Abstract
OBJECTIVES
Coronavirus Disease 2019 (COVID-19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Serological testing for anti-SARS-CoV-2 nucleocapsid (N) antibodies (Abs) and anti-SARS-CoV-2 spike (S) Abs is performed to detect prior COVID-19 infection. It is still controversial which antibodies are the most sensitive and specific, and which can be detected earliest after infection. Here, we evaluated the results of serological tests of anti-SARS-CoV-2 N and S Abs in Japan.
METHODS
Symptomatic COVID-19 patients (n = 84) and control patients with rheumatoid arthritis (n = 93) were recruited at Tokyo National Hospital. Anti-SARS-CoV-2 N and S Abs were measured by commercial electrochemiluminescence immunoassays.
RESULTS
The fraction of patients positive for anti-SARS-CoV-2 N and S Abs was highest >14 days after symptom onset. The frequency of anti-SARS-CoV-2 S Ab positivity at this time (80.4%) tended to be slightly but not significantly lower than anti-SARS-CoV-2 N Ab positivity (84.8%). Optimized cut-off levels for anti-SARS-CoV-2 N and S Ab positivity were lower than the manufacturer's recommended cut-off levels. Using multiple linear regression analyzes with anti-SARS-CoV-2 N and S Abs, we created an Ab-index with high sensitivity.
CONCLUSION
To increase the sensitivity of serological diagnostic tests for COVID-19, it is suggested that both anti-SARS-CoV-2 N and S Abs should be measured and cut-off levels decreased.
Keywords: COVID-19, anti-SARS-CoV-2 antibody, electrochemiluminescence immunoassay, nucleocapsid, spike
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19) which emerged in December 2019 in Wuhan, China. 1 Real-time reverse transcription polymerase chain reaction detection of SARS-CoV-2 in nasopharyngeal swabs, saliva, sputum or bronchoalveolar lavage samples is the standard method of COVID-19 virological diagnosis. 2 Serological detection of anti-SARS-CoV-2 antibodies (Abs) is necessary for the detection of prior infection or evaluation of vaccine efficacy. The major structural proteins of SARS-CoV-2 include spike (S), nucleocapsid (N), envelope, and membrane proteins. 3 The S and N proteins possess immunogenic epitopes and are target antigens for serological assays4–11 which have accordingly been developed against both. It is unknown how long these antibodies persist after COVID-19 infection and is controversial which antibodies are the most sensitive, specific, or first to be detected after infection. Hence, we evaluated the results of serological tests for anti-SARS-CoV-2 N and S Abs in Japan using commercial electrochemiluminescence immunoassays (ECLIA).
Materials and Methods
Patients and sera
Symptomatic COVID-19 patients recruited at Tokyo National Hospital from April 2020 to February 2021 (n = 84) were diagnosed by real-time reverse transcription polymerase chain reaction or loop-mediated isothermal amplification methods. 12 Severity of COVID-19 was defined as follows: patients not requiring oxygen (severity 1), patients requiring oxygen (severity 2), and patients requiring mechanical ventilation or deceased due to respiratory failure (severity 3). Control patients with rheumatoid arthritis (n = 93) or healthy controls (n = 498) were also recruited at Tokyo National Hospital, not diagnosed as having COVID-19 before serum collection. No participants were vaccinated against SARS-CoV-2 before serum collection. Sera from these groups of patients were analyzed for anti-SARS-CoV-2 Abs. This study was approved by The Research Ethics Committee of Tokyo National Hospital (469), which has waived written informed consent for emerging infectious diseases. Oral informed consent was obtained from the patients with COVID-19 and written informed consent was obtained from the control patients. This study was conducted in accordance with the principles expressed in the Declaration of Helsinki.
