Abstract
Chemokines are a family of cytokines that mediate leukocyte trafficking and are involved in tumor cell migration, growth, and progression. Although there is emerging evidence that multiple chemokines are expressed in tumor tissues and that each chemokine induces receptor‐mediated signaling, their collaboration to regulate tumor invasion and lymph node metastasis has not been fully elucidated. In this study, we examined the effect of CXCL12 on the CCR7‐dependent signaling in MDA‐MB‐231 human breast cancer cells to determine the role of CXCL12 and CCR7 ligand chemokines in breast cancer metastasis to lymph nodes. CXCL12 enhanced the CCR7‐dependent in vitro chemotaxis and cell invasion into collagen gels at suboptimal concentrations of CCL21. CXCL12 promoted CCR7 homodimer formation, ligand binding, CCR7 accumulation into membrane ruffles, and cell response at lower concentrations of CCL19. Immunohistochemistry of MDA‐MB‐231–derived xenograft tumors revealed that CXCL12 is primarily located in the pericellular matrix surrounding tumor cells, whereas the CCR7 ligand, CCL21, mainly associates with LYVE‐1+ intratumoral and peritumoral lymphatic vessels. In the three‐dimensional tumor invasion model with lymph networks, CXCL12 stimulation facilitates breast cancer cell migration to CCL21‐reconstituted lymphatic networks. These results indicate that CXCL12/CXCR4 signaling promotes breast cancer cell migration and invasion toward CCR7 ligand–expressing intratumoral lymphatic vessels and supports CCR7 signaling associated with lymph node metastasis.
Keywords: breast cancer, chemokine, invasion, lymph node, metastasis
Chemokines are a family of cytokines that mediate leukocyte trafficking and are involved in tumor cell migration, growth, and progression. We here demonstrated that CXCL12/CXCR4 signaling enhances breast cancer cell response at lower concentrations of CCR7 ligand chemokine. In the tumor microenvironment, CXCL12 may indirectly regulate lymph node metastasis by promoting CCR7 signaling in cancer cells.
1. INTRODUCTION
Lymphoid chemokines, CXCL12, CCL21, and CCL19, are expressed abundantly at luminal sites of the high endothelial venules (HEVs), stromal cells in the parenchyma of secondary lymphoid tissues, and lymphatic vessels in peripheral tissues. They regulate directional lymphocyte trafficking from HEVs into the lymphoid tissue parenchyma, intranodal motility of T cells, and dendritic cell migration into lymphatic vessels. 1 , 2 , 3 In addition to these physiological roles, studies have demonstrated that these lymphoid chemokines play a central role in cancer cell metastasis to lymph nodes (LNs). 4 , 5 , 6 CXCL12/SDF‐1α functions as a chemoattractant for lymphocytes through its receptor, CXCR4. 7 A variety of cancer types such as breast, prostate, gastric, and pancreatic cancers are CXCR4 positive, 8 and the CXCR4 is positively correlated with the metastatic potential to the organs with high CXCL12 expression. 9 , 10 Various mechanisms have been proposed concerning the function of CXCL12 in cancer proliferation and metastasis. For example, the CXCL12/CXCR4 axis promotes the growth of brain tumors and breast cancer metastasis by increasing cell survival. 11 , 12 It contributes to tumor growth by promoting angiogenesis in concert with VEGF, 13 and it also indirectly promotes tumor angiogenesis by recruiting CXCR4‐positive endothelial progenitor cells into carcinomas. 14
CCL21 and CCL19 are constitutively expressed in the lymphatic vessels, and their cognate receptor CCR7 is expressed in certain cancer types including breast cancer, melanoma, non–small cell lung cancer, stomach cancer, colorectal cancer, and T‐cell leukemia. 15 Clinical studies have demonstrated a positive correlation between CCR7 expression and the metastatic potential of cancer cells to the LNs, 16 , 17 , 18 suggesting that CCR7 expression promotes LN metastasis. In mouse metastasis models, CCR7 expression confers metastatic potential to B16 murine melanoma and breast cancer cells to regional LNs. 19 , 20 Additionally, several studies have demonstrated that CCL21 produced by lymphatic endothelial cells guides CCR7‐expressing metastatic malignant melanoma cell lines and that the CCL21 concentration gradient mediates CCR7‐dependent breast cancer chemotaxis under interstitial flow. 21 , 22
In the tumor microenvironment, a variety of chemokines are concomitantly expressed by cancer and stromal cells, and they can orchestrate cancer progression. 23 We have previously demonstrated that T‐cell reactivity to CCR7 ligands is enhanced when cells are pretreated with CXCL12 at higher concentration than the optimal one for migration. 24 CXCL12 treatment activates CXCR4‐mediated signaling and subsequently induces CCR7 homodimerization that promotes CCR7‐dependent migration and reactivity to CCR7 ligands. 25 , 26 , 27 In clinical samples, the relative expression levels of CXCR4 and CCR7 predicts LN metastasis in breast cancer. 28 These results support the idea that a collaboration between CXCR4‐ and CCR7‐mediated signaling promotes cancer cell metastasis to the LNs. To gain insight into the synergy between CXCL12 and CCR7 ligands during cancer progression, we determined whether CXCR4‐mediated signaling affects the CCR7‐dependent response in breast cancer cell migration, invasion, and LN metastasis. Here, we demonstrate that CXCL12‐CXCR4 engagement enhances CCR7‐dependent intracellular signaling and thus increases CCR7‐dependent cell migration to lymphatic tissues in a three dimensional (3D) reconstituted model.
2. MATERIALS AND METHODS
2.1. Cells
The human breast cancer cell line, MDA‐MB‐231 (MDA231), was kindly provided by Dr. K. Itoh of the Osaka Medical Center for Cancer and Cardiovascular Diseases. The cells were cultured in RPMI1640 medium supplemented with 10% (v/v) FCS, 2 mmol/L l‐glutamine, 1 mmol/L sodium pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin, 50 μmol/L 2‐ME, 0.1 mmol/L nonessential amino acids, and 10 mmol/L HEPES. The MDA231‐derived cell lines, Meta‐1 and Meta‐2, were established as follows. The puromycin‐resistant cells established by transfection of pPGKpuro plasmid 29 into MDA231 cells were inoculated into the thoracic fat pads of CB17/Icr‐Prkdc SCID mice (Charles River Laboratories Japan). Twenty weeks after injection, axillary LNs from the MDA231 tumor–bearing mice were dissected, cut into small pieces, washed, and digested in HBSS supplemented with 0.1% (w/v) collagenase type I (Worthington Biochemicals), 0.01% (w/v) hyaluronidase, and 20% (w/v) DNase (Sigma‐Aldrich) for 3 hours with stirring at 37°C. The tissue digest was allowed to form colonies in RPMI1640 supplemented with 10% FCS and puromycin. The puromycin‐resistant cells were reinoculated into SCID mice. Another round of in vivo selection and in vitro expansion yielded Meta‐1 and Meta‐2, which were cultured in a selective medium containing 0.1 μg/mL puromycin.
