Abstract
Emerging evidence has been reported to support the involvement of the gut microbiota in the host’s blood lipid and hyperlipidemia (HLP). However, there remains unexplained variation in the host’s blood lipid phenotype. Herein a nonhuman primate HLP model was established in cynomolgus monkeys fed a high-fat diet (HFD) for 19 months. At month 19%, 60% (3/5) of the HFD monkeys developed HLP, but surprisingly 40% of them (2/5) exhibited strong tolerance to the HFD (HFD-T) with their blood lipid profiles returning to normal levels. Metagenomic analysis was used to investigate the compositional changes in the gut microbiota in these monkeys. Furthermore, the relative abundance of Megasphaera remarkably increased and became the dominant gut microbe in HFD-T monkeys. A validation experiment showed that transplantation of fecal microbiota from HFD-T monkeys reduced the blood lipid levels and hepatic steatosis in HLP rats. Furthermore, the relative abundance of Megasphaera significantly increased in rats receiving transplantation, confirming the successful colonization of the microbe in the host and its correlation with the change of the host’s blood lipid profiles. Our results thus suggested a potentially pivotal lipid-lowering role of Megasphaera in the gut microbiota, which could contribute to the variation in the host’s blood lipid phenotype.
Keywords: cynomolgus monkey, hyperlipidemia, high-fat diet, fecal microbiota transplantation, Megasphaera
Introduction
Hyperlipidemia (HLP) is considered one of the main contributing factors in developing metabolic syndromes and increasing the risk of atherosclerosis, stroke, coronary heart disease, and myocardial infarction (Karr, 2017; Panahi et al., 2018; Libby et al., 2019). In addition, high blood cholesterol (total cholesterol, TC) is a strong risk factor for hypertension, fatty liver, and diabetes (Elkins and Friedrich, 2018).
Recently, emerging evidence suggests the role of gut microbiota in human health and disease (Nicholson et al., 2012; Qin et al., 2012; Guo et al., 2016; Tong et al., 2018). Gut dysbiosis, defined as an imbalance in gut microbial communities, has been linked to a variety of diseases, including malnutrition, inflammatory bowel disease, neurological disorders, and cancer, as well as diabetes and obesity (Blanton et al., 2016; DeGruttola et al., 2016). For example, in genetically obese (ob/ob) mice, compared with wild-type and heterozygous lean counterparts, the abundance of Firmicutes was significantly increased and Bacteroidetes were decreased (Turnbaugh et al., 2006, 2009).
Probiotics could confer health benefits on the host by protecting them from gut dysbiosis. Akkermansia muciniphila is currently recommended as a new potential complementary therapy for obesity, diabetes, and liver diseases in clinics (Cani and de Vos, 2017; Plovier et al., 2017). It has been demonstrated that there are metabolic benefits of Parabacteroides distasonis (P. distasonis) on decreasing weight gain, hyperglycemia, and hepatic steatosis in ob/ob and high-fat diet (HFD)-fed mice. Administration of P. distasonis into obese mice dramatically changed the profile of secondary bile acids (Wang et al., 2019). In addition, Faecalibacterium prausnitzii and the peptides secreted by it showed anti-inflammatory effects on chemically induced colitis in mice (Breyner et al., 2017). Lactobacillus was reported to have a lipid-lowing effect on hypercholesterolemia and hyperlipidemic rat or mice (Zhou et al., 2013; Singh et al., 2015; Jiang et al., 2019; Qian et al., 2019). Therefore, it is promising to improve such diseases and gut dysbiosis by targeting the gut microbiota using probiotics.
As a typical chronic disease, HLP is closely related to gut microbiota dysbiosis and could also be potentially alleviated and cured through gut microbiota regulation. Given the high similarity to humans in terms of genetics, anatomy, reproduction, development, and metabolism, non-human primates (NHPs) are used as distinctive and indispensable model organisms in various areas of biomedical research and disease studies (Manara et al., 2019). Furthermore, it has been found that NHPs in captivity have a similar gut microbiota composition to that in humans (Clayton et al., 2016).
To understand the role of the gut microbiota in HLP, we used NHPs (cynomolgus monkeys) fed a high-fat diet (HFD) to establish an HLP model in the current study. HFD monkeys exhibited HLP by Month 7. After 12 months of HFD feeding, some individuals of the monkeys developed tolerance to HFD with their blood lipid profiles returning to normal levels. To explore the dynamics of the gut microbiota associated with such changes in lipid profiles, we conducted metagenomic sequencing in the HFD monkeys. Furthermore, transplantation of fecal microbiota from HFD-tolerant cynomolgus monkeys alleviated HLP and hepatocyte lesion in HFD rats.
Materials and Methods
Animals
Ten male cynomolgus monkeys were purchased from Guangdong Landau Biotechnology Co. Ltd. (Guangzhou, China) which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International (AAALAC). All the animals were confirmed to be in healthy condition by records and veterinary examination before the experiment.
All monkeys were kept in a well-controlled and comfortable environment with temperature (16°C–28°C) and relative humidity of 40%–70%, as well as a 12/12-h light–dark cycle. All the animals were free to access food and drinking water. The protocol for this study was approved by the Institutional Animal Care and Use Committee of Guangdong Landau Biotechnology Co., Ltd. (Code: LD20150518).
All monkeys were randomly divided into two groups, including an HFD group (n = 5) and a normal chow diet group (NCD, n = 5). The NCD group was fed normal chow, while the HFD group was fed normal chow daily plus emulsion containing 10% sucrose, 10% lard, 1% cholesterol, and 0.5% cholate (5 ml/kg body weight) via nasogastric gavage 6 days a week. All the monkeys in the HFD group were fed HFD for 19 months.