Anti-SARS-CoV-2 Ab analyzes
IgM and IgG classes of anti-SARS-CoV-2 N Abs were detected using the ECLIA system (Elecsys Anti-SARS-CoV-2, Roche Diagnostics, Mannheim, Germany), according to the manufacturer's instructions, and cut-off indices (COI) were calculated. Samples with COI < 1.0 were considered negative for anti-SARS-CoV-2 N Abs; samples with COI ≥ 1.0 were considered positive. Results of anti-SARS-CoV-2 N Ab assays for the control patients with rheumatoid arthritis have already been reported. 13 IgM and IgG classes of anti-SARS-CoV-2 S Abs were also detected using the ECLIA system (Elecsys Anti-SARS-CoV-2 S, Roche Diagnostics), according to the manufacturer's instructions, and COI were calculated. Samples with COI < 0.8 were negative and those with COI ≥ 0.8 were positive.
Statistical analysis
Differences between characteristics of the patients with COVID-19 or control patients were analyzed by Student's t-test or Fisher's exact test using 2 × 2 contingency tables. The area under the curve (AUC) values of the receiver operating characteristic (ROC) curves for anti-SARS-CoV-2 N Abs and for anti-SARS-CoV-2 S Abs were compared with the AUC value of 0.5 by Chi-square analysis. From ROC curves, optimized cut-off levels, sensitivities, and specificities were calculated based on the highest Youden's index. Multiple linear regression analyzes of anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs were performed to create Ab-indices as new biomarkers.
Results
Characteristics of the patients
The characteristics of the patients are shown in Table 1. The mean age of the patients with COVID-19 was lower than controls and the proportion of men was higher.
Table 1.
Characteristics of COVID-19 patients and controls.
| COVID-19 PATIENTS | RA CONTROLS | P | HEALTHY CONTROLS | P | |
|---|---|---|---|---|---|
| Number | 84 | 93 | 498 | ||
| Mean age, years (SD) | 58.8 (19.5) | 73.4 (10.7) | 3.96 × 10−8 | 39.5 (11.7) | 9.39 × 10−32 |
| Male, n (%) | 56 (66.7) | 23 (24.7) | *3.13 × 10−8 | 132 (26.5) | *2.69 × 10−12 |
| Sevirity 2 or 3, n (%) | 24 (28.6) |
Numbers or average values are shown. Standard deviations or percentages are shown in parentheses. Significance of the differences were tested by Fisher's exact test using 2 × 2 contingency tables or Student's t-test. *Fisher's exact test was employed. COVID-19, coronavirus disease 2019; RA, rheumatoid arthritis; SD, standard deviation.
Anti-SARS-CoV-2 antibodies in sera from COVID-19 patients
Anti-SARS-CoV-2 N Abs were assessed in patients with COVID-19 (Table 2). The frequency of patients positive for anti-SARS-CoV-2 N Abs was low 0-6 days after symptom onset (3.7%), but reached 84.8% >14 days later. Anti-SARS-CoV-2 S Abs were also measured in patients with COVID-19. The frequency was also low 0-6 days after symptom onset (5.6%), rising to 66.7% 7-13 days thereafter. This was slightly higher than the frequency of anti-SARS-CoV-2 N Abs (56.5%), although this difference did not achieve statistical significance (P = .2936). The frequency of patients with anti-SARS-CoV-2 S Abs after day 14 (80.4%) tended to be lower than for anti-SARS-CoV-2 N Abs (84.8%), but this was again not significant (P = .7841). The frequency of COVID-19 patients with one or the other anti-N or anti-S Abs was highest after day 14 (93.5%), suggesting that measurement of both Abs should be used for diagnosis of COVID-19. The frequency of anti-SARS-CoV-2 N and S Abs in COVID-19 patients stratified by age, sex, or disease severity is shown in Supplementary Table S1, indicating no influence of these variables. Thus, the frequency of all patients positive for anti-SARS-CoV-2 N Abs or anti-SARS-CoV-2 S Abs was highest after day 14. In contrast, no SARS-CoV-2 N Abs or anti-SARS-CoV-2 S Abs were detected in control patients with rheumatoid arthritis, when the manufactureŕs recommended cut-off levels were used.
