utilized to unequivocally detect ferroptosis versus general oxidative stress in tissue samples is of high demand and currently a barrier towards creating ferroptosis-based medicines. Identification of novel biomarkers with high ferroptosis specificity over other cell death modalities, as well as contexts that trigger oxidative stress without inducing ferroptosis, and vice versa are of high demand. Such biomarkers can be of diverse nature, and would include mRNAs, metabolites, lipids, proteins, enzymes as well as membrane proteins and receptors. Therefore, to comprehensively address the challenge of selective ferroptosis detection, a two-step process may be helpful to (i) identify specific biomarkers and to (ii) create reagents to detect these markers.
As a first step, a set of exploratory studies could be initiated that dissect the nature and molecular details of cell death modalities, as well as overlapping and non-overlapping features with oxidative stress pathways to identify unambiguous ferroptosis markers. In particular, side-by-side comparisons of omic methods could specifically map critical components of and responses to individual cell death pathways and cell stresses. Each omics technology could be measured using diverse cell death modalities and other stressors in parallel experiments to enable direct comparison of markers among cell states. This concept is supported by a recent comparative study demonstrating that excessive phospholipid peroxidation distinguishes ferroptosis from apoptosis, necroptosis and pyroptosis19. Thus, only with a broad picture of the ingredients of distinct cell death and oxidative stress pathways will we be able to choose unambiguous biomarkers of ferroptosis. In addition, biomarker discovery may yield markers for different stages of ferroptosis induction (early phase, peroxidation phase and late phase) to be able to develop ferroptosis-preventive and ferroptosis-inducing medicines, and where these fit within the ferroptosis process.
Once a collection of selective ferroptosis biomarkers is defined, subsequent studies need to explore their consistency in detecting ferroptosis within different cell and tissue populations. For example, inducing ferroptosis in CD8+ T-cells would have a very different functional impact than inducing ferroptosis in tumor cells--responses of different cell types in a tissue is critical. Thus, it is important to analyze their presence on a single-cell level to choose highly consistent markers. Notably, identification of markers for different ferroptosis stages relies even more on single cell resolution, as individual cells within populations will inevitably be at different stages. Single-cell studies will include single-cell transcriptomics, single-cell phenotyping by high-content image analysis, as well as recently developed ultra-sensitive single-cell proteomics20 and single-cell lipidomics/metabolomics21. In addition to reagent development for ferroptosis detection, the implementation of mass spectrometry imaging to visualize metabolic biomarkers in tissue samples could be an important technology to detect ferroptosis in human and animal samples. The field should be able to nominate highly selective biomarkers of ferroptotic cell death that are operative in different cell and tissue types within the next five-year span of research.
As a second step, suitable clinical-grade reagents that selectively recognize ferroptosis biomarkers in tissue samples should be created. This will form the next generation of tools to precisely detect ferroptosis in animal models and human patient samples. It will be important to develop a broad spectrum of detection technologies in order to be able to monitor ferroptosis in fixed animal as well as human tissues, frozen samples, and for live patient imaging. Such tools include (i) chemical probes that specifically detect previously specified ferroptosis markers in mouse and human FFPE tissue sections, (ii) probes suitable for patient imaging by magnetic resonance tomography (MRI), optoacoustic imaging, computer tomography (CT), or position emission tomography (PET) to detect lipid peroxides or other relevant biomarkers in vivo, and (iii) antibodies, as well as fluorescently tagged single-chain antibodies and nanobodies to detect ferroptosis biomarkers on the surface of cells, which can be used for tissue imaging in immunohistological studies. These three lines of reagent development should be a key future research area to advance the transition of ferroptosis biology into medicines.
Application of probes, fluorescent agents, or antibodies to detect ferroptosis biomarkers in tissues and primary cells generates large quantities of imaging data that need proper analysis in order to be able to classify samples as containing or lacking ferroptosis. Analyses of such images can be carried out manually, which is limited in terms of throughput, and, most importantly, prone to bias by individual researchers. In contrast, artificial intelligence (AI) tools can help analyzing images in an unbiased way to extract image features that human eyes would not perceive or select, and use this information to distinguish ferroptosis from other cell death modalities (e.g., apoptosis, necroptosis, and pyroptosis) and oxidative stress. We present a brief description of general AI concepts for image analysis in Box 1. Together, the combination of novel tools (probes, reagents, antibodies) to detect ferroptosis-selective biomarkers in tissues, together with unbiased image analysis by AI methods could be critical to specifically detect ferroptosis in animal and human tissues (Fig. 2).