Twenty seven male Sprague–Dawley (SD) rats used for validation experiments were purchased from Guangzhou University of Chinese Medicine. All rats were kept in a specific pathogen-free environment with free access to food and water. The protocol for this study was approved by Institute of Zoology, Guangdong Academy of Sciences (GIABR20200908).
The rats were randomly divided into three groups (n = 9 for each group): an NCD group (NCD), an HFD group, and a fecal microbiota transplantation group (HFD + M). All rats were fed with an NCD. The HFD and HFD + M groups were fed normal chow daily plus the same emulsion used in the monkeys (5 ml/kg body weight) by gavage (six days a week for 12 weeks.
Fecal Microbiota Transplantation
Fresh stool samples from monkeys that showed tolerance to the HFD were pooled, suspended in sterile phosphate buffer saline (PBS, pH = 7.0), and centrifuged at 500 g for 5 min. 200 μl of the prepared supernatant (108 CFU) was given to the HFD + M group of rats via oral gavage 6 days per week at week 7–12 of HFD feeding.
Biochemical Analysis
Blood samples of rats and monkeys were periodically collected from the ocular vein and the upper limb saphenous vein, respectively. Serum total cholesterol (TC), triacylglycerol (TG), low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C) were measured using commercially available kits from Guangdong Lewwin Pharmaceutical Research Institute Co., Ltd. (Guangzhou, China).
Histological Analysis
Liver tissues of rats were collected, fixed in 4% paraformaldehyde, paraffin-embedded, and sectioned at 5 μm. Hematoxylin and eosin (H&E) and Oil Red O staining of paraffin sections were conducted following a standard method.
Metagenomic Sequencing and Analysis
DNA of macaque fecal samples was collected at month 19 of HFD feeding and sequenced on the HiSeq-X10 platform (Illumina) using a paired-end 150 bp configuration. Cleaning data were obtained by filtering raw data using KneadData (v0.7.4).1 Karaken2 (v2.0.8) was used for taxonomic analysis (Wood and Lu, 2019), and vegan 2.5–7 was used for diversity indices analysis.
16S rRNA Gene Sequencing and Analysis
SD rat fecal samples were collected at Week 12 for gut microbial analysis. Bacterial genomic DNA was extracted from frozen fecal samples stored at −80°C using TIANamp Stool DNA kit (Cat.#DP328, Tiangen, China) according to the manufacturer’s instructions.
The hypervariable V4 regions of bacterial 16 s rRNA genes were amplified using the polymerase chain reaction and V4-specific primers as described previously (Wei et al., 2021). After quality control, PCR products were purified and sequenced on an Ion S5XL sequencer (Thermo Fisher Scientific, Waltham, Massachusetts) with a single-end 400-bp read length configuration.
Bioinformatic analysis of the 16S rRNA gene sequencing data was conducted using the QIIME2 (version 2018.6.0) analysis pipeline (Bolyen et al., 2019). Briefly, the dada2 program was used to filter and remove low-quality and chimeric sequences, and generate unique feature tables equivalent to operational taxonomic unit (OTU) tables at exact match or 100% sequence similarity. Taxonomic identifications were then assigned to these features using the q2-feature-classifier and the full-length SILVA database (release r138) at a 99% similarity cutoff (Parks et al., 2014). PICRUSt (version 1.1.4) was used to predict microbial functions from the 16S rRNA gene sequencing data, which were further categorized using the BRITE hierarchy of the KEGG database (Langille et al., 2013). Differences in the microbial functions were then analyzed by the Linear discriminant analysis (LDA) Effect Size (LEfSe) algorithm with a log (LDA) score cutoff of 2 and default settings (Segata et al., 2011).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism V.7.0a (GraphPad Software, United States) and the R statistical language (version 3.6.0). The levels of serum lipid, the relative abundance of OTUs, and alpha diversity indices among groups were compared using the Student’s t-test and evaluated for pair-wise inter-group differences with Tukey’s post-hoc test if overall significance was found.
Results
Tolerance to HFD in Monkeys After Long-Term Dietary Induction
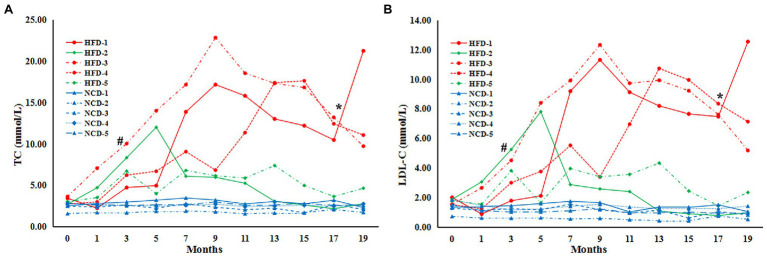
To explore the characteristics of HLP, the levels of TC and LDL-C were significantly higher in monkeys fed HFD than NCD controls (n = 5) by 3 months (TC, HFD vs NCD, p = 0.001; LDL-C, HFD vs. NCD, p = 0.003) (Figures 1A,B). After 7 months, all HFD monkeys showed HLP (TC > 6.20 mmol/L and LDL-C > 3.64 mmol/L) with higher levels of TC and LDL-C compared to the NCD controls (TC, HFD vs. NCD, p = 0.006; LDL-C, HFD vs. NCD, p = 0.007). Intriguingly, after 17 months of diet induction, 2 monkeys (HFD-2 and HFD-5) exhibited obvious tolerance to HFD with their blood lipid profiles returning to normal levels (TC, HFD-2/5 vs. NCD control, p > 0.05; HFD-2/5 vs. HFD-1/3/4, p = 0.004. LDL-C, HFD-2/5 vs. NCD control, p > 0.05; HFD-2/5 vs. HFD-1/3/4, p < 0.0001). In the subsequent analysis, we refer to HFD-2 and HFD-5 as HFD-tolerant (HFD-T), and HFD-1, HFD-3, and HFD-4 as HFD-intolerant (HFD-I). However, long-term HFD did not cause significant body weight changes in HFD monkeys compared to NCD monkeys by month 19 (data not shown).