The cells expressing human CCR7‐EGFP and CXCR4‐mCherry were established as follows: The entire open reading frames of human CCR7 and CXCR4 were amplified by PCR, and subcloned into the pEGFP‐N3 and pmCherry‐N1 plasmids (Takara Bio USA), respectively. The DNA fragment of CCR7‐EGFP was inserted into the retroviral expression vector pCXbsr 30 to generate pCXbsr‐CCR7‐EGFP, and that of CXCR4‐mCherry was inserted into the pCAGI‐puro vector (kindly provided by Dr. J. Miyazaki, Osaka University). The pCXbsr‐CCR7‐EGFP plasmid was transiently introduced into HEK293T cells and the supernatant containing retroviral particles were added to MDA231 cells. The infected MDA231 cells were cultured in a growth media containing 0.5 μg/mL blasticidin, and drug‐resistant cells were pooled. The blasticidin‐resistant cells were then transfected with a plasmid encoding CXCR4‐mCherry by MultiFectam (Promega) and cultured in a selective culture medium containing 0.5 μg/mL puromycin and blasticidin to establish MDA‐R7/X4 cells. To establish CCR7 overexpressing cell lines and control cells, Meta‐1 cells were treated with recombinant retroviral particles including pCXbsr‐CCR7‐EGFP or pCXbsr‐EGFP and subsequently cultured in a growth medium containing 0.5 μg/mL blasticidin. Single‐cell–derived colonies were selected by the colony‐picking method from the culture plate of pCXbsr‐CCR7‐EGFP retroviral infection to establish four Meta‐1 subclones, CCR7‐1 to CCR7‐4. As a control, a polyclonal cell pool of pCXbsr‐EGFP retroviral infection was obtained. For the establishment of CCR7‐knockdown cells, Meta‐1 cells were transfected with a human CCR7‐specific shRNA expression plasmid (CCR7i; 348‐TCTCTGGTCGTGTTGACCTATATCTGCAG‐473, SureSilencing shRNA plasmid, SABiosciences) or a negative control plasmid (shRNA‐NC; 348‐TCTCGGAATCTCATTCGATGCATACGCAG‐473) using ESCORT V transfection reagent (Sigma‐Aldrich) and were subsequently cultured in a growth medium containing 100 μg/mL hygromycin to establish drug‐resistant cells.
2.2. Cell migration assay
Recombinant human CXCL12 and CCL21 were purchased from R&D Systems. A transwell cell migration assay was performed using a 24‐well chemotaxis chamber with 5‐µm pore inserts precoated with 20 µg/mL fibronectin (Sigma‐Aldrich) for 1 hour at room temperature. The cells (1 × 105 cells/mL) were resuspended in DMEM/F12 supplemented with 2% FCS and added to the upper chambers, whereas CXCL12 or CCL21 to the lower chambers. After incubating for 18 hours, the cells on the lower surface of the membrane were fixed with methanol and stained with hematoxylin and eosin. Cell number was counted under a light microscope in eight fields per membrane at a 200‐fold magnification. The relative chemotactic index was determined as the ratio of the proportion of cells that migrated in response to multiple chemokines to the proportion that migrated in response to a single chemokine. The assay was performed in triplicate. Data were expressed as the mean ± SD and were analyzed by Student's t test. A two‐tailed P‐value of 0.05 or less was considered significant.
3. COLLAGEN GEL INVASION ASSAY
A 48‐well tissue culture plate was treated with 0.8 mg/mL type I collagen from rat tail (Merck Millipore) and incubated at 37°C for 3 hours. After washing with RPMI1640 for 10 minutes, 100 μL of 10 ng/mL CCL21 was added to each well. The cells (1 × 104 cells/well, 0.5 mL) were treated with or without 2.5 µg/mL CXCL12 for 30 minutes in the presence of anti‐human CCR7 mAb (clone 150503, R&D Systems), anti‐human CXCR4 mAb (clone 44717, R&D Systems), or control mouse IgG (R&D Systems), plated onto the surface of the gel, and then incubated for 15 hours. The number of cells on the surface of the collagen matrix and those that migrated into the matrix were counted microscopically at a 400‐fold magnification in three random fields per well for triplicate gel cultures. Data are expressed as the percentage of cell invasion calculated as cells in the gel relative to the total cell number (in gel + on gel) × 100.
3.1. In vivo bioluminescence imaging
The pCBR‐Control vector (Promega) was double‐digested with HindIII and XbaI, and the 1667‐bp DNA fragment corresponding to the click beetle luciferase gene (CBRluc) was subcloned into the mammalian expression vector pCR3.1‐Uni (Invitrogen). The MDA231 variants transfected with pCR3.1‐Uni containing CBRluc were grown in standard media containing 1 mg/mL G418, and the drug‐resistant cells were analyzed by a luciferase assay using the Chroma‐Glo Luciferase Assay System (Promega). The luciferase‐positive cells (5 × 106 cells) in 50 μL PBS containing 40% Matrigel (BD Biosciences) were injected into the right inguinal mammary fat pad of 6‐9‐week‐old female SCID mice. A sham operation was performed with PBS on the contralateral side. The mice were anesthetized with isoflurane, and D‐luciferin (VivoGlo, Promega) was injected intraperitoneally (150 mg/kg). Cells were imaged using an IVIS Imaging System (Caliper Life Sciences). The presence of LN metastasis was determined when the bioluminescence signal was first detected in the right axillary LN with a 5‐second exposure time by IVIS.
3.2. Flow cytometry
Cells harvested in 0.02% EDTA were incubated with 10 µg/ml phycoerythrin‐ or 20 µg/mL Alexa Fluor647–conjugated mouse anti‐human CCR7 mAb (clone MAB197, R&D Systems), biotinylated mouse anti‐human CXCR4 (clone 44717, R&D Systems), or biotinylated control mouse IgG for 30 minutes on ice, and subsequently with 5 μg/mL allophycocyanin‐conjugated streptavidin (BD Biosciences). The cells were washed, centrifuged at 1200 rpm for 5 minutes at 4°C, and resuspended in PBS containing 0.1% BSA. Flow cytometric analysis was performed on a Gallios flow cytometer (Beckman Coulter).
