Abstract
Androgens and the androgen receptor (AR) play crucial roles in the biology of normal and diseased prostate tissue, including prostate cancer (PCa). This dependence is evidenced by the use of androgen depletion therapy (ADT) as the primary treatment for locally advanced, metastatic, or relapsed PCa. This dependence is further evidenced by the various mechanisms employed by PCa cells to re-activate the AR to circumvent the growth-inhibitory effects of ADT. Re-activation of the AR during ADT is central to the disease evolving into the lethal castration resistant PCa (CRPC) phenotype, which is responsible for nearly all PCa mortality. Thus, understanding the regulation of AR and AR signaling is important for understanding the development and progression of PCa. This understanding provides the foundation for development of newer approaches for targeting CRPC therapeutically.
AR Structure and Function
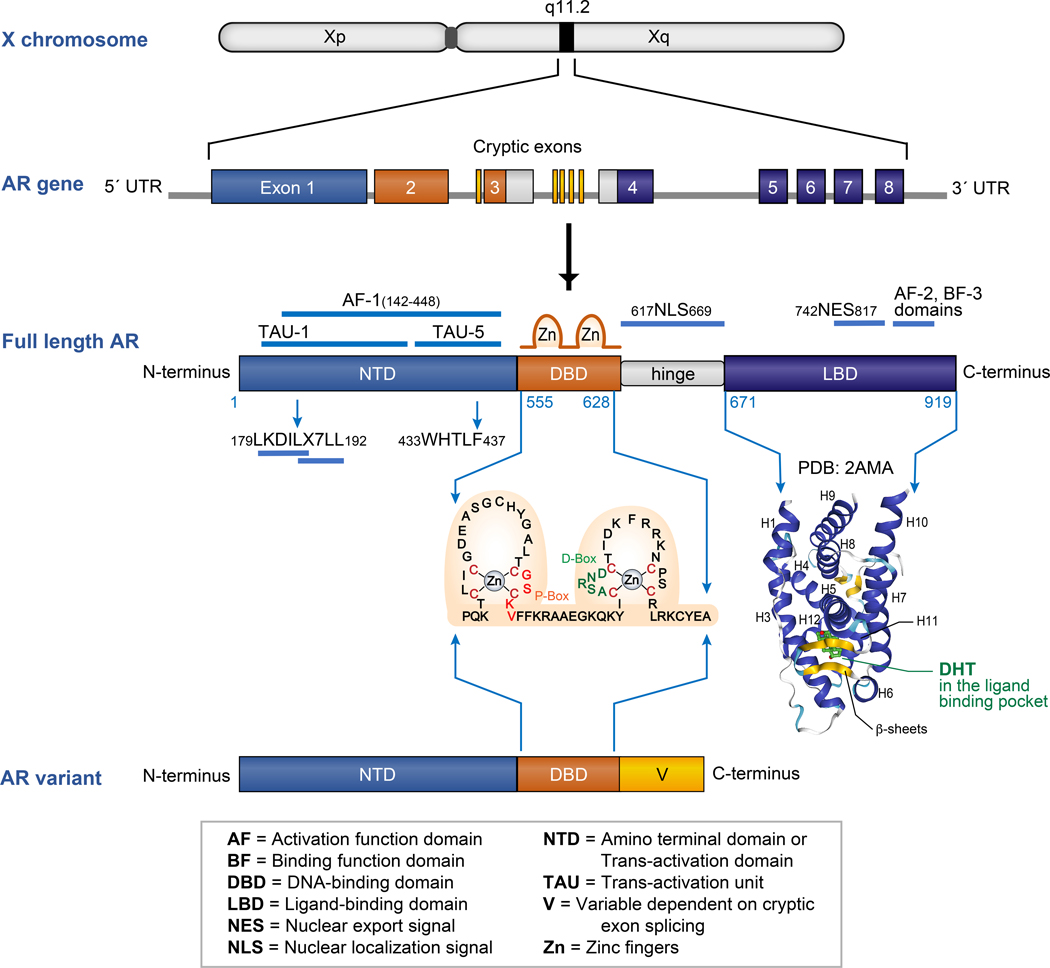
AR is a member of the class I nuclear receptor transcription factor family, which includes the steroid receptors glucocorticoid receptor (GR), mineralocorticoid receptor (MR), estrogen receptor (ER), and progesterone receptor (PR). It is a 110 kDa phospho-protein encoded by the AR gene located on chromosome X at Xq11–12; hence XY males have 1 copy of AR. The AR gene comprises eight exons which encode four distinct functional domains of the full-length AR protein: (i) an intrinsically-disordered NH2-terminal domain (NTD) encoded by exon 1; (ii) a 2-zinc finger DNA-binding domain (DBD) encoded by exon 2 and the 5’ end of exon 3; (iii) a short flexible hinge region harboring the nuclear localization signal (NLS) encoded by the 3’ end of exon 3 and 5’ end of exon 4; and (iv) a ligand-binding domain (LBD) encoded by the 3’ end of exon 4 along with exons 5–8 (Figure 1) [1, 2].
Figure 1:
AR gene and protein structure: AR is located on the X chromosome at position q11.2. The AR gene is encoded by 8 exons that are color coded to represent the domains of the full-length AR protein they encode. The full-length AR is comprised of an amino terminal domain (NTD, in blue), DNA binding domain (DBD, in orange), a short hinge region (in grey) and a ligand binding domain (LBD, in purple). The amino acid sequence of the two zinc finger units containing the P-box and D-box of the AR DBD are shown. The structure of human AR LBD domain with a DHT bound in its ligand binding pocket is represented (PDB: 2AMA). AR variants contain the AR NTD and DBD but lack the LBD. The C-termini of AR variants have variable lengths (V, in yellow) and sequences based on the splicing of the cryptic exons in the AR gene.
The physiological ligands for AR include testosterone and dihydrotestosterone (DHT), which bind to the steroid binding site in the LBD. Like the other steroid receptors, there are two distinct transcriptional activation regions in AR: a strong activation function domain (AF-1) in the NTD and a weak activation function domain (AF-2) in the LBD, both of which can recruit various co-regulators of AR (Figure 1). The relative roles of these two transcriptional activation domains have been studied extensively for AR as well as other steroid receptors. In the case of AR, it is AF-1 that appears to be necessary and sufficient for transcriptional activity [3–6]. This knowledge has generated considerable interest in dissecting the mechanisms of AF-1 function, and has led to the finding that AF-1 can be further sub-divided into two discrete transcriptional activation units, termed TAU-1 and TAU-5 [7–9]. TAU-1 (amino acids 101–360) contains two motifs: (i) an LKDIL motif, which is similar to the nuclear receptor box sequence found in nuclear receptor co-regulator proteins; and (ii) an LX7LL motif, which is evolutionarily conserved in AR, ERα and PR (Figure 1). Deletion of the LKDIL motif causes significant loss in transcriptional activity of AR, whereas the LX7LL motif is required for de-repression of a cohort of genes in response to inflammatory cytokine signaling [10, 11]. TAU-5 (amino acids 361–490) contains the WHTLF motif, which appears to play a selective transactivation role under conditions of no/low androgens [12, 13]. Additionally, as elaborated below, this WHTLF motif mediates an intramolecular interaction between the AR amino and carboxyl termini by binding the AR AF-2 domain, indicating that accessibility of this transactivation motif is regulated, whether or not it is bound to AF-2 [14, 15].
The AR DBD is cysteine-rich and highly conserved among steroid receptors. There are 2 clusters of four cysteine residues, each of which coordinate a single zinc ion to make up the two zinc fingers of the DBD. As shown in Figure 1, the first zinc finger contains the P or proximal box (amino acids 577–581), which specifically recognizes DNA androgen response elements (ARE). The second zinc finger contains the D or distal box (amino acids 596–600), which mediates dimerization between two AR monomers [16–18].
Like LBDs of other nuclear receptors, the structure of the AR LBD is arranged in a three-layer, antiparallel α-helical sandwich fold that surrounds an interior hydrophobic ligand binding pocket (Figure 1). The AF-2 domain in the LBD is a shallow, hydrophobic groove formed by helices H12, H3 and H4 in the agonist-bound conformation. A domain proximal to AF-2, which is composed of a hydrophobic cleft made at the junction of H1 with the H3-H4 loop and H9 on the surface of the AR LBD, is referred to as binding function-3 domain (BF-3). BF-3 can allosterically regulate the binding of coactivators at AF-2 [19, 20]. The shallow AF-2 groove functions to bind LXXLL and LX7LL motifs found in nuclear receptor co-activator proteins, which are referred to as NR boxes [21]. As illustrated by AR LBD crystal structure 2AMA [22], deposited in The Protein Data Bank [23], agonists like testosterone or DHT, upon binding to the LBD re-position H12 to act as a lid and lock the agonist in the ligand-binding pocket. In contrast, when an antagonist binds the AR ligand binding pocket, it pushes H12 outwards to subsequently cause conformational changes in AF-2, thus rendering it incapable of binding co-activators [2, 19, 22]. In addition to binding NR boxes of co-activator proteins, the AF-2 domain also mediates interactions with the AR NTD, an intramolecular interaction referred to as the N/C interaction. The WHTLF motif of TAU-5 and the FXXLF motif both bind to the AF-2 domain of AR [14, 24].
