Abstract
Human papillomavirus (HPV) is a significant risk factor for head and neck squamous cell carcinoma (HNSCC). HPV+ HNSCC patients have a higher survival rate, which may be related to its unique tumor microenvironment. Exosomes are emerging as a communication tool between tumor cells and the tumor microenvironment, including cancer‐associated fibroblasts (CAFs). In this study, 111 clinical samples tissues and public sequencing data were analyzed. Our study found fewer CAFs infiltrated in HPV+ HNSCC, and poor CAF infiltration level was associated with a good prognosis. HPV+ HNSCC cell‐derived exosomes can significantly reduce the phenotypic transformation of fibroblasts. miR‐9‐5p, as a miRNA enriched in HPV+ HNSCC cell‐derived exosomes, can be transferred to fibroblasts. miR‐9‐5p mimic transfection decreased the expression of NOX4 and the level of intracellular reactive oxygen species (ROS), which inhibited the transforming growth factor beta 1(TGF‐β1)‐induced increase of αSMA levels. Therefore, these results indicated that HPV+ HNSCC‐derived exosomal miR‐9‐5p inhibits TGF‐β signaling‐mediated fibroblast phenotypic transformation through NOX4, which is related to the excellent prognosis of HPV patients.
Keywords: cancer‐associated fibroblasts, exosome, head and neck squamous cell carcinoma, HPV, miR‐9‐5p
In conclusion, we first demonstrated that CAF infiltration is reduced in HPV+ HNSCC, which may be related to the inhibition of TGF‐β1‐mediated fibroblast phenotypic transformation by HPV+ HNSCC‐derived miR‐9‐rich exosomes. miR‐9 reduced ROS levels in fibroblasts by targeting NOX4, which is necessary for TGF‐β1 signaling transduction.
![]()
Abbreviations
- CAFs
cancer‐associated fibroblasts
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DCFH‐DA
dichlorofluorescein diacetate
- EV
extracellular vesicle
- FBS
fetal bovine serum
- GO
gene ontology
- HGF‐1
human gingival fibroblast cell line
- HNSCC
head and neck squamous cell carcinoma
- HPV
human papillomavirus
- IF
immunofluorescence
- IHC
immunohistochemistry
- MFI
mean fluorescence intensity
- MUT
mutation‐type
- NF
normal fibroblast
- NTA
nanoparticles tracking analysis
- PCA
principal component analysis
- ROS
reactive oxygen species
- TGF‐β1
transforming growth factor beta 1
- TME
tumor microenvironment
- UMAP
uniform manifold approximation and projection
- UTR
untranslated region
1. INTRODUCTION
Head and neck squamous cell carcinoma is the seventh most common cancer worldwide, and nearly 890 000 cases are newly diagnosed annually, making it become a significant social health burden. 1 , 2 Human papillomavirus (HPV) is an essential risk factor for HNSCC. 3 HPV+ HNSCC has unique tumor biology and clinical characteristics from HPV− HNSCC, which may be related to the difference in the tumor microenvironment (TME). 4 , 5
In the past decade, the role of the TME in tumor progression has attracted significant attention. 6 Cancer‐associated fibroblasts (CAFs), one of the main components of the TME, establish dangerous relationships with cancer cells and other components of TME and form an environment supporting tumor victory in many types of cancers. Previous studies have proved that CAFs are an essential part of the tumor matrix and play a key role in promoting the progression of HNSCC. 7 CAFs can encourage the development of HNSCC by secreting inflammatory factors, remodeling the extracellular matrix, and stimulating angiogenesis. 8 , 9 , 10 , 11
Exosome, a kind of EV, is a disk‐shaped vesicle with a diameter of 50‐200 nm. 12 , 13 , 14 Exosomes can transport their contents, such as mRNAs, miRNAs, and proteins, to other cells, which plays a crucial role in the communication among various cells. 15 Recently, several studies have demonstrated that exosomes derived from tumor cells play an essential role in cell communication. 16 miRNA is a short single chain non‐encoding RNA that can play a post‐transcriptional regulatory role, resulting in translation inhibition or gene silencing. 17 miRNA plays various roles in tumors, such as regulating cell proliferation, affecting tissue differentiation, promoting angiogenesis, metabolism, and so on. 18 , 19 , 20 Since the discovery of miRNA, miRNA‐related research and medical methods have thrived. 21 , 22 In HNSCC, miRNA has been widely studied, but there are still gaps in some fields. 23 , 24
In this study, we demonstrated for the first time that HPV+ HNSCC‐derived exosomal miR‐9‐5p mediated fibroblast phenotypic transformation by affecting the level of ROS and inhibited transforming growth factor beta 1 (TGF‐β1) signaling in fibroblasts. This provides a new direction for the study of HPV‐related tumor microenvironments.
2. MATERIALS AND METHODS
2.1. Patient samples
In total, 111 HNSCC samples were collected from the Third Affiliated Hospital of Harbin Medical University (Harbin, China) from 2008‐2018. All participants provided written informed consent regarding this study, and ethical approval for the study was obtained from the Ethics Committee of Harbin Medical University.
2.2. Immunohistochemical staining
Tumor tissues were fixed, embedded in paraffin, and sectioned at a thickness of 4 μm. After deparaffinization and rehydration, sections were covered with Tris‐EDTA (pH 9.0) buffer for antigen retrieval and blocked with 3% H2O2 for 20 min. Next, sections were blocked with 5% goat serum for 30 min and incubated with αSMA antibody (1:1500) (Proteintech, MA, USA) and p16INK4a (ZSGB‐Bio, China) at 4°C overnight, followed by incubation with horseradish peroxidase‐labeling secondary antibody at 37°C for 30 min. Blots were visualized with 3,3′‐diaminobenzidine (DAB) solution and counterstained with hematoxylin. Images were taken under an Olympus BX51 microscope with ×100 and ×200 objectives. Stained slides were reviewed and scored using ImageJ software.
2.3. Cell culture
The human HNSCC cell line SCC090 was purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences. SCC47, SAS cell lines were kindly provided by Dr. Henning Willers (Harvard University, Boston, USA). CAL27 was a kind gift from Professor Songbin Fu (Harbin Medical University, Harbin, China). Human gingival fibroblast cell line HGF‐1 was purchased from Otwo Biotech (Shenzhen, China). HEK293T was obtained from the authors’ stock at Harbin Medical University. All cells were cultured in DMEM supplemented with 10% exosome‐free FBS and maintained at 37°C in a humidified atmosphere of 5% CO2. To obtain EV‐depleted FBS, FBS was ultracentrifuged at 4℃, 120 000 g for 20 h and the serum far away from the precipitation in the upper layer was filtered through a 0.22‐µm filter and used for subsequent cell culture. 25 Human Recombinant TGF‐β1 (PeproTech, Inc. USA) was reconstituted follow the manufacturer’s instructions and administered to cells at a dose of 5 ng/ml. GKT137831 (Meilun, China) was reconstituted in DMSO and administered at 20 µM. GW4869 (Meilun, China) was reconstituted in DMSO and added to the cell culture medium at 10 µM.