3.3. Immunohistochemistry
Tumor xenografts were dissected twelve weeks after cell injection, and serial sections of fresh‐frozen tumor specimens were subjected to immunohistochemical staining. CXCL12 and p53 were detected with goat anti‐human/mouse CXCL12 pAb (sc‐6193, Santa Cruz), biotinylated mouse anti‐human p53 mAb (clone DO‐7, Dako), Alexa Fluor488–conjugated donkey anti‐goat IgG (Molecular Probes), and Alexa Fluor 568–conjugated streptavidin (Thermo Fisher Scientific). LYVE‐1 was detected with 2 μg/ml rat anti‐mouse LYVE‐1 mAb (clone 223322, R&D Systems) and Alexa Fluor488–conjugated mouse anti‐rat IgG. CCL21 was detected with biotinylated rabbit anti‐mouse CCL21 pAb (500‐P114, Peprotech) and by a tyramide signal amplification system with horseradish peroxidase–conjugated streptavidin (NEL750, PerkinElmer) and Alexa Fluor568 Tyramide (B40933, Thermo Fisher Scientific). Alpha‐smooth muscle actin (α‐SMA) was detected with FITC‐labeled anti‐antibody (F3777, Sigma; 1: 250 dilution). As a control, 1 μg/mL FITC‐labeled mouse IgG2a (eBioscience) was used. For detection of CCL21 and LYVE‐1 in 3D lymphatic networks, 1 μg/mL biotinylated goat anti‐human CCL21 pAb (BAF366, R&D Systems), horseradish peroxidase–conjugated streptavidin, Alexa Fluor488 Tyramide (B40953, Thermo Fisher Scientific), and 5 μg/mL eFluor660‐conjugated anti‐LYVE‐1 mAb (clone ALY7, eBioscience) were used. The tissues were stained with 2 μg/mL Hoechst 33342 (Invitrogen) for 10 minutes, mounted with Fluoromount‐G (SouthernBiotech), and observed with a confocal laser scanning microscope (Fluoview1000‐D, Olympus).
3.4. RT‐PCR and DNA sequencing
Total RNA was purified using the RNeasy Mini kit (Qiagen). PCR amplification was performed using DNA polymerase (KOD‐FX, Takara) and 0.2 μg of template cDNA synthesized by the ReverTra Ace qPCR RT Kit (TOYOBO Co., Ltd.). The PCR reaction was performed with a pair of oligonucleotide primers (0.3 µmol/L) with gene‐specific sequences described below:
CXCL12 forward: 5′‐CGCTCTGCATCAGTGACGGTA‐3′
CXCL12 reverse: 5′‐GTTCTTCAGCCGTGCAACAATC‐3′
CCL21 forward: 5′‐GGACCCAAGGCAGTGATGGAGG‐3′
CCL21 reverse: 5′‐TTGGAGCCCTTTCCTTTCTT‐3′
GAPDH forward: 5′‐GGAAAGCTGTGGCGTTGGCGTGAT‐3′.
GAPDH reverse: 5′‐CTGTTGCTGTAGCCGTATTC‐3′
The PCR product was separated by electrophoresis on a 2% agarose gel and purified using the Wizard SV Gel and PCR Clean‐Up System (Promega), and the nucleotide sequence was determined by using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific).
3.5. Cell invasion model using 3D multilayered tissues with lymphatic networks
The 3D multilayered tissues including lymphatic networks were constructed as described previously. 31 , 32 Briefly, primary human dermal lymphatic endothelial cells (Lonza) with or without 100 ng/mL CCL21 were embedded in multilayered human dermal fibroblasts constructed on a cell culture insert with 0.4‐μm pores. After 2 days of culture, which allows lymphatic networks to extend into the 3D tissues, CCR7‐1 cells pretreated with or without 5 μg/mL CXCL12 were added to the top surface. The tissues were fixed with 4% paraformaldehyde after a 3‐day culture and stained with anti‐human CD31 antibody. Images of the tissues were obtained by fluorescence microscopy (Fluoview FV10i, Olympus) and analyzed by Volocity 3D image analysis software (PerkinElmer).
3.6. Luciferase complementation assay
pEF1‐CXCR4‐Nluc and pN1‐β‐arrestin2‐Cluc plasmids 33 were gifted by Drs. K. E. Luker and G. D. Luker (University of Michigan Medical School). pEF1‐CXCR4‐Nluc was digested with the NotI/SalI restriction enzymes to remove CXCR4 from the plasmid. The human CCR7 gene amplified by PCR was inserted into the vector to generate the pEF1‐CCR7‐Nluc plasmid. MDA231 cells were cotransfected with pEF1‐CCR7‐Nluc and pN1‐beta‐arrestin2‐Cluc plasmids using Nucleofector technology (Lonza) according to the manufacturer's instructions. One day after transfection, the cells were seeded at a density of 1 × 104/well in a 96‐well plate coated with collagen I (BD Biosciences) and treated with or without 2.5 µg/mL recombinant human CXCL12 and 150 µg/ml D‐luciferin for 30 minutes at 37°C. The cells were then treated with 1 µg/mL recombinant human CCL21 and 150 µg/mL D‐luciferin in growth medium supplemented with 0.5% FCS and analyzed by IVIS Imaging System.
For the detection of CCR7 homodimer, cells were transiently transfected with the plasmids, CCR7‐NGLuc and CCR7‐CGLuc, encoding a chimeric N‐terminal or C‐terminal protein of gaussia luciferase and human CCR7. 26 The cells were then treated with 0.5 µg/mL recombinant human CXCL12 or 0.1% BSA‐containing PBS for 30 minutes at 37°C. Following the addition of 10 µg/mL coelenterazine, luminescence signals were integrated for 1 second using a Glomax 20/20 luminometer (Promega). Four samples were used for all experimental conditions.
3.7. CCL19‐Fc binding
For the CCR7 ligand‐binding assay, cells were seeded into collagen type I–coated wells and treated with 0.1% BSA in PBS or 100 ng/mL CXCL12 for 30 minutes at 37°C, followed by 1 μg/mL CCL19‐Fc (eBioscience) for 30 minutes at 4°C. After staining with 5 μg/mL biotin‐conjugated anti‐human IgG (American Qualex) and 5 μg/mL Alexa Fluor 647–conjugated streptavidin (Thermo Fisher), the cells were imaged by DeltaVision Imaging System (Applied Precision).