Androgen Regulation of AR Nuclear Translocation and DNA Binding
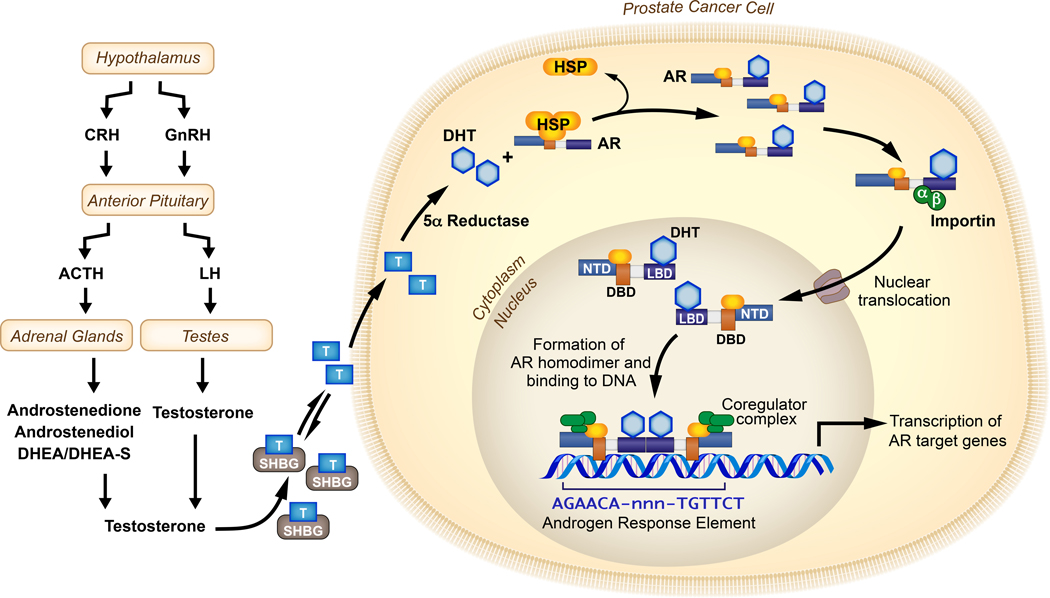
AR shuttles between the cytoplasm and the nucleus in a manner that is regulated by binding to androgen ligand. In the un-liganded state, chaperones and co-chaperones, like members of the heat shock protein family, including Hsp23, Hsp40, Hsp56, Hsp70 and Hsp90, associate with the AR LBD and sequester AR in the cytoplasm in a conformation that is competent for ligand binding [1, 25, 26]. The principal androgen circulating in the blood is testosterone, mostly produced by the Leydig cells in testes with a minor contribution from the adrenal cortex [27]. Synthesis of testosterone is regulated by the hypothalamus-pituitary-gonad and hypothalamus-pituitary-adrenal axes of the endocrine system. Several steroidogenic enzymes and isoenzymes are required to generate testicular and adrenal androgens from cholesterol in the canonical pathway. The hypothalamus secretes gonadotropin releasing hormone (GnRH) which acts upon the anterior pituitary to release the luteinizing hormone, subsequently signaling the release of testosterone from the testes [27, 28]. The anterior pituitary also releases the adrenocorticotropin hormone (ACTH) that acts on the adrenal cortex where the action of CYP17A1 and other enzymes produces dihydroepiandrosterone (DHEA), androstenedione and androstenediol. These weak adrenal androgens can then be converted to testosterone or DHT in peripheral tissues through various pathways such as the 5α−dione pathway or backdoor pathway [28, 29]. Although most of the testosterone in circulation is bound to sex hormone binding globulin (SHBG), ≤2% testosterone is free. When testosterone enters normal or cancerous prostate cells, it gets converted by 5α-reductase enzyme activity into DHT, which is a more potent androgen by virtue of it stabilizing the AR protein to a greater degree than testosterone and having a slower dissociation rate from the AR LBD. The binding of androgens to the AR LBD induces a conformational change in AR, thereby exposing the NLS and promoting translocation to the nucleus via direct interactions with the importin-α adapter protein and importin-β carrier protein, leading to transit through the nuclear pore complex [30–32]. In the nucleus, AR binds as a dimer via DBDs to androgen response elements. These AR dimers provide a platform for recruitment of a variety of co-regulators that govern the transcriptional program of AR. Androgen synthesis regulated by the hypothalamus-pituitary-gonad and hypothalamus-pituitary-adrenal axes and ultimate transmission of this hormonal signal via AR to the nucleus and genome of target cells is broadly referred to as the AR signaling axis (Figure 2). This AR signaling axis provides the foundation for the biological property of androgen-dependence of PCa cells.
Figure 2:
The AR signaling axis: The production of androgens (e.g. Testosterone) by the hypothalamus-pituitary-adrenal axis or hypothalamus-pituitary-gonadal axis is shown (left). In the bloodstream, testosterone is bound by sex-hormone binding globulin (SHBG), which releases free testosterone to enter cells where it is metabolized to DHT by 5-α reductase. AR bound to heat shock proteins (HSPs) in the cytoplasm binds DHT and translocates to the nucleus. In the nucleus, DHT-bound AR binds androgen response elements as dimers. The recruitment of various coactivators and corepressors determines the transcription profile of AR target genes.
AR Interactions with Chromatin
To understand the functional consequences of AR binding to AREs, researchers have focused their efforts on deciphering the AR transcriptome (the sets of mRNAs regulated by transcriptional activity of AR) and AR cistrome (the cis-regulatory elements in the genome to which AR binds). Genome-wide studies that have evaluated AR binding to AREs in various PCa models using ChIP-seq has provided fundamental information, although the exact number of AR binding events in PCa cells has not been clearly established. For instance, comparison of the number of AR-binding events in LNCaP cells (11,053) versus VCaP cells (51,811) demonstrated vastly different numbers. However, this is likely due to much higher expression of AR in VCaP cells due to AR gene amplification in this cell line. Nevertheless, despite this difference in number of AR binding events, the AR binding events observed in LNCaP cells displayed 90% overlap with the binding events observed in VCaP cells. In androgen-activated LNCaP cells, ChIP-seq studies have further revealed that recruitment of RNA Polymerase II to AR binding sites correlated with transcription of AR-upregulated genes. These AR-upregulated targets include genes involved in glucose uptake and glycolysis, biosynthetic pathways, regulators of cell cycle, and cellular metabolism [33].
Comparing and contrasting cistrome data from genome-wide ChIP-seq studies with structure/function studies of AR DNA binding has advanced the concept that there is flexibility with which AR binds ARE sites. For instance, global ChIP-seq studies have confirmed that the canonical ARE motif is a 15-mer sequence comprising of inverted repeats of a 6 base pair half-site ( 5’-AGAACA-3’) separated by 3 bases [34]. Structural studies have demonstrated that AR monomers engage with these ARE sequences as a homodimer arranged in a head-to-head symmetrical conformation. This leads to one AR monomer bound with high affinity to one ARE half-site, but the other AR monomer bound with lower affinity to the adjacent half-site. By reducing the stringency requirements for this adjacent half-site, AR can selectively bind its AREs [35]. This suboptimal binding of AR to its target DNA suggests an efficient way for AR to distinguish its various target genes and a mechanism to modulate transcription of ARE-driven AR target genes based on the strength of this binding interaction [17, 18]. Therefore, the way AR influences its target genes is non-uniform and heterogeneous, yet specific and strong.
Differential expression of AR target genes and variable occupancy of AR binding sites have been observed under different cellular contexts. For example, more than 50% of AR binding sites observed in CRPC tissue were not present in PCa cell lines, highlighting the divergence in AR signaling pathways under these conditions [36]. Further, comparative analysis of ChIP-seq data from 13 PCa tissue specimens versus 7 histologically normal prostate tissue specimens (6 of which were pair-matched from the same patient) revealed that prostate epithelial cells undergo re-programming of the AR cistrome to achieve a neoplastic phenotype [37].
These genome wide studies reinforce the idea that under different cellular contexts and through different stages of PCa progression, AR displays alterations in the repertoire of transcriptional targets to which it binds and regulates. There are multiple mechanistic explanations for these alterations, including changes in AR gene expression levels, AR protein structure, changes in expression or activity of AR co-regulators, and global changes in the epigenome that affect the chromatin environment around AR binding sites [38]. Thus, global profiling of androgen-AR-ARE-co-regulator complexes in clinical specimens provides an important framework for understanding the role of the AR cistrome and transcriptome in disease progression and identifying new therapeutic avenues that could be exploited.