Figure 1.
Serum lipids in cynomolgus monkeys during HFD feeding. Serum levels (mmol/L) are shown for (A) total cholesterol (TC), (B) low-density lipoprotein cholesterol (LDL-C). HFD: high-fat diet monkeys; NCD: normal chow diet. Significant differences are indicated as #, at month 3, HFD monkeys vs NCD monkeys, p < 0.05; *, at Month 17, HFD-1/HFD-3/HFD-4 (HFD-intolerant) monkeys vs HFD-2/HFD-5 (HFD-tolerant) monkeys, p < 0.05.
Gut Microbiota Profiles Associated With HFD Tolerance in Monkeys
To explore the dynamics of gut microbiota associated with such changes in lipid profiles, we conducted metagenomic sequencing using fecal DNA from both HFD-T and HFD-I monkeys after HFD-induction for 19 months. Our results showed that long-term HFD induction changes the composition of the gut microbiota and slightly reduced alpha diversity indices (Chao1, Shanno’s index, and Simpson’s index) in HFD-I monkeys, while HFD-T monkeys had similar alpha diversity compared to NCD controls (Figure 2).
Figure 2.
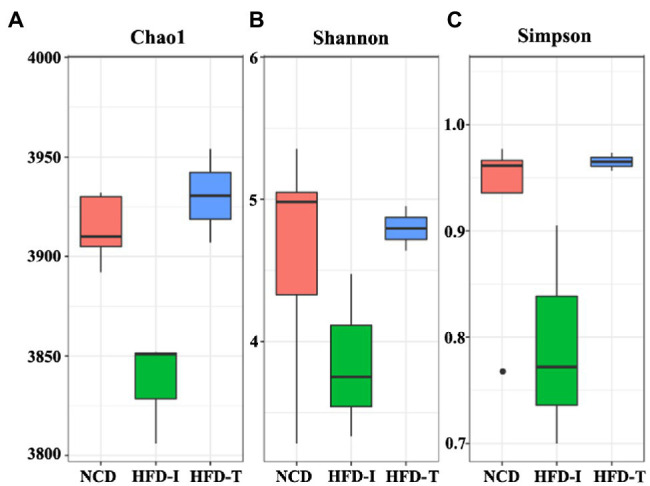
Boxplots showing changes in alpha diversity of the gut microbiota in NCD, HFD-I, and HFD-T monkeys. Alpha diversity indices including Chao1 (A), Shannon’s (B) index, and Simpson’s index (C) are shown.
Analysis at the phylum level showed that HFD markedly decreased the relative abundance of Firmicutes, and increased the relative abundance of Bacteroidetes. In addition, Spirochaetes was increased in HFD-I monkeys. However, the relative abundance of Firmicutes was found to be partially increased in HFD-T monkeys, and Bacteroidetes also increased in HFD-I monkeys (Supplementary Figure S1).
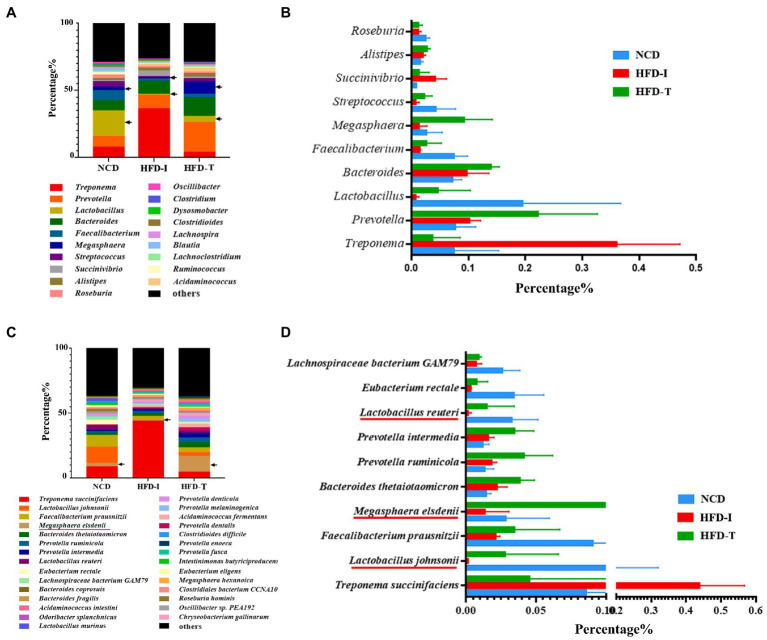
A detailed analysis of the bacterial genera detected in each group is shown in Figure 3A. The relative abundance of Lactobacillus was the highest in NCD monkeys but was markedly reduced in HFD monkeys (HFD-I and HFD-T groups). Intriguingly, the relative abundance of Prevotella, Bacteroides, Lactobacillus, and especially Megasphaera were increased in HFD-T monkeys (Figures 3A,B).
Figure 3.
The most abundant genera and species in the gut microbiota of NCD, HFD-I, and HFD-T monkeys. Relative abundances of the gut microbiota at the genus level (A), top 10 abundant genera (B), relative abundances of the gut microbiota at the species level (C), and top 10 abundant species (D) are shown.