Table 2.
Frequencies of anti-SARS-CoV-2 N and S Abs in serum samples of COVID-19 patients after symptom onset.
| DAYS SINCE SYMPTOM ONSET | |||
|---|---|---|---|
| DAY 0–6 | DAY 7–13 | DAY 14– | |
| Median (days) | 3 | 10 | 15 |
| Range (days) | (0–6) | (7–13) | (14–32) |
| Anti-SARS-CoV-2 N Ab positive, n (%) | 2 (3.7) | 39 (56.5) | 39 (84.8) |
| Anti-SARS-CoV-2 S Ab positive, n (%) | 3 (5.6) | 46 (66.7) | 37 (80.4) |
| Anti-SARS-CoV-2 N Ab and anti-SARS-CoV-2 S Ab positive, n (%) | 1 (1.9) | 34 (49.3) | 33 (71.7) |
| Anti-SARS-CoV-2 N Ab or anti-SARS-CoV-2 S Ab positive, n (%) | 4 (7.4) | 51 (73.9) | 43 (93.5) |
Number of positive patients in each group is shown, with percentages in parentheses. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARS-CoV-2 N, SARS-CoV-2 nucleocapsid protein; SARS-CoV-2 S, SARS-CoV-2 spike protein; Ab, antibody.
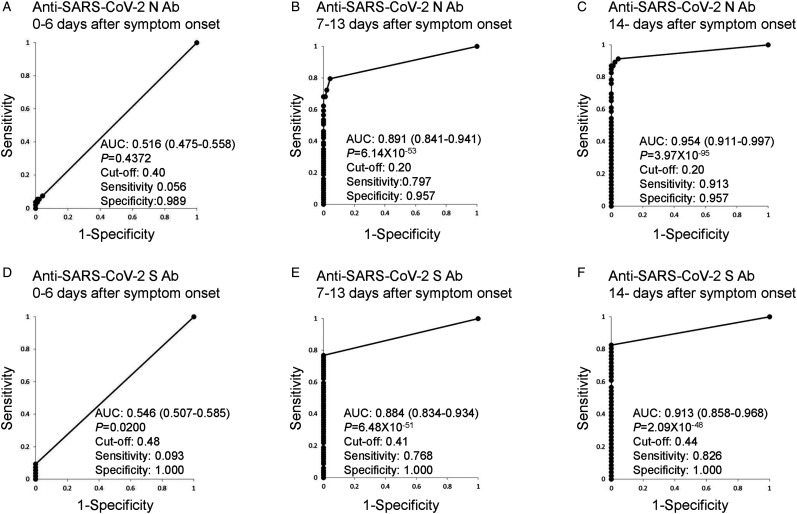
The AUC values of the ROC curves for anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs are shown in Figure 1. The AUC values on day 0-6 after symptom onset were lowest, and those after day 14 were highest (anti-SARS-CoV-2 N Abs: AUC, 0.954; 95% confidence intervals [CI] 0.911-0.997, P = 3.97 × 10−95, and for anti-SARS-CoV-2 S Abs: AUC, 0.913; 95% CI 0.858-0.968, P = 2.09 × 10−48). From these ROC curves, optimized cut-off levels, sensitivities, and specificities were calculated based on the highest Youden's index. The optimized cut-off level of anti-SARS-CoV-2 N Abs was 0.20; this is lower than the manufactureŕs recommended cut-off level of 1.0. The optimized cut-off level of anti-SARS-CoV-2 S Abs was 0.44, also lower than the recommended cut-off level of 0.8. When optimized cut-off levels were used, SARS-CoV-2 N Abs were detected in two patients with rheumatoid arthritis (2.2%), though no anti-SARS-CoV-2 S Abs were detected in control patients with rheumatoid arthritis. These data suggest that lower cut-off values should be used for these Abs for a better sensitivity.
Figure 1.
