TO THE EDITOR:
Chronic lymphocytic leukemia (CLL) is characterized by a clonal expansion of mature CD19+CD5+ B cells, which are highly dependent on microenvironmental cues for their survival.1 This common adult leukemia is preceded by a precursor phase termed monoclonal B-cell lymphocytosis (MBL),2,3 which has been characterized as indistinguishable from CLL at the genetic, transcriptomic, and epigenomic level.4-6 However, how leukemia cells coevolve with immune cells in their circulating microenvironment during the onset of MBL and upon progression to CLL remains incompletely characterized.7
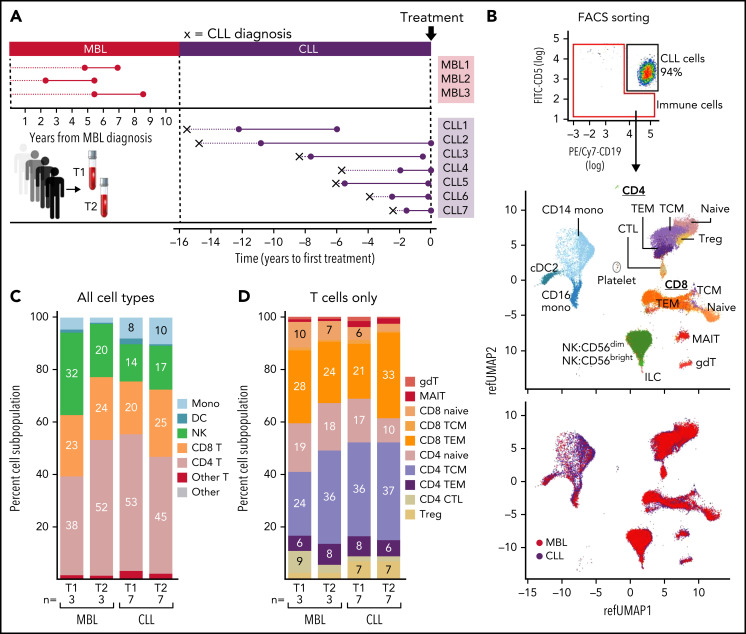
Recently, single-cell transcriptome sequencing (scRNA-seq) approaches have transformed our ability to gain a comprehensive evaluation of the spectrum of immune cells within the tumor microenvironment and of their potential cross talk with cancer cells.8-14 In our study, we applied scRNA-seq to broadly characterize circulating immune cells coexisting with leukemic cells during natural CLL progression. Although we acknowledge the critical role of the bone marrow and lymph node microenvironments on CLL cells, the lack of feasibility for procuring serial specimens from these tissue compartments led us to focus our study on circulating immune cells. We therefore collected serial peripheral blood mononuclear cell (PBMC) samples from 3 individuals with high-count MBL who did not progress to CLL after a median follow-up of 7.0 years and 7 patients with CLL, whose genetic characterization of CD19+CD5+ cells over time by whole-exome sequencing, has been reported15 (Figure 1A). We processed paired samples from all patients: the first samples were collected at time point 1 (T1), at a median of 4.96 years (range, 2.44-5.46) after MBL diagnosis or 2.54 years (range, 0.5-4.2) after CLL diagnosis; whereas the second group were collected at T2, a median of 2.97 years (range, 2.01-2.99) after T1 for the MBL patients and 4.75 years (range, 1.3-10.6) for the CLL patients. T2 samples for CLL patients were collected at a median of 0.2 years (range, 0-5.9) before the first treatment (supplemental Table 1, available on the Blood Web site).
Figure 1.
scRNA-seq analysis of immune cells from nonprogressive MBL patients and CLL patients. (A) PBMCs from 2 serial samples were collected for 3 patients with MBL and 7 with CLL. (B) Non-CD19+CD5+ cells were isolated by fluorescence-activated cell sorting. UMAP visualization of all immune cells colored by immune cell type (top) and CLL or MBL assignment (bottom). (C) Proportion of immune cell types per time point in patients with MBL or CLL. (D) Proportion of T-cell types per time point in patients with MBL or CLL. Cell percentages were calculated after the number of cell from all samples. CTL, cytotoxic T lymphocyte; DC, dendritic cell; gdT, γ-δ T (cells); ILC, innate lymphoid cell; MAIT, mucosa-associated invariant T (cells); Mono, monocyte; NK, natural killer (cell); pDC, plasmacytoid dendritic cell; T, T cell; TCM, central memory T (cell); TEM, effector memory T (cell); Treg, regulatory T (cells).