2.4. Exosome isolation and identification
Ultracentrifugation was used for exosome isolation. Cell culture medium was collected and centrifuged at 300 g for 10 min, then 2000 g for 10 min to remove cells and debris. Next the supernatant was centrifuged on an ultracentrifuge (Beckman XPN‐100) using a SW 32 Ti rotor at 10 000 g for another 40 min. The supernatant was then ultracentrifuged at 120 000 g for 70 min to collect pellet exosomes. To purify the exosomes again, the pellet exosomes were resuspended using PBS and then centrifuged again at 120 000 g for 70 min. All steps above were performed at 4°C. Finally, exosomes dissolved in 50 μl PBS buffer were filtered through a 0.22‐µm filter.
The morphology of the exosomes was then detected. Resuspended exosomes were dropped into a carbon‐coated copper grid, and stained with 2% uranyl acetate. Exosomes were observed using a Hitachi transmission electron microscopy (TEM) system. The size and concentration of exosomes were measured by NTA at VivaCell Biosciences with ZetaView PMX 110. Western blot analysis was then performed to detect exosome markers, including CD9, CD63, and TSG101, as well as the endoplasmic reticulum‐derived calnexin protein. 12
2.5. Exosome labeling and tracking
In accordance with the manufacturer's instructions, exosomes isolated from the culture medium of HNSCC cells were collected and stained with PKH26 red fluorescent membrane linker dye (Umibio, China). Labeled exosomes were collected and added to the fibroblasts growing on glass slides for exosome uptake analysis. After incubation at 37°C for 2 h, the cytoplasm of fibroblasts was stained with vimentin (1:100) (Proteintech, MA, USA) and Coralite 488‐conjugated anti‐rabbit IgG (H+L) (1:200) (Proteintech, MA, USA) as secondary antibodies. The nucleus was stained with DAPI. Then treated fibroblasts on a glass slide were examined under an Olympus BX51 microscope at the appropriate excitation wavelength for the fluorophore.
2.6. Western blotting
Cells or exosomes were collected and denatured. Proteins were separated by HEPES‐Tris gel and transferred onto polyvinylidene difluoride membranes. After blocking with 5% skimmed milk for 90 min, membranes were probed with CD9 antibody (1:500) (Proteintech, MA, USA), CD63 antibody (1:200) (Proteintech, MA, USA), TSG101 antibody (1:1000) (Proteintech, MA, USA), calnexin antibody (1:1000) (ABclonal, China), αSMA antibody (1:5000) (Proteintech, MA, USA), NOX4 antibody (1:1000) (Proteintech, MA, USA), Smad2 + Smad3 antibody (1:1000) (Abcam, USA), Phospho‐Smad2‐S465/467 + Smad3‐S423/425 antibody (1:1000) (ABclonal, China) and β‐actin antibody (1:5000) (ZSGB‐Bio, China) at 4°C overnight, followed by incubation with horseradish peroxidase‐linked secondary antibodies at room temperature for 1.5 h. Protein bands were visualized using the BeyoECL Plus kit (Beyotime, China). Finally, the membranes were exposed to a gel imaging system ChemiDox XRS + System. Gray values of protein blots were calculated using ImageJ software v.1.8.0 software. β‐actin served as the internal control.
2.7. Immunofluorescence
HGF‐1 cells were seeded onto 12‐well plates containing 15‐mm diameter glass coverslips. HGF‐1 cells were transfected with 100 nM miR‐9‐5p mimic or mimic NC. Then cells were incubated with 5 ng/ml TGF‐β1 for 48 h. Then treated cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.25% Triton X‐100 in PBS for 20 min at room temperature. Cells were washed with PBS, blocked with 1% BSA in PBS for 1 h, and incubated overnight at 4°C with αSMA antibody (1:200) (Proteintech, MA, USA) and NOX4 antibody (1:100) (Proteintech, MA, USA). Nuclear staining was performed with DAPI (Beyotime, China). Then, Coralite 594‐conjugated anti‐mouse IgG (H+ L) (1:200) (Proteintech, MA, USA) and Coralite 488‐conjugated anti‐rabbit IgG (H+L) (1:200) (Proteintech, MA, USA) were used as secondary antibodies. The coverslips were mounted on slides using an anti‐fluorescence quenching sealing agent (Beyotime, China). An Olympus BX51 microscope was used to visualize cell fluorescence at the appropriate excitation wavelength for the fluorophore.
2.8. ELISA assay
The cell culture medium for the HNSCC cell lines was collected. Secretion levels of human IL‐6, TGF‐β1, and PDGF‐BB were measured using ELISA kits (ABclonal, China).
2.9. Real‐time PCR
TRIzol reagent was used to extract total RNA from cells (TaKaRa, JPN). Exosomal RNA was extracted using TRIzol LS (Ambion, USA). PrimeScript™ RT Master Mix (Perfect Real Time) (TaKaRa, JPN) was used to perform reverse transcription. Real‐time PCR was performed with TB Green® Premix Ex Taq™ II (Tli RNase H Plus) (TaKaRa, JPN). TBP and U6 served as internal controls. Expression changes were determined using the 2−ΔΔCt method in comparison with the negative control.
2.10. Dual‐luciferase reporter gene activity assay
The NOX4 3ʹ untranslated region (UTR) WT/mutation‐type (MUT) constructs were cloned into the pmirGLO vector, and HEK293T cells were transfected with pmirGLO‐NOX4‐WT/MUT along with the miR‐9‐5p mimic or mimic NC. The luciferase activity of the cells was determined 48 h later using a dual‐luciferase activity assay kit (Beyotime, China).
2.11. ROS assay
Intracellular ROS were measured using flow cytometry analysis of dichlorofluorescein diacetate (DCFH‐DA) fluorescence. Cells were trypsinized, centrifuged, and incubated for 25 min at 37°C with 10 μM DCFH‐DA to load cells with the dye. Then cells were resuspended in cold PBS and kept on ice, protected from light. Fluorescence was measured on the FL‐1 channel of a BD Accuri C6, and the MFI was calculated using FlowJo software.
2.12. Transfection of miRNA mimics and inhibitors
Cells at 70% confluency were transfected with 100 nM of either miRNA mimic of miR‐9‐5p (micrON hsa‐miR‐9‐5p mimic) (Ribobio, China) or miR‐9 inhibitor (micrOFF hsa‐miR‐9‐5p inhibitor) (Ribobio, China) using Lipofectamine 3000 (Invitrogen, USA). In all experiments, an equal concentration of a non‐targeting sequence miRNA mimic negative control (micrON mimic NC #22) (Ribobio, China) or miRNA inhibitor negative control (micrOFF inhibitor NC #22) (Ribobio, China) was used.