3.8. Analysis of membrane ruffling
For phalloidin staining near the cell membrane, MDA‐R7/X4 cells were cultured in eight‐well Lab‐Tek Permanox chamber slides coated with fibronectin and were treated with or without 25 μg/mL AMD3100 (Sigma‐Aldrich) in 0.1% BSA/HBSS for 30 minutes at 37°C, followed by the indicated concentrations of CXCL12 for an additional 30 minutes at 37°C. The cells were permeabilized with 0.1% Triton X‐100 for 3 minutes, fixed with 4% PFA for 10 minutes at room temperature, and incubated with 4 μg/mL Alexa647‐phalloidin (Molecular Probes) for 5 minutes at room temperature. The cell images were captured by a confocal laser microscope. The ruffling index was calculated as described previously with minor modifications 34 ; 0 = no ruffles, 1 = isolated areas of ruffling covering less than 25% of the peripheral area, 2 = extensive ruffling covering more than 25% of the peripheral area. At least 50 cells were analyzed, and the average score per cell was calculated. Statistical analysis was done using a two‐tailed Mann‐Whitney U test.
3.9. Detection of GM1 and CD44
The cells were cultured overnight on an eight‐well slide chamber coated with type I collagen (BD biosciences). After 4% paraformaldehyde fixation, the cells were treated with 4 μg/mL Alexa Fluor 647–labeled cholera toxin subunit B (CTxB, Molecular Probes) for 20 minutes at room temperature or 10 μg/mL biotinylated anti‐CD44 mAb (clone 5F12) for 30 minutes at 4°C, and subsequently with 10 μg/mL Alexa Fluor 647–labeled streptavidin (Molecular Probes). The samples were analyzed using a DeltaVision microscope system.
3.10. Ethics approval and consent to participate
The experimental protocols for use of laboratory animals were approved by the Ethics Review Committee of Kindai University. All experiments were conducted in accordance with the approved guidelines from Kindai University.
4. RESULTS
4.1. CXCL12 promotes CCL21‐induced cancer cell migration
We previously reported that the CXCR4‐mediated signaling promotes CCR7 ligand–induced lymphocyte chemotaxis and cell migration to LNs. 24 , 25 To determine whether the chemokine cooperation also contributes to breast cancer cell chemotaxis and migration, we used MDA231 cells, which express both CXCR4 and CCR7 at the mRNA and protein level. We confirmed that MDA231 exhibits CXCL12‐ and CCL21‐mediated cell migration, reaching its highest values at 40 ng/mL CXCL12 and at 100 ng/mL CCL21, respectively (Figure 1A and B). When MDA231 cells were mixed with 1−5 μg/mL CXCL12 in the upper wells, they exhibited enhanced cell migration toward CCL21 in the lower wells, which was not observed with CXCL12 alone or CCL21 alone, suggesting that CXCL12 and CCL21 functionally cooperated to induce cell migration (Figure 1C). The enhancing effect of CXCL12 was almost completely reduced to the basal levels by adding anti‐CXCR4 neutralizing antibody (Figure 1D), which indicates that the effect of CXCL12 was primarily dependent on CXCR4.
FIGURE 1.
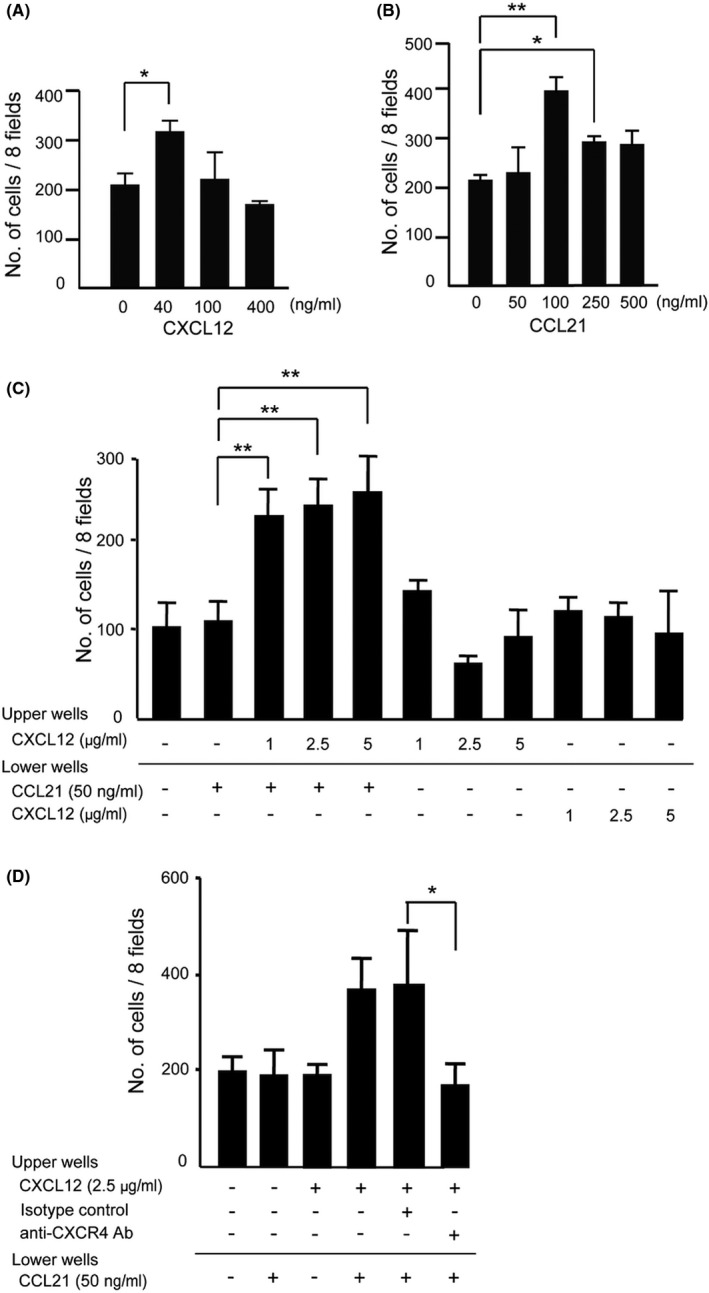
The effects of CXCL12 on CCR7 ligand–dependent MDA231 cell migration. A, CXCL12‐induced cell migration assay was performed using MDA231 cells in the presence or absence of CXCL12 at indicated concentrations. The number of migrated cells to the lower wells in response to CXCL12 was analyzed. Graphs represent means ± SD of the number of cells in eight random fields per membrane in triplicate assay. * p < 0.05 by Student's t test. B, CCL21‐induced cell migration of MDA‐MB‐231. The assay was performed in the presence of CCL21 in the lower wells at indicated concentrations. One‐way ANOVA; *p < 0.05, **p < 0.01. C, The effect of CXCL12 on MDA231 cell migration induced by a suboptimal concentration of CCL21 (50 ng/mL). The assay was performed under the indicated concentrations of CXCL12 and CCL21 added to the upper or the lower wells. One‐way ANOVA; **p < 0.01. D, The effect of CXCL12 on CCL21‐induced cell migration in the presence of anti‐CXCR4 neutralizing antibody. CXCL12 (2.5 μg/mL) was added to the upper wells with or without anti‐CXCR4 mAb or an isotype control immunoglobulin (10 μg/mL). Mean ± SD (Student's t test; *p < 0.05, **p < 0.01, n = 3)