Further analysis at the species level showed similar results to those observed at the genus level. As shown in Figure 3C, the relative abundance of Lactobacillus johnsonii, Lactobacillus reuteri and Megasphaera elsdenii were highest in NCD monkeys. However, the relative abundance of these bacteria was markedly reduced after long-term HFD feeding (Figure 3D). However, the relative abundance of L. johnsonii and L. reuteri were found to be partially increased in HFD-T monkeys (Figures 3D, 4A,B). However, M. elsdenii was dramatically increased and became predominant in HFD-T monkeys (Figures 3C,D, 4C). In addition, another two species of Megasphaera (including Megasphaera hexanoica and Megasphaera stantonii) were also increased in HFD-T monkeys (Figures 4D,E).
Figure 4.
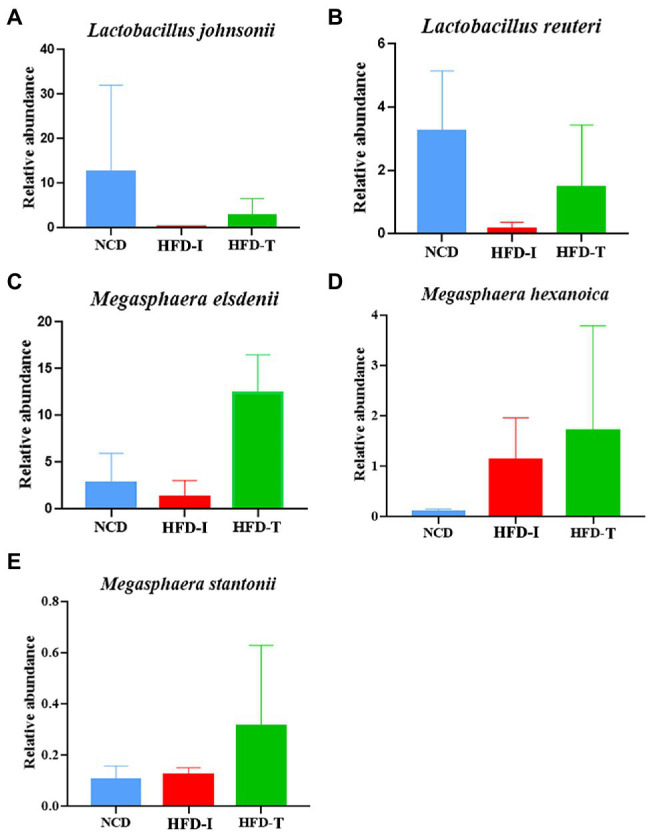
The relative abundance of selected species in the gut microbiota in NCD, HFD-I, and HFD-T monkeys. Lactobacillus johnsonii (A), Lactobacillus reuteri (B), Megasphaera elsdenii (C), Megasphaera hexanoica (D), and Megasphaera stantonii (E).
Transplantation of Fecal Microbiota From HFD-T Monkeys to HFD Rats Alleviated HLP and Hepatic Steatosis
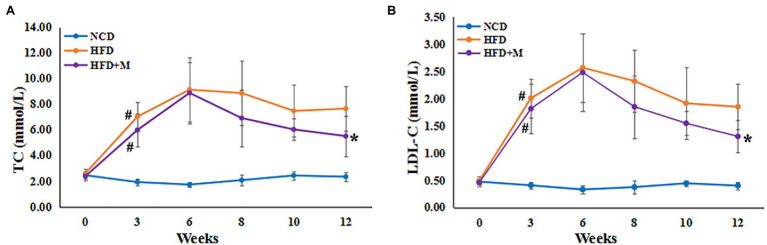
To validate the possible lipid-lowering effects of the gut microbiota in HFD-T monkeys, fecal microbiota from HFD-T monkeys was transplanted to HLP rats fed with an HFD. All rats showed significantly higher levels of TC and LDL-C compared to NCD rats after 3 weeks of HFD feeding. Transplantation of fecal microbiota from HFD-T monkeys alleviate HLP in the recipient HFD rats with significantly lower levels of TC (NCD vs HFD, p < 0.0001; NCD vs HFD + M, p < 0.0001; HFD vs HFD + M, p = 0.0227) and LDL-C (NCD vs HFD, p < 0.0001; NCD vs HFD + M, p < 0.0001; p = 0.0094; Figures 5A,B). Furthermore, HFD-fed rats showed enhanced lipid accumulation, which was markedly inhibited with improved hepatic steatosis by the fecal microbiota transplantation in the HFD + M mice (Figure 6).
Figure 5.
Serum lipids in SD rats following fecal microbiota transplantation from HFD-T monkeys. Serum levels (mmol/L) are shown for (A) total cholesterol (TC), (B) low-density lipoprotein cholesterol (LDL-C). NCD, Normal Chow Diet rats; HFD, HFD rats; HFD + M, HFD rats with transplantation from the fecal microbiota from HFD-T monkeys. #, at Week 3 of HFD feeding, HFD and HFD + M rats vs NCD rats, p < 0.05; *, at Week 12, HFD rats vs HFD + M rats, p < 0.05.
Figure 6.
