Abstract
The outbreak of the SARS-CoV-2 pandemic created an unprecedented requirement for diagnostic testing, challenging not only healthcare workers and laboratories, but also providers. Quantitative RT-PCR of various specimen types is considered the diagnostic gold standard for the detection of SARS-CoV-2, both in terms of sensitivity and specificity. The pre-analytical handling of patient specimens is a critical factor to ensure reliable and valid test results. Therefore, the effect of storage duration and temperature on SARS-CoV-2 RNA copy number stability was examined in various commercially available specimen collection, transport and storage devices for naso/oropharyngeal swabs and saliva. The swab specimen transport and storage devices tested showed no significant alteration of viral RNA copy numbers when stored at room temperature, except for one system when stored for up to 96 h. However, at 37 °C a significant reduction of detectable RNA was found in 3 out of 4 of the swab solutions tested. It was also found that detectability of viral RNA remained unchanged in all 7 saliva devices as well as in unstabilized saliva when stored for 96 h at room temperature, but one device showed marked RNA copy number loss at 37 °C. All tested saliva collection devices inhibited SARS-CoV-2 infectivity immediately, whereas SARS-CoV-2 remained infectious in the swab transport systems examined, which are designed to be used for viral or bacterial growth in cell culture systems.
Abbreviations: BE, Artificial body excretion; CPE, Cytopathic effect; PAHO, Pan American Health Organization; UTM, universal transport media; VTM, viral transport media
Keywords: SARS-CoV-2, Pre-analytic, Swab systems, Saliva collection devices, Infectivity, RNA stability
Introduction
Diagnostic testing for the presence of SARS-CoV-2 in the upper respiratory tract has become a main strategy to combat COVID-19 [1], [2]. The broad application of SARS-CoV-2 diagnostic tests, however, raised a series of challenges such as shortages of personal protective equipment (PPE), sampling devices and diagnostic kits, as well as the compliance with specific biosafety procedures to handle diagnostic specimens of individuals with a suspected COVID-19 infection. To ensure a precise definition of terms throughout this article in compliance with ISO Standards and ISO Technical Specifications, a “specimen” is defined as the primary specimen directly collected from a patient or test person into a collection device, and a “sample” is derived from a specimen after every kind of processing of a primary specimen, e.g. aliquots or isolates (nucleic acids, proteins, etc.). The policies of the leading public health institutions (WHO, CDC, PAHO, PHE, FDA, UCSF) were summarized in [3], emphasizing that COVID-19 specimens, such as nasopharyngeal swabs, saliva, tissues, blood or urine, must be collected, processed and preserved by trained personnel equipped with the right PPE to prevent the transmission of the coronavirus and ensure the specimen quality for testing and research. The WHO prescribes a biosafety level (BSL)-2 facility for non-propagative and BSL-3 for virus propagative work and staff trained in relevant procedures [4]. These requirements limit the number of qualified laboratories and can ultimately increase transport duration from the specimen collection facility to the analyzing laboratory. The WHO and CDC strongly recommend a storage temperature of 2–8 °C for all specimens intended for SARS-CoV-2 testing for up to 72 h. For long-term storage, specimens should be kept at − 70 °C [5], [6]. However, maintaining an appropriate cooling chain according to international guidelines could be endangered during hot seasons due to widely scattered test centers, a high number of specimens, storage over many hours and mostly end-of-day transports to a diagnostic laboratory. Pre-analytical and analytical vulnerabilities in the laboratory diagnosis of COVID-19 have been described [7]. They are aimed at raising the awareness of personnel who are organizationally and practically responsible for specimen logistics towards the importance of pre-analytical influences and on how the specimens should be collected, managed and stored before analysis [7].
Correct sampling and specimen handling is of utmost importance to obtain high-quality RNA and ensure reliable and valid test results. Various manufacturers offer nasopharyngeal swabs (hereafter only referred to as swabs) which are most commonly used. They are made of different materials, such as nylon or cotton, individually packed in bags and delivered in combination with different transport media, e.g. Amies (a modification of Stuart’s medium to prevent drying and death of bacteria), viral transport media (VTM) or universal transport media (UTM). The media used by various companies in various compositions are usually indicated with these abbreviations, mostly not further defined, and can be either virus-inactivating or suitable for virus cultivation. These details are relevant for the selection of biosafety measures for personnel and specimen transport and storage.
A valuable alternative to nasopharyngeal swab sampling is the collection of saliva when aiming for simplification and acceleration of COVID-19 diagnosis. Since saliva can be collected by non-professional personnel it may better be suited for self-collection. Saliva collection devices intended for molecular testing are usually delivered together with nucleic acid stabilization solutions. Easy-to-handle instructions for use should be safe so as to enable so-called home collection. Several studies have compared the performance of saliva and nasopharyngeal specimens for RT-qPCR analysis [11], [12], [13], [14]. It is important to assess whether the virus RNA-stabilizing and virus-inactivating substances interfere with molecular analysis such as RT-qPCR or sequencing.
RT-qPCR is currently the diagnostic gold standard for detecting SARS-CoV-2 RNA because of its high sensitivity and specificity, no matter how the specimen was collected, either using swabs or saliva. However, RT-qPCR testing requires transport of specimens to laboratories, trained staff for analysis and a one to three day timeframe to provide the test result.
As of yet, there are only few studies assessing the pre-analytical effects of different sampling systems although billions of specimens have been processed worldwide. The performance of MRSA swabs (Copan eSwabs 480C flocked nasal swab in Amies medium) and viral swabs (Becton Dickenson, BD H192(07) flocked swabs in UTM) was compared for parallel SARS-CoV-2 testing and revealed a concordance of more than 96% after expeditious transport at room temperature (RT) and freezing at − 20 °C until PCR analysis [8]. High intra-individual and inter-individual reliability was reported for the detection of three SARS-CoV-2 genes in endotracheal secretion specimens transported in phosphate-buffered saline (PBS) and VTM stored for 18 h at RT [9]. In another study comparing five different specimen transport systems similar differences in the average viral RNA Cq-values were reported across the tested media and temperature storage conditions [10]. As these studies only spotlight certain pre-analytical aspects, it is important to systematically examine the impact of pre-analytic conditions. The type and claimed purpose of sampling devices and the effect of suboptimal transport and storage conditions, which might be beyond the manufacturer’s specification for SARS-CoV-2 RNA stability, may have an impact on the reliability and validity of test results.
In this study four commonly used swab systems and seven saliva collection devices for SARS-CoV-2 RNA testing were evaluated systematically. The widely used PBS and VTM, prepared according to protocols published by the US Centers for Disease Control (CDC), were used for comparison. First, specimen types and devices were tested for their ability to stabilize SARS-CoV-2 RNA under distinct storage conditions and durations to allow for direct comparisons when analyzed by RT-qPCR. Additionally, the virus inactivation properties of the sampling solutions were examined.