Non-CD19+CD5+ cells were isolated by fluorescence-activated cell sorting, and samples from each patient were processed on the same day to minimize the batch effect. Cell suspensions were loaded on a GemCode Single-Cell Instrument (10× Genomics), and libraries were prepared as previously described16 (supplemental Methods). Analysis was conducted using Seurat V4.0.0 selecting cells with gene count between 500 and 3000 and less than 10% mitochondrial reads. Using the trimmed data set, we isolated the nontumor population and assigned immune cell types by performing multimodal reference mapping, using a CITE-seq (cellular indexing of transcriptomes and epitope–sequencing) reference of 162 000 PBMCs measured with 228 antibodies.17 B cells were excluded because of potential CLL contamination. After quality control, we obtained 67 333 single-cell transcriptomes (median number of cells per sample, 3711; range, 491-6633; Figure 1B; supplemental Table 1). For each sample, we evaluated the potential for processing and batch artifacts between samples and cohorts, and we selected cohorts with similar “cold-shock signature”18 for comparison (supplemental Figure 1A). In total, we identified 16 clusters across 3 distinct lineages: T cells, natural killer cells, and myeloid cells (Figure 1B; top, UMAP [uniform manifold approximation and projection]). The distribution of immune cell types from MBL and CLL samples and across patients appeared to be balanced across the cell clusters (Figure 1B; bottom, UMAP; supplemental Figure 1B). Analysis of the proportions of immune cell types, including various T-cell subsets, between MBL and CLL samples revealed no differences, even across time points (T1 vs T2; Figure 1C-D; supplemental Table 2A).
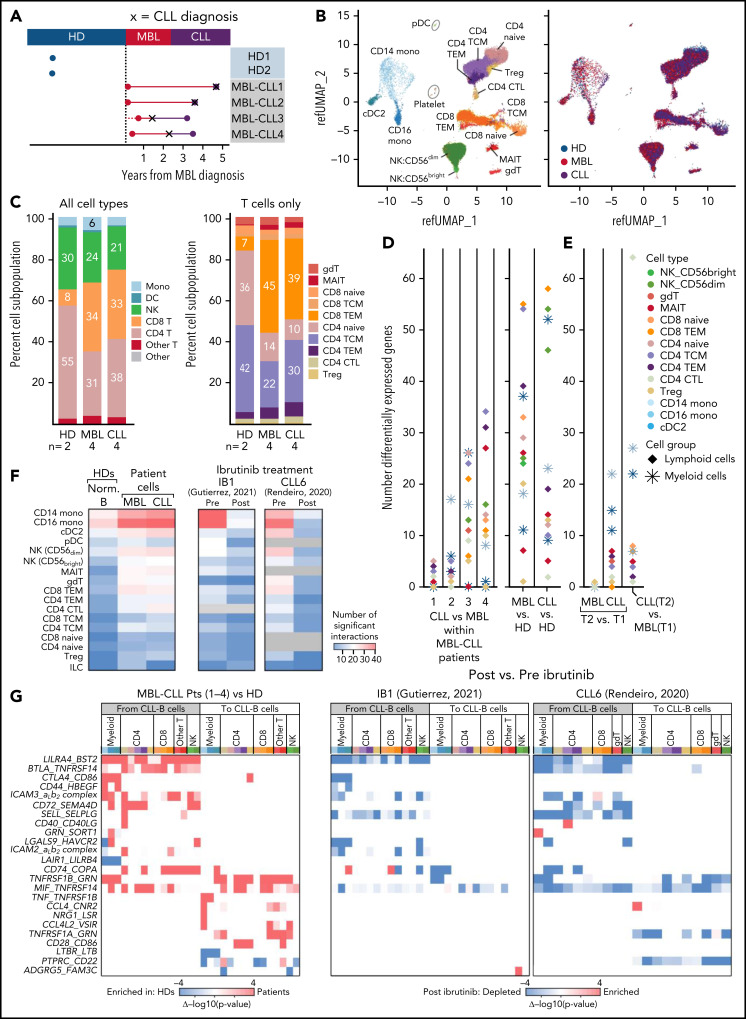
To confirm the absence of major differences in immune cell proportions between MBL and CLL, we performed scRNA-seq on PBMCs collected from a separate cohort of 4 patients with high-count MBL that progressed to CLL (MBL-CLL1-4); the median time from MBL (T1) to CLL diagnosis was 2.68 years (range, 0.7-4.6) and from CLL diagnosis to T2 was 0.6 years (range, 0-1.8). We also evaluated 2 age-matched healthy donors (HDs, median number of cells per sample, 4400; range, 2630-7596 cells) using the same approach described above (Figure 2A-B). Again, we observed an absence of major compositional or phenotypic changes in immune cell populations in the transition from MBL to CLL, whereas marked differences in the composition in immune cell types were evident in patients with CLL compared with HDs. In particular, the proportion of CD8+ T cells was higher in patients with CLL than in HDs (33% vs 8%, P = .037), with a corresponding decrease in CD4+ T cells (Figure 2C, left; supplemental Table 2B). The CD4+ and CD8+ T-cell subtypes that contributed to these differences were naive, central memory CD4+ and terminal effector memory CD8+ cells (Figure 2C; right). A higher number of differentially expressed genes (adjusted P < .05 and |avg_log2 FC| >0.6) was observed between HDs and patients with MBL/CLL than between MBL and CLL at the time