Receiver operating characteristic (ROC) curves using anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs. ROC curves for anti-SARS-CoV-2 N Ab 0-7 (A), 8-13 (B), and >14 (C) days after symptom onset. ROC curves for anti-SARS-CoV-2 S Ab 0-7 (D), 8-13 (E), and >14 (F) days after symptom onset. The area under the curve (AUC) values of the ROC curves with 95% confidence intervals and the optimized cut-off levels with specificities and sensitivities are depicted. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARS-CoV-2 N, SARS-CoV-2 nucleocapsid protein; SARS-CoV-2 S, SARS-CoV-2 spike protein; ROC, receiver operating characteristic; AUC, area under the curve.
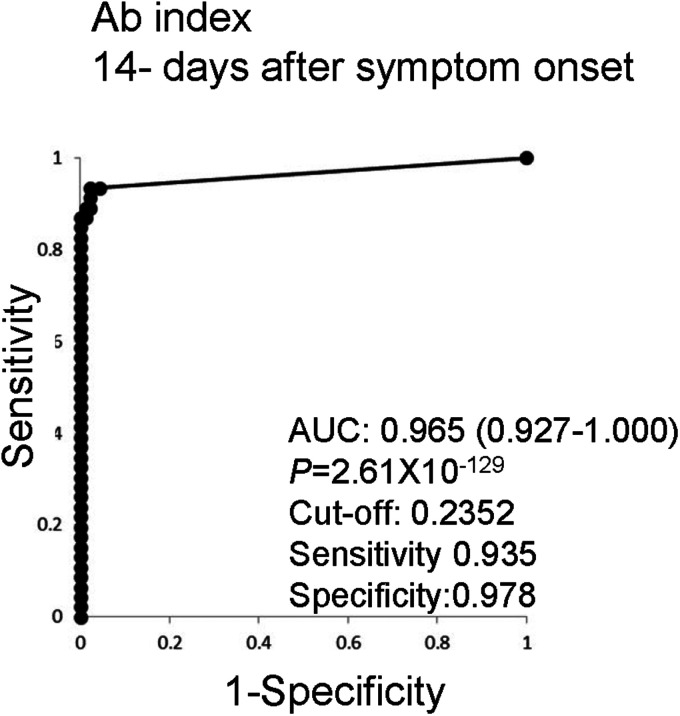
Multiple linear regression analyzes of anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs >14 days after symptom onset were performed and an Ab-index was generated as (COI of anti-SARS-CoV-2 N Abs) × 0.0120 + (COI of anti-SARS-CoV-2 S Abs) × 0.0003 + 0.2325. A ROC curve was calculated with an AUC of 0.965 (95% CI 0.927-1.000, P = 2.61 × 10−129, Figure 2), which is higher than for anti-SARS-CoV-2 N Abs or anti-SARS-CoV-2 S Abs alone. Sensitivities and specificities were calculated and the sensitivity with the highest Youden's index (0.935) was found to be higher than for either anti-SARS-CoV-2 N Abs or anti-SARS-CoV-2 S Abs. Thus, multiple linear regression analyzes with anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs resulted in the creation of an Ab-index with high sensitivity.
Figure 2.
Receiver operating characteristic (ROC) curves using multiple regression analysis with the anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs. Multiple linear regression analyzes of anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs >14 days after symptom onset were performed and an Ab-index was generated to create a complex biomarker for COVID-19. The ROC curve of the Ab-index, the area under the curve (AUC) value of the ROC curve with 95% confidence intervals, and the optimized cut-off level with the specificity and sensitivity are depicted. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SARS-CoV-2 N, SARS-CoV-2 nucleocapsid protein; SARS-CoV-2 S, SARS-CoV-2 spike protein; ROC, receiver operating characteristic; AUC, area under the curve.
Anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S >14 days after symptom onset were also compared with the results of healthy controls (Supplementary Figure S1A and S1B). Similar results were obtained to those from controls with rheumatoid arthritis. The Ab-index was validated in the comparison with healthy controls (Supplementary Figure S1C); the sensitivity of the Ab-index was still high.