Material and methods
Saliva collection devices and transport swab systems
Information on devices and recommended storage conditions and claims towards inactivation properties listed in Table 1 and Table 2 were cited from the suppliers’ homepages including relevant instructions for use.
Table 1.
Specimen collection, transport and storage devices and respective manufacturers’ claims.
| Device | Abbreviation | Storage condition | Regulatory status |
|---|---|---|---|
| Copan eSwab™ 480C + Single Regular Size Nylon® Flocked Swab | eSwab | INTENDED USE Copan Liquid Amies Elution Swab (eSwab®) Collection and Transport System is intended for the collection and transport of clinical specimens containing aerobes, anaerobes and fastidious bacteria from the collection site to the testing laboratory. Swab specimens for bacterial investigations collected using eSwab® should be transported directly to the laboratory, preferably within 2 h of collection (2–4) to maintain optimum organism viability. If immediate delivery or processing is delayed, then specimens should be refrigerated at 4–8 °C or stored at room temperature (20–25 °C) and processed within 48 h. | EU: CE IVD Outside EU: FDA cleared |
| Copan UTM-RT® Universal Transport Medium 359C + Single Regular Size Nylon® Flocked Swab | UTM | INTENDED USE UTM® is an FDA cleared collection and transport system suitable for collection, transport, maintenance and long-term freeze storage of clinical specimens containing viruses, including COVID-19, chlamydia, mycoplasma or urea plasma organisms. Using the UTM-RT® System, collected specimens can be stored for up to 72 h at 2–8 °C (according to CDC recommendations https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html) | EU: CE IVD Outside EU: FDA cleared |
| IMPROVIRAL™ Viral Preservative Medium (8110111) + ImproSwab® (550040A) | VPM | Widely used for the collection, preservation and transportation of nasopharyngeal pathogen specimens such as influenza, pneumonia, avian influenza, hand-foot-mouth disease, measles and other. Storage and transport under 0–8 ℃ and no more than 30 days at room temperature. | EU: CE IVD |
| Viral transport mediuma | VTM | As recommended by CDC: Store respiratory specimens at 2–8 °C for up to 72 h after collection. If a delay in testing or shipping is expected, store specimens at − 70 °C or below. | n. a. |
(HBSS (Life Technologies Europe, Bleiswijk, Netherlands) + 2% FCS (Thermo Fisher Scientific, Waltham, USA) + 0,2% PenStrep (Thermo Fisher Scientific, Waltham, USA).
Table 2.
Saliva collection devices and respective manufacturers’ claims.
| Device | Abbreviation | Storage condition | Regulatory status |
|---|---|---|---|
| PAXgene Saliva Collector (769040) – (PreAnalytiX GmbH, Switzerland) | PreAnalytiX | Saliva collected and stabilized with the PAXgene Saliva Collector can be stored for DNA isolation for at least 24 months at temperatures up to 25 °C (study ongoing). In addition, PAXgene Saliva can be frozen long term at − 20 or − 80 °C when transferred into a suitable cryovial. SARS-CoV-2-derived RNA copy numbers are stabilized in saliva collected into PAXgene Saliva Collector for at least 4 days (96 h) at 20 °C [15]. | MBA (for molecular biology applications) world wide |
| Zeesan Saliva RNA Sample Collection Kit (401115) – (Zeesan, P.R. China) | Zeesan | Collected sample should be stored and transported under 37 °C. It is recommended to extract RNA within 1 month or be stored below 8 °C for long-term storage (Package insert, see Supplementary information) | EU: CE IVD Outside EU: CFDA approval for clinical use in China |
| DNA/RNA Shield™ Saliva/sputum collection kit (R1210) – (Zymo Research Corp., U.S.A) | Zymo | Each collection tube is pre-filled with 2 mL of DNA/RNA Shield™ that preserves nucleic acids in samples at ambient temperature (DNA > 1 year; RNA at least 1 month). Samples in DNA/RNA Shield™ can be frozen (− 20/− 80 °C) for prolonged periods [16]. | EU: CE IVD Outside EU: FDA 510(k) Number K202641 (substantial equivalence determination decision summary) |
| GeneFiX RNA Saliva collection (RFX-01) – (Isohelix DNA/RNA Sampling and Purification, UK) | Isohelix | RNA is stabilized at room temperature for at least 4 weeks [17]. | For research use only (Patient instructions, Version September 2018) |
| Saliva RNA Collection and Preservation Device Dx (53810) – (Norgen Biotek Corp. Canada) | Norgen | Once collected, saliva RNA is stable for up to 2 months when kept tightly sealed and stored at room temperature [18]. | EU: CE IVD Outside EU: authorized for diagnostic use in Canada by Health Canada |
| Spectrum SDNA-2000 Saliva Collection device – (Spectrum Solutions, USA) | Spectrum | No required post-collection temperature-controlled storage or transport of saliva samples. Provides over 10 days of post-collection stability with no degradation in sample efficacy [19]. | EU: CE Approved and authorized for COVID-19 testing Outside EU: Health Canada Approved and authorized for COVID-19 testing. FDA accelerated emergency use authorization for SARS-CoV-2 testing. |
| OMNIgene ORAL (OME-505) – (DNA Genotek Inc, Canada) | DNAGenotek | Saliva samples collected in OMNIgene ORAL (OM-505) can be stored at room temperature for up to 21 days. Storing at 4 °C is NOT recommended for saliva samples collected in OMNIgene ORAL [20]. |
EU: CE IVD |
Cell culture
African green monkey kidney epithelial cells (VeroE6) were from Biomedica (VC-FTV6, Vienna, Austria) and grown in Minimal Essential Medium (MEM) containing Earle’s Salts and L-Glutamine (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 2% fetal calf serum (FCS) (Thermo Fisher Scientific) and 1% Penicillin-Streptomycin (PenStrep) (Thermo Fisher Scientific).
Virus propagation
All experimental procedures with SARS-CoV-2 were performed in a BSL-3 laboratory [21]. Human 2019-nCoV Isolate (Ref-SKU: 026 V-03883, Charité, Berlin, Germany) was propagated in VeroE6 cells at 37 °C and 5% CO2 for 72 h. Prior to harvesting, cells were lysed by a freeze and thaw cycle, followed by a centrifugation step (10 min, 3000 × g) to remove cellular debris and the supernatant was filtered with 0.2 µm syringe filters (Thermo Fisher Scientific). Virus stocks were stored at − 80 °C until use.