4.2. CXCL12 promotes CCL21‐dependent cancer cell invasion
Considering that CXCL12 promotes CCR7‐dependent signaling in cancer cells, we determined whether the CXCL12/CXCR4 axis contributes to CCR7‐driven cell invasion and LN metastasis. Therefore, we established the MDA231‐derived variants, Meta‐1 and Meta‐2, both of which metastasize to LNs in contrast to the parental MDA231 cells (Table 1). The two variants showed comparable growth and invasive properties into collagen gels in the presence or absence of CCL21 or CXCL12 in vitro (Figure S1A and B, Figure S2). On the other hand, these variant cells exhibited more aggressive growth and LN‐specific metastatic properties in vivo (Table 1) and a slight increase in cell surface CXCR4 expression and decrease in CCR7 expression (Figure S1C). CXCL12 promoted CCR7‐dependent cell invasion in the two metastatic variants but not parental cells at a minimal CCL21 concentration (10 ng/mL) required for invasion (Figure 2A). One of the variants, Meta‐1, metastasized to LNs in a CCR7‐dependent manner, as confirmed by a shRNA‐mediated knockdown of CCR7, whereas the cells with >94% silencing efficiency exhibited a tendency to decrease in tumor growth (Figure S3A) and LN metastasis (Table 1 and Figure S3B). The effect of CXCL12 on the cell invasion frequency of Meta‐1 was significantly inhibited by anti‐CXCR4 or anti‐CCR7 monoclonal antibody (Figure 2B), which is in agreement with the idea that the CXCL12/CXCR4 axis promotes CCR7‐dependent cell invasion.
TABLE 1.
Lymph node metastasis frequency of MDA231‐derived cells
| 2 (w) | 3 (w) | 4 (w) | 7 (w) | |
|---|---|---|---|---|
| Parental cell | 0/3 a | 0/3 | 0/3 | NM b |
| Meta‐1 | 1/4 | 2/4 | 3/4 | NM |
| Meta‐2 | 0/3 | 0/3 | 1/3 | NM |
| Control shRNA (Meta‐1) | 0/5 | 2/5 | 2/5 | 5/5 |
| CCR7 shRNA (Meta‐1) | 0/6 | 0/6 | 0/6 | 5/5 |
Luciferase‐expressing cells were orthotopically injected into the right inguinal mammary fat pad of 6‐9‐w‐old female SCID mice on day 0, and bioluminescence signal was detected by IVIS.
Number of mice with detectable signals of axillary lymph node metastasis/number of mice with tumor at injection site.
Not measured.
FIGURE 2.
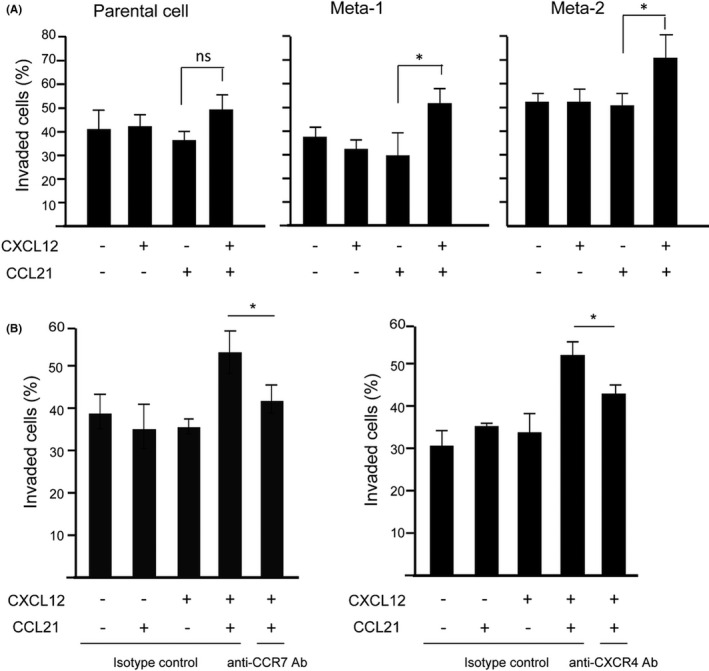
The effects of CXCL12 on CCR7‐mediated MDA231 cell invasion to the collagen gel. A, Parental MDA231 cells, Meta‐1, and Meta‐2 cells were pretreated with 1 μg/mL CXCL12 and subjected to a collagen gel cell invasion assay under a suboptimal concentration (10 ng/mL) of CCL21. After 15 h incubation, the number of cells on the surface of the collagen matrix and those migrated into the matrix was counted, and the percentage of cell invasion was analyzed. Data represent the mean ± SD of three independent experiments, each performed in triplicate wells (Student's t test; *p < 0.05). B, Meta‐1 cells were pretreated with CXCL12 in the presence of 10 μg/mL anti‐CCR7 mAb (left panel), anti‐CXCR4 mAb, or isotype control and were then applied to a collagen gel containing CCL21
Further support for this idea came when we used different MDA231‐derived variant cell lines, CCR7‐1 to ‐4, that overexpress human CCR7 and EGFP‐tag fusion proteins (Figure S4A). Using an invasion assay with a minimum concentration of CCL21 for parental cells, most CCR7‐overexpressing cell lines were more invasive compared with mock‐transfected cells (Figure S4B), indicating that CCR7‐overexpression confers a higher sensitivity to CCL21. When the CCR7‐overexpressing cells were orthotopically injected, they tended to metastasize to the ipsilateral axillary LNs more rapidly compared with mock‐transfected cells (Table 2), which supports the idea that increased CCR7 signaling promotes efficient cell invasion and metastasis in vivo. CCR7‐overexpressing cells grew at comparable rates to the mock‐transfected cells over a 2 to 4‐week period, indicating that primary tumor growth is not significantly affected by CCR7 signaling (Figure S4C).