Discussion
In the present study, the diagnostic performance of anti-SARS-CoV-2 N Ab and anti-SARS-CoV-2 S Ab according to their detection by Elecsys was investigated. It was found that the frequency of COVID-19 patients with anti-SARS-CoV-2 S Abs during the 7-13 days after symptom onset was slightly higher than for anti-SARS-CoV-2 N Abs. However, after day 14, the frequency of anti-SARS-CoV-2 S Abs was lower than that of anti-SARS-CoV-2 N Abs (Table 2). These data suggest earlier production, higher specificity, and lower sensitivity of anti-SARS-CoV-2 S relative to N Abs. However, the sensitivity of anti-SARS-CoV-2 S Abs was higher, although the levels of these Abs were increased earlier in a previous study. 14 It was reported that anti-SARS-CoV-2 S Abs were more specific due to the lower cross-reactivity with S proteins of other coronaviruses,15–18 supporting the results obtained in our study (Figure 1).
The frequency of patients with anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs was not sufficiently high and the rate of “anti-SARS-CoV-2 N Abs or anti-SARS-CoV-2 S Abs” positivity after day 14 was higher than for either of these Abs alone (Table 2). The sensitivities of these Abs were relatively lower after day 14 compared with the Ab-index (Figures 1 and 2). These data suggest that both anti-SARS-CoV-2 N and S Abs should be measured in order to increase the sensitivity of serological diagnostic methods for COVID-19. On the other hand, specificities of anti-SARS-CoV-2 N Abs or anti-SARS-CoV-2 S Abs are high enough and the false positive rates of these Abs low. In the ROC curve analyzes in our study, the optimized cut-off levels of anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs were lower compared than the manufacturer's recommended cut-off levels (Figure 1). Although the condition of the setting of manufacturer's recommended cut-off levels were not reported, they were higher than our optimized cut-off levels, suggesting that these conditions was set in order to decrease false positive rates. We therefore suggest to decrease the cut-off levels of anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs for improvement of the serological diagnostic tests of COVID-19. Anti-SARS-CoV-2 N (n = 7, 15.2%) or S (n = 9, 19.6%) Abs were not detected in some COVID-19 patients after day 14 (Table 2). These results suggested the limitation of the serological analyzes for the discrimination of prior infection.
The vaccination against SARS-CoV-2 produces anti-SARS-CoV-2 S Abs in sera and anti-SARS-CoV-2 S Abs cannot be used for the discrimination of prior infection in vaccinated subjects. However, both anti-SARS-CoV-2 N and S Abs can be measured in unvaccinated subjects. Anti-SARS-CoV-2 N Abs can be used as an indicator of prior infection in vaccinated subjects, though they were not detected in 15.2% of COVID-19 patients after day 14 (Table 2).
It was reported that serum IgM levels of anti-SARS-CoV-2 N and S Abs decreased after day 18, though IgG levels were maintained.19,20 In the method used for detection in the present study, serum IgM and IgG levels were measured for anti-SARS-CoV-2 N Abs or anti-SARS-CoV-2 S Abs. Therefore, these characteristics of IgM and IgG separately could not be evaluated here. Because the sample size of this study is modest, further large-scale studies are necessary to validate the results presented here. When rheumatoid arthritis patients were vaccinated, antibody levels were increased. However, it was known that the treatment with some disease modifying anti-rheumatic drugs decreased the antibody levels.21,22 When anti-SARS-CoV-2 Abs in COVID-19 patients were compared with healthy controls, similar results were obtained (Supplementary Figure S1). Thus, the antibody levels could be compared with the controls with rheumatoid arthritis and the ROC curve analyzes could be performed. Antibody profiles in asymptomatic patients with COVID-19 were not analyzed in this study. These are the limitations of the present study. Antibody profiles should be measured in future studies with this in mind.