SARS-CoV-2 virus titer was determined by a focus forming assay. For titration of virus, 48-well plates containing 100% confluent VeroE6 cells were infected with 200 µL of serial 10-fold diluted virus for 1 h at 37 °C and 5% CO2. Subsequently, cells were washed with medium once and overlayed with 1.5% carboxymethyl cellulose (Sigma-Aldrich, St. Louis, MO, USA) in MEM supplemented with 2% FCS and 1% PenStrep. After 72 h incubation, cells were fixed with 4% neutral-buffered formalin and stained immunohistochemically (see section immunohistochemistry).
SARS-CoV-2 RNA isolation and quantification via RT-qPCR
Viral RNA was extracted from 140 µL of the cell culture supernatant using QIAamp® Viral RNA Mini Kit (QIAGEN GmbH, Hilden, Germany), according to the manufacturer’s instructions. RNA samples were eluted in 40 µL ultra-pure water. 5 µL of the eluate were used for RT-qPCR using N1 or N2 primers (Eurofins Genomics Ebersberg, Germany) and a probe set from 2019-Novel Coronavirus (2019-nCoV) Real-time rRT-qPCR Panel [22] in combination with the QuantiTect® Multiplex RT-qPCR Kit (QIAGEN GmbH) on a Rotor Gene® Q cycler (QIAGEN GmbH). Amplification was performed in a total volume of 25 µL. Primer and probe sequences and thermal profile are shown in Table 3 and Table 4.
Table 3.
Primers and probe sequences for SARS-CoV-2 N1 region, adapted by Centers for Disease Control and Prevention (CDC).
| Name | Description | Sequence (5′ > 3′) | Label |
|---|---|---|---|
| 2019-nCoV_N1-F | Forward Primer | GAC CCC AAA ATC AGC GAA AT | none |
| 2019-nCoV_N1-R | Reverse Primer | GAC CCC AAA ATC AGC GAA AT | none |
| 2019-nCoV_N1-P | Probe | ACC CCG CAT TAC GTT TGG TGG ACC | FAM, BHQ-1 |
| 2019-nCoV_N2-F | Forward Primer | TTA CAA ACA TTG GCC GCA AA | none |
| 2019-nCoV_N2-R | Reverse Primer | GCG CGA CAT TCC GAA GAA | none |
| 2019-nCoV_N2-P | Probe | ACA ATT TGC CCC CAG CGC TTC AG | FAM, BHQ-1 |
Table 4.
Thermal profile for RT-qPCR using a Rotor-Gene Q thermal cycler.
| Step | Temperature (°C) | Time | |
|---|---|---|---|
| Reverse transcription | 50 | 30 min | |
| Initial denaturation | 95 | 15 min | |
| Amplification | 95 | 3 s | 45 cycles |
| 55 | 30 s |
A commercially available SARS-CoV-2 RNA copy number standard (VR-1986D genomic RNA from 2019 Novel Coronavirus, Lot: 70035624, ATCC, Glasgow, UK) was serially diluted and analyzed by RT-qPCR. The resulting Cq-values were plotted against ln[copy numbers] and the equation obtained from a simple linear regression analysis was used to calculate the copy numbers from the Cq-values (Eq. (1)).
Standard curve to calculate between Cq-values and viral copy numbers (Eq. (1))
| (1) |
Based on the standard curves, cut-offs were set to Cq values of 35.1 and 38.4 for N1 and N2, respectively. Cq-values exceeding the cut-off were set to the value 1 for graphical representation and further calculations.
Immunohistochemistry
For the immunohistochemical detection of SARS-CoV-2 in infected cells, 48-well plates were fixed for at least 30 min with 4% neutral-buffered formalin (SAV Liquid Production GmbH, Flintsbach, Germany). Subsequently, cells were washed 3 times with phosphate buffered saline (PBS) (Gatt-Koller GmbH, Absam, Austria), permeabilized for 10 min with 0.1% Triton X-100 (Merck KGaA, Darmstadt, Germany) in PBS and washed again 3 times. Endogenous peroxidases were suppressed by a 30 min incubation step with 3% H2O2 (Merck KGaA) in methanol (Merck KGaA). Thereafter, cells were incubated with anti-coronavirus nucleocapsid antibody (Sino Biological, Beijing, China, AB_2827973, diluted 1:1000) for 1 h. Cells were washed 3 times and incubated for 30 min with a HRP-conjugated secondary antibody (Dako REAL EnVision, HRP Rabbit/Mouse; Agilent Dako, Glostrup, Denmark). Bound secondary antibodies were visualized with the substrate AEC + High Sensitivity Substrate Chromogen Ready-to-use (Agilent Dako,). Images were taken by light microscope (Nikon, Eclipse, TS100; Nikon Europe BV, Amsterdam, The Netherlands) equipped with a JENOPTIK GRYPHAX® camera (Breitschopf, Innsbruck, Austria).
Viral RNA stability in various transport swab systems at different storage conditions
The definition used for room temperature ranged from 18 to 25 °C as defined by the ISO standard ISO 4307:2021 “Molecular in vitro diagnostic examinations – Specifications for pre-examination processes for saliva – Isolated human DNA” and the actual range measured was 20.6–22.4 °C.
Four different swab collection systems were selected (Table 1) to compare the SARS-CoV-2 RNA stability at different storage durations and temperatures. Artificial body excretion (BE) was prepared with 2.5 mg/mL BSA (Carl Roth GmbH, Karlsruhe, Germany), 3.5 mg/mL tryptone (Becton Dickinson, Le Pont de Claix, France) and 0.8 mg/mL mucin (Merck KGaA) [23] and spiked with 10,000 (= high spike-in), 1000 (= medium spike-in) and 100 (= low spike-in) copies of SARS-CoV-2 per swab device. 100 µL of spiked BE were applied onto each swab which was directly transferred into the respective transport medium. Spiked swab collection devices were stored at RT and at 37–37.4 °C for zero (t0), 24 h (t24) and 96 h (t96), respectively. At each time point, swabs were vortexed and a sample was taken for viral RNA quantification. In total, every swab collection device was examined in 9 replicates for each condition. Viral RNA was isolated and quantified via RT-qPCR (as described above). Detected viral copy numbers were extrapolated to the total copy numbers per device according to Eq. (1).