AR Interactions with Co-Regulators and other Transcription Factors
The co-regulators recruited as a result of AR-ARE interactions serve different roles in normal prostate function and PCa by fine-tuning AR transcriptional output. There is strong evidence that certain co-regulators display expression changes during PCa development and progression, and that these changes in expression re-direct or re-program AR chromatin binding and/or transcriptional output [39]. Therefore, there has been great interest in identifying the roles and regulatory mechanisms of AR co-regulators to better understand similarities and differences in regulation of AR action between normal and cancerous prostate tissue. This is an ambitious undertaking, since more than 200 co-activators (enhance transcription) and co-repressors (inhibit transcription) affect AR transcriptional activity and/or chromatin binding, and at least 50 have expression patterns that correlate with important clinical parameters in PCa specimens [40]. Mechanistically, co-regulators can affect stability and complex formation of AR, influence AR nuclear or cytoplasmic localization, DNA occupancy, chromatin remodeling, chromatin looping, interactions with other transcription factors and complexes, as well as priming and assembly of the overall transcription complex [41].
Some of the best-defined classes of AR co-regulators play important roles in regulating transcriptional output of many transcription factors. These co-regulators include molecular chaperones like FKBP1 (FKBP1A), FKBP2 (FKBP2), FKBP5 (FKBP5) and HSP90 (HSP90AA1) , the p160 family of steroid receptor coactivators like SRC-1 (NCOA1), SRC-2/TIF-2/GRIP1 (NCOA2), SRC-3/AIB1 (NCOA3), p300 (EP300), CBP (CREBBP), ARA70 (NCOA4), ARA54 (RNF14) and ARA55 (TGFB1I1), as well as pioneer transcription factors like Oct1 (POU2F1) and GATA-2 (GATA-2) [42, 43]. AR-associated co-regulators are crucial to AR dependence in PCa. Several such co-regulators affect AR binding to DNA and/or AR-gene regulation in genome-wide associated studies (GWAS) or AR-cistrome analysis. BAF57 (SMARCE1), an accessory subunit of the SWI/SNF chromatin-remodeling complex is one such cofactor, which is dramatically upregulated in metastatic PCa. Increased expression of BAF57 directed AR and the SWI/SNF complex to a distant intragenic region of the ITGA2 gene, which encodes integrin alpha 2. In vitro studies confirmed that elevated levels of integrin alpha 2 protein results in an increased migratory and invasive phenotype in cells, supporting a prometastatic role for BAF57 [44].
FOXA1 and HOXB13 are key factors associated with growth and development of PCa through their binding interactions with AR [45, 46]. Physical interactions between AR-FOXA1 [47] and AR-HOXB13 [48] have been known for some time, but more recent global analyses have revealed that these interactions occur as a result of overlap with, and significant crosstalk between, the respective cistromes of AR, FOXA1 and HOXB13 to alter the transcriptional landscape of PCa cells. Furthermore, in a comparative analysis of FOXA1 and HOXB13 dependency across 102 cell lines from various tissue types, the PCa cell line LNCaP scored very high (second for HOXB13 and fifth for FOXA1), underscoring the relative importance of these factors in PCa cells [37]. For example, ectopic expression of FOXA1 and HOXB13 in immortalized LHSAR cells was sufficient to reprogram the AR cistrome to a state that was similar to that in a PCa cell line [37, 39, 49]. Additionally, FOXA1 is important for proliferation and cell cycle regulation in PCa, and knock down of FOXA1 expression in a PCa cell line led to an overall increase in other AR binding events. It is noteworthy that mutations in the coding sequence of FOXA1 occur in clinical PCa specimens, which are predicted to disrupt the forkhead DNA binding domain and thereby alter the affinity or specificity of FOXA1 for FOXA1 binding sites across the genome [50–52]. The role of these FOXA1 mutations in regulating the AR cistrome is an ongoing area of investigation.
In a recent study that used an unbiased proteomics technique termed RIME (rapid immunoprecipitation and mass spectrometry of endogenous proteins), Grainyhead-like 2 (GRHL2) was identified as a co-activator of AR with dichotomous roles in PCa development and progression. GRHL2 is pro-tumorigenic in early stages of PCa growth, but suppresses stromal invasion, intravasation of tumor cells, and survival of circulating tumor cells to reduce epithelial-mesenchymal transition and hence progression to metastatic PCa [53]. Another study used RIME and ChIP-seq to identify 66 known and novel interacting proteins of AR in LNCaP cells stimulated by a synthetic androgen R1881. These interaction partners were found to be members of the DNA repair machinery, chromatin remodeling factors, cell cycle regulators, cytoskeletal remodelers, and other transcriptional factors. These proteomics findings were subsequently followed by ChIP-seq studies to reveal that certain AR binding sites are co-occupied by AR and these interacting partners, including ARID1A, BRG1, FOXA1, HOXB13, TLE3, TRIM28 and WDHD1[54].
Within PCa cells, co-regulators can modulate distinct sets of genes to affect AR regulated pathways. This is illustrated by a study wherein 18 clinically important AR co-regulators were selectively inhibited in a PCa cell line using siRNA knock-down. Inhibition of specific co-regulators was found to selectively activate or repress discrete sets of genes within a 452-AR-target gene panel. This demonstrated specific, context-dependent effects of individual AR co-regulators, providing a mechanistic basis for intracellular heterogeneity in AR gene regulation [55]. A precise definition of the mechanisms by which co-regulators affect AR target gene expression based on the availability of androgens, presence of different drugs, cell line under investigation, and other factors influencing PCa growth and progression, could ultimately enable a better assessment of this disease through various stages of PCa progression and enable the development of more effective therapeutics.
Therapeutic Targeting of the AR Signaling Axis
The concept of AR-dependence was first introduced by Charles Huggins and Clarence V. Hodges almost 75 years ago [56]. Since then, androgen depletion therapy (ADT) has remained the principal treatment strategy for locally advanced, metastatic, or relapsed PCa. ADT targets various points of the AR signaling axis, with the goal of inhibiting transcriptional activity of the AR, which is the most widely accepted driver of PCa development and progression [57]. The earliest implementation of ADT included orchiectomy to eliminate the testicular source of androgens, or treatment with the oral synthetic estrogen diethylstilbestrol. These castration-based ADT modalities, and benefit for advanced PCa patients, formed the basis for the 1966 Nobel Prize in Medicine being awarded to Charles Huggins. Gonadotropin-releasing hormone (GnRH) agonists and antagonists like leuprolide, goserelin, triptorelin, and histrelin have replaced diethylstilbestrol as the main castration-based therapies, due to increased risk of cardiovascular mortality with estrogen therapy. Additionally, AR antagonists including bicalutamide, flutamide, and nilutamide function as competitive antagonists by binding the testosterone binding site in the AR LBD [58]. These drugs, collectively referred to as “first-generation” ADT, lead to suppression of circulating testosterone levels and blockade of AR signaling. This is best exemplified by the ensuing reduction in serum levels of prostate specific antigen (PSA), an AR transcriptional gene target in PCa cells. The main limitation of ADT is that it is not curative, and the duration of the therapeutic response of patients varies from a few months to several years. This stage of the disease, where patients have stopped responding to ADT, is referred to as CRPC. This stage of the disease is lethal and often progresses quickly due to a lack of durable treatment options [58]. Progression to CRPC is usually indicated by rising serum PSA levels despite ADT, suggesting re-engagement of the AR signaling axis. This has driven efforts to understand the mechanisms by which AR signaling can resume under conditions of ADT, and develop new therapies that can counteract these mechanisms in patients with CRPC [58, 59].
AR Gene Amplification in CRPC
An early comparative genomic hybridization study with matched PCa tissues from patients collected pre-ADT and post-ADT demonstrated that 30% of patients displayed AR gene amplification, specifically in post-ADT tissues [60]. A follow-up study using fluorescence in situ hybridization confirmed these initial findings, and also demonstrated that AR mRNA expression was higher in tumors displaying AR gene amplification [61]. Comparing the global gene expression profiles of seven isogenic pairs of hormone sensitive and castration-resistant PCa xenografts revealed that the CRPC phenotype is consistently associated with increased expression of AR [62]. Mechanistically, this study further showed that higher expression of AR is sufficient for transition from hormone-sensitive PCa to a CRPC phenotype. For example, hormone sensitive LNCaP cells engineered to express a 2–3 fold higher level of AR display increased growth under castrate conditions, as well as bicalutamide-stimulated growth Consistent with these functional data, more contemporary DNA sequencing studies of localized PCa and CRPC-stage tumors demonstrated that AR gene amplification is the most frequent event in CRPC genomes, occurring in approximately 55–60% of CRPC cases but almost never in localized PCa [63]. Whole genome sequencing of multiple metastases from CRPC patients revealed that persistent selective pressure of ADT drives separate cancer cell clones within the same patient to undergo distinct AR amplification events in distinct metastatic lesions. This study reinforces the importance of AR amplification as a key mechanism of resistance to ADT in CRPC [64].