2.13. Bioinformatic analysis
RNA HT‐Seq counts data and miRNA profiling data on 494 HNSCC tissue samples and the corresponding clinical information were downloaded from TCGA (https://cancergenome.nih.gov). TPM normalization was used to remove technical biases in sequenced data. In total, 70 patients who were HPV positive and 424 patients who were HPV negative were divided by their HPV status in accordance with previous studies. 26 One additional, independent HNSCC data set (GSE74927) containing gene expression profiles was retrieved from the NCBI GEO platform (https://www.ncbi.nlm.nih.gov/gds/), including 18 HPV‐positive and 18 HPV‐negative samples. Gene expression data from 3 pairs of CAFs and normal fibroblasts from the same individual were retrieved from GSE83314. CAF infiltration levels and scores were analyzed using the EPIC R package (https://github.com/GfellerLab/EPIC) and MCPcount R package (https://github.com/ebecht/MCPcounter). The expression of ACTA2 and has‐miR‐9‐5p in HNSCC patients was compared using the DEseq2 R package. In total, 489 samples were divided into miR‐9‐5p high expression group (n = 144) and low expression group (n = 345) in accordance with the cutoff of miR‐9‐5p expression.
The online microRNA databases and target prediction tools miRDB (http://mirdb.org/), RNAInter (http://www.rnainter.org/), and miRwalk (http://mirwalk.umm.uni‐heidelberg.de/) were used to identify potential miR‐9‐5p targets. The intersection of target genes predicted by 3 databases was selected for functional annotation clustering in the bioinformatics tool DAVID v.6.8 (http://david.abcc.ncifcrf.gov/) using the GO database. Enrichment score was reported as the minus log transformation of the geometric mean of P‐values. Terms with P‐values < .05 were considered significant.
2.14. Single‐cell RNA‐seq data and analysis
HNSCC single‐cell RNA‐seq data (GSE103322) was download from the NCBI GEO platform. The gene expression matrix was converted to Seurat objects using the Seurat R package (v.3.0.2) (https://github.com/topics/seurat). HNSCC cells were robustly grouped by PCA and UMAP was applied for visualization of the data. Then 9 clusters were separately grouped and annotated based on the cell signature markers. 27 Survival analyses of the top 50 differentially expressed genes of CAFs were processed using GEPIA2 (http://gepia2.cancer‐pku.cn/#general).
2.15. Statistical analysis
All statistical analyses, survival analyses, and correlation analyses in this study were conducted using GraphPad Prism 7 software (GraphPad Software, Inc.), and data are presented as means ± SD. The significant difference between 2 groups or more was determined using Student t test and 1‐way ANOVA. Pearson chi‐square tests were performed to assess the statistical correlation between 2 variables. A P‐value < .05 was considered as statistically significant.
3. RESULTS
3.1. CAFs differentially infiltrated in HPV+ and HPV− HNSCC and prognostic significance
To examine the infiltration characteristics of CAFs in HPV‐related HNSCC, 71 HNSCC patient tissues (34 HPV+ tissues and 37 HPV− tissues) with complete prognostic information were chosen for this study. Patient information is summarized in Table S1. All HNSCC tissues were stained with αSMA, the most widely accepted CAF marker. In accordance with CAF density analyzed by ImageJ software, tissues from patients with HNSCC were scored for CAF infiltration. HPV+ HNSCC were infiltrated with less CAFs compared with HPV− HNSCC, as shown in Figure 1A,B. In addition, HPV+ HNSCC patients and CAF‐low patients had higher survival rates (Figure 1C,D). To further explore the influence of CAFs on survival rates of patients with HNSCC, single‐cell sequencing data from HNSCC samples were analyzed, and the infiltration of 9 types of cells was clustered. Top differentially expressed genes for CAFs compared with other cell types were distinguished, in which low expression of 5 genes significantly correlated with the better prognosis of patients with HNSCC (Figure S1A‐C). Another 40 HNSCC tissue samples were divided into HPV+ and HPV− groups based on the expression of p16 protein. CAF infiltration level was lower in the HPV+ HNSCC group compared with the HPV− HNSCC group, as expected (Table S2, Figure S1D,E).
FIGURE 1.
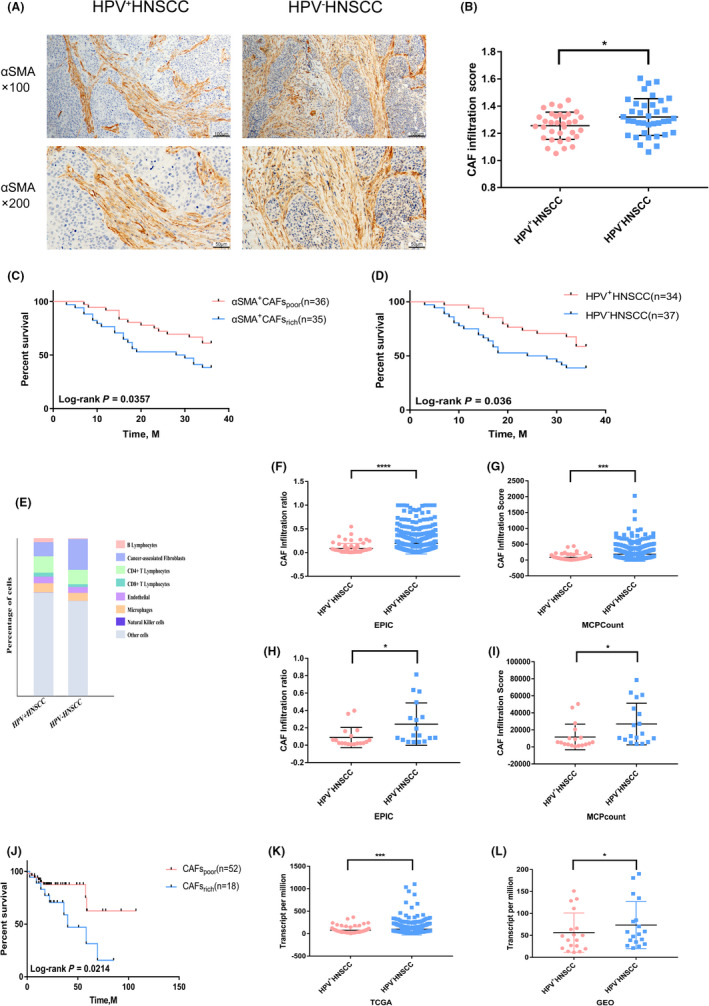
Differential infiltration and prognostic significance of CAFs in HPV+ and HPV− HNSCC. A, Representative immunohistochemistry (IHC) images of αSMA+ CAFs in HPV+ and HPV− HNSCC (n = 71). B, Quantitative analysis of IHC scores of αSMA+ CAFs in HNSCC with HPV status (n = 71). C, D, Survival analysis of αSMA+ CAFs and HPV in 71 HNSCC patients. E, Average fractions of various cells in HPV+/− HNSCC in TCGA data sets evaluated by EPIC. F, Infiltration ratio of CAFs in HPV+/− HNSCC in TCGA data sets evaluated by EPIC. G, Infiltration score of CAFs in HPV+/− HNSCC in TCGA data sets evaluated by MCPcount. H, Infiltration ratio of CAFs in HPV+/− HNSCC in GSE74927 evaluated by EPIC. I, Infiltration score of CAFs in HPV+/− HNSCC in GSE74927 evaluated by MCP count. J, Survival analysis of CAFs evaluated by EPIC in HPV+ HNSCC in TCGA data sets. K, L, Differential expression of ACTA2 analyzed by DEseq2 in HPV+/− HNSCC in TCGA data sets and GSE74927. *P < .05; ***P < .001; ****P < .0001
Next, we collected HNSCC sequencing data from the TCGA database and GEO database (GSE73917) for further analysis. The infiltration of CAFs in HPV+ HNSCC samples was lower than that in HPV− HNSCC, evaluated by 2 R packages with different principles, consistent with our immunohistochemical results (Figure 1E‐I). In addition, patients with lower CAF infiltration had a better prognosis in HPV+ HNSCC (Figure 1J). The relative expression levels of ACTA2, which encodes αSMA, was analyzed in HNSCC sequencing data from TCGA and GSE73917. There was a lower expression level of ACTA2 in HPV+ HNSCC than in HPV− HNSCC (Figure 1K,L). In addition, the expression of ACTA2 in CAFs isolated from HNSCC was significantly higher than that of NF in the corresponding normal tissues (Figure S1F). These data implied that the HPV infection plays a crucial role in influencing CAF infiltration in HNSCC. Moreover, CAFs levels were closely related to the prognosis of patients with HNSCC.