TABLE 2.
Lymph node metastasis frequency of CCR7‐expressing subclones
| 1 (w) | 2 (w) | 3 (w) | 4 (w) | |
|---|---|---|---|---|
| Mock | 0/5 a | 0/5 | 0/5 | 2/5 |
| CCR7‐1 | 0/5 | 2/5 | 3/5 | 3/5 |
| CCR7‐2 | 0/5 | 2/5 | 3/5 | 4/5 |
| CCR7‐3 | 0/5 | 1/5 | 2/5 | 4/5 |
| CCR7‐4 | 0/5 | 1/5 | 2/5 | 3/5 |
Luciferase‐expressing cells were orthotopically injected into the right inguinal mammary fat pad of SCID mice on day 0.
Number of mice with detectable signals of axillary lymph node metastasis/number of mice with tumor at injection site.
4.3. CXCL12 is expressed in the pericellular matrix, whereas CCL21 associates with lymphatic vessels at the primary tumor site
To investigate the possible interplay between CXCL12 and CCR7 ligands in the tumor microenvironment, we determined the localization of CXCL12 and CCL21 in MDA231‐derived tumor tissue. We found that CXCL12 was primarily expressed in the pericellular matrix surrounding p53‐positive tumor cells (Figure 3), whereas CCL21 was selectively expressed in the matrix surrounding and within the lumen of LYVE‐1+ intratumoral lymphatic vessels (Figure 3A). These results are consistent with the idea that CXCL12 signals CXCR4‐expressing tumor cells to modulate their response to perilymphatically expressed CCL21.
FIGURE 3.
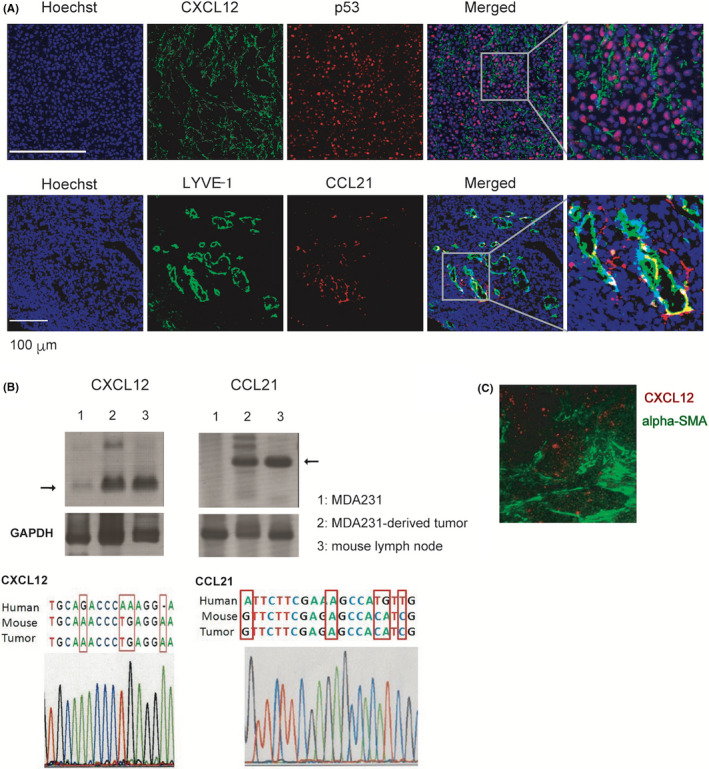
Expression of CXCL12 and CCL21 in MDA231‐derived tumor. A, Parental MDA231 cells (1 × 106) were injected into the thoracic mammary fat pad of female SCID mice at 9 weeks of age (n = 3). A sham operation was performed on the contralateral side. Twelve weeks after injection, primary tumors were dissected. Serial sections of fresh‐frozen tumor specimens from tumor xenograft were subjected to immunohistochemical staining with anti‐CXCL12 (green, upper panel) and anti‐human p53 mAb (red, upper panel) or anti‐LYVE‐1 (green, lower panel) and anti‐CCL21 (red, lower panel) antibodies. Scale bar, 100 µm. B, Upper row: RT‐PCR analysis of CXCL12 and CCL21 in the parental MDA231 cell line, MDA231‐derived tumor tissue, and C57BL/6 mouse lymph node using oligonucleotide primer pairs common to mouse and human sequences. GAPDH were used as an endogenous control. Bottom: the tumor‐derived nucleotide sequence of the RT‐PCR products was determined and compared with the species‐specific nucleotide positions of the CXCL12 and CCL21 genes. C, Localization of CXCL12 expression (red) and alpha‐smooth muscle actin (α‐SMA)‐positive cancer‐associated fibroblast (green) in MDA231‐derived tumor tissue
We examined whether CXCL12 and CCL21 expressed in tumor tissue were derived from transplanted MDA231 cells or from recipient mouse tissue. Total RNA was extracted from the tumor tissue, and RT‐PCR was performed using primers containing human and mouse common nucleotide sequences for CXCL12 and CCL21. As shown in Figure 3B, signals of expected size (152‐bp fragment for CXCL12 and 301‐bp for CCL21) were detected in MDA231‐derived tumor and mouse LNs. On the other hand, no prominent signal was detected in MDA231 cells cultured in vitro. The DNA sequencing of the amplified band from the tumor tissue sample revealed that they exhibited the mouse CXCL12 and CCL21 nucleotide sequences (Figure 3B, lower row), suggesting that CXCL12 and CCL21 expressed in tumor tissues are predominantly derived from mouse cells.
It has been reported that CXCL12 is produced by cancer stromal cells including cancer‐associated fibroblasts (CAFs), which are involved in cancer cell growth. 14 , 35 In human breast tumor xenografts, α‐SMA–expressing CAFs with myofibloblastic properties (myCAFs) secreted CXCL12. 14 Considering the possibility that CXCL12 is produced from myCAFs in MDA231‐derived tumor tissue, we analyzed the localization of CXCL12 and α‐SMA expression in the tumor tissue. Both CXCL12 and α‐SMA were detected in the interstitial part of the cancer tissue, but CXCL12 and α‐SMA were detected mainly in different locations (Figure 3C), suggesting that CXCL12 in the MDA231‐derived tumor tissue is produced by mouse cells other than myCAFs.