Viral RNA stability in saliva collection devices at different storage conditions
Seven saliva collection devices from different manufacturers were selected (Table 2) to compare the viral RNA copy number stability at different storage durations and temperatures. In addition to the saliva collection devices, PBS and unstabilized saliva were examined. The collection of saliva from self-reporting healthy volunteers was approved by the research ethics committee of the Medical University Graz (approval number EK 32-666). After obtaining informed consent, saliva from 4 to 6 healthy donors was pooled and spiked with three different virus copy loads, 100,000 (= high spike-in), 10,000 (= medium spike-in) and 1000 (= low spike-in) copies per device. One mL of each of the saliva stabilization solutions was added to 2 mL spiked saliva. Spiked saliva collection devices were stored at RT and at 37–37.4 °C for zero (t0), 24 h (t24) and 96 h (t96), respectively. At each time point, the devices were vortexed and 3 replicates were aliquoted from each device. In total, 3 saliva collection devices were examined for each tested condition. Viral RNA was isolated and quantified via RT-qPCR (as described above). Detected viral copy numbers were extrapolated to the total copy numbers per device according to Eq. (1).
Testing viral inactivation properties of saliva collection devices and swab transport systems
VeroE6 cells were seeded in 48-well cell culture plates (30,000 cells per well) in MEM supplemented with 2% FCS and 1% PenStrep overnight to reach approximately 80% confluence on the day of infection. Saliva collection devices were spiked with 277,000 plaque forming units (pfu)/mL device and diluted 1:2000 in serum free-medium, to avoid cell cytotoxic effects. Thus, cells were infected with 28 pfu/well. PBS and medium were treated the same way. After 1 h incubation (37 °C and 5% CO2) cells were washed once with medium and incubated in MEM supplemented with 2% FCS and 1% PenStrep for 24 and 72 h, respectively. Cytopathic effects (CPE) were documented after 72 h post infection via bright field microscopy. Additionally, 140 µL of cell culture supernatant were collected, viral RNA was isolated as described above and quantified via RT-qPCR to determine the ratio of virus replication between the control and the saliva collection devices. Furthermore, in a second independent experiment, cells were fixed 24 h post infection and stained via immunohistochemistry.
Swab transport systems were spiked with 277,000 pfu/mL and diluted 1:200 in serum free-medium, to avoid cell cytotoxic effects. Thus, cells were infected with 277 pfu/well. After 1 h incubation (37 °C, 5% CO2) cells were washed once with medium and incubated in MEM supplemented with 2% FCS and 1% PenStrep. 24 h post infection viral RNA obtained from 140 µL supernatant were quantified via RT-qPCR to determine the ratio of virus replication between the control and swab transport systems. Additionally, infected cells were stained immunohistochemically as an independent read-out.
Statistical analysis
The copy numbers were calculated with Excel 2016 and statistics and graphical presentations were performed with GraphPad Prism 8. Statistical differences between time points were determined by repeated measurement ANOVA test. t0 was defined as control column and the means of t24 and t96 were compared to the mean of the control column. Definition of Asterisks: ns = P > 0.05; * = P 0.01–0.05; ** = P 0.001–0.01; *** = P 0.0001–0.001; **** = P ≤ 0.0001.
Results
Commonly used transport swab systems differ in their ability to stabilize SARS-CoV-2 RNA
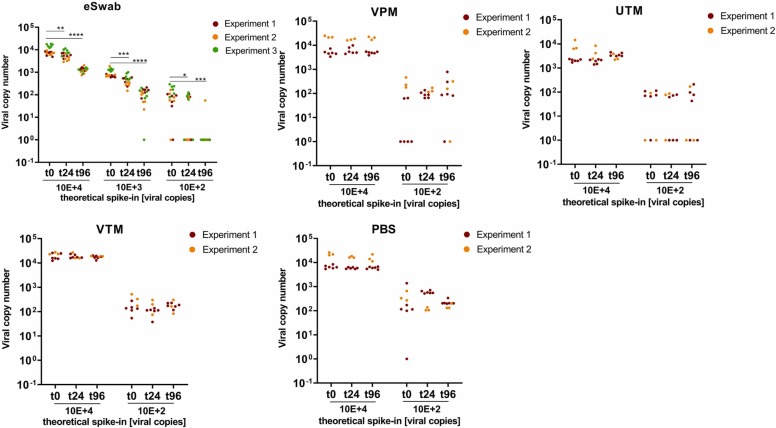
Various transport media were spiked with different virus loads (10,000 - 100 copies/mL transport medium) and stored at RT and 37 °C, respectively. RNA concentrations were examined directly after spiking (= t0) and after 24 h and 96 h of storage to monitor viral RNA detectability. Viral RNA levels in VPM, UTM, VTM and PBS remained constant over 96 h at RT ( Fig. 1). In contrast, viral copies in eSwab significantly decreased over time, when stored at RT. When eSwabs were spiked with a copy number concentration close to the detection limit, only 16 out of 18 samples showed detectable SARS-CoV-2 RNA at t0. After 96 h storage at RT, virus was detectable only in 1 out of 18 samples spiked with low copy numbers. At time point zero (t0) only 5 of the low spike-in samples of VPM were detectable, though after 24 h and 96 h, 9 and 7 samples respectively gave a signal above the threshold. A similar situation was observed with UTM with the low spike-in samples where independent of the storage duration virus was detectable in 6 of 9, 5 of 9, and 5 of 9 samples, respectively.
Fig. 1.
Stabilization of SARS-CoV-2 RNA copy numbers by collection and transport devices at room temperature. Data shown refer to two independent experimental series with either 6 or 3 replicates, with the exception of eSwabs, which were examined in 3 independent experimental series with 6 replicates each. Columns represent mean ± standard deviation (SD). Copy numbers were extrapolated to the total amount of copies per device. ns = P > 0.05; * = P 0.01–0.05; ** = P 0.001–0.01; *** = P 0.0001–0.001; **** = P ≤ 0.0001.
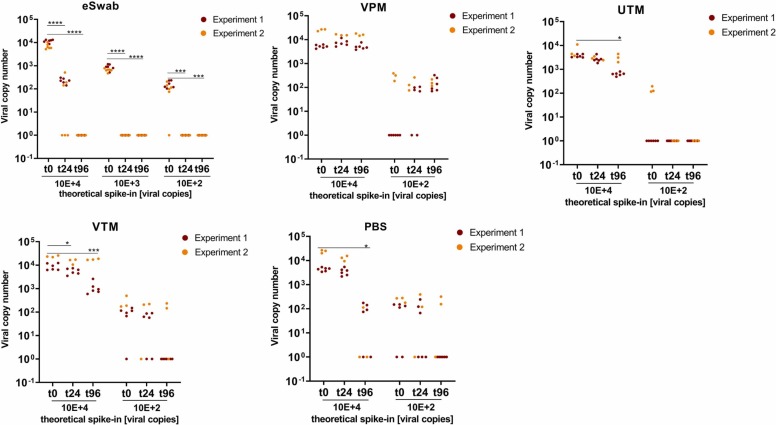
Increasing storage temperature to 37 °C had a profound impact on the stability of viral RNA in 3 of 4 systems ( Fig. 2). Significant reduction of SARS-CoV-2 RNA was found with eSwabs already after 24 h storage, and no RNA was detectable at any of the spike-in concentrations after 96 h. A significant but less pronounced decrease of viral RNA was also observed in UTM, VTM and PBS at 96 h storage. In contrast, SARS-CoV-2 RNA remained stable in VPM for up to 96 h at 37 °C, similar to RT (compare Fig. 1 and Fig. 2).