of progression (patients MBL-CLL-1 and -2; Figure 2D; supplemental Table 3). More differences in gene expression were seen in those paired CLL samples obtained at a time more distant from transition to CLL (patients MBL-CLL-3 and -4), suggesting further evolution of the immune response over time with CLL progression. Effector memory CD8+ T cells and CD56dim natural killer cells consistently showed more differentially expressed genes in patients with MBL and CLL than in HDs (Figure 2D, right), which we also observed in a pseudobulk reanalysis of the same data (supplemental Figure 2). Comparable shifts in immune cell expression profiles were observed in the evaluation of independent MBL (MBL1-3, T1) vs CLL (CLL1-7, T2), but only minimal differences were observed in nonprogressing MBL (Figure 2E). We acknowledge that the low number of replicates (n = 2) did not provide sufficient power to detect the biological variability among HDs and that individual-specific variations may have confounded the observed differences between HD and MBL/CLL samples, but we minimized that risk by selecting age-matched HDs and applied uniform processing to all samples.
Figure 2.
scRNA-seq analysis of immune cells from healthy donors and disease progression from MBL to CLL. (A) scRNA-seq was performed on PBMCs collected from 4 patients with MBL (red dots) that progressed to CLL (purple dots), and from 2 HDs (blue dots). X, the time of diagnosis of CLL. (B) UMAP visualization of all immune cells colored by immune cell types (left) and by sample types (right). (C) Proportion of immune cell types (left) and T-cell subtypes (right). (D) Number of significant differentially expressed genes for each cell type by performing a comparison of paired samples within patients (left) or comparison between MBL samples or CLL samples vs healthy donors (right). Cells were categorized based on lymphoid and myeloid cells. (E) Same analysis for significant differentially expressed genes was performed on 3 independent patients with nonprogressive MBL and 7 with CLL (Figure 1). (F) Heat maps with the number of the significant ligand-receptor interactions for each cell type under different conditions using CellPhoneDB v2.1.7. Heat map comparing the number of significant interactions between healthy donors and patient samples from either MBL stage or CLL stage (left). Heat maps including samples before and after ibrutinib for 2 additional patients (right).20,21 Gray boxes indicate an insufficient number of cells to perform interactome analysis. (G) Heat maps representing the difference of P values for each ligand-receptor pair regarding specific cell types (x-axis). Interactions that are enriched in patients (red) or enriched in healthy donors (blue) were calculated by subtracting −log10 (P value) in healthy donors from −log10 (P value) in patients (left). The same interactions that are either enriched (red) or depleted (blue) after ibrutinib (right)20,21 are calculated by subtracting −log10 (P value) in preibrutinib from −log10 (P value) in postibrutinib. Pts, patients; cell type abbreviations are the same as in Figure 1.