AR Somatic Mutations in CRPC
Primary PCa typically shows less mutational burden than other solid tumors, but upon progression of the disease, about 20% of patients progressing with CRPC show somatic mutations in the AR gene [65, 66]. Similar to AR gene amplification, AR point mutations are exceedingly rare in ADT-naïve PCa. The best-described AR mutations are T878A, H875Y, W742C and L702H in the AR LBD, which play a key role in promoting resistance to ADT. For example, T878A confers resistance to ADT by enabling AR activation in response to alternative ligands, including progesterone and the antiandrogen flutamide. Similarly, H875Y and W742C mutations enable AR activation in response to the antiandrogens bicalutamide and flutamide [52, 67, 68]. The L702H mutation, alone or in combination with T878A, also broadens the agonist repertoire of AR, enabling AR activation by glucocorticoids [69]. The frequency of these somatic AR point mutations appears to be enriched in CRPC patients treated with antiandrogens, indicating this is a major mechanism of resistance in patients under continuous selective pressure from AR antagonists.
Amplification of an Upstream AR Enhancer in CRPC
Three recent studies integrated whole genome sequencing datasets or copy number microarrays with epigenetic datasets to reveal an important enhancer region regulating expression of the AR in CRPC. One study analyzed genome-wide copy number alterations from 149 tumors and identified an amplification hotspot encompassing the AR gene body, and another amplification hotspot located 650 kb centromeric to the AR gene body [70]. This upstream genomic region coincides with a region of DNaseI hypersensitivity in LNCaP cells that is essential for LNCaP cell viability. Further analysis of H3K27ac ChIP-seq data in this study revealed that this upstream genomic region resembles a developmental enhancer that is selectively acetylated in CRPC, indicating potential reactivation [70]. In a related study using linked read WGS, 70%−87% of metastatic CRPC patient samples showed tandem duplication events leading to amplification of this upstream AR enhancer region compared to only 2% of ADT naïve PCa cases [71]. Another study employed integrative deep whole genome sequencing coupled with RNA-seq to find that 81% of 101 CRPC specimens displayed increased AR gene expression correlated with amplification of this enhancer region [72]. Collectively, these studies have demonstrated that amplification of an enhancer located ∼650 kb upstream of the AR gene plays an important role in increasing the expression of AR mRNA in CRPC-stage tumors.
AR Variants in CRPC
Alternative splicing of AR mRNA to create AR variant (AR-V) proteins that lack the LBD represents a resistance mechanism where AR can function independent of androgen ligands to bypass ADT [73]. To date, several AR-Vs have been discovered and reported in PCa cell lines, xenograft tumors, primary tumors, metastatic lesions, and circulating tumor cells [74, 75]. However, the most widely-studied AR-V is termed AR-V7, composed of contiguously-spliced AR exons 1, 2, 3 and cryptic exon 3 (CE3). Development of antibodies specific to AR-V7 led to the finding that AR-V7 protein is rarely expressed (<1%) in primary PCa but detectable in >75% of CRPC cases. Expression of AR-V7 was homogenous within a tumor sample but was heterogeneous between different metastatic lesions from the same patient [76]. These studies aimed at evaluating the expression profiles of AR-V7 have suggested the potential to develop AR-Vs as biomarkers for resistance [77–79]. For example, detection of AR-V7 mRNA or protein in circulating tumor cells from patients with CRPC has been evaluated as a treatment selection biomarker that predicts poor treatment outcomes with second-generation AR targeted therapies abiraterone and enzalutamide, but better treatment outcomes with taxane chemotherapy [80–82]. Another AR-V expressed in clinical tissues that has been correlated with resistance to abiraterone acetate is AR-V9, composed of contiguously spliced AR exons 1/2/3/CE5 [79, 83]. Importantly, many AR-Vs are co-expressed in clinical CRPC [84, 85], raising the question of whether AR-Vs function alone, or cooperatively with other AR-Vs to promote resistance. More than 20 such variants have been reported in PCa models and clinical tissues in the last several years [86]. It also remains unresolved whether the functional effects of AR-V7 in CRPC cells requires the activity of full-length AR. For instance, knock-down of full-length AR in LNCaP cells engineered to overexpress AR-V7 inhibited androgen-independent growth [87]. Similarly, antisense oligonucleotides that blocked expression of full-length AR inhibited the growth of an AR-V7 positive LNCaP model of acquired resistance to enzalutamide [88]. Conversely, antisense oligonucleotides that blocked the expression of AR-V7 had no effect on growth of this enzalutamide-resistant LNCaP model. In light of these findings, it is important to note that AR-V7 is co-expressed with full-length AR, and the main mechanism underlying AR-V7 expression appears to be amplification of the AR gene [89]. These findings underscore the context-dependent roles of AR-Vs in PCa and point to a need to understand the interplay between full length AR and AR-Vs in disease staging, developing predictive biomarkers, and devising strategies for new therapies.
AR-V transcriptome and cistrome studies have provided important insights into the system-wide influence of these numerous AR isoforms in PCa. Gene expression profiling has shown that AR-Vs can activate many of the same transcriptional targets as full-length AR, while also displaying unique and distinct transcriptional targets. However, these differences may reflect different thresholds of activation between AR-Vs and full-length AR, and not absolute differences in transcriptional targets [87]. For example, AR-Vs were reported to uniquely activate genes involved in G2/M phase cell cycle progression like UBE2C and CCNA2 [90]. However, a subsequent study demonstrated that UBE2C and CCNA2 were also full-length AR targets that were induced depending on whether cells were maintained under conditions of low or high androgens [87]. In addition to differences in cell cycle regulation, differences in metabolic programs have been noted in cells expressing full-length AR vs. AR-V7 [91], with AR-V7-expressing cells displaying increased dependence on glutaminolysis and reductive carboxylation. One mechanism explaining differential regulation of transcriptional targets is differences in chromatin binding affinity, with AR-Vs having lower affinity for canonical AREs than full-length AR [92, 93].
AR Cross-Talk with Other Signaling Pathways
The AR signaling axis displays extensive crosstalk with other oncogenic pathways that are highly relevant in PCa. One such relevant pathway is the PI3K/AKT/PTEN pathway. About 20% of primary PCa samples display loss-of-function genomic alterations in PTEN, which increases to over 40% in CRPC. These PTEN alterations are in addition to somatic mutations or gene amplification of PIK3CA and PIK3CB in PCa [63, 68]. AR-mediated non-genomic activation of PI3K in the cytosol promotes cell survival and inhibits apoptosis in androgen-sensitive cells [94]. Mouse xenografts of LNCaP cells overexpressing AKT show accelerated tumor growth relative to control xenografts [95]. Mechanistically, AKT mediates direct phosphorylation of AR at Ser-213 and Ser-791, although the clinical relevance of these post-translational modifications has not yet been deciphered [96]. Collectively, these studies indicate that the PI3K signaling pathway positively regulates AR activity in PCa. However, PI3K signaling can negatively regulate AR and AR can negatively regulate PI3K. For example, FOXO3a binds to the AR promoter to upregulate AR expression, while FOXO1 recruits histone deacetylase 3 to decrease AR activity [97–99]. Further, PTEN loss results in suppression of androgen responsive transcription, while active expression of AR results in increased expression of FKBP5 and dephosphorylation of AKT, thereby suppressing AKT activity [100]. Using a PTEN- deficient murine PCa model, it was shown that this negative crosstalk between PI3K and AR is reciprocal, such that inhibition of one pathway leads to the activation of another to maintain tumor cell survival [101]. All these studies suggest that a combined therapeutic regimen targeting both AR and PI3K signaling would be more effective than targeting either pathway alone.
The role of AR in directing PCa cells towards distinct microenvironments like bone in advanced PCa provide an insight into the role of AR in tumor metastasis. Regulation of chemokine signaling via the Kruppel-Like Factor 5 (KLF5) transcription factor, chemokine receptor 4 (CXCR4), and the CXCR4 ligand CXCL12 is one such proposed mechanism in PCa cells [102]. The normal prostate gland expresses CXCR4, which becomes upregulated in response to androgens. Expression of CXCR4 is further elevated in bone metastatic lesions of PCa [103]. The ligand CXCL12 is a soluble chemoattractant highly enriched in bones. Upregulation of CXCR4 at the surface of LNCaP cells promotes cellular migration towards a CXCL12 gradient. Mechanistically, CXCR4 is indirectly regulated by AR via KLF5, which is an androgen-induced transcription factor necessary and sufficient for upregulation of CXCR4 and subsequent cellular functions in LNCaP cells [102]. The concept of increased androgen signaling, leading to increased CXCR4 expression to cause cellular migration to distant bony sites provides a foundation for future work to explore the roles and therapeutic vulnerabilities of chemokine signaling in aggressive metastatic PCa [46].