3.2. HPV influenced fibroblast phenotypic transformation in the tumor microenvironment of HNSCC
An in vitro co‐culture system was used to examine the role of HNSCC cells in fibroblast phenotypic transformation. After co‐culture with the HNSCC cell line for 48 h, immunofluorescence showed that αSMA levels were increased in HGF‐1. Next, we further detected αSMA levels in HGF‐1 after co‐culture using western blotting. The results showed that the expression of αSMA after co‐culture with HPV+ HNSCC cells was lower than that of HPV− HNSCC cells. What is more, HPV+ HNSCC cells induced lower ACTA2 expression at the mRNA level than HPV− HNSCC cells after 24 h of co‐culture (Figure 2A‐E). Fibroblast phenotypic transformation was related to cytokines; therefore, we detected the levels of widely reported soluble factors secreted by HNSCC cells involved in this progress. IL‐6, PDGF‐BB, and TGF‐β1 levels were detected in HPV+ and HPV− HNSCC cell supernatants. We found IL‐6 levels to be higher in the supernatants of HPV+ cells compared with HPV− cells. However, a high level of IL‐6 did not drive fibroblast phenotypic transformation in the HPV+ cell co‐culture environment. In addition, no significant relationship was seen between the other 2 cytokines and HPV (Figure S2A‐C). In a word, HPV infection can influence fibroblast phenotypic transformation through a non‐cytokine approach.
FIGURE 2.
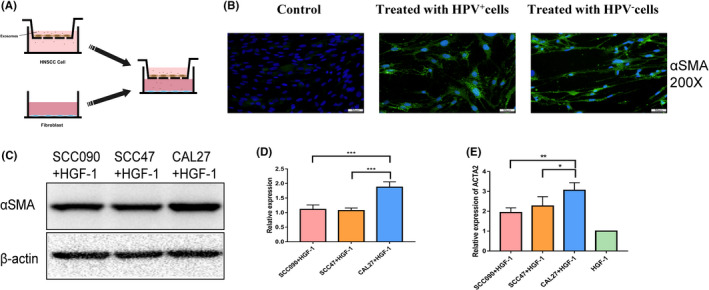
HPV influences fibroblast phenotypic transformation in the tumor microenvironment of HNSCC. A, Schema for the in vitro co‐culture system. B, Immunofluorescence (IF) analysis of αSMA in HGF‐1 for 48 h after co‐culture with HPV+/− HNSCC cells. C, D, Protein levels of αSMA were assessed and quantified by the western blot analysis. E, qRT‐PCR analysis of ACTA2 expression in HGF‐1 cells after co‐culture with HPV+/− HNSCC cells as described in (A) for 24 h. *P < .05; **P < .01; ***P < .001
3.3. HPV+ HNSCC‐derived exosomes reduce fibroblast phenotypic transformation
To examine the impact of HPV+ HNSCC cell‐derived exosomes on fibroblast phenotypic transformation, exosomes derived from HPV+ and HPV− HNSCC cells were isolated from the cell culture medium and quantified. The exosomes used for later studies were typical rounded particles ranging from 50 to 200 nm in diameter (Figure 3A). NTA showed that exosomes harvested from HPV+ HNSCC cells had significantly higher nanoparticle concentrations compared with those derived from HPV− HNSCC cells (Figure 3B). Exosome identity was confirmed by 3 exosomal markers CD9, CD63, and TSG101. These exosomal markers were all expressed, whereas the endoplasmic reticulum marker calnexin was absent in the isolated exosomes (Figure 3C). We next examined whether exosomes could be transported to fibroblasts. Exosomes, labeled with red fluorescence dyes, were co‐cultured with unstained fibroblasts. Then, the cytoplasm and the nucleus of fibroblasts were stained. Fluorescence microscope analysis showed that fibroblasts internalized exosomes over time (Figure 3D). In addition, the production of exosomes in HNSCC cells was blocked with GW4869. Then fibroblasts were cultured with exosome‐removed HNSCC cell culture supernatant and untreated HNSCC cells culture supernatant. Western blot results showed that HPV+ HNSCC cell‐derived exosomes could reduce the phenotypic transformation of fibroblasts to CAFs (Figure 3E). These results suggested that a substance enriched in HPV+ cell‐derived exosomes may play a role in the phenotypic transformation of fibroblasts.
FIGURE 3.