Fig. 2.
Stabilization of SARS-CoV-2 RNA by transport devices at 37 °C. Data shown refer to 2 independent experimental series with either 6 or 3 replicates, with the exception of eSwabs. Here 3 independent experimental series with 6 replicates each were carried out. Columns represent mean ± SD. Copy numbers were extrapolated to the total amount of viral copies per device. ns = P > 0.05; * = P 0.01–0.05; ** = P 0.001–0.01; *** = P 0.0001–0.001; **** = P ≤ 0.0001.
SARS-CoV-2 remains infective in all tested transport swab systems
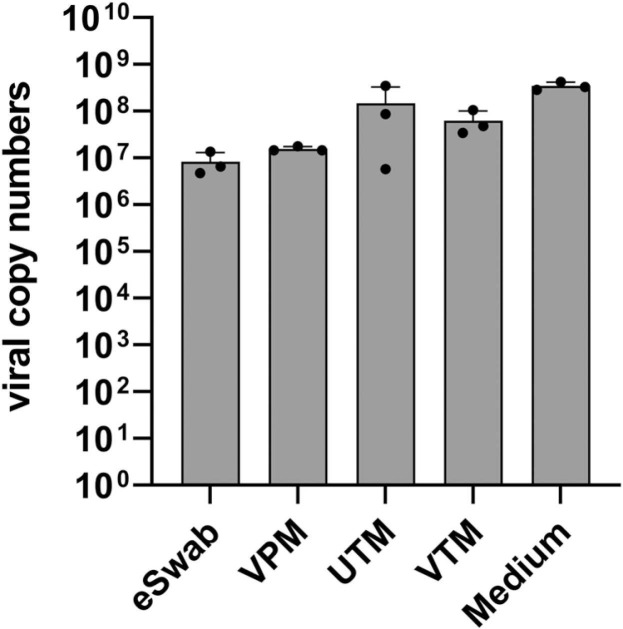
Virus particles recovered from transport media were used to test their infectivity in VeroE6 cells. Viral replication was assessed at 72 h after infection by isolating viral RNA from the VeroE6 cell culture supernatant and subsequent quantification via RT-qPCR. Cell culture medium infected with the same amount of virus as used to spike the devices served as positive control. Results obtained show some reduction of virus infectivity by the tested transport swab systems but none of them inactivated the SARS-CoV-2 infectivity ( Fig. 3).
Fig. 3.
Infectivity of SARS-CoV-2 in transport swab systems. Four different transport swab systems were spiked with SARS-CoV-2. Cell culture medium (Medium) infected with the same amount of medium as used for spiking the swab systems, which served as positive control, Viral replication in VeroE6 cells was determined after 72 h by using RT-qPCR.
Viral RNA stability in various saliva collection devices at room temperature
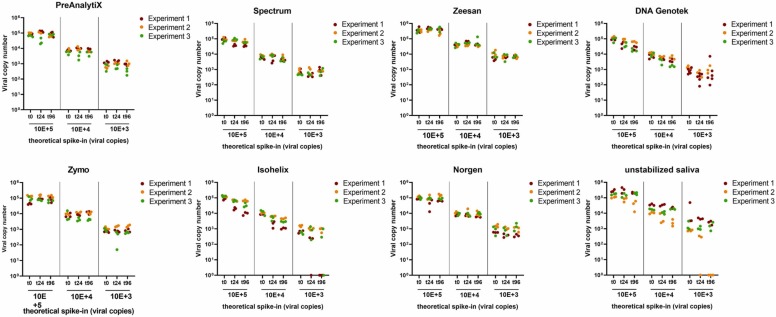
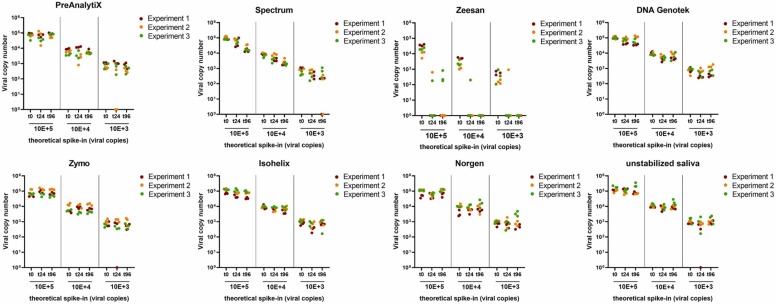
Saliva spiked with a defined amount of virus copies was used to evaluate the stability of viral RNA in saliva collection devices. PBS and unstabilized saliva served as references. Samples were drawn in triplicates after spike-in (t0), 24 h and 96 h, respectively. Even at the lowest virus load tested, corresponding to 1000 viral copies per device, no alteration of viral RNA was detected in the PreAnalytiX, Spectrum, Zeesan, Genotek, Zymo and Norgen devices as well as in PBS for up to 96 h at RT ( Fig. 4). In Isohelix, a trend towards reduction of viral copies was observed in one of the experimental series, the same was observed with unstabilized saliva. Further, in one experimental series, none of the replicates of unstabilized saliva with low virus input yielded detectable viral RNA after 96 h.
Fig. 4.
Comparison of SARS-CoV-2 RNA copy number stability in saliva collection devices and unstabilized saliva at room temperature over time. Three different viral copy numbers were spiked and monitored at (t0), after 24 h and 96 h. Data represent 3 independent experimental series each consisting of 3 technical replicates per spike-in concentration and time point. Copy numbers were extrapolated to the total amount of copies per device.
Viral RNA stability in saliva collection devices at 37 °C
In order to test RNA stability at elevated temperature, saliva collection devices were incubated at 37 °C. As shown in Fig. 5, levels of viral RNA levels remained unchanged in PreAnalytiX, Isohelix, Norgen, DNA Genotek, and unstabilized saliva for 96 h °C. A tendency to viral RNA decrease was observed after 24 h and 96 h for Spectrum. In Zeesan, only 2 samples gave a signal above the threshold after 24 h and no viral RNA was detectable after 96 h when stored at 37 °C.
Fig. 5.