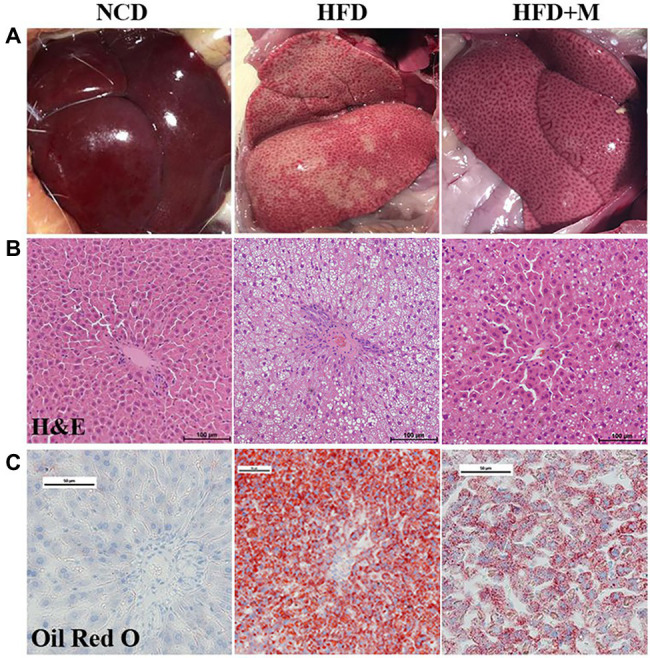
Changes in hepatic steatosis after fecal microbiota transplantation. (A) Photos showing liver morphology. (B) H and E staining of liver section. Scale bars, 100 μm. (C) Liver Oil Red O staining. NCD, Normal Chow Diet rats; HFD, HFD rats; HFD + M, HFD rats received fecal microbiota transplant from HFD-T monkeys. Scale bars, 50 μm.
Fecal Microbiota Transplantation Changed Gut Microbiota of HFD Rats.
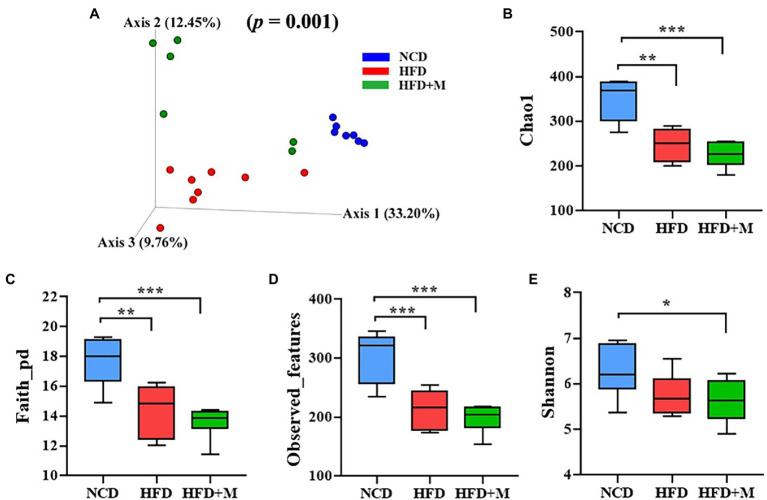
To further investigate the lipid-lowering mechanism of the gut microbiota from the HFD-T monkeys, we also analyzed the gut microbiota composition of rats in the NCD, HFD, and HFD + M groups using 16S rRNA gene high-throughput sequencing. Principal component analysis (PCA), the NCD, HFD, and HFD + M groups of rats each presented a separated clustering of microbiota composition (Figure 7A). Such results demonstrate that transplantation of fecal microbiota significantly changed the gut microbiota profiles in rats (p = 0.001).
Figure 7.
The beta diversity, structure, and richness of the gut microbiota in three groups of rats. (A) Nonmetric multidimensional scaling (NMDS) score plot based on principal component analysis (PCA) score plot based on microbiota composition. (B–E) The gut microbiota diversity as calculated using Shannon’s index (B), Chao1 (C), Faith-pd (D), and observed-features (E) index of each rat group: NCD, Normal Chow Diet (control) rats; HFD, HFD rats; HFD + M, HFD rats received fecal microbiota transplant from HFD-T monkeys. Values are presented as mean ± SE. (n = 7). *, p < 0.05; **, p < 0.01; ***, p < 0.001, compared with NCD rats.
In addition, it has been demonstrated that HFD could significantly reduce the richness and diversity of the gut microbiota in HFD fed rats (including both the HFD and HFD + M groups) by using a range of alpha diversity indices (Shannon, Chao1, Faith-pd, and Observed-features; Figures 7B–E). However, there was no significant difference in the richness and alpha diversity of the gut microbiota between the HFD and HFD + M groups (Figures 7B–E).
As shown in Supplementary Figure S2, at the phylum level analysis HFD significantly decreased the relative abundance of Firmicutes and increased the relative abundance of Bacteroidetes. These changes are consistent with findings in monkeys. However, the relative abundance of Firmicutes and Bacteroidetes in HFD + M rats showed no significant changes compared to HFD rats.
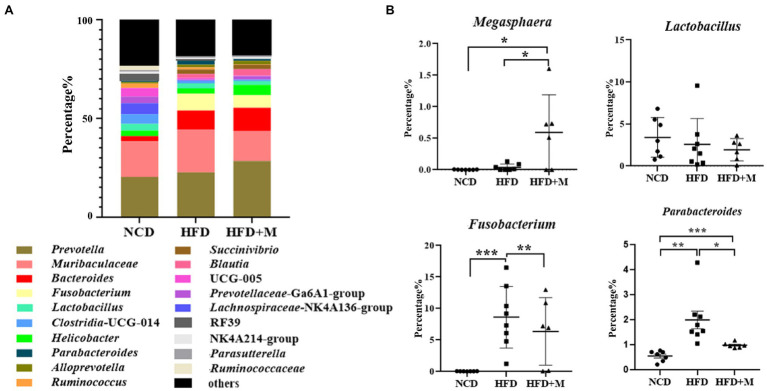
Furthermore, we analyzed the differences in the gut microbiota at the genus level. Genera Prevotella, Bacteroides, and Lactobacillus were predominant in the rat gut microbiota and Bacteroides was significantly increased in HFD and HFD + M rats (NCD vs HFD, p < 0.001; NCD vs HFD + M, p < 0.0001; Figure 8A). However, Fusobacterium (NCD vs HFD, p < 0.001; HFD vs HFD + M, p < 0.01) and Parabacteroides (NCD vs HFD, p < 0.01; HFD vs. HFD + M, p < 0.01) were increased in HFD rats but were significantly decreased after fecal microbiota transplantation (Figure 8B). The abundance of Lactobacillus showed no significant difference among the three groups of rats (p > 0.05; Figure 8B). Interestingly, Megasphaera was significantly increased in HFD + M rats (p < 0.05), in line with its changes observed in HFD-T monkeys, implicating its role in the reduction of the host’s blood lipids.