Box 1:
Machine Learning (ML) analysis routines can be performed with supervised learning strategies, where the model is first fit on a training data set with defined labels, which can subsequently be used to assign test sample images to previously classified labels. Notably, this analysis method needs (i) extracted features from labeled training data sets, and (ii) a priori knowledge of the outcome (e.g., morphological understanding of different cell death pathways). Thus, this analysis strategy can only be used in settings where suitable controls are available. However, in many cases the researcher is not aware of the outcome, e.g., which type of cell death modality is the cause of a certain disease. Hence, supervised ML limits the ability to gain novel and unexpected biological insights. Deep learning (DL) algorithms can overcome the need for a priori knowledge by using neural networks (e.g., convolutional neural networks) to extract features from raw images of various conditions without seeing the labels. Subsequently, it learns differences between these conditions and classifies the output23. This type of analysis routine is perfectly suited to distinguish larger as well as subtle differences for instance between diverse cell death and oxidative stress pathways. Current image-based ML/DL methods largely concentrate on analysis of cell culture models in high-content imaging approaches. However, recent advances have also applied ML/DL tools to analyzing tissue images, especially by applying DL algorithms24. Finally, the field is growingly developing tools that can be used by AI-inexperienced researchers to train and apply DL networks for their image analysis. A recent example for user-friendly DL application is the ZeroCostDL4Mic platform25.
Figure 2:
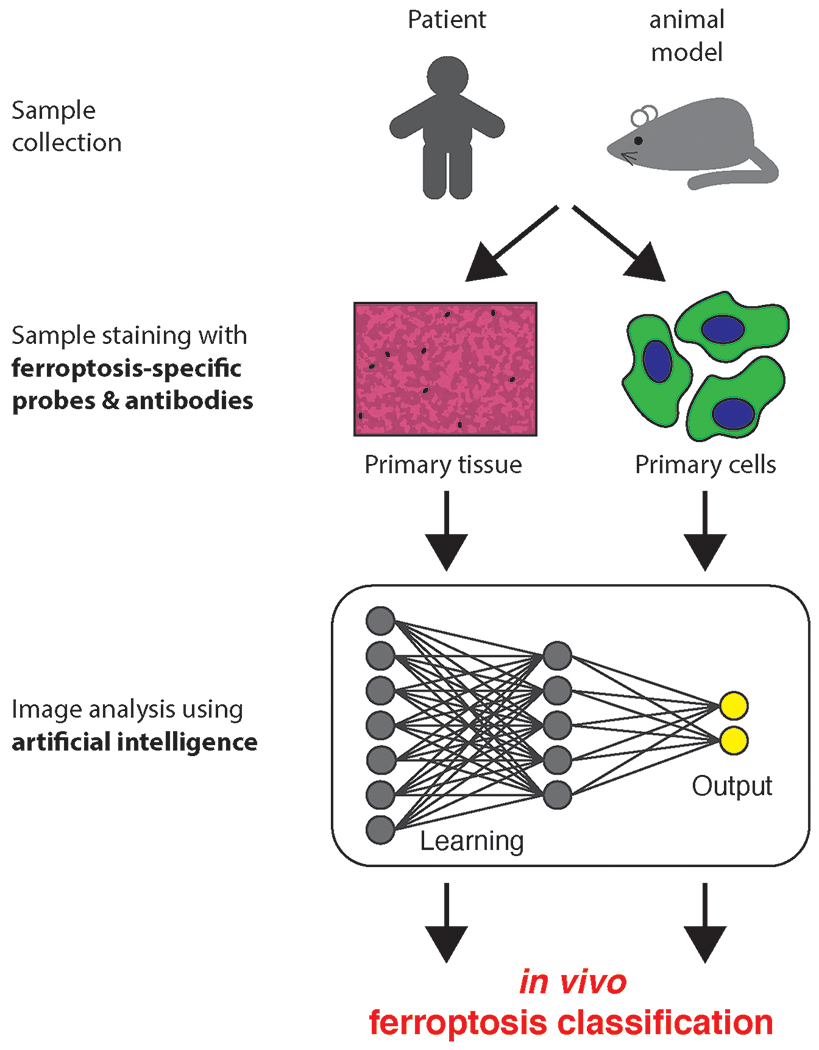
Ferroptosis detection and classification. Sample staining with ferroptosis specific reagents and subsequent artificial intelligence methods for image analysis to specifically classify ferroptosis in vivo.
Advances in ferroptosis-based therapeutics
Introduction of the term of ferroptosis in 2012 fueled the investigation of diverse small molecule modulators of this pathway. These modulators include ferroptosis inhibitors that may be used to treat degenerative diseases and ferroptosis inducers that may be applied to treatment of specific cancer subtypes.