Conclusion
This study characterized the diagnostic performance of anti-SARS-CoV-2 N Abs and anti-SARS-CoV-2 S Abs in patients with COVID-19 in Japan. To increase the sensitivity of serological diagnostic tests for COVID-19, it is suggested to measure both anti-SARS-CoV-2 N Abs or anti-SARS-CoV-2 S Abs and to decrease the cut-off levels for assigning positivity for these Abs. These amendments could extend the application of serological diagnostic tests for COVID-19.
Supplemental Material
Supplemental material, sj-rtf-1-cra-10.1177_11795484221075492 for Detection of Anti-SARS-CoV-2 Nucleocapsid and Spike Antibodies in Patients with Coronavirus Disease 2019 in Japan by Hiroshi Furukawa, Shomi Oka, Takashi Higuchi, Miho Yamaguchi, Shota Uchiyama, Tomohiro Koiwa, Moriyuki Nakama, Masaaki Minegishi, Hideaki Nagai and Shigeto Tohma in Clinical Medicine Insights: Circulatory, Respiratory and Pulmonary Medicine
Supplemental material, sj-tiff-2-cra-10.1177_11795484221075492 for Detection of Anti-SARS-CoV-2 Nucleocapsid and Spike Antibodies in Patients with Coronavirus Disease 2019 in Japan by Hiroshi Furukawa, Shomi Oka, Takashi Higuchi, Miho Yamaguchi, Shota Uchiyama, Tomohiro Koiwa, Moriyuki Nakama, Masaaki Minegishi, Hideaki Nagai and Shigeto Tohma in Clinical Medicine Insights: Circulatory, Respiratory and Pulmonary Medicine
Abbreviations
- CI
confidence interval
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- N
nucleocapsid
- S
spike
- COVID-19
coronavirus disease 2019
- SD
standard deviation
- ECLIA
electrochemiluminescence immunoassay
- Ab
antibody
- COI
cut-off index
- AUC
area under the curve
- ROC
receiver operating characteristic.
Footnotes
Author Contributions: Conceived and designed the experiments: HF and ST, Performed the experiments: SO, TH, and HF, Analyzed the data: HF and ST, Contributed reagents/materials/analysis tools: HF, SO, TH, MY, SU, TK, HN, and ST. Wrote the manuscript: HF and ST.
Conflict of Interest: HF was supported by research grants from Bristol-Myers Squibb Co. ST was supported by research grants from 9 pharmaceutical companies: Abbott Japan Co., Ltd., Astellas Pharma Inc., Chugai Pharmaceutical Co., Ltd., Eisai Co., Ltd., Mitsubishi Tanabe Pharma Corporation, Merck Sharp and Dohme Inc., Pfizer Japan Inc., Takeda Pharmaceutical Company Limited, Teijin Pharma Limited. The other authors declare no financial or commercial conflict of interest.
Funding: This work was supported by Grants-in-Aid for Clinical Research from the National Hospital Organization, and research grants from the following pharmaceutical companies: Bristol-Myers Squibb Co., Abbott Japan Co., Ltd, Astellas Pharma Inc., Chugai Pharmaceutical Co., Ltd, Eisai Co., Ltd, Mitsubishi Tanabe Pharma Corporation, Merck Sharp and Dohme Inc., Pfizer Japan Inc., Takeda Pharmaceutical Company Limited, Teijin Pharma Limited. The funders had no role in study design, data collection and analysis, decision to publish, or preparing the manuscript.