Recent studies have reported bidirectional cross-talk between AR and the nuclear receptor super family member peroxisome proliferator activated receptor gamma (PPAR-γ). PPAR-γ can either activate or repress the activity of AR, and AR can also repress the activity of PPAR-γ [104, 105]. These interactions between AR and PPAR-γ are mediated through PPAR coactivator 1 alpha (PGC1α) or through fatty acid binding proteins 4 and 5 (FABP4, FABP5), but also other, yet to be defined mechanisms [106–108]. Thus, AR-dependent control of metabolic pathways appears to be central to PCa development and progression. PPAR-γ expression varies among PCa cell lines, with lower PPAR-γ expression in castration-sensitive cell lines like LNCaP and, higher PPAR-γ expression in castration resistant cell lines like C4–2 [109]. Although previous studies using PPAR-γ agonists suggested its role as a tumor suppressor in PCa [110], later transposon-based ‘sleeping beauty’ screen found that increased expression of PPAR-γ coupled with loss of PTEN promotes prostate tumorigenesis [111]. Further studies showed that PPAR-γ agonists increase AR signaling through an androgen-dependent and PPAR-γ-dependent mechanism [112, 113]. In larger studies using tissue microarray, RT-PCR and immunohistochemistry, PPAR-γ expression was found to be positively correlated with advanced PCa suggesting a more oncogenic role for PPAR-γ and its ligands [114, 115]. Gene set enrichment analysis of AR target genes regulating metabolism and biosynthetic pathways, showed enrichment for carbohydrate metabolism and PGC1α gene sets, further underscoring the relevance of this pathway in regulating AR and metabolic pathways in PCa cells [33]. As more ligands of AR and PPAR-γ enter clinical development, the intricacy of the bidirectional crosstalk between AR and PPAR-γ needs to be fully characterized in castration-sensitive PCa and CRPC.
Therapeutic Advances in AR Targeting for CRPC-Stage Disease
Studies of a cohort of CRPC tissues collected from PCa patients indicated that intra-tumoral levels of androgens were persistently high, despite castrate levels of androgens in the blood. This suggested that intracrine steroidogenesis in tumors could bypass the low levels of circulating androgens [116–118]. Understanding the mechanisms of AR re-activation in response to ADT in CRPC led to the development of second-generation AR-targeted therapies abiraterone acetate and enzalutamide [119–121]. Abiraterone acetate targets CYP17A1, an enzyme involved in conversion of cholesterol to the androgen precursor pregnenolone by blocking its 17, 20 lyase and 17α-hydroxylase activities, thus inhibiting synthesis of DHT and hence reducing de novo production of androgens in the tumor tissue. Additionally, abiraterone acetate inhibits these CYP17A1 activities in the adrenal cortex, thus preventing the synthesis of adrenal androgens. More recently, AR antagonist activity was reported for a metabolite of abiraterone, Δ4-abiraterone, which provides further basis for its anti-tumor activity [122]. Enzalutamide (MDV-3100) acts as a competitive antagonist of the AR LBD, which reduces AR nuclear translocation and chromatin binding, and thereby blocks expression of AR target genes. As with first-generation ADT, development of resistance represents a major limitation of therapy with both abiraterone and enzalutamide. As discussed earlier, expression of AR-V7 and perhaps other AR-Vs is associated with resistance to both of these agents. Additionally, mechanisms like increased expression of steroidogenic enzyme AKR1C3 and activation of the 5α-dione pathway have been implicated in developing resistance to abiraterone [119, 123, 124]. Somatic mutations such as F876L in the AR LBD are associated with resistance to enzalutamide in models of CRPC progression, although the prevalence of F876L AR in clinical specimens appears to be low [125, 126].
Emerging Therapeutic Strategies to Target AR in CRPC
The ongoing durability of AR signaling in CRPC, which includes patients that have been treated with potent inhibitors such as enzalutamide and abiraterone, indicates an ongoing need to develop novel AR-targeted therapies. Broadly speaking, the current arsenal of AR-targeted therapies for PCa patients all exerts their action by preventing androgen production, or by binding to the AR LBD. Given the importance of additional functional domains of the AR protein, one emerging strategy is to develop therapeutics that targets the AR NTD or the AR DBD. Additionally, there are currently no approved PCa therapies that degrade or block expression of AR protein, which may be important for counteracting the widespread overexpression of AR observed in CRPC tumors harboring AR amplification. Below, we highlight experimental therapies that are being developed to target alternative domains on the AR protein, or block AR expression in PCa cells.
One strategy for targeted degradation of AR is using proteolysis targeting chimeric (PROTAC) technology. A PROTAC that has been developed to target AR is a bifunctional drug-like small molecule with one chemical moiety that binds the Von-Hippel-Lindau (VHL) E3 ubiquitin ligase complex and the other chemical moiety representing DHT, which binds the AR LBD [127, 128]. In treated PCa cells, these PROTACs bind to AR and recruited VHL E3 ligase, which induces AR polyubiquitination and degradation, leading to reduced levels of AR protein in cells and G1 growth arrest. Although these compounds are cell permeable and specific to AR, prolonged treatment with these PROTACs leads to cytotoxicity [128]. Recently, a more potent enzalutamide-based PROTAC called ARCC-4 has been developed and compared to enzalutamide under different cellular conditions. ARCC-4 selectively degraded about 95% of cellular AR in LNCaP cells. ARCC-4 was also very effective in LNCaP cells overexpressing AR point mutations F876L and T877A, as measured by reduced PSA levels in these cells upon treatment with ARCC-4. Unlike enzalutamide, ARCC-4 was able to block proliferation of VCaP cells under high androgen conditions, further demonstrating the advantage of this PROTAC over its parent compound [129]. The development of these AR degraders for therapeutic benefit in CRPC offers a new treatment strategy that can be tailored to create additional PROTACs targeting other proteins like bromodomain and extraterminal (BET) family proteins. Recently, the BET degrader ARV-771 was shown to indirectly target expression and activity of the AR-V7 splice variant [130].
A pressing challenge in the CRPC field is the development of agents that selectively target expression or activity of AR-Vs. Recently, selective AR degraders (SARDs) were developed that lead to efficient reduction in the activity of full length AR and AR-Vs even at sub-micromolar doses. SARDs UT-69 and UT-155 reduce AR expression and downstream transcription in LNCaP cells more effectively than enzalutamide. These SARDs are competitive antagonists of the AR LBD, but also bind the AR NTD domain at the AF-1 region. Further modification of UT-155 led to the development of R-UT-155, which could directly bind the AF-1 domain, but did not bind the AR LBD. Consistent with an AF-1-directed mechanism of action, R-UT-155 inhibited expression of AR and AR-Vs in the AR-V7-positive CRPC cell line 22Rv1. Moreover, R-UT-155 inhibits the growth of 22Rv1 xenografts in mice. These SARD compounds may provide a new avenue to inhibit AR by binding to and reducing expression of AR and AR-V proteins in CRPC cells [131].
In addition to the development of novel molecular entities for blocking AR expression, recent efforts have involved screening FDA-approved drugs for efficacy in CRPC cells. This led to the identification of niclosamide, an anti-helminthic drug, as a possible therapeutic that could be re-purposed for inhibition of AR in PCa [132]. Functional studies with niclosamide showed this drug could re-sensitize CRPC cells to treatment with both abiraterone and enzalutamide [133, 134]. Further, niclosamide was able to overcome the ability of AR-V7 to promote resistance to bicalutamide [135]. Based on these encouraging pre-clinical findings, niclosamide is being tested in combination with enzalutamide in a phase I clinical trial (NCT02532114).
Mutational hot spots that reside near the AR LBD, such as the binding function-3 (BF-3) pocket located near the AF-2 domain, have also been explored as targets in PCa cells resistant to enzalutamide. The BF-3 domain has functional significance in nuclear translocation of AR through interactions with cytoplasmic (like SGTA) and nuclear (e.g. FKBP52, BAG1L) co-chaperones [136–139]. VPC-13566 was developed as a potent and selective small molecule inhibitor of AR that binds specifically to the AR BF-3 domain [139]. In cells treated with VPC-13566, reduced AR transcriptional activity was observed. Mechanistically, this appears to be due in part to impaired translocation of AR to the nucleus. Because this compound inhibits AR BF-3 binding to cytoplasmic SGTA and nuclear BAG1L factors, it could be perceived to affect two separate pathways and therefore have less likelihood of promoting resistance. In xenograft studies, mice treated with VPC-13566 showed reduced tumor growth [139]. However, due to pharmacokinetic limitations, VPC13566 needs to be optimized for better in vivo stability and bioavailability before it can advance in clinical development [139].