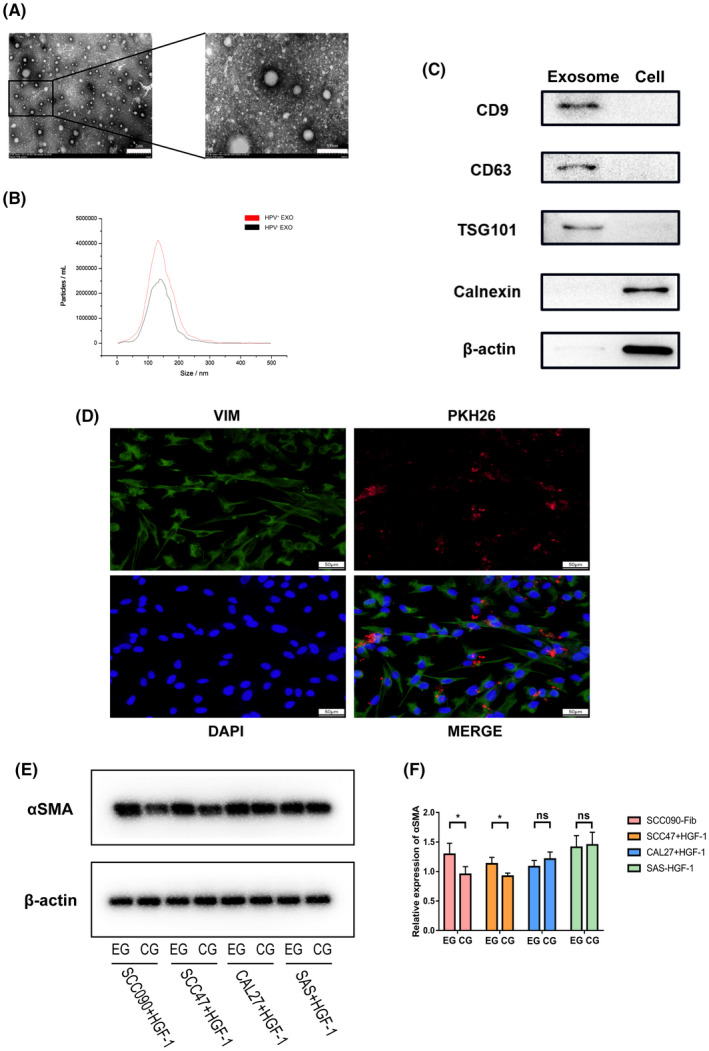
HPV+ HNSCC‐derived exosomes reduce fibroblast phenotypic transformation. A, Representative TEM micrographs of exosomes derived from HNSCC cells. B, NTA of exosomes isolated from HPV+ and HPV− HNSCC cells. C, Western blot analysis for exosomal proteins CD9, CD63, and TSG10, endoplasmic reticulum marker calnexin and β‐actin. D, Immunofluorescence (IF) staining showing the internalization of PKH26‐labeled HNSCC‐derived exosomes (red) by HGF‐1 (green). E, F Protein levels of αSMA were assessed and quantified by the western blot analysis after incubated culture supernatant of HPV+/− HNSCC cells treated with GW4869 (EG) or DMSO (CG). *P < .05
3.4. Exosomal miR‐9‐5p influences fibroblast phenotypic transformation through NOX4/TGF‐β1 signaling
Exosomes contain bioactive molecules, such as miRNAs, which are involved in intercellular communication. miR‐9‐5p has been reported to be the miRNA most related to HPV. TCGA data suggested that miR‐9 was highly expressed in HPV+ HNSCC (Figure 4A). Therefore, we detected miR‐9‐5p expression levels in HPV+ HNSCC and HPV− HNSCC cell lines. qRT‐PCR results showed that miR‐9‐5p was highly expressed in HPV+ HNSCC cells but not in HPV− HNSCC cells (Figure 4B). miR‐9‐5p expression in exosomes derived from HNSCC cells was then further confirmed. As shown in Figure 4C, miR‐9‐5p was significantly enriched in HPV+ HNSCC‐derived exosomes compared with the HPV− group. As exosomes can deliver bioactive molecules to other cells, we used SCC47‐ and CAL27‐derived exosomes to treat fibroblasts and then evaluated whether miR‐9‐5p could be delivered to fibroblasts. The result showed that fibroblasts treated with HPV+ HNSCC‐derived exosomes had a higher level of miR‐9‐5p compared with those treated with HPV− HNSCC‐derived exosomes (Figure 4D). Therefore, miR‐9‐5p was highly expressed in HPV+ HNSCC‐derived exosomes, and could be transferred to fibroblasts. The focus of the following exploration was whether an increased level of miR‐9 was correlated with improved prognosis in HNSCC. A log‐rank test was used to determine the prognostic value in accordance with miR‐9‐5p expression level. Survival analysis suggested that an increased level of miR‐9‐5p was positively correlated with an improved prognosis of HNSCC (Figure 4E). Correlation between CAFs infiltration ratio analyzed by EPIC and miR‐9‐5p expression levels was also explored. miR‐9‐5p was negatively correlated with the ratio of CAFs infiltration in HPV+ HNSCC data and total HNSCC data from TCGA (Figure 4F,G). miR‐9‐5p may be closely related to the better prognosis of HNSCC through influencing CAFs.
FIGURE 4.

Exosomal miR‐9‐5p influences fibroblast phenotypic transformation through NOX4/TGF‐β1 signaling. A, Expression profiling data of miR‐9‐5p between HPV+ and HPV− HNSCC from TCGA. B, Expression of miR‐9‐5p between HPV+ and HPV− HNSCC cells. C, Expression of miR‐9‐5p between HPV+ and HPV− HNSCC cell‐derived exosomes. D, Expression of miR‐9‐5p between HPV+ and HPV− HNSCC cell‐derived exosome‐treated HGF‐1. E, Survival analysis of miR‐9 in HNSCC from TCGA. F, G, Correlation analysis between miR‐9‐5p and CAF ratio evaluated by EPIC in HPV+ HNSCC and HNSCC from TCGA. **P < .01, ***P < .001, ****P < .0001
To explore the mechanism underlying exosomal miR‐9‐induced fibroblast phenotypic transformation, we searched for miR‐9‐5p target genes using miRDB, miRWalk, and RNAInter (Figure 5A). Then the intersection of target genes screened from the 3 databases was enriched using the GO database (Figure 5B). Finally, in the cellular response to the TGF‐β stimulus biological pathway, NOX4 turned out to be a potential target (Figure 5C). Increase in NOX4 protein expression will increase the level of ROS in cells, which is necessary for Smad2/3 phosphorylation. Dual‐luciferase assay showed that miR‐9‐5p strikingly inhibited the activity of pmirGLO‐NOX4‐WT‐3′UTR, while having a minimal effect on the negative control (Figure 5D). Then, we evaluated the expression of NOX4 of HNSCC transcriptome sequencing data in TCGA database and GEO data. NOX4 expression was lower in HPV+ NNSCC (Figure 5E,F), and NOX4 expression was correlated with ACTA2 expression (Figure 5G,H).
FIGURE 5.