To investigate which dysfunctional immune mechanisms may impact CLL biology, we interrogated major molecular interactions between immune and normal B or CLL-B cells in HDs or patients, respectively, using CellPhoneDB v2.1.7, which predicts potential interactions between ligand-receptor pairs based on elevated expression in the corresponding cell types.19 In so doing, we observed an increased total number of potential interactions in subjects with MBL compared with those in HDs. This increase remained stable with progression to CLL and was evident across diverse immune cell types but was most distinctly observed in monocytes (Figure 2F, left heat map). To examine the effects of B-cell receptor signaling inhibition with ibrutinib on the cellular interactions between immune and leukemia cells, we reanalyzed 4 additional scRNA-seq samples previously generated from PBMCs before and during ibrutinib treatment (cells collected 30-240 days after treatment) from 2 patients with CLL.20,21 We again observed that the number of cellular interactions in pretreatment CLL samples was higher across immune cell types, especially in monocytes in both patients. Consistently, the number of interactions decreased after ibrutinib treatment to levels similarly observed in HDs (Figure 2F, right heat maps). Most of the interactions upregulated in patients with MBL/CLL involved inhibitory signals of immune cell function proceeding from CLL cells across to various immune cell types, such as BTLA/MIF-TNFRSF14 (HVEM, observed in MBL-CLL1, -3, and -4), CTLA4-CD86 (observed in MBL-CLL-4), and LGALS9-HAVCR2 (TIM3, observed in MBL-CLL1-4; Figure 2G, left; supplemental Figure 3). Notably, only a proportion of cancer cells express these inhibitory signals: BTLA (17.4%), MIF (41.6%), LGALS9 (18.2%), and CTLA4 (10.4%) (supplemental Figure 4). We observed that all these interactions were downregulated after ibrutinib treatment (Figure 2G, right).
Altogether, we observed that the composition and state of immune cells was markedly different between HDs and patients with MBL, whereas no major additional transcriptional changes manifested during natural progression from MBL to CLL. These observations suggest that the key drivers of transcriptional immune dysfunction in CLL may be present early during the course of the disease and are in keeping with the early transcriptomic, genomic, and epigenetic changes already present in MBL, as well as the known increased risk of infections, even at the earliest stages of the disease.22 Among the features that distinguished immune and leukemia cells interactions in patients with CLL were an increased number of cellular interactions compared with HDs, especially within myeloid cells, that predominantly involved multiple inhibitory immune signals and that were no longer detected after ibrutinib treatment. Thus, although T-cell deficits in CLL have been well investigated,23,24 the contribution of myeloid cells to inhibitory signals has been far less well characterized and warrants further assessment.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Jerome Ritz and the DFCI Pasquarello Tissue Bank in Hematologic Malignancies for prospective collection and processing of blood samples from healthy donors.
This work was supported, in part, by National Institutes of Health (NIH), National Cancer Institute (NCI) grants 5P01CA081534-14, P01CA206978, R01CA216273, and UG1CA233338. C.J.W. acknowledges support from the CLL Global Research Foundation. S.H.G. was supported by a Kay Kendall Leukaemia Fund Fellowship. E.M.P. acknowledges research funding from the Doris Duke Charitable Foundation (Physician-Scientist Fellowship), a Conquer Cancer (The ASCO Foundation) Young Investigator Award, and Dana-Farber Flames FLAIR. J.F. received support from the NIH, National Institute of General Medical Science under award R35GM142889. P.G. was supported by Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy (Special Program on Metastatic Disease; 5 per mille #21198), and ERA NET TRANSCAN-2 Joint Transnational Call for Proposals: JTC 2016 (project #179 NOVEL), project code (MIS) 5041673. S.L. is supported by an NCI Research Specialist Award (R50CA251956).
Footnotes
All single-cell data reported in this article have been deposited in the dbGAP repository (accession number Pha002705.vi). Questions regarding methods and protocols will be answered in response to e-mail request to the corresponding author.
There is a Blood Commentary on this article in this issue.
Authorship
Contribution: N.P., J.F., S.H.G., and C.J.W. designed and conceived the study; N.P., L.Z.R., T.J.K., S.L.S., N.E.K., C.L., T.D.S., P.G., and L.S. collected samples and clinical annotations; S.L. generated the scRNA-seq libraries and processed the raw sequencing data; N.P., Y.E.T., C.K.L., N.C., E.M.P., W.Z., K.J.L., P.V.K., D.S.N., L.R.O., J.F., and S.H.G. analyzed and interpreted data; J.F., S.H.G., and C.J.W. supervised the project; and N.P., Y.E.T., S.H.G., and C.J.W. wrote the paper with assistance from all other authors.