Given that the AR NTD is responsible for the majority of AR transcriptional activity, the AR NTD represents an attractive therapeutic target to block activity of full length AR as well as AR-Vs. However, the AR NTD represents a challenging therapeutic target, because it is an intrinsically disordered domain of the AR [13]. Two classes of molecules, the EPI-series of bisphenol-like compounds, as well as Sintokamides, bind the AR NTD directly [140–142]. The compounds EPI-001 and EPI-002 engage and covalently bind to the AR NTD in treated cells, and thereby block the ability of the AR NTD to recruit co-activators such as CBP [140]. In NMR studies, EPI-001 was shown to bind to the AR TAU5 domain in the AR NTD, which is presumed to precede formation of a covalent bond between TAU5 via a chlorhydrin moiety on EPI-001. However, the specificity of EPI-series compounds for binding the AR NTD is debatable, given that the highly-reactive chlorhydrin moiety of EPI-series compounds is required for the anti-AR action in cell models [140]. Indeed, EPI-001 was shown to have general non-specific alkylating activity in a pH-dependent manner, and also have PPAR-γ agonist activity, two properties which could also account for the anti-AR action of these compounds [143]. A pro-drug formulation of EPI-002, termed EPI-506, recently advanced to a Phase I/II clinical trial for metastatic PCa (NCT02606123) [99], but this trial was recently discontinued.
An additional domain of AR that could provide a therapeutic targeting opportunity is the DBD. Small molecule inhibitors have been designed to target a small pocket exposed at the surface of the AR DBD and block the ability of the AR DBD to bind DNA. One such molecule termed VPC-14449 inhibits activity of full length AR and induced regression of LNCaP xenografts in mouse studies [144]. Mechanistically, VPC-14449 affects the chromatin binding interactions of wild-type and mutant forms of full length AR as well as AR-Vs. As a result, transcriptional programs mediated by full length AR, AR-Vs, or AR mutants such as F876L are all repressed. Interestingly, additive effects of VP-14449 and enzalutamide co-administered simultaneously suggest an attractive pre-clinical rationale for the development of combination therapies [145]. These studies led to the development of another lead compound termed VPC-17005, which binds selectively to the D-box of the AR DBD, thereby blocking AR dimerization. Consequently, this compound inhibits transcription of AR target genes [146].
Conclusions
AR is a master regulator in PCa that is crucial to disease development, progression and treatment. The presence of full-length AR along with generation of multiple AR-Vs creates intra-tumoral and intra-cellular heterogeneity of AR expression and activity in CRPC. There are myriad complexities to these heterogeneous transcriptomes and cistromes that are important for the field to decipher and understand. The biphasic nature of androgen signaling, escape from ADT, and rapid progression of aggressive CRPC present many variables that impact the androgen dependence and therapeutic responsiveness of PCa. The failure of several single-agent drug targets and pathway inhibitors in clinical trials that showed promising results in pre-clinical studies could be attributable to this vast heterogeneity. Efforts aimed at carefully selecting patients based on the presence of AR gene mutations, AR amplification, expression of AR-Vs, and status of related pathways including PTEN, could all impact the success of novel AR-targeted therapies in clinical trials. The myriad challenges also bring new and interesting solutions to target AR, AR-Vs, and AR target genes with potent and selective inhibitors that work alone or in combination with current anti-androgens.
References
- 1.Heinlein CA and Chang C, Androgen receptor in prostate cancer. Endocr Rev, 2004. 25(2): p. 276–308. [DOI] [PubMed] [Google Scholar]
- 2.Tan MH, et al. , Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin, 2015. 36(1): p. 3–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callewaert L, Van Tilborgh N, and Claessens F, Interplay between two hormone-independent activation domains in the androgen receptor. Cancer Res, 2006. 66(1): p. 543–53. [DOI] [PubMed] [Google Scholar]
- 4.Bevan CL, et al. , The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol Cell Biol, 1999. 19(12): p. 8383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenster G, et al. , Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem, 1995. 270(13): p. 7341–6. [DOI] [PubMed] [Google Scholar]
- 6.Christiaens V, et al. , Characterization of the two coactivator-interacting surfaces of the androgen receptor and their relative role in transcriptional control. J Biol Chem, 2002. 277(51): p. 49230–7. [DOI] [PubMed] [Google Scholar]
- 7.Dehm SM and Tindall DJ, Alternatively spliced androgen receptor variants. Endocr Relat Cancer, 2011. 18(5): p. R183–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warnmark A, et al. , Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol, 2003. 17(10): p. 1901–9. [DOI] [PubMed] [Google Scholar]
- 9.McEwan IJ, Molecular mechanisms of androgen receptor-mediated gene regulation: structure-function analysis of the AF-1 domain. Endocr Relat Cancer, 2004. 11(2): p. 281–93. [DOI] [PubMed] [Google Scholar]
- 10.Chamberlain NL, Whitacre DC, and Miesfeld RL, Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J Biol Chem, 1996. 271(43): p. 26772–8. [DOI] [PubMed] [Google Scholar]
- 11.Zhu P, et al. , Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell, 2006. 124(3): p. 615–29. [DOI] [PubMed] [Google Scholar]
- 12.Dehm SM, et al. , Selective role of an NH2-terminal WxxLF motif for aberrant androgen receptor activation in androgen depletion independent prostate cancer cells. Cancer Res, 2007. 67(20): p. 10067–77. [DOI] [PubMed] [Google Scholar]
- 13.De Mol E, et al. , Regulation of Androgen Receptor Activity by Transient Interactions of Its Transactivation Domain with General Transcription Regulators. Structure, 2018. 26(1): p. 145–152 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He B, Kemppainen JA, and Wilson EM, FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J Biol Chem, 2000. 275(30): p. 22986–94. [DOI] [PubMed] [Google Scholar]
- 15.He B, et al. , Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J Biol Chem, 2002. 277(28): p. 25631–9. [DOI] [PubMed] [Google Scholar]
- 16.Xu D, et al. , Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer Res, 2015. 75(17): p. 3663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer PL, et al. , Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci U S A, 2004. 101(14): p. 4758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadal M, et al. , Structure of the homodimeric androgen receptor ligand-binding domain. Nat Commun, 2017. 8: p. 14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estebanez-Perpina E, et al. , A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A, 2007. 104(41): p. 16074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosdidier S, et al. , Allosteric conversation in the androgen receptor ligand-binding domain surfaces. Mol Endocrinol, 2012. 26(7): p. 1078–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao W, Bohl CE, and Dalton JT, Chemistry and structural biology of androgen receptor. Chem Rev, 2005. 105(9): p. 3352–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira de Jesus-Tran K, et al. , Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity. Protein Sci, 2006. 15(5): p. 987–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman HM, et al. , The Protein Data Bank. Nucleic Acids Res, 2000. 28(1): p. 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson EM, Analysis of interdomain interactions of the androgen receptor. Methods Mol Biol, 2011. 776: p. 113–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saporita AJ, et al. , Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem, 2003. 278(43): p. 41998–2005. [DOI] [PubMed] [Google Scholar]
- 26.Haelens A, et al. , The hinge region regulates DNA binding, nuclear translocation, and transactivation of the androgen receptor. Cancer Res, 2007. 67(9): p. 4514–23. [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama T, Serum testosterone levels after medical or surgical androgen deprivation: a comprehensive review of the literature. Urol Oncol, 2014. 32(1): p. 38 e17–28. [DOI] [PubMed] [Google Scholar]
- 28.Dai C, Heemers H, and Sharifi N, Androgen Signaling in Prostate Cancer. Cold Spring Harb Perspect Med, 2017. 7(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharifi N and Auchus RJ, Steroid biosynthesis and prostate cancer. Steroids, 2012. 77(7): p. 719–26. [DOI] [PubMed] [Google Scholar]
- 30.Kaku N, et al. , Characterization of nuclear import of the domain-specific androgen receptor in association with the importin alpha/beta and Ran-guanosine 5’-triphosphate systems. Endocrinology, 2008. 149(8): p. 3960–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni L, et al. , Androgen induces a switch from cytoplasmic retention to nuclear import of the androgen receptor. Mol Cell Biol, 2013. 33(24): p. 4766–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cutress ML, et al. , Structural basis for the nuclear import of the human androgen receptor. J Cell Sci, 2008. 121(Pt 7): p. 957–68. [DOI] [PubMed] [Google Scholar]
- 33.Massie CE, et al. , The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J, 2011. 30(13): p. 2719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claessens F, Joniau S, and Helsen C, Comparing the rules of engagement of androgen and glucocorticoid receptors. Cell Mol Life Sci, 2017. 74(12): p. 2217–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahu B, et al. , Androgen receptor uses relaxed response element stringency for selective chromatin binding and transcriptional regulation in vivo. Nucleic Acids Res, 2014. 42(7): p. 4230–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma NL, et al. , The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell, 2013. 23(1): p. 35–47. [DOI] [PubMed] [Google Scholar]
- 37.Pomerantz MM, et al. , The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet, 2015. 47(11): p. 1346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills IG, Maintaining and reprogramming genomic androgen receptor activity in prostate cancer. Nat Rev Cancer, 2014. 14(3): p. 187–98. [DOI] [PubMed] [Google Scholar]