Comparison of SARS-CoV-2 RNA stability in saliva collection devices and unstabilized saliva at 37 °C over time. Three different viral copy numbers were spiked and analyzed at t0, after 24 h and 96 h. Data represent 3 independent experimental series each consisting of 3 technical replicates per spike-in concentration and time point. Copy numbers were extrapolated to the total amount of copies per device.
Inactivation of SARS-CoV-2 by saliva collection devices
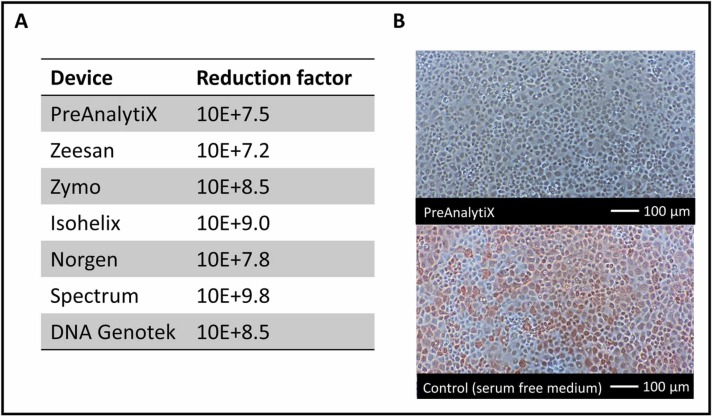
The saliva devices were spiked with SARS-CoV-2 and subsequently diluted 1:2000 prior to infection. The dilution step was required, because cell lysis was observed using bright field microscopy after exposure of cells to stabilization solutions. Cells infected with spiked, diluted saliva collection devices formed a confluent monolayer with no evidence of a CPE at 72 h post infection. In contrast, cells infected with virus diluted in serum-free medium showed a CPE after 72 h, which manifested as cell rounding and detachment (not shown). Further confirmation of the virus inactivating property of the saliva collection devices was shown by the capacity of viral replication, which was determined by quantifying viral RNA in the cell culture supernatant 72 h post infection. As shown in Fig. 6A, no viral replication was detected when the virus was treated with any of the saliva collection devices. Compared to the control, this corresponds to reduction factors ranging from 10E + 7.2 to 10 + E9.8. Additionally, infected cells were visualized by immunohistochemistry staining with a SARS-CoV-2 specific antibody. The majority of cells infected with SARS-CoV-2 diluted in serum free medium are stained red, indicating an infection with SARS-CoV-2. In contrast, cells incubated with spiked saliva devices remained unstained, suggesting viral inactivation (Fig. 6B).
Fig. 6.
SARS-CoV-2 inactivation by saliva collection and transport devices. A: Data show the log10 reduction of virus copy numbers after treatment of virus with collection device as compared to non-treated control from one experimental series with 3 technical replicates each. B: Immunohistochemistry with antibodies to SARS-CoV-2 n protein of VeroE6 cell at 24 h post infection. Top: Example of cells incubated with a spiked saliva collection device. Bottom: Cells incubated with virus diluted in cell culture medium. Red stained cells indicate SARS-CoV-2 infection.
Discussion
In this study, the impact of pre-analytical conditions on the stabilization and detection of SARS-CoV-2 RNA was analyzed in order to offer experimental data–based information on appropriate nasopharyngeal swab and saliva preservation solutions. Conditions tested were temperature (RT and 37 °C), transport/storage duration (up to 96 h) and transport/storage medium to simulate critical pre-analytical conditions that may occur in COVID-19 diagnostics. The spike-in amounts were deliberately chosen at the lower detection limit to show also the pre-analytical effect due to borderline amounts as they occur in natural specimens.
In the first part of the study 5 different transport swab systems, which are commonly used for nasopharyngeal and oropharyngeal specimen collection, were tested with respect to their viral RNA stabilization properties at RT (Fig. 1) and 37 °C (Fig. 2) for up to 96 h.
Some of the transport swab systems (e.g. eSwab, UTM) are designed for collecting and recovering bacteria from patient specimens, and therefore do not contain antibacterial substances. Transport systems, intended for sampling of virus specimens (e.g., VPM and UTM) are typically substituted with antibiotics, to suppress bacterial growth and protect viral nucleic acids e.g. from degradation by bacterial nucleases. As a sterile spike matrix was used in this study, no conclusions towards the impact of bacterial growth on SARS-CoV-2 RNA stability can be drawn.
At RT, the viral RNA copy number in VPM, UTM, VTM as well as in PBS remained stable over 96 h. According to the manufacturer’s instructions, eSwab samples can be stored at RT (20–25 °C) for up to 48 h. However, the results demonstrate that within 24 h the level of SARS-CoV-2 RNA significantly decreased in eSwabs. The latter observation was reproduced in 3 independent experimental series, each consisting of 6 replicates for each tested condition (Fig. 1).
UTM samples stored at 37 °C were spiked with a slightly lower viral load, because of variations in the preparation of the spike-in dilutions, compared to the other transport systems, which in turn led to undetectable signals at low-virus inputs. Surprisingly, in VPM spiked with low virus loads, an increase of viral copies was observed after prolonged storage. This might be due to so-called matrix effects of the swab device, which describes the release of adsorbed viral particles from the swab material. This effect was observed at RT as well as at 37 °C (Fig. 1, Fig. 2).
In one study [10], different transport systems including eSwabs and UTM were spiked with approximately 1500 SARS-CoV-2 copies/mL, covering comparable amounts of virus as used for the medium spike-in (1000 copies/mL) in the present study. For eSwabs, a Cq-value of 31.7 ± 0.4 was reported at time point zero and a Cq-value of 31.9 ± 0.4 after 5 days storage at RT. In contrast to these findings, the mean Cq-value in the present study at time point zero increased from 31.5 ± 0.4 to 34.4 ± 0.9 after 96 h, which corresponds to a ΔCq = 2.9 ± 0.3 and a 7.5-fold reduction of viral copy numbers.
Others reported on RNA stability of several respiratory viruses including SARS-CoV-2 in saline, VTM and UTM devices at temperatures up to 28 °C and storage durations of up to 28 days [27]. This is essentially in agreement with the present observations for UTM and VTM medium stored at RT for up to 4 days. However, a significant RNA reduction was observed in VTM at prolonged storage at 37 °C (Fig. 2).
In the second part of this study the SARS-CoV-2 RNA stabilization properties of seven commercially available saliva collection devices were examined.Saliva was spiked with 3 different SARS-CoV-2 copy number loads (low, medium, high) and preserved in saliva collection devices. Subsequently, they were stored at RT and at 37 °C for up to 96 h. The extent of viral RNA degradation was indicated by decreased viral genome copy numbers, which correlates with an increase of Cq-values in RT-qPCR.