Figure 8.
Relative abundances of gut microbiota at genus level in different rat groups. (A) The most abundant genera (top 20) of the gut microbiota in SD rats of different groups. (B) The relative abundance of Megasphaera, Lactobacillus, Parabacteroides, and Fusobacterium in fecal microbiota. NCD, Normal Chow Diet rats; HFD, HFD rats; HFD + M, HFD rats received fecal microbiota transplant from HFD-T monkeys. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
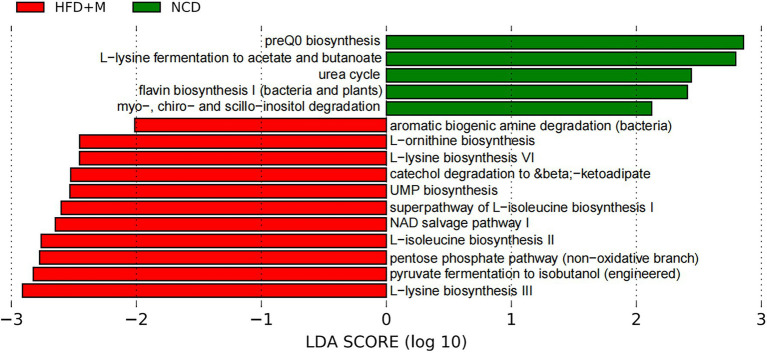
In addition, as shown in LEfSe analysis shown in Figure 9, PICRUSt prediction results indicated that transplantation of fecal microbiota from HFD-T monkeys changed various functions of gut microbiota in rats, particularly biosynthesis of amino acids, including L-lysine and L-isoleucine.
Figure 9.
LEfSe analysis of microbial functions predicted by PICRUSt in different rat groups. NCD, Normal Chow Diet rats; HFD, HFD rats; HFD + M, HFD rats received fecal microbiota transplant from HFD-T monkeys; LDA, linear discriminant analysis (LDA); LEfSe, linear discriminant analysis effect size.
Discussion
HLP is considered an important risk factor for cardiovascular diseases (CVDs). More than 17.9 million people died from CVDs in 2019, representing 32% of all deaths worldwide (Roth et al., 2020). However, emerging evidence shows that gut microbiota plays an important role the host’s energy metabolism and blood lipid level modulation (Velagapudi et al., 2010; Mestdagh et al., 2012). Therefore, we developed an NHP HLP model with high similarity to humans in genetics, physiology, and gut microbial composition, for analysis of the effect of gut microbiota on the host’s blood lipids. A unique and interesting feature of such an NHP model was that 40% of the monkeys in our study developed tolerance to HFD with normal lipid profiles after prolonged exposure. Such an NHP model provided a unique tool to investigate changes in the gut microbiota related to the development of HFD tolerance.
Previous studies have demonstrated that multiple species of Lactobacillus can reduce serum cholesterol, TG, and LDL-C, and have been used in clinical practice as potential probiotics (Zhou et al., 2013; Singh et al., 2015; Jiang et al., 2019; Qian et al., 2019). In the current study, we found that Lactobacillus (such as Lactobacillus johnsoni and L. reuteri) was significantly increased in HFD-T monkeys with TC and LDL-C returning to the normal level. However, after transplantation of fecal microbiota from HFD-T monkeys to HFD rats, Lactobacillus showed no significant change in HFD + M rats. Therefore, Lactobacillus might not be the major microbial microbe responsible for the reduction of serum lipids in the HFD + M rats.
In previous studies, Parabacteroides showed a beneficial effect against weight gain, hyperglycemia, hepatic steatosis, and HLP (Wang et al., 2019). However, the relative abundance of Parabacteroides was also significantly decreased in HFD + M rats. HFD rats showed the highest relative abundance of Parabacteroides, suggesting that Parabacteroides might not play a lipid-lowering effect role in HFD + M rats.
In our previous study, Megasphaera was found to be a dominant genus in the primate gut microbiota throughout different life stages and a key driver that contributes to long-term gut microbiota development (Wei et al., 2021). The genus belongs to the family Veillonellaceae under the order Veillonellales, class Negativicutes, and phylum Firmicutes (Maki and Looft, 2018). Previous studies have demonstrated that M. elsdenii could metabolize lactate and buffer fat and produce acetate, propionate, and butyrate (Chen and Wang, 2016; Chen et al., 2019). These short-chain fatty acids may act through down-regulating cholesterol biosynthesis and increasing the bile acid excretion to regulate the lipid levels (Illman et al., 1988; Marcil et al., 2003; Falcinelli et al., 2015, 2018). Our results found that Megasphaera, especially M. elsdenii, was a predominant component of the microbiota, and showed a dramatic increase in abundance in HFD-T monkeys. Surprisingly, the relative abundance of Megasphaera was also dramatically increased in HFD + M rats and this increasing trend was consistent with changes in the abundance of Megasphaera species in monkeys. In contrast, Megasphaera is almost undetectable in NCD and HFD rats without fecal microbiota transplantation. Therefore, it is reasonable to speculate that the Megasphaera (more likely M. elsdenii) successfully colonized in HFD + M rats after fecal microbiota transplantation, and Megasphaera might play a critical role in lowering blood lipids in these HLP animals.