Notable ferroptosis inhibitors are radical-trapping antioxidants (RTAs), deuterated PUFAs, or iron-chelating molecules, which mostly lack specific cellular targets, but act through general mechanisms to remove hallmarks of ferroptosis, namely lipid peroxides and iron. In addition, inhibition of ACSL4 by glitazone compounds has shown to be effective for ferroptosis inhibition. Inducers of ferroptosis have largely been developed against components of the system-xc-/glutathione/GPX4 axis (e.g., imidazole ketone erastin, buthionine sulfoximine, RSL3, and ML210). While these are promising targets to induce ferroptosis, they have only utilized the potential of a small part of the known ferroptosis regulatory network. Thus, additional targets would be desirable to enlarge the repertoire of small molecules modulating ferroptosis to be used perhaps in combination with system-xc-/glutathione/GPX4 axis inhibition or in contexts where system-xc-/glutathione/GPX4 inhibition is not viable. Moreover, adaptations of cancer cells to loss of ferroptosis-protective enzymes, such as GPX4, may benefit from combinatorial treatments.
A recent review summarized the progress of the most widely used ferroptosis modulators towards becoming medicines10. Although some of these small molecules have shown useful in vivo effects, most of them have not (yet) passed the stage of preclinical development and few have entered clinical testing. Hence, to extend the number of ferroptosis-modulating clinical candidates, current and future research efforts may employ more target-based therapeutic strategies for small molecule combinatorial treatment along with inhibitors of the system-xc-/glutathione/GPX4 axis. A target-based approach should be beneficial because having defined targets will facilitate lead optimization as well as preclinical validation to accelerate the path of novel candidate molecules towards clinical evaluation and in vivo target engagement and pharmacodynamics studies. Based on a number of excellent discoveries of recent years, the ferroptosis community is able to nominate promising targets from diverse ferroptosis-regulating pathways to induce or block ferroptosis, with selected examples highlighted in the following.
Until recently, the system-xc-/glutathione/GPX4 axis was considered to be the only ferroptosis inhibitory pathway, which resulted in many inhibitors of this axis to drive ferroptosis. However, it is now understood that three other gatekeepers counteract ferroptosis in a GPX4-independent manner, namely the FSP1/ubiquinol axis3,4, the GCH1/DHFR/tetrahydrobiopterin axis5,6, and the DHODH/ubiquinol axis7. Thus, targeting these enzymes and their associated pathways with selective inhibitors in combination with system-xc-/glutathione/GPX4 axis inhibitors holds promise for effective cancer treatment.
In order to inhibit ferroptosis, the process of iron-dependent lipid peroxidation needs to be blocked. As an alternative to established RTAs and iron-chelators, target-based strategies to attenuate or block lipid peroxidation are the inhibition of Acyl-CoA Synthetase Long Chain Family Member 4 (ACSL4) or cytochrome P450 oxidoreductase (POR). Here, inhibition of defined targets to block lipid peroxidation may increase specificity of ferroptosis-related medicine. Undoubtedly, future research will illuminate additional targets from genetic and chemical screens to counteract ferroptosis in degenerative diseases.
Besides small molecule drug development, the field of ferroptosis medicines could explore the generation of large molecule therapeutics, including antibodies, nanobodies, and single-chain antibodies to activate and repress ferroptosis. Although the number of large molecule therapeutics in cancer and immune therapies has skyrocketed in recent years, the ferroptosis field has barely entered this therapeutic space to explore the potential of an alternative set of promising targets. Notably, large molecule approaches will have their potential in specifically targeting extracellular-oriented surface proteins and receptors. The biggest advantage of large molecule ferroptosis medicines may be the high specificity of these drug candidates towards their targets. As an example, antibodies against TfR1, which recently were developed to visualize ferroptosis15, may be used to block transferrin-mediated iron import and hence inhibit ferroptosis. An additional example would be the generation of antibodies against system xc-to induce ferroptosis. Given the success of large molecule drug discovery in the past decade, the ferroptosis field could benefit from this opportunity to incorporate a novel modality. In addition to targeting cell surface receptors and membrane proteins by antibodies, administration of specific enzymes may be an additional strategy to modulate ferroptosis as recently demonstrated by the use of cyst(e)inases in pancreatic cancer12. Together, large molecule drug discovery may become an essential component of ferroptosis-based medicines.