Conflict-of-interest disclosure: N.P. is currently an employee of AstraZeneca; C.J.W. holds equity in BioNTech, Inc. and receives research funding from Pharmacyclics. S.H.G. has received speaker fees from Janssen UK and travel and honoraria from AbbVie and provides research consultancy for Novalgen Limited. P.G. has received honoraria from AbbVie, AstraZeneca, ArQule MSD, BeiGene, Celgene/Juno/BMS, Janssen, Loxo/Lilly, and Roche and research funding from AbbVie, AstraZeneca, Janssen, and Sunesis. P.V.K serves on the scientific advisory boards of Celsius Therapeutics Inc. and Biomage Inc. N.E.K. serves on the advisory boards of AbbVie, Astra Zeneca, Behring, Cytomx Therapy, Dava Oncology, Janssen, Juno Therapeutics, Oncotracker, Pharmacyclics and Targeted Oncology and on the data safety monitoring committees of Agios Pharm, AstraZeneca, BMS-Celgene, Cytomx Therapeutics, Janssen, Morpho-sys, and Rigel and research funding from AbbVie, Acerta Pharma, Bristol Meyer Squib, Celgene, Genentech, MEI Pharma, Pharmacyclics, Sunesis, TG Therapeutics, and Tolero Pharmaceuticals. L.S. has received honoraria from AbbVie, AstraZeneca, Janssen and travel funding from Janssen. T.D.S. received institutional research support from Genentech and Phamacyclics. The remaining authors declare no competing financial interests.
Correspondence: Catherine J. Wu, Dana-Farber Cancer Institute, Dana 520C, 450 Brookline Ave, Boston, MA 02215; e-mail: cwu@partners.org.
REFERENCES
- 1.Burger JA. The CLL cell microenvironment. Adv Exp Med Biol. 2013;792:25-45. [DOI] [PubMed] [Google Scholar]
- 2.Dagklis A, Fazi C, Scarfo L, Apollonio B, Ghia P. Monoclonal B lymphocytosis in the general population. Leuk Lymphoma. 2009;50(3):490-492. [DOI] [PubMed] [Google Scholar]
- 3.Rawstron AC, Bennett FL, O’Connor SJM, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359(6):575-583. [DOI] [PubMed] [Google Scholar]
- 4.Puente XS, Beà S, Valdés-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526(7574):519-524. [DOI] [PubMed] [Google Scholar]
- 5.Agathangelidis A, Ljungström V, Scarfò L, et al. Highly similar genomic landscapes in monoclonal B-cell lymphocytosis and ultra-stable chronic lymphocytic leukemia with low frequency of driver mutations. Haematologica. 2018;103(5):865-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kretzmer H, Biran A, Purroy N, et al. Preneoplastic alterations define CLL DNA methylome and persist through disease progression and therapy. Blood Cancer Discov. 2021;2(1):54-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purroy N, Wu CJ. Coevolution of leukemia and host immune cells in chronic lymphocytic leukemia. Cold Spring Harb Perspect Med. 2017;7(4):a026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plass M., Solana J., Wolf F.A., et al. Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science. 2018; 360(6391):eaaq1723. [DOI] [PubMed] [Google Scholar]
- 9.Villani AC, Satija R, Reynolds G, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335):eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014; 344(6190):1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirosh I, Izar B, Prakadan SM, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gohil SH, Iorgulescu JB, Braun DA, Keskin DB, Livak KJ. Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat Rev Clin Oncol. 2021;18(4):244-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roerink SF, Sasaki N, Lee-Six H, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556(7702):457-462. [DOI] [PubMed] [Google Scholar]
- 15.Gruber M, Bozic I, Leshchiner I, et al. Growth dynamics in naturally progressing chronic lymphocytic leukaemia. Nature. 2019;570(7762):474-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8(1):14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573-3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massoni-Badosa R, Iacono G, Moutinho C, et al. Sampling time-dependent artifacts in single-cell genomics studies. Genome Biol. 2020;21(1):112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15(4):1484-1506. [DOI] [PubMed] [Google Scholar]
- 20.Rendeiro AF, Krausgruber T, Fortelny N, et al. Chromatin mapping and single-cell immune profiling define the temporal dynamics of ibrutinib response in CLL. Nat Commun. 2020;11(1):577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez C, Al’Khafaji AM, Brenner E, et al. Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment. Nat Can. 2021;2(7):758-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreira J, Rabe KG, Cerhan JR, et al. Infectious complications among individuals with clinical monoclonal B-cell lymphocytosis (MBL): a cohort study of newly diagnosed cases compared to controls. Leukemia. 2013;27(1):136-141. [DOI] [PubMed] [Google Scholar]
- 23.Ramsay AG, Johnson AJ, Lee AM, et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008;118(7):2427-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long M, Beckwith K, Do P, et al. Ibrutinib treatment improves T cell number and function in CLL patients. J Clin Invest. 2017;127(8):3052-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.