Our results also indicated the functional impact of fecal microbiota transplantation from HFD-T monkeys on the gut microbiota in HFD rats, pointing to a potential role of amino acid biosynthesis. Effects of lysine and isoleucine on lipid metabolism have been implicated in previous studies. Dietary lysine restriction in rats was reported to cause lipid accumulation in skeletal muscle (Goda et al., 2021). Branched-chain amino acids leucine and isoleucine were found to reduce lipid accumulation in HFD-induced obese mice (Ma et al., 2020). It reminds a possible mechanism via altered biosynthesis of such amino acids in the gut microbiota in improving the host’s lipid metabolism.
Our current study demonstrates that changes in the gut microbiota are associated with variation of blood lipid phenotype in both monkeys and rats under HFD feeding. Transplantation of the gut microbiota from HFD-T monkeys alleviated HLP and hepatic steatosis in rats. Further studies are thus warranted to confirm the lipid-lowering and liver-protecting effects of Megasphaera, which could be potentially used as a probiotic to lower blood lipids, and improve hepatic steatosis in the treatment of HLP.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Guangdong Landau Biotechnology and Institute of Zoology, Guangdong Academy of Sciences.
Author Contributions
J-MG, J-HR, Z-YW, and J-HC designed the study. J-MG, LH, M-TT, T-TZ, J-HL, G-AZ, and Y-XS conducted the experiments. J-MG, Z-YW, and S-YX conducted the data analysis data or performed the statistical analysis. J-MG, GH, J-HC, and J-HR drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported in part by grants from the National Natural Science Foundation of China (no. 31671311), the Guangdong Key Laboratory of Non-human Primate Research (2020B121201006), GDAS’ Project of Science and Technology Development (2022GDASZH-2022010110), Guangdong Basic and Applied Basic Research Foundation (2019A1515012062 and 2020A1515010480), Natural Science Foundation of Guangdong Province (2018A030313307), the Program for High-Level Entrepreneurial and Innovative Talents Introduction of Jiangsu Province, the Taihu Lake Talent Plan, and Wuxi Institute of Translational Medicine.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.876043/full#supplementary-material
References
- Blanton L. V., Charbonneau M. R., Salih T., Barratt M. J., Venkatesh S., Ilkaveya O., et al. (2016). Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351:10.1126. doi: 10.1126/science.aad3311, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyner N. M., Michon C., de SoUnited States C. S., Vilas Boas P. B., Chain F., Azevedo V. A., et al. (2017). Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-κB pathway. Front. Microbiol. 8:114. doi: 10.3389/fmicb.2017.00114, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P. D., de Vos W. M. (2017). Next-generation beneficial microbes: The case of Akkermansia muciniphila. Front. Microbiol. 8:1765. doi: 10.3389/fmicb.2017.01765, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Shen Y., Wang C., Ding L., Zhao F., Wang M., et al. (2019). Megasphaera elsdenii lactate degradation pattern shifts in rumen acidosis models. Front. Microbiol. 10:162. doi: 10.3389/fmicb.2019.00162, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang H. (2016). Advances in the metabolism and regulation of lactic acids in the rumen. Pratacultural Sci. 33, 972–980. [Google Scholar]
- Clayton J. B., Vangay P., Huang H., Ward T., Hillmann B. M., Al-Ghalith G. A., et al. (2016). Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. U. S. A. 113, 10376–10381. doi: 10.1073/pnas.1521835113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGruttola A. K., Low D., Mizoguchi A., Mizoguchi E. (2016). Current understanding of Dysbiosis in disease in human and animal models. Inflamm. Bowel Dis. 22, 1137–1150. doi: 10.1097/MIB.0000000000000750, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins C., Friedrich D. (2018). Hypertriglyceridemia: A review of the evidence. Nurse Pract. 43, 22–29. doi: 10.1097/01.NPR.0000544997.22887.0b [DOI] [PubMed] [Google Scholar]
- Falcinelli S., Picchietti S., Rodiles A., Cossignani L., Merrifield D. L., Taddei A. R., et al. (2015). Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Sci. Rep. 5:9336. doi: 10.1038/srep09336, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcinelli S., Rodiles A., Hatef A., Picchietti S., Cossignani L., Merrifield D. L., et al. (2018). Influence of probiotics administration on gut microbiota core: a review on the effects on appetite control, glucose, and lipid metabolism. J. Clin. Gastroenterol. 52, S50–S56. doi: 10.1097/MCG.0000000000001064, PMID: [DOI] [PubMed] [Google Scholar]
- Goda Y., Yamanaka D., Nishi H., Masuda M., Kamei H., Kumano M., et al. (2021). Dietary lysine restriction induces lipid accumulation in skeletal muscle through an increase in serum threonine levels in rats. J. Biol. Chem. 297:101179. doi: 10.1016/j.jbc.2021.101179, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Zhang J., Wang Z., Ang K. Y., Huang S., Hou Q., et al. (2016). Intestinal microbiota distinguish gout patients from healthy humans. Sci. Rep. 6, 20602. doi: 10.1038/srep20602, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illman R. J., Topping D. L., McIntosh G. H., Trimble R. P., Storer G. B., Taylor M. N., et al. (1988). Hypocholesterolaemic effects of dietary propionate: studies in whole animals and perfused rat liver. Ann. Nutr. Metab. 32, 95–107. doi: 10.1159/000177414, PMID: [DOI] [PubMed] [Google Scholar]