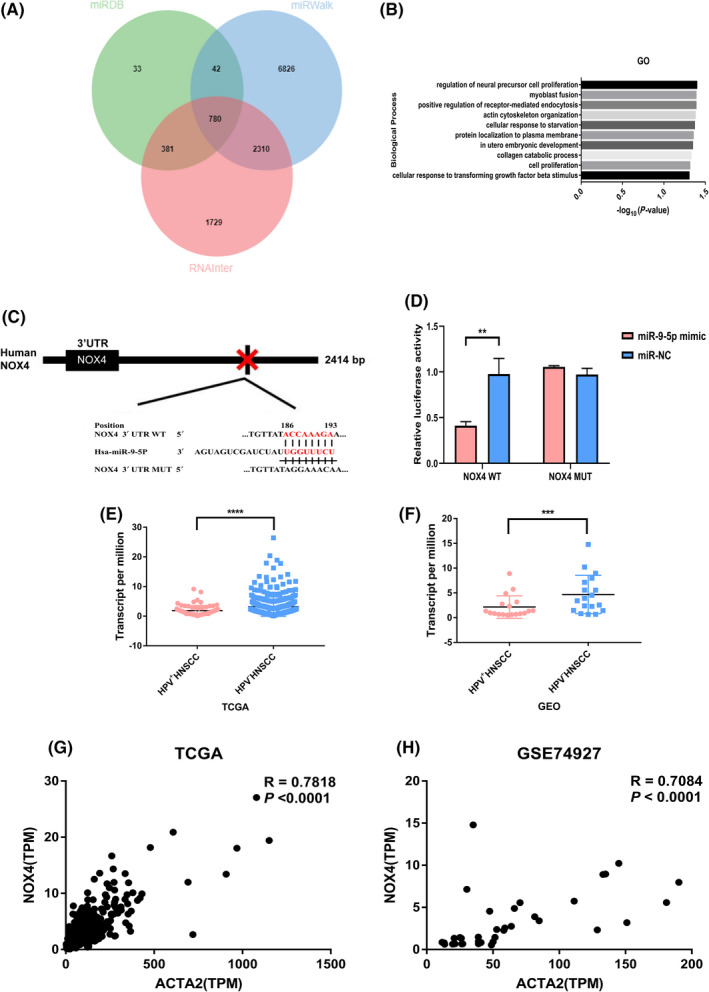
NOX4 is potential target gene of miR‐9‐5p. A, Intersection of potential target genes predicted by miRDB, RNAInter, and miRWalk. B, Enrichment of potential target genes of miR‐9‐5p in biological processes. C, Binding site of miR‐9‐5p in the 3′UTR of NOX4. D, Dual‐luciferase activity as described in (C) with the WT or mutated 3′UTR sequences of human NOX4 gene. E, F, Differential expression of NOX4 analyzed by DEseq2 in HPV+/− HNSCC in TCGA data sets and GSE74927. G, H, Correlation analysis between ACTA2 and NOX4 in HNSCC from TCGA and GSE74927. **P < .01, ***P < .001, ****P < .0001
TGF‐β1 is the most potent stimulus for the differentiation of fibroblasts into myofibroblasts. To determine whether TGF‐β1 was necessary for expression of αSMA in HGF‐1, we examined the expression of αSMA at the mRNA and protein levels after stimulation with TGF‐β1 for different times. As shown in Figure S3A‐F, TGF‐β1 upregulated αSMA at the mRNA and protein levels, and ROS levels also increased. Additionally, a NOX4 inhibitor, GKT137831, was used to inhibit NOX4 activity. The suppression of NOX4 activity suppressed TGF‐β1‐induced myofibroblast differentiation (Figure 6A,B). To further investigate if miR‐9‐5p was involved in the phenotypic transformation of fibroblasts by TGF‐β1, miR‐9‐5p transfection levels were first examined (Figure S3G). HGF‐1 cells were transfected with miR‐9‐5p mimic or mimic NC and immediately treated with TGF‐β1. Flow cytometry results showed that miR‐9‐5p could significantly reduce the increase in intracellular ROS due to TGF‐β1 stimulation. In addition, miR‐9‐5p inhibitor transfection could increase ROS levels (Figure 6C,D). Consequently, miR‐9‐5p transfection significantly affected the expression of NOX4 and αSMA (Figures 6E‐G and 7A,B). Inhibition of miR‐9‐5p effectively increased protein abundance of NOX4 and αSMA (Figure S4A‐C). Phospho‐smad2/3, necessary signal transduction factors in the TGF‐β1 signaling pathway, were also reduced by miR‐9‐5p mimic transfection (Figure S4D,E).
FIGURE 6.
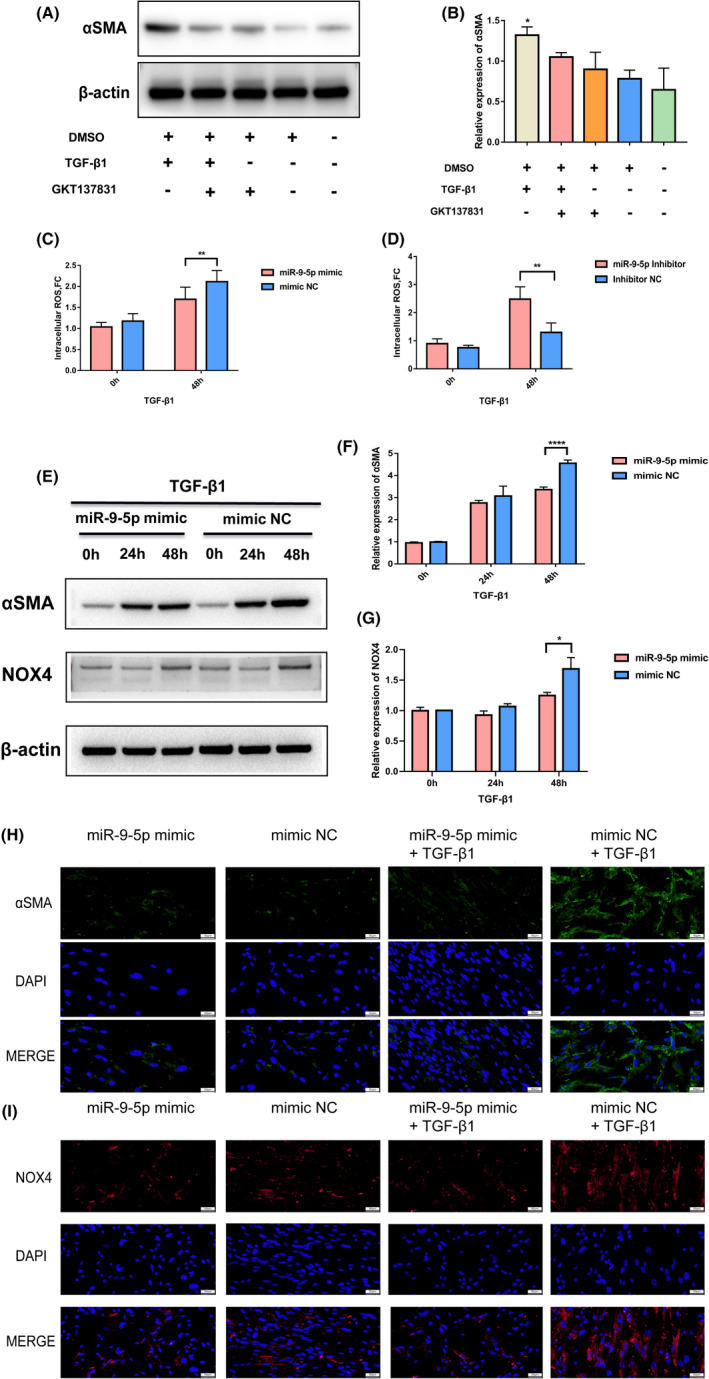
Exosomal miR‐9‐5p influences fibroblast phenotypic transformation through NOX4/TGF‐β1 signaling. A, B, Protein levels of αSMA were assessed and quantified by western blot analysis. C, Flow cytometry analysis of DCFH‐DA fluorescence in HGF‐1 treated with TGF‐β1 for 48 h after transfection with miR‐9‐5p mimic or mimic NC. D, Flow cytometry analysis of DCFH‐DA fluorescence in HGF‐1 treated with TGF‐β1 for 48 h after transfection with miR‐9‐5p inhibitor or inhibitor NC. E‐G, Protein levels of αSMA and NOX4 were assessed and quantified by western blot analysis in HGF‐1 treated with TGF‐β1 for 48 h after transfection with miR‐9‐5p mimic or mimic NC. H, I, Immunofluorescence analysis of αSMA and NOX4 in HGF‐1 treated with TGF‐β1 for 48 h after transfection with miR‐9‐5p mimic or mimic NC. *P < .05, **P < .01, ****P < .0001
FIGURE 7.