A further exciting drug discovery avenue that has not been explored yet by the ferroptosis field is the use of PROTACs and LYTACs. Mechanistically, PROTACs have the ability to degrade proteins via the proteasome by hijacking E3 ligases and LYTACs induce endocytosis of receptors or external proteins for lysosomal degradation22. One obvious benefit of PROTACs or LYTACs for the ferroptosis field in distinct instances over conventional inhibitory small molecules may be that targets without desirable pockets for compound inhibition can be marked for degradation; thus, expanding ferroptosis medicines to less conventionally druggable regulators. Moreover, eradication of a given target, in contrast to inhibition, might be more effective to modulate ferroptosis in some contexts. In general, PROTACs/LYTACs are additional drug modalities in suitable target contexts and not an overall replacement for inhibitory compounds. As a proof-of-concept, PROTACs may be directed against GPX4 and HMGCR for ferroptosis induction, or against ACSL4 for ferroptosis inhibition. With regards to LYTACs, degradation of SLC7A11 may induce ferroptosis, while degradation of TfR1 would inhibit ferroptosis. Such exploratory studies could be initiated to evaluate the suitability of these drug classes as valid ferroptosis medicines and upon success expand to additional targets.
In summary, to accomplish clinical translation of ferroptosis, the community needs to develop selective small and large molecule therapeutics against ferroptosis-regulating targets that have a favorable set of pharmacokinetics and ADME parameters, alongside demonstrated efficacy and safety.
Conclusion
In this commentary, we argue for the need for specifically detecting ferroptosis in vivo as well as therapeutics that act through ferroptosis-regulating targets in order to ultimately generate ferroptosis-based medicines. Thus, exploring the following two strategies as the core of future efforts may facilitate translating ferroptosis research from bench to bedside:
Identification of ferroptosis BIOMARKERS and development of REAGENTS to detect ferroptosis in (human) primary tissues.
Development of small molecule and large molecule THERAPEUTICS capable of modulating ferroptosis in animal models and human patients.
It is also important to mention that the community may want to nominate distinct animal models as standards for understanding ferroptosis biology as well as studying distinct diseases, where ferroptosis may be involved. This will facilitate the discovery and detection of specific ferroptosis biomarkers and the effective testing of ferroptosis-based drugs. Defined animal models will ease comparison of studies between research labs and clinicians.
The goals suggested in this commentary are interconnected and this endeavor is only feasible if the field brings together researcher from multiple disciplines and scientific as well as medical backgrounds to cooperatively tackle the critical tasks needed to create ferroptosis-based medicines.
Footnotes
Competing financial interest
Kamyar Hadian declares no competing financial interest. Brent R. Stockwell is an inventor on patents and patent applications involving ferroptosis, co-founded and serves as a consultant to Inzen Therapeutics and Nevrox Limited, and serves as a consultant to Weatherwax Biotechnologies Corporation.
References:
- 1.Tang D, Kang R, Berghe TV, Vandenabeele P & Kroemer G Cell Res 29, 347–364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dixon SJ et al. Cell 149, 1060–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bersuker K et al. Nature 575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doll S et al. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Kraft VAN et al. ACS Cent Sci 6, 41–53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soula M et al. Nat Chem Biol 16, 1351–1360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao C et al. Nature 593, 586–590 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou Y et al. Nat Chem Biol 16, 302–309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadian K & Stockwell BR Cell 181, 1188–1188.e1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad M, Lorenz SM & Proneth B Trends Mol Med 27, 113–122 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Stockwell BR, Jiang X & Gu W Trends Cell Biol 30, 478–490 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Badgley MA et al. Science 368, 85–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ubellacker JM et al. Nature 585, 113–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang WM et al. Nature 569, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng H et al. Cell Rep 30, 3411–3423 e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y et al. Cell Chem Biol 26, 623–633 e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedmann Angeli JP et al. Nat Cell Biol 16, 1180–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon SJ & Stockwell BR Annual Review of Cancer Biology, Vol 3 3, 35–54 (2019). [Google Scholar]
- 19.Wiernicki B et al. Cell Death Dis 11, 922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunner A-D et al. bioRxiv, 2020.12.22.423933 (2021). [Google Scholar]
- 21.Capolupo L et al. bioRxiv, 2021.02.23.432420 (2021). [Google Scholar]
- 22.Ding Y, Fei Y & Lu B Trends Pharmacol Sci 41, 464–474 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin S, Schorpp K, Rothenaigner I & Hadian K Drug Discov Today 25, 1348–1361 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Binder A et al. Nature Machine Intelligence 3, 355–366 (2021). [Google Scholar]
- 25.von Chamier L et al. Nat Commun 12, 2276 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