4.4. CXCL12 promotes CCL21‐induced cell migration toward artificial lymphatic vessels
To verify that CXCL12 supports the guidance of cancer cell migration toward CCL21‐expressing lymphatic vessels, we determined the effect of CXCL12 on cell migration using a 3D tumor invasion model, in which cancer cells were added to lymphatic vessels grown in the 3D multilayered artificial tissues (Figure S5A). Because these tissues did not express detectable levels of CCL21 as confirmed by quantitative PCR and immunohistochemical analysis, we exogenously added recombinant human CCL21 to the lymphatic networks so that CCL21 would be locally present in lymphatic tissues, and subsequently added CXCL12‐ or PBS‐treated cells on top of the 3D tissues. CCL21 is known to bind to podoplanin on lymphatic endothelial cell surface 36 and extracellular matrix proteins; 37 therefore, exogenously added CCL21 is potentially captured by lymphatic networks grown in the 3D tissues. The CCL21‐reconstituted lymphatic endothelial cells were verified by colocalization of CCL21 and LYVE‐1 (Figure S5B). As shown in Figure 4A, CXCL12 treatment enhanced the interaction between cancer cells and lymphatics (Figure 4A). Further analysis of these images revealed that CXCL12‐treated cells were more intimately associated with CCL21‐reconstituted lymphatic vessels, as determined by measuring the distance between individual cancer cells and the nearest lymphatics (Figure 4B). Furthermore, when the data were analyzed at a 10‐μm cutoff, CXCL12 treatment exhibited a higher percentage of cells present within this distance compared with mock treatment, which supports the idea that CXCL12 promotes cancer cell migration toward CCL21‐expressing lymphatic vessels (Figure 4C).
FIGURE 4.
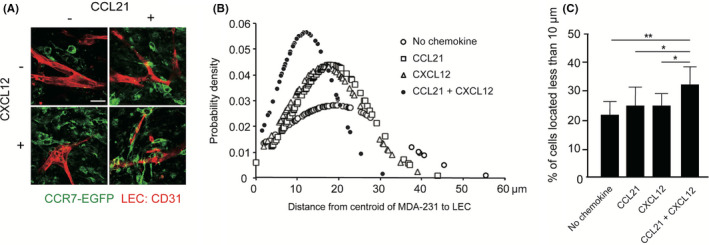
The effects of CXCL12 on MDA231 cell migration toward lymphatic vessels. A, The fluorescent images of CCR7‐1 cells (green) invading into tissues are shown. Lymphatic networks were visualized by immunostaining with anti‐CD31 antibody (red). Scale bar, 50 µm. B, Distribution of CCR7‐1 cells’ distance from the nearest lymphatic network is shown. In each sample, images including 100–150 cells (n = 5) were captured and subjected to the analysis. The results shown are representative of three independent experiments. C, The percentages of CCR7‐1 cells located less than 10 µm away from the nearest network are analyzed. Data represent the mean ± SD percentage from triplicate images of three independent experiments (one‐way ANOVA; *p < 0.05, **p < 0.01)
4.5. CXCL12 promotes CCR7 homodimer formation, ligand binding, and CCR7 localization to membrane ruffles
We previously demonstrated that CCR7 homodimer formation promotes CCR7‐mediated cell migration and CCR7 ligand binding. 26 To determine whether CXCL12 affects CCR7 homodimer formation in MDA231 cells, we conducted a split luciferase assay as previously described. 26 Consistent with our previous work, CXCL12 significantly increased luminescence signals compared with the control treatment of MDA231 cells, indicating that CXCL12 promoted CCR7 homodimer formation (Figure 5A). As expected, CXCL12 treatment resulted in an approximately twofold increase in CCL19‐Fc fusion protein binding to membrane ruffles in CCR7‐EGFP–expressing cells compared with mock treatment (Figure 5B). To determine whether CXCL12 potentiates CCR7‐mediated signaling, we performed a firefly luciferase–based fragment complementation assay, in which CCR7‐mediated signaling was evaluated by β‐arrestin 2 binding to the receptor, a method used for detecting CXCR4 signaling. 33 After CXCL12 pretreatment, the cells showed significantly increased levels (approximately 2.5‐fold) of a luminescence signal that peaked at 10–15 minutes in response to CCL21 as compared with the vehicle control, whereas CCL21 alone exhibited an approximately 1.5‐fold induction (Figure 5C). These results indicate that CXCL12 enhances CCR7‐mediated signaling.
FIGURE 5.
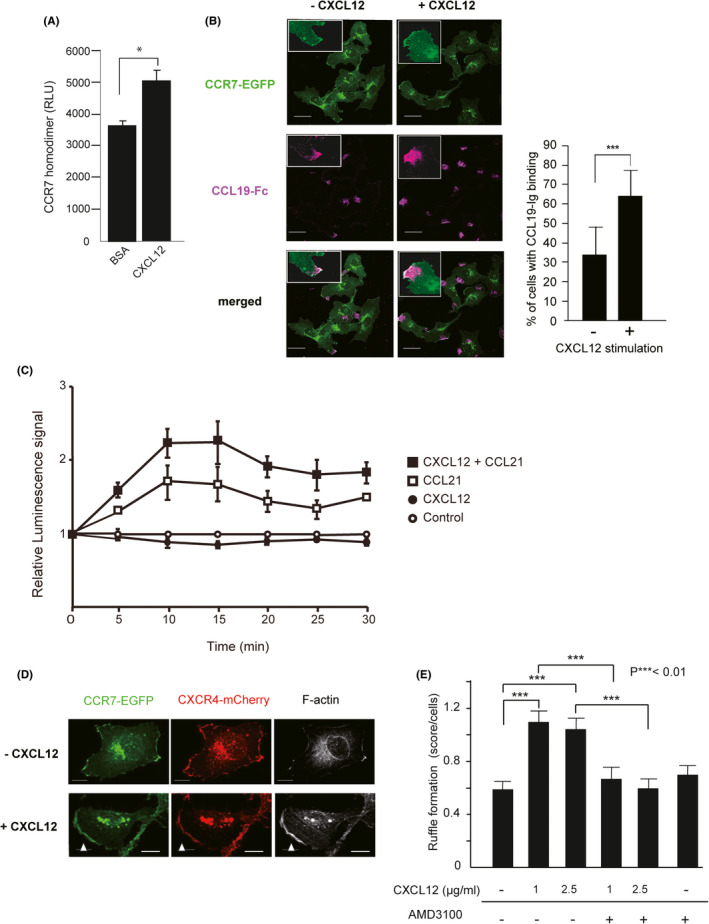