- 39.Copeland BT, et al. , The androgen receptor malignancy shift in prostate cancer. Prostate, 2018. 78(7): p. 521–531. [DOI] [PubMed] [Google Scholar]
- 40.Heemers HV and Tindall DJ, Unraveling the complexities of androgen receptor signaling in prostate cancer cells. Cancer Cell, 2009. 15(4): p. 245–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heemers HV and Tindall DJ, Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev, 2007. 28(7): p. 778–808. [DOI] [PubMed] [Google Scholar]
- 42.Foley C and Mitsiades N, Moving Beyond the Androgen Receptor (AR): Targeting AR-Interacting Proteins to Treat Prostate Cancer. Horm Cancer, 2016. 7(2): p. 84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dehm SM and Tindall DJ, Ligand-independent androgen receptor activity is activation function-2-independent and resistant to antiandrogens in androgen refractory prostate cancer cells. J Biol Chem, 2006. 281(38): p. 27882–93. [DOI] [PubMed] [Google Scholar]
- 44.Balasubramaniam S, et al. , Aberrant BAF57 signaling facilitates prometastatic phenotypes. Clin Cancer Res, 2013. 19(10): p. 2657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dadaev T, et al. , Fine-mapping of prostate cancer susceptibility loci in a large meta-analysis identifies candidate causal variants. Nat Commun, 2018. 9(1): p. 2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Augello MA, Den RB, and Knudsen KE, AR function in promoting metastatic prostate cancer. Cancer Metastasis Rev, 2014. 33(2–3): p. 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao N, et al. , The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol, 2003. 17(8): p. 1484–507. [DOI] [PubMed] [Google Scholar]
- 48.Jung C, et al. , HOXB13 induces growth suppression of prostate cancer cells as a repressor of hormone-activated androgen receptor signaling. Cancer Res, 2004. 64(24): p. 9185–92. [DOI] [PubMed] [Google Scholar]
- 49.Whitington T, et al. , Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat Genet, 2016. 48(4): p. 387–97. [DOI] [PubMed] [Google Scholar]
- 50.Robinson JL, Holmes KA, and Carroll JS, FOXA1 mutations in hormone-dependent cancers. Front Oncol, 2013. 3: p. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbieri CE, et al. , Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet, 2012. 44(6): p. 685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grasso CS, et al. , The mutational landscape of lethal castration-resistant prostate cancer. Nature, 2012. 487(7406): p. 239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paltoglou S, et al. , Novel Androgen Receptor Coregulator GRHL2 Exerts Both Oncogenic and Antimetastatic Functions in Prostate Cancer. Cancer Res, 2017. 77(13): p. 3417–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stelloo S, et al. , Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene, 2018. 37(3): p. 313–322. [DOI] [PubMed] [Google Scholar]
- 55.Liu S, et al. , A comprehensive analysis of coregulator recruitment, androgen receptor function and gene expression in prostate cancer. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huggins C and Hodges CV, Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin, 1972. 22(4): p. 232–40. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt LJ and Tindall DJ, Androgen receptor: past, present and future. Curr Drug Targets, 2013. 14(4): p. 401–7. [DOI] [PubMed] [Google Scholar]
- 58.Watson PA, Arora VK, and Sawyers CL, Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer, 2015. 15(12): p. 701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knudsen KE and Kelly WK, Outsmarting androgen receptor: creative approaches for targeting aberrant androgen signaling in advanced prostate cancer. Expert Rev Endocrinol Metab, 2011. 6(3): p. 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Visakorpi T, et al. , In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet, 1995. 9(4): p. 401–6. [DOI] [PubMed] [Google Scholar]
- 61.Koivisto P, et al. , Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res, 1997. 57(2): p. 314–9. [PubMed] [Google Scholar]
- 62.Chen CD, et al. , Molecular determinants of resistance to antiandrogen therapy. Nat Med, 2004. 10(1): p. 33–9. [DOI] [PubMed] [Google Scholar]
- 63.Network TCGAR, The Molecular Taxonomy of Primary Prostate Cancer. Cell, 2015. 163(4): p. 1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gundem G, et al. , The evolutionary history of lethal metastatic prostate cancer. Nature, 2015. 520(7547): p. 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azad AA, et al. , Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin Cancer Res, 2015. 21(10): p. 2315–24. [DOI] [PubMed] [Google Scholar]
- 66.Carreira S, et al. , Tumor clone dynamics in lethal prostate cancer. Sci Transl Med, 2014. 6(254): p. 254ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taplin ME, et al. , Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res, 1999. 59(11): p. 2511–5. [PubMed] [Google Scholar]
- 68.Taylor BS, et al. , Integrative genomic profiling of human prostate cancer. Cancer Cell, 2010. 18(1): p. 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao XY, et al. , Glucocorticoids can promote androgen-independent growth of prostate cancer cells through a mutated androgen receptor. Nat Med, 2000. 6(6): p. 703–6. [DOI] [PubMed] [Google Scholar]
- 70.Takeda DY, et al. , A Somatically Acquired Enhancer of the Androgen Receptor Is a Noncoding Driver in Advanced Prostate Cancer. Cell, 2018. 174(2): p. 422–432 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Viswanathan SR, et al. , Structural Alterations Driving Castration-Resistant Prostate Cancer Revealed by Linked-Read Genome Sequencing. Cell, 2018. 174(2): p. 433–447 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Quigley DA, et al. , Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell, 2018. 175(3): p. 889. [DOI] [PubMed] [Google Scholar]
- 73.Dehm SM, et al. , Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res, 2008. 68(13): p. 5469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wadosky KM and Koochekpour S, Androgen receptor splice variants and prostate cancer: From bench to bedside. Oncotarget, 2017. 8(11): p. 18550–18576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paschalis A, et al. , Alternative splicing in prostate cancer. Nat Rev Clin Oncol, 2018. 15(11): p. 663–675. [DOI] [PubMed] [Google Scholar]
- 76.Sharp A, et al. , Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest, 2019. 129(1): p. 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antonarakis ES, et al. , Androgen receptor variant-driven prostate cancer: clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis, 2016. 19(3): p. 231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scher HI, et al. , Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol, 2016. 2(11): p. 1441–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kohli M, et al. , Androgen Receptor Variant AR-V9 Is Coexpressed with AR-V7 in Prostate Cancer Metastases and Predicts Abiraterone Resistance. Clin Cancer Res, 2017. 23(16): p. 4704–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Antonarakis ES, et al. , AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med, 2014. 371(11): p. 1028–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Antonarakis ES, et al. , Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol, 2015. 1(5): p. 582–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Scher HI, et al. , Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol, 2018. 4(9): p. 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mostaghel EA, et al. , Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: therapeutic implications for castration-resistant prostate cancer. Cancer Res, 2007. 67(10): p. 5033–41. [DOI] [PubMed] [Google Scholar]
- 84.Robinson D, et al. , Integrative clinical genomics of advanced prostate cancer. Cell, 2015. 161(5): p. 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyamoto DT, et al. , RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science, 2015. 349(6254): p. 1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van der Steen T, Tindall DJ, and Huang H, Posttranslational modification of the androgen receptor in prostate cancer. Int J Mol Sci, 2013. 14(7): p. 14833–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Watson PA, et al. , Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A, 2010. 107(39): p. 16759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamamoto Y, et al. , Generation 2.5 antisense oligonucleotides targeting the androgen receptor and its splice variants suppress enzalutamide-resistant prostate cancer cell growth. Clin Cancer Res, 2015. 21(7): p. 1675–87. [DOI] [PubMed] [Google Scholar]
- 89.Henzler C, et al. , Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nat Commun, 2016. 7: p. 13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu R, et al. , Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res, 2012. 72(14): p. 3457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shafi AA, et al. , Differential regulation of metabolic pathways by androgen receptor (AR) and its constitutively active splice variant, AR-V7, in prostate cancer cells. Oncotarget, 2015. 6(31): p. 31997–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan SC, et al. , Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res, 2015. 43(12): p. 5880–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan SC, Li Y, and Dehm SM, Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem, 2012. 287(23): p. 19736–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baron S, et al. , Androgen receptor mediates non-genomic activation of phosphatidylinositol 3-OH kinase in androgen-sensitive epithelial cells. J Biol Chem, 2004. 279(15): p. 14579–86. [DOI] [PubMed] [Google Scholar]
- 95.Graff JR, et al. , Increased AKT activity contributes to prostate cancer progression by dramatically accelerating prostate tumor growth and diminishing p27Kip1 expression. J Biol Chem, 2000. 275(32): p. 24500–5. [DOI] [PubMed] [Google Scholar]