According to the manufacturer’s instructions of the Zymo, Isohelix, Norgen and DNA Genotek devices, viral RNA should be maintained for weeks at ambient temperature. The manufacturer’s specification of the PreAnalytiX device states SARS-CoV-2 stability for up to 4 days at RT. The instructions for use of the Spectrum device do not indicate any post-collection temperature-controlled storage but claim analytic stability for up to 10 days (for reference see Table 2). In the present study, no major alteration in viral RNA copies was observed for any of the tested devices when stored at RT and 37 °C, with exception of the Zeesan device.
Xiamen Zeesan Biotech points out that samples should be kept below 37 °C and analyzed within one month. In the present setting, viral RNA was stable at RT, but a drastic RNA degradation was observed already after 24 h at 37 °C. After 96 h no positive signals were obtained for any of the spike-in concentrations by RT-qPCR analysis. However, Xiamen Zeesan Biotech also markets another saliva collection device (product no.: 401109), for which stability data for SARS-CoV-2 for up to 25 °C are shown.
Surprisingly, viral RNA in unstabilized saliva seemed to be less stable at RT than at 37 °C in one of the experimental series. This might be an effect of heterogeneity of the sample with regard to viscosity. Since saliva was less viscous at 37 °C, this might reduce heterogeneity caused by pipetting compared to RT conditions. Others quantified SARS-CoV-2 RNA in spiked saliva samples from 9 donors after a 7-day storage period at RT and reported no significant differences for the ORF1 and E viral RNA [25]. These authors also reported negative ΔCq-values of spiked saliva samples after a storage time of 7 days at RT, which was explained by potential virus replication in residual host cells within the sample or assay variability.
In another study, unchanged Cq-values for unstabilized saliva over a seven-day storage period at RT and 4 °C were reported [26]. Saliva was spiked with 300- and 60-fold higher virus load compared to the present work. It can be proposed that a degradation of SARS-CoV-2 RNA was detected in unstabilized saliva because a very low virus load was used, which is close the detection limit of the RT-qPCR assay; RNA copy numbers falling below the detection limit were thus more easily detectable.
For SARS-CoV-2 diagnostics, no virus cultivation from patient specimens is required in most cases. Therefore, virus-inactivating devices should be used preferably to minimize the risk of infection of healthcare workers. All saliva devices examined showed SARS-CoV-2 inactivation properties in cell culture experiments and reduced the viral load in the supernatant by a factor of at least 10E + 7 compared to the positive control. At the same time the SARS-CoV-2 RNA copy number was surprisingly stable in unstabilized saliva. Thus, additives for stabilization of viral RNA are not mandatory under the conditions tested in this study. The inactivation properties of the saliva collection devices are beneficial for the application in home collection based testing. In contrast, all swab-based viral transport systems spiked with SARS-CoV-2 remained infectious in cell culture experiments (Fig. 6).
The European Centre for Disease Prevention and Control (ECDC) considers saliva to be a suitable alternative specimen type to the widely used swabs derived from nose and pharynx, because saliva sampling is not only well suited for self-sampling in large population screenings, but is also non-invasive and well accepted [13]. Some studies compared the detection of SARS-CoV-2 RNA in saliva and nasopharyngeal specimen and reported similar sensitivities for samples with high viral loads [14], [24]. Even though the number of tests based on saliva are increasing strongly, nasopharyngeal swabs remain the gold standard for SARS-CoV-2 diagnosis up to now, although these specimens have to be collected by trained healthcare workers [28]. Various studies have produced partially contradictory results. One report showed that SARS-CoV-2 in saliva samples has a higher detection rate (88.09%) compared to throat swabs (45.24%) and nasopharyngeal swabs (76.19%) [29]. Other studies determined that COVID-19 tests from saliva are as sensitive as tests based on nasopharyngeal swabs [30], [31], [32], [33] while lower sensitivities were reported in other studies [26], [34]. To the best of our knowledge, the approach used here is one of the most comprehensive systematic studies, examining the impact of pre-analytical conditions such as storage and transport on SARS-CoV-2 RNA copy number stability in swab and saliva collection devices. The results demonstrate that different real life-relevant storage conditions of various sampling devices differentially affect the capability of SARS-CoV-2 detection in RT-qPCR. Some manufacturer's data on RNA stability could not be confirmed. Particularly at low virus spike-in amounts, the dispersion of the Cq values became larger, which can already have an effect on whether a test result is positive or negative. Additionally, false negative results may be caused by incorrect pre-analytical processes. The data demonstrate that pre-analytical variables, such as the choice of the specimen collection device, the transport storage and transport conditions and durations, should be determined during the development of SARS-CoV-2 RNA amplification based tests. An ISO Technical Specification on SARS-CoV-2 detection by nucleic acid amplification methods is currently under development (ISO/PRF TS 5798 Quality practice for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by nucleic acid amplification methods; https://www.iso.org/standard/81712.html).
It is hoped that this study can contribute to raising awareness that reliable laboratory diagnostics depends strongly on defined pre-analytical specimen handling. This requires adequate performance studies, examining a broad spectrum of pre-analytic conditions that may occur when using the device in different countries and highlighting the importance of strict adherence to manufacturers’ instructions, as well as thorough validation of the actual workflow.
Funding
This work was supported by the ERINHA-Advance project (funding from the European Union’s Horizon 2020 Research & Innovation Program, Grant agreement no. 824061).
CRediT authorship contribution statement
MH: performed experiments, analyzed and interpreted data, contributed to study design, writing and proof reading of the manuscript. EFH: performed experiments, analyzed and interpreted data, contributed to study design, writing and proof reading of the manuscript. SF: contributed to sample preparation and proof reading of the manuscript. AK: contributed to sample preparation and proof reading of the manuscript. ML contributed to study design, writing and proof reading of the manuscript. KZ: contributed to study design, writing and proof reading of the manuscript. All authors have approved the final article.
Declaration of Competing Interest
The authors declare no conflict of interest in relation to this study.
Acknowledgements
We thank the team of the BSL-3 laboratory at the Medical University Graz, for excellent technical assistance. Thanks to Penelope Kungl for help in proof reading of the manuscript. We acknowledge QIAGEN’s valuable support on the study protocol design, data interpretation and critical reading of the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.nbt.2022.04.001.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Raffle A.E., Pollock A.M., Harding-Edgar L. Covid-19 mass testing programmes. BMJ. 2020;370:m3262. doi: 10.1136/bmj.m3262. [DOI] [PubMed] [Google Scholar]
- 2.Peto J. Covid-19 mass testing facilities could end the epidemic rapidly. BMJ. 2020;368:m1163. doi: 10.1136/bmj.m1163. [DOI] [PubMed] [Google Scholar]
- 3.Gao F., Tao L., Ma X., Lewandowski D., Shu Z. A study of policies and guidelines for collecting, processing, and storing coronavirus disease 2019 patient biospecimens for biobanking and research. Biopreserv Biobank. 2020;18:511–516. doi: 10.1089/bio.2020.0099. [DOI] [PubMed] [Google Scholar]
- 4.WHO . Coronavirus (COVID-19) Dashboard 〈https://covid19.who.int/〉.