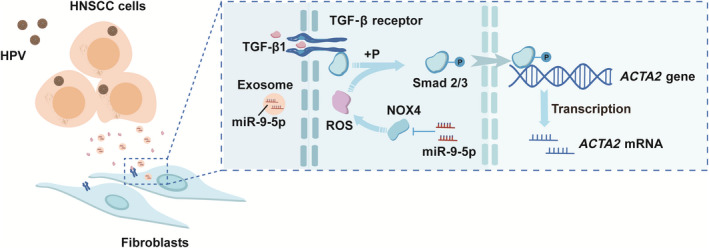
Schematic model shows that HPV+ HNSCC‐derived exosomal miR‐9‐5p inhibits TGFβ signaling‐mediated fibroblast phenotypic transformation through NOX4
4. DISCUSSION
In conclusion, we first demonstrated that CAF infiltration is reduced in HPV+ HNSCC, which may be related to the inhibition of TGF‐β1‐mediated fibroblast phenotypic transformation by HPV+ HNSCC‐derived miR‐9‐rich exosomes. miR‐9 reduced ROS levels in fibroblasts by targeting NOX4, which is necessary for TGF‐β1 signaling transduction.
Cancer‐associated fibroblasts may be transformed from different cells in different tumor environments under various stimuli. Still, most commonly, they are thought to originate from TGF‐β‐dependent phenotypic transformation of local fibroblasts. 28 , 29 The formation and development of tumors are long‐term processes. Cell level experiments in vitro can only restore the process at a few levels. The transformation and influence of CAFs also depend on the communication with other tumor microenvironment cells. CAF‐containing tumors often have low levels of lymphocytes. 30 Previous studies have shown that CAFs are associated with CD8+ T‐cell depletion. 31 The disturbance of immune cell function may be the reason why CAF affects the survival rate of patients. Large numbers of studies have focused on the effect of CAFs on tumor cells. For instance, exosomal miR‐196a, derived from tumor‐associated fibroblasts, can endow tumor cells with cisplatin resistance. 32 In contrast, it is not clear how tumor cells in different infection states affect fibroblasts.
HPV is a critical risk factor for HNSCC. It has been reported that HPV can affect the tumor microenvironment of HNSCC, 16 , 33 , 34 but the effect of HPV on CAFs in the tumor microenvironment has not been deeply studied. This study suggested that HPV infection was associated with CAF infiltration. In addition, lower levels of CAFs indicated a higher survival probability of HNSCC patients. These results are consistent with the clinical characteristics in which patients who are HPV positive usually have a better prognosis.
Exosomes have attracted extensive attention as an essential means of communication between cells. 35 , 36 Exosomes can be released from different cell types, especially tumor cells. 37 , 38 Accumulating shreds of evidence suggest that exosomes are involved in cellular communication between tumor cells and immune cells, stromal cells, and epithelial cells in the tumor microenvironment. 39 miRNAs are small non‐coding RNAs that can play post‐transcriptional regulation via binding to the 3′UTR of target mRNAs. 40 Previous studies have shown that miRNAs could be isolated from host cells and transferred into the surrounding target cells through exosomes. 41 miR‐9 is crucial for cancer growth, metastasis, immunity, angiogenesis, and radiosensitivity. 42 , 43 , 44 , 45 Accumulated studies have shown HPV mediated the activation by miR‐9 in cervical cancer and HNSCC. 46 , 47 , 48 This study demonstrated that miR‐9 was enriched in exosomes secreted from HPV+ HNSCC cells and transferred into fibroblasts via exosomes. The miR‐9 level is associated with CAF infiltration in HPV+ HNSCC. A high level of miR‐9 indicates a higher patient survival rate.
DISCLOSURE
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
BZW, LLW, and FJT conceived and designed the project. BZW performed all experiments. BZW performed the statistical analysis and analyzed and interpreted all the data. BZW and SWZ prepared the figures and tables. BZW wrote the paper. BZW, YW, and LLW reviewed and revised the manuscript. All authors approved the final manuscript.
Supporting information
Fig S1‐4
Table S1‐2
ACKNOWLEDGMENTS
We are grateful to the Third Affiliated Hospital of Harbin Medical University in Heilongjiang Province for providing technical guidance and HNSCC patients tissues. We give thanks to Wu Lien‐Teh Institute for providing the experimental platform.