- 96.Xin L, et al. , Progression of prostate cancer by synergy of AKT with genotropic and nongenotropic actions of the androgen receptor. Proc Natl Acad Sci U S A, 2006. 103(20): p. 7789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang L, et al. , Induction of androgen receptor expression by phosphatidylinositol 3-kinase/Akt downstream substrate, FOXO3a, and their roles in apoptosis of LNCaP prostate cancer cells. J Biol Chem, 2005. 280(39): p. 33558–65. [DOI] [PubMed] [Google Scholar]
- 98.Liu P, et al. , A transcription-independent function of FOXO1 in inhibition of androgen-independent activation of the androgen receptor in prostate cancer cells. Cancer Res, 2008. 68(24): p. 10290–9. [DOI] [PubMed] [Google Scholar]
- 99.Leung JK and Sadar MD, Non-Genomic Actions of the Androgen Receptor in Prostate Cancer. Front Endocrinol (Lausanne), 2017. 8: p. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mulholland DJ, et al. , Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell, 2011. 19(6): p. 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carver BS, et al. , Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell, 2011. 19(5): p. 575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Frigo DE, et al. , Induction of Kruppel-like factor 5 expression by androgens results in increased CXCR4-dependent migration of prostate cancer cells in vitro. Mol Endocrinol, 2009. 23(9): p. 1385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun YX, et al. , Expression of CXCR4 and CXCL12 (SDF-1) in human prostate cancers (PCa) in vivo. J Cell Biochem, 2003. 89(3): p. 462–73. [DOI] [PubMed] [Google Scholar]
- 104.Olokpa E, Moss PE, and Stewart LV, Crosstalk between the Androgen Receptor and PPAR Gamma Signaling Pathways in the Prostate. PPAR Res, 2017. 2017: p. 9456020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elix C, Pal SK, and Jones JO, The role of peroxisome proliferator-activated receptor gamma in prostate cancer. Asian J Androl, 2018. 20(3): p. 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shiota M, et al. , Peroxisome proliferator-activated receptor gamma coactivator-1alpha interacts with the androgen receptor (AR) and promotes prostate cancer cell growth by activating the AR. Mol Endocrinol, 2010. 24(1): p. 114–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Olokpa E, Bolden A, and Stewart LV, The Androgen Receptor Regulates PPARgamma Expression and Activity in Human Prostate Cancer Cells. J Cell Physiol, 2016. 231(12): p. 2664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bao Z, et al. , A novel cutaneous Fatty Acid-binding protein-related signaling pathway leading to malignant progression in prostate cancer cells. Genes Cancer, 2013. 4(7–8): p. 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mueller E, et al. , Effects of ligand activation of peroxisome proliferator-activated receptor gamma in human prostate cancer. Proc Natl Acad Sci U S A, 2000. 97(20): p. 10990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hisatake JI, et al. , Down-Regulation of prostate-specific antigen expression by ligands for peroxisome proliferator-activated receptor gamma in human prostate cancer. Cancer Res, 2000. 60(19): p. 5494–8. [PubMed] [Google Scholar]
- 111.Ahmad I, et al. , Sleeping Beauty screen reveals Pparg activation in metastatic prostate cancer. Proc Natl Acad Sci U S A, 2016. 113(29): p. 8290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tew BY, et al. , Vitamin K epoxide reductase regulation of androgen receptor activity. Oncotarget, 2017. 8(8): p. 13818–13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moss PE, Lyles BE, and Stewart LV, The PPARgamma ligand ciglitazone regulates androgen receptor activation differently in androgen-dependent versus androgen-independent human prostate cancer cells. Exp Cell Res, 2010. 316(20): p. 3478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rogenhofer S, et al. , Enhanced expression of peroxisome proliferate-activated receptor gamma (PPAR-gamma) in advanced prostate cancer. Anticancer Res, 2012. 32(8): p. 3479–83. [PubMed] [Google Scholar]
- 115.Segawa Y, et al. , Expression of peroxisome proliferator-activated receptor (PPAR) in human prostate cancer. Prostate, 2002. 51(2): p. 108–16. [DOI] [PubMed] [Google Scholar]
- 116.Nishiyama T, Hashimoto Y, and Takahashi K, The influence of androgen deprivation therapy on dihydrotestosterone levels in the prostatic tissue of patients with prostate cancer. Clin Cancer Res, 2004. 10(21): p. 7121–6. [DOI] [PubMed] [Google Scholar]
- 117.Montgomery RB, et al. , Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res, 2008. 68(11): p. 4447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ho Y and Dehm SM, Androgen Receptor Rearrangement and Splicing Variants in Resistance to Endocrine Therapies in Prostate Cancer. Endocrinology, 2017. 158(6): p. 1533–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chandrasekar T, et al. , Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl Androl Urol, 2015. 4(3): p. 365–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scher HI, et al. , Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med, 2012. 367(13): p. 1187–97. [DOI] [PubMed] [Google Scholar]
- 121.de Bono JS, et al. , Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med, 2011. 364(21): p. 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li Z, et al. , Conversion of abiraterone to D4A drives anti-tumour activity in prostate cancer. Nature, 2015. 523(7560): p. 347–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chang KH, et al. , Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc Natl Acad Sci U S A, 2011. 108(33): p. 13728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu C, et al. , Inhibition of AKR1C3 Activation Overcomes Resistance to Abiraterone in Advanced Prostate Cancer. Mol Cancer Ther, 2017. 16(1): p. 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu H, et al. , Molecular Dynamics Studies on the Enzalutamide Resistance Mechanisms Induced by Androgen Receptor Mutations. J Cell Biochem, 2017. 118(9): p. 2792–2801. [DOI] [PubMed] [Google Scholar]
- 126.Culig Z, Molecular Mechanisms of Enzalutamide Resistance in Prostate Cancer. Curr Mol Biol Rep, 2017. 3(4): p. 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schneekloth JS Jr., et al. , Chemical genetic control of protein levels: selective in vivo targeted degradation. J Am Chem Soc, 2004. 126(12): p. 3748–54. [DOI] [PubMed] [Google Scholar]
- 128.Rodriguez-Gonzalez A, et al. , Targeting steroid hormone receptors for ubiquitination and degradation in breast and prostate cancer. Oncogene, 2008. 27(57): p. 7201–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Salami J, et al. , Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun Biol, 2018. 1: p. 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Raina K, et al. , PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A, 2016. 113(26): p. 7124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ponnusamy S, et al. , Novel Selective Agents for the Degradation of Androgen Receptor Variants to Treat Castration-Resistant Prostate Cancer. Cancer Res, 2017. 77(22): p. 6282–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu C, et al. , Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res, 2014. 20(12): p. 3198–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu C, et al. , Niclosamide enhances abiraterone treatment via inhibition of androgen receptor variants in castration resistant prostate cancer. Oncotarget, 2016. 7(22): p. 32210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu C, et al. , Niclosamide suppresses cell migration and invasion in enzalutamide resistant prostate cancer cells via Stat3-AR axis inhibition. Prostate, 2015. 75(13): p. 1341–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu C, et al. , Niclosamide and Bicalutamide Combination Treatment Overcomes Enzalutamide- and Bicalutamide-Resistant Prostate Cancer. Mol Cancer Ther, 2017. 16(8): p. 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.De Leon JT, et al. , Targeting the regulation of androgen receptor signaling by the heat shock protein 90 cochaperone FKBP52 in prostate cancer cells. Proc Natl Acad Sci U S A, 2011. 108(29): p. 11878–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Buchanan G, et al. , Control of androgen receptor signaling in prostate cancer by the cochaperone small glutamine rich tetratricopeptide repeat containing protein alpha. Cancer Res, 2007. 67(20): p. 10087–96. [DOI] [PubMed] [Google Scholar]
- 138.Philp LK, et al. , SGTA: a new player in the molecular co-chaperone game. Horm Cancer, 2013. 4(6): p. 343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lallous N, et al. , Targeting Binding Function-3 of the Androgen Receptor Blocks Its Co-Chaperone Interactions, Nuclear Translocation, and Activation. Mol Cancer Ther, 2016. 15(12): p. 2936–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Andersen RJ, et al. , Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell, 2010. 17(6): p. 535–46. [DOI] [PubMed] [Google Scholar]
- 141.Myung JK, et al. , An androgen receptor N-terminal domain antagonist for treating prostate cancer. J Clin Invest, 2013. 123(7): p. 2948–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sadar MD, et al. , Sintokamides A to E, chlorinated peptides from the sponge Dysidea sp. that inhibit transactivation of the N-terminus of the androgen receptor in prostate cancer cells. Org Lett, 2008. 10(21): p. 4947–50. [DOI] [PubMed] [Google Scholar]
- 143.Brand LJ, et al. , EPI-001 is a selective peroxisome proliferator-activated receptor-gamma modulator with inhibitory effects on androgen receptor expression and activity in prostate cancer. Oncotarget, 2015. 6(6): p. 3811–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dalal K, et al. , Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J Biol Chem, 2014. 289(38): p. 26417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dalal K, et al. , Bypassing Drug Resistance Mechanisms of Prostate Cancer with Small Molecules that Target Androgen Receptor-Chromatin Interactions. Mol Cancer Ther, 2017. 16(10): p. 2281–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dalal K, et al. , Selectively targeting the dimerization interface of human androgen receptor with small-molecules to treat castration-resistant prostate cancer. Cancer Lett, 2018. 437: p. 35–43. [DOI] [PubMed] [Google Scholar]