- 5.U.S. Department of Health & Human Services: USA. Interim guidelines for collecting and handling of clinical specimens for COVID-19 testing 〈https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html〉, [Accessed 11 October 2021].
- 6.WHO. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance. Geneva PP - Geneva: World Health Organization; 2020.
- 7.Lippi G., Simundic A.-M., Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) Clin Chem Lab Med. 2020;58:1070–1076. doi: 10.1515/cclm-2020-0285. [DOI] [PubMed] [Google Scholar]
- 8.Federman D.G., Gupta S., Stack G., Campbell S.M., Peaper D.R., Dembry L.M., et al. SARS-CoV-2 detection in setting of viral swabs scarcity: are MRSA swabs and viral swabs equivalent? PLoS One. 2020;15 doi: 10.1371/journal.pone.0237127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radbel J., Jagpal S., Roy J., Brooks A., Tischfield J., Sheldon M., et al. Detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is comparable in clinical samples preserved in saline or viral transport medium. J Mol Diagnostics. 2020;22:871–875. doi: 10.1016/j.jmoldx.2020.04.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers A.A., Baumann R.E., Borillo G.A., Kagan R.M., Batterman H.J., Galdzicka M.M., et al. Evaluation of transport media and specimen transport conditions for the detection of SARS-CoV-2 by use of real-time reverse transcription-PCR. J Clin Microbiol. 2020:58. doi: 10.1128/JCM.00708-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czumbel L.M., Kiss S., Farkas N., Mandel I., Hegyi A., Nagy Á., et al. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front Med. 2020;7:465. doi: 10.3389/fmed.2020.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrera L.A., Hidalgo-Miranda A., Reynoso-Noverón N., Meneses-García A.A., Mendoza-Vargas A., Reyes-Grajeda J.P., et al. Saliva is a reliable and accessible source for the detection of SARS-CoV-2. Int J Infect Dis. 2021;105:83–90. doi: 10.1016/j.ijid.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler-Laporte G., Lawandi A., Schiller I., Yao M., Dendukuri N., McDonald E.G., et al. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Intern Med. 2021;181:353–360. doi: 10.1001/jamainternmed.2020.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.PAXgene® (25). Saliva Collectors Handbook; 2021. 〈file:///C:/Users/FoederlHE/Downloads/HB-2926-001_HB_PX_SalivaCol_Kit_0821_WW(2).pdf〉.
- 16.DNA/RNA Shield™ Saliva Collection Kit Catalog no. R1210 Quick Protocol 〈https://files.zymoresearch.com/quick-protocol/_r1210_shield-saliva_tube.pdf〉.
- 17.GENEFIX RNA saliva collection & stabilization 〈https://isohelix.com/products/rfx/〉.
- 18.Saliva RNA Collection and Preservation Device (50) Product Insert 〈https://norgenbiotek.com/sites/default/files/resources/Saliva-RNA-Collection-and-Preservation-Devices-50-Research-Use-PIRU53800-1.pdf〉.
- 19.Spectrum Solution – SDNA-2000 〈https://spectrumsolution.com/sdna-whole-saliva-dna-collection-devices/〉.
- 20.Storage recommendations for samples collected using OMNIgene products from DNA Genotek 〈https://www.dnagenotek.com/ROW/pdf/PD-PR-01036.pdf〉.
- 21.Loibner M., Langner C., Regitnig P., Gorkiewicz G., Zatloukal K. Biosafety requirements for autopsies of patients with COVID-19: example of a BSL-3 autopsy facility designed for highly pathogenic agents. Pathobiology. 2021;88:37–45. doi: 10.1159/000513438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.2019-Novel Coronavirus (2019-nCoV) Real-time rRT-qPCR Panel 〈https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html〉.
- 23.Riddell S., Goldie S., Hill A., Eagles D., Drew T.W. The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol J. 2020;17:145. doi: 10.1186/s12985-020-01418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kojima N., Turner F., Slepnev V., Bacelar A., Deming L., Kodeboyina S., et al. Self-collected oral fluid and nasal swab specimens demonstrate comparable sensitivity to clinician-collected nasopharyngeal swab specimens for the detection of SARS-CoV-2. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandel C., Zheng J., McCready J., Serbanescu M., Racher H., Desaulnier M., et al. Detection of SARS-CoV-2 from saliva as compared to nasopharyngeal swabs in outpatients. Viruses. 2020;12:1314. doi: 10.3390/v12111314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griesemer S.B., Van Slyke G., Ehrbar D., Strle K., Yildirim T., Centurioni D.A., et al. Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing. J Clin Microbiol. 2021;59:e01418–e01420. doi: 10.1128/JCM.01418-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Summer S., Schmidt R., Herdina A.N., Krickl I., Madner J., Greiner G., et al. Detection of SARS-CoV-2 by real-time PCR under challenging pre-analytical conditions reveals independence of swab media and cooling chain. Sci Rep. 2021;11:13592. doi: 10.1038/s41598-021-93028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Control EC for DP and. Considerations for the use of saliva as sample material for COVID-19 testing; 2021. 〈https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-use-saliva-sample-material-testing.pdf〉.
- 29.Zheng S., Yu F., Fan J., Zou Q., Xie G., Yang X., et al. Saliva as a diagnostic specimen for SARS-CoV-2 by a PCR-based assay: a diagnostic validity study. SSRN Electron J. 2020 doi: 10.2139/ssrn.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.To K.K.-W., Tsang O.T.-Y., Yip C.C.-Y., Chan K.-H., Wu T.-C., Chan J.M.-C., et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bastos M.L., Perlman-Arrow S., Menzies D., Campbell J.R. The sensitivity and costs of testing for SARS-CoV-2 infection with saliva versus nasopharyngeal swabs. Ann Intern Med. 2021;174:501–510. doi: 10.7326/M20-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alizargar J., Etemadi Sh M., Aghamohammadi M., Hatefi S. Saliva samples as an alternative for novel coronavirus (COVID-19) diagnosis. J Formos Med Assoc. 2020;119:1234–1235. doi: 10.1016/j.jfma.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamal A.J., Mozafarihashjin M., Coomes E., Powis J., Li A.X., Paterson A., et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2. Clin Infect Dis. 2021;72:1064–1066. doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material