ORCID iD: Hiroshi Furukawa https://orcid.org/0000-0003-1353-8056
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pascarella G, Strumia A, Piliego C, et al. COVID-19 diagnosis and management: a comprehensive review. J Intern Med. 2020;288(2):192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Che XY, Qiu LW, Pan YX, et al. Sensitive and specific monoclonal antibody-based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J Clin Microbiol. 2004;42(6):2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo PC, Lau SK, Wong BH, et al. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J Clin Microbiol. 2005;43(7):3054–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer B, Drosten C, Müller MA. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rikhtegaran Tehrani Z, Saadat S, Saleh E, et al. Performance of nucleocapsid and spike-based SARS-CoV-2 serologic assays. PLoS One. 2020;15(11):e0237828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Algaissi A, Alfaleh MA, Hala S, et al. SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients. Sci Rep. 2020;10(1):16561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariën J, Ceulemans A, Michiels J, et al. Evaluating SARS-CoV-2 spike and nucleocapsid proteins as targets for antibody detection in severe and mild COVID-19 cases using a Luminex bead-based assay. J Virol Methods. 2021;288(114025):114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama R, Kurano M, Morita Y, et al. Validation of a new automated chemiluminescent anti-SARS-CoV-2 IgM and IgG antibody assay system detecting both N and S proteins in Japan. PLoS One. 2021;16(3):e0247711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noda K, Matsuda K, Yagishita S, et al. A novel highly quantitative and reproducible assay for the detection of anti-SARS-CoV-2 IgG and IgM antibodies. Sci Rep. 2021;11(1):5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitagawa Y, Orihara Y, Kawamura R, et al. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol. 2020;129(104446):104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka S, Higuchi T, Furukawa H, et al. Detection of anti-SARS-CoV-2 antibodies in patients with rheumatoid arthritis. Res Sq. 2020. DOI: 10.21203/rs.3.rs-112294/v1 [DOI] [Google Scholar]
- 14.Liu W, Liu L, Kou G, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6):00461–00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kontou PI, Braliou GG, Dimou NL, Nikolopoulos G, Bagos PG. Antibody tests in detecting SARS-CoV-2 infection: a meta-analysis. Diagnostics (Basel). 2020;10(5):319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng M, Song L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell Mol Immunol. 2020;17(5):536–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang P, Gao Q, Wang T, et al. Development and evaluation of a serological test for diagnosis of COVID-19 with selected recombinant spike proteins. Eur J Clin Microbiol Infect Dis. 2021;40(5):921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano Y, Kurano M, Morita Y, et al. Time course of the sensitivity and specificity of anti-SARS-CoV-2 IgM and IgG antibodies for symptomatic COVID-19 in Japan. Sci Rep. 2021;11(1):2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Migita K, Akeda Y, Akazawa M, et al. Effect of Abatacept on the immunogenicity of 23-valent pneumococcal polysaccharide vaccination (PPSV23) in rheumatoid arthritis patients. Arthritis Res Ther. 2015;17(357):357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Migita K, Akeda Y, Akazawa M, et al. Opsonic and antibody responses to pneumococcal polysaccharide in rheumatoid arthritis patients receiving golimumab plus methotrexate. Medicine (Baltimore). 2015;94(52):e2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-rtf-1-cra-10.1177_11795484221075492 for Detection of Anti-SARS-CoV-2 Nucleocapsid and Spike Antibodies in Patients with Coronavirus Disease 2019 in Japan by Hiroshi Furukawa, Shomi Oka, Takashi Higuchi, Miho Yamaguchi, Shota Uchiyama, Tomohiro Koiwa, Moriyuki Nakama, Masaaki Minegishi, Hideaki Nagai and Shigeto Tohma in Clinical Medicine Insights: Circulatory, Respiratory and Pulmonary Medicine
Supplemental material, sj-tiff-2-cra-10.1177_11795484221075492 for Detection of Anti-SARS-CoV-2 Nucleocapsid and Spike Antibodies in Patients with Coronavirus Disease 2019 in Japan by Hiroshi Furukawa, Shomi Oka, Takashi Higuchi, Miho Yamaguchi, Shota Uchiyama, Tomohiro Koiwa, Moriyuki Nakama, Masaaki Minegishi, Hideaki Nagai and Shigeto Tohma in Clinical Medicine Insights: Circulatory, Respiratory and Pulmonary Medicine