Wang B, Zhang S, Tong F, Wang Y, Wei L. HPV+ HNSCC‐derived exosomal miR‐9‐5p inhibits TGF‐β signaling‐mediated fibroblast phenotypic transformation through NOX4. Cancer Sci. 2022;113:1475–1487. doi: 10.1111/cas.15281
Funding information
National Natural Science Foundation of China, Grant/Award no. 82102822; Shenzhen Longgang Fund for Science and Technology Project, Grant no. LGKCYLWS2020101; Shenzhen Science and Technology R&D Fund, Grant no. JCYJ20210324131607019
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Chow LQM. Head and neck cancer. N Engl J Med. 2020;382(1):60‐72. [DOI] [PubMed] [Google Scholar]
- 3. Psyrri A, DiMaio D. Human papillomavirus in cervical and head‐and‐neck cancer. Nat Clin Pract Oncol. 2008;5(1):24‐31. [DOI] [PubMed] [Google Scholar]
- 4. Ward MJ, Thirdborough SM, Mellows T, et al. Tumour‐infiltrating lymphocytes predict for outcome in HPV‐positive oropharyngeal cancer. Br J Cancer. 2014;110(2):489‐500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Economopoulou P, Kotsantis I, Psyrri A. Tumor microenvironment and immunotherapy response in head and neck cancer. Cancers. 2020;12(11):3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423‐1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qin X, Yan M, Zhang J, et al. TGFbeta3‐mediated induction of Periostin facilitates head and neck cancer growth and is associated with metastasis. Sci Rep. 2016;6:20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qin X, Yan M, Wang XU, et al. Cancer‐associated fibroblast‐derived IL‐6 promotes head and neck cancer progression via the osteopontin‐NF‐kappa B signaling pathway. Theranostics. 2018;8(4):921‐940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bergers G, Hanahan D. Modes of resistance to anti‐angiogenic therapy. Nat Rev Cancer. 2008;8(8):592‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monteran L, Erez N. The dark side of fibroblasts: cancer‐associated fibroblasts as mediators of immunosuppression in the tumor microenvironment. Front Immunol. 2019;10:1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeltz C, Primac I, Erusappan P, et al. Cancer‐associated fibroblasts in desmoplastic tumors: emerging role of integrins. Semin Cancer Biol. 2020;62:166‐181. [DOI] [PubMed] [Google Scholar]
- 12. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88: 487‐514. [DOI] [PubMed] [Google Scholar]
- 13. Greenberg AS, Reeves AR. The good and bad of adipose tissue macrophage exosomes in obesity. Cell Metab. 2021;33(4):700‐702. [DOI] [PubMed] [Google Scholar]
- 14. Thery C, Witwer KW, Aikawaet E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Record M, Carayon K, Poirot M, et al. Exosomes as new vesicular lipid transporters involved in cell‐cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841(1):108‐120. [DOI] [PubMed] [Google Scholar]
- 16. Tong F, Mao X, Zhang S, et al. HPV + HNSCC‐derived exosomal miR‐9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer Lett. 2020;478:34‐44. [DOI] [PubMed] [Google Scholar]
- 17. Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15(1):1‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alamoudi AA, Alnoury A, Gad H. miRNA in tumour metabolism and why could it be the preferred pathway for energy reprograming. Brief Funct Genomics. 2018;17(3):157‐169. [DOI] [PubMed] [Google Scholar]
- 19. Tiwari A, Mukherjee B, Dixit M. MicroRNA key to angiogenesis regulation: MiRNA biology and therapy. Curr Cancer Drug Targets. 2018;18(3):266‐277. [DOI] [PubMed] [Google Scholar]
- 20. Sun Z, Shi KE, Yang S, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17(1):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451‐5465. [DOI] [PubMed] [Google Scholar]
- 22. Mishra S, Yadav T, Rani V. Exploring miRNA based approaches in cancer diagnostics and therapeutics. Crit Rev Oncol Hematol. 2016;98:12‐23. [DOI] [PubMed] [Google Scholar]
- 23. Zhao X, Cui L. A robust six‐miRNA prognostic signature for head and neck squamous cell carcinoma. J Cell Physiol. 2020;235(11):8799‐8811. [DOI] [PubMed] [Google Scholar]
- 24. Wang J, Jiang C, Li N, et al. The circEPSTI1/mir‐942‐5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis. 2020;11(8):682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 26. Campbell JD, Yau C, Bowlby R, et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018;23(1):194‐212.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang X, Lan Y, Xu J, et al. Cell Marker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721‐D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park D, Sahai E, Rullan A. SnapShot: cancer‐associated fibroblasts. Cell. 2020;181(2):486‐486.e1. [DOI] [PubMed] [Google Scholar]
- 29. Desmoulière A, Geinoz A, Gabbiani F, et al. Transforming growth factor‐beta 1 induces alpha‐smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol. 1993;122(1):103‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chakravarthy A, Khan L, Bensler NP, Bose P, De Carvalho DD. TGF‐beta‐associated extracellular matrix genes link cancer‐associated fibroblasts to immune evasion and immunotherapy failure. Nat Commun. 2018;9(1):4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ford K, Hanley CJ, Mellone M, et al. NOX4 inhibition potentiates immunotherapy by overcoming cancer‐associated fibroblast‐mediated CD8 T‐cell exclusion from tumors. Cancer Res. 2020;80(9):1846‐1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qin X, Guo H, Wang X, et al. Exosomal miR‐196a derived from cancer‐associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019;20(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen X, Fu E, Lou H, et al. IL‐6 induced M1 type macrophage polarization increases radiosensitivity in HPV positive head and neck cancer. Cancer Lett. 2019;456:69‐79. [DOI] [PubMed] [Google Scholar]
- 34. Zhang S, Wang B, Ma F, et al. Characteristics of B lymphocyte infiltration in HPV(+) head and neck squamous cell carcinoma. Cancer Sci. 2021;112(4):1402‐1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen J, Zhou R, Liang Y, et al. Blockade of lncRNA‐ASLNCS5088–enriched exosome generation in M2 macrophages by GW4869 dampens the effect of M2 macrophages on orchestrating fibroblast activation. FASEB J. 2019;33(11):12200‐12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Czernek L, Duchler M. Functions of cancer‐derived extracellular vesicles in immunosuppression. Arch Immunol Ther Exp. 2017;65(4):311‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907‐1920. [DOI] [PubMed] [Google Scholar]
- 39. Wu Q, Zhou L, Lv D, et al. Exosome‐mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol. 2019;12(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Valadi H, Ekström K, Bossios A, et al. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654‐659. [DOI] [PubMed] [Google Scholar]
- 42. Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR‐9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci U S A. 2009;106(13):5282‐5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen XU, Yang F, Zhang T, et al. MiR‐9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J Exp Clin Cancer Res. 2019;38(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arora H, Qureshi R, Jin S, Park AK, Park WY. miR‐9 and let‐7g enhance the sensitivity to ionizing radiation by suppression of NFkappaB1. Exp Mol Med. 2011;43(5):298‐304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma LI, Young J, Prabhala H, et al. miR‐9, a MYC/MYCN‐activated microRNA, regulates E‐cadherin and cancer metastasis. Nat Cell Biol. 2010;12(3):247‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Park S, Eom K, Kim J, et al. MiR‐9, miR‐21, and miR‐155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer. 2017;17(1):658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Božinović K, Sabol I, Dediol E, et al. Genome‐wide miRNA profiling reinforces the importance of miR‐9 in human papillomavirus associated oral and oropharyngeal head and neck cancer. Sci Rep. 2019;9(1):2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shiiba M, Uzawa K, Tanzawa H. MicroRNAs in Head and Neck Squamous Cell Carcinoma (HNSCC) and Oral Squamous Cell Carcinoma (OSCC). Cancers. 2010;2(2):653‐669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐4
Table S1‐2