- Jiang J., Feng N., Zhang C., Liu F., Zhao J., Zhang H., et al. (2019). Lactobacillus reuteri A9 and lactobacillus mucosae A13 isolated from Chinese superlongevity people modulate lipid metabolism in a hypercholesterolemia rat model. FEMS Microbiol. Lett. 366:fnz254. doi: 10.1093/femsle/fnz254, PMID: [DOI] [PubMed] [Google Scholar]
- Karr S. (2017). Epidemiology and management of hyperlipidemia. Am. J. Manag. Care 23, S139–S148. PMID: [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology 31, 814–821. doi: 10.1038/nbt.2676, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby P., Buring J. E., Badimon L., Hansson G. K., Deanfield J., Bittencourt M. S., et al. (2019). Atherosclerosis. Nat. Rev. Dis. Primers. 5, 56. doi: 10.1038/s41572-019-0106-z [DOI] [PubMed] [Google Scholar]
- Ma Q., Zhou X., Hu L., Chen J., Zhu J., Shan A. (2020). Leucine and isoleucine have similar effects on reducing lipid accumulation, improving insulin sensitivity and increasing the browning of WAT in high-fat diet-induced obese mice. Food Funct. 11, 2279–2290. doi: 10.1039/c9fo03084k, PMID: [DOI] [PubMed] [Google Scholar]
- Maki J. J., Looft T. (2018). Megasphaera stantonii sp. nov., a butyrate-producing bacterium isolated from the cecum of a healthy chicken. Int. J. Syst. Evol. Microbiol. 68, 3409–3415. doi: 10.1099/ijsem.0.002991, PMID: [DOI] [PubMed] [Google Scholar]
- Manara S., Asnicar F., Beghini F., Bazzani D., Cumbo F., Zolfo M., et al. (2019). Microbial genomes from non-human primate gut metagenomes expand the primate-associated bacterial tree of life with over 1000 novel species. Genome Biol. 20, 299. doi: 10.1186/s13059-019-1923-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcil V., Delvin E., Garofalo C., Levy E. (2003). Butyrate impairs lipid transport by inhibiting microsomal triglyceride transfer protein in Caco-2 cells. J. Nutr. 133, 2180–2183. doi: 10.1093/jn/133.7.2180, PMID: [DOI] [PubMed] [Google Scholar]
- Mestdagh R., Dumas M. E., Rezzi S., Kochhar S., Holmes E., Claus S. P., et al. (2012). Gut microbiota modulate the metabolism of brown adipose tissue in mice. J. Proteome Res. 11, 620–630. doi: 10.1021/pr200938v, PMID: [DOI] [PubMed] [Google Scholar]
- Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- Panahi Y., Ahmadi Y., Teymouri M., Johnston T. P., Sahebkar A. (2018). Curcumin as a potential candidate for treating hyperlipidemia: A review of cellular and metabolic mechanisms. J. Cell. Physiol. 233, 141–152. doi: 10.1002/jcp.25756, PMID: [DOI] [PubMed] [Google Scholar]
- Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plovier H., Everard A., Druart C., Depommier C., Van Hul M., Geurts L., et al. (2017). A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113. doi: 10.1038/nm.4236, PMID: [DOI] [PubMed] [Google Scholar]
- Qian Y., Li M., Wang W., Wang H., Zhang Y., Hu Q., et al. (2019). Effects of lactobacillus Casei YBJ02 on lipid metabolism in Hyperlipidemic mice. J. Food Sci. 84, 3793–3803. doi: 10.1111/1750-3841.14787, PMID: [DOI] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450, PMID: [DOI] [PubMed] [Google Scholar]
- Roth G. A., Mensah G. A., Fuster V. (2020). The global burden of cardiovascular diseases and risks: A compass for global action. J. Am. Coll. Cardiol. 76, 2980–2981. doi: 10.1016/j.jacc.2020.11.021 [DOI] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T. P., Malik R. K., Katkamwar S. G., Kaur G. (2015). Hypocholesterolemic effects of lactobacillus reuteri LR6 in rats fed on high-cholesterol diet. Int. J. Food Sci. Nutr. 66, 71–75. doi: 10.3109/09637486.2014.953450, PMID: [DOI] [PubMed] [Google Scholar]
- Tong X., Xu J., Lian F., Yu X., Zhao Y., Xu L., et al. (2018). Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by metformin and a traditional Chinese herbal formula: a multicenter, randomized, open label clinical trial. mBio. 9, e02392–e02317. doi: 10.1128/mBio.02392-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414, PMID: [DOI] [PubMed] [Google Scholar]
- Velagapudi V. R., Hezaveh R., Reigstad C. S., Gopalacharyulu P., Yetukuri L., Islam S., et al. (2010). The gut microbiota modulates host energy and lipid metabolism in mice. J. Lipid Res. 51, 1101–1112. doi: 10.1194/jlr.M002774, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liao M., Zhou N., Bao L., Ma K., Zheng Z., et al. (2019). Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 26, 222–235.e5. doi: 10.1016/j.celrep.2018.12.028, PMID: [DOI] [PubMed] [Google Scholar]
- Wei Z. Y., Rao J. H., Tang M. T., Zhao G. A., Li Q. C., Wu L. M., et al. (2021). Characterization of changes and driver microbes in gut microbiota during healthy aging using a captive monkey model. Genomics Proteomics Bioinformatics S1672-0229:00258. doi: 10.1016/j.gpb.2021.09.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D. E., Lu J., Langmead B. (2019). Improved metagenomic analysis with kraken 2. Genome Biol. 20, 257. doi: 10.1186/s13059-019-1891-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Inoue N., Ozawa R., Maekawa T., Izumo T., Kitagawa Y., et al. (2013). Effects of heat-killed lactobacillus pentosus S-PT84 on postprandial hypertriacylglycerolemia in rats. Biosci. Biotechnol. Biochem. 77, 591–594. doi: 10.1271/bbb.120830, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.