The effects of CXCL12 on CCR7 homodimerization, ligand binding, and the plasma membrane localization. A, CCR7 homodimer formation after treatment with CXCL12 in MDA231 cells. The levels of bioluminescence signals are shown for cells transfected with combinations of CCR7‐CGLuc and CCR7‐NGLuc in the presence of BSA or CXCL12. A representative experiment from at least three independent experiments is shown. Data represent mean ± SD (n = 4). *p < 0.05 by Student's t test. B, Confocal microscopic images of MDA‐R7/X4 cells treated with or without CXCL12 for 30 min, fixed, and stained with recombinant CCL19‐Fc, biotin‐anti‐human IgG, and Alexa Fluor 647–conjugated streptavidin. The expressions of CCR7‐EGFP (green) and CCL19‐Fc (magenta) are shown. Insets show high magnification of membrane ruffles. The images were analyzed to obtain the percentage of the cells with CCL19‐Fc binding. Scale bar, 30 µm. ***p < 0.01 by Student's t test. C, Luciferase complementation assay using CCR7‐Nluc and β‐arrestin 2‐Cluc. CCL21 was added 30 min after treatment of cells with CXCL12 or a solvent, and the luminescence signal was measured at each time point indicated. Error bars indicate standard error of the mean. D, MDA‐R7/X4 cells were stimulated with or without CXCL12 for 30 min, fixed, and stained with Alexa647‐phalloidin. Fluorescence images were captured by a confocal microscopy. The expression of CCR7 (green), CXCR4 (red), and F‐actin (white) are shown. White arrowheads show CXCR4‐ and CCR7‐enriched membrane ruffles. Scale bar, 10 µm. E, The average score of CXCR4/CCR7‐enriched membrane ruffles of each cell. The cells were treated with the indicated concentrations of CXCL12 in the presence or absence of CXCR4‐specific antagonist AMD3100. The ruffling index was evaluated as 0 = no ruffles, 1 = ruffles covering less than 25% of the peripheral area, and 2 = ruffles covering more than 25% of the peripheral area. The statistical difference was determined by one‐way ANOVA and depicted with ***p < 0.01
To further investigate the mechanism responsible for the enhanced CCR7‐dependent signaling, we determined the effects of CXCL12 treatment on the subcellular distribution of CCR7 in plasma membrane. In MDA231 cells stably expressing CCR7‐EGFP and CXCR4‐mCherry molecules (MDA‐CCR7/CXCR4), the majority of CCR7 and CXCR4 were located in partially overlapping cellular compartments (Figure 5D). Without CXCL12 stimulation, large portions of CCR7‐EGFP and CXCR4‐mCherry were observed intracellularly, and the majority of CCR7 were found in close association with the plasma membrane, as previously reported. 38 Upon CXCL12 stimulation, CCR7 was recruited to the plasma membrane ruffles and localized closely with CXCR4 and F‐actin (Figure 5D, white arrowheads). The analysis of CXCR4/CCR7‐enriched membrane ruffles in each cell revealed that CXCL12 stimulation increased membrane ruffle formation in MDA231 cells (Figure 5E). The effect was restored to basal levels by treatment with the selective CXCR4 antagonist, AMD3100, suggesting that the effect of CXCL12 is mediated by CXCR4. These data suggest that CXCL12/CXCR4 signaling facilitates the formation of CCR7‐enriched plasma membrane ruffles.
Previous studies have demonstrated that chemokine‐dependent downstream signaling is initiated by molecules recruited to lipid rafts in the ruffling membrane. 39 We confirmed that both CCR7 and CXCR4 are localized closely at this site with GM1‐ and CD44‐positive lipid rafts in MDA231 cells (Figure 6).
FIGURE 6.
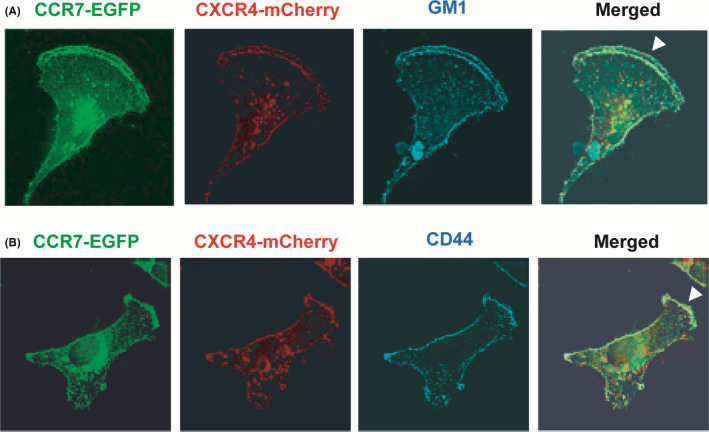
Localization of CCR7 and CXCR4 in the lipid rafts of ruffling membrane. A, CCR7 (green) and CXCR4 (red) were detected in cells expressing CCR7‐EGFP and CXCR4‐mCherry. GM1 (blue) was detected with Alexa Fluor647‐labeled CTxB. B, CD44 (blue) was detected with a biotinylated anti‐human CD44 mAb followed by Alexa Fluor647–conjugated streptavidin. White arrowheads represent ruffling membrane
5. DISCUSSION
We have previously demonstrated that CXCR4 and CCR7, the major chemokine receptors expressed in naïve lymphocytes, are critically required for T‐cell entry into LNs and Payer's patches that express chemokine ligands and act cooperatively during these events. 24 , 25 In the present study, we have proposed that CXCL12/CXCR4 signaling enables cancer cells to react efficiently to the CCR7 ligands, which promotes cancer cell interaction with intratumoral lymphatic vessels that eventually travel to tumor‐draining LNs.
Consistent with our previous findings in mouse and human T cells, 24 , 25 our data suggest that CXCL12 and CCR7 ligands act synergistically in human breast cancer cells and promote cell migration and invasion in vitro. We have previously reported that the efficacy of CCR7 ligand–dependent T‐cell migration correlates with CCR7 homo‐ and CCR7/CXCR4 heterodimer formation, 25 and that there is a direct contribution of CCR7 homodimerization to CCR7‐dependent T‐cell migration and signaling. 26 We speculate that CXCL12‐induced CCR7 homodimerization contributes to the CCR7‐dependent cell migration and signaling in MDA231 cells, although we cannot exclude a possible involvement of CCR7/CXCR4 heterodimer, which leads to the acquisition of invasive phenotype in MDA231 cells. 40 We also demonstrated that CXCL12 induces not only CCR7 homodimer formation but also CCR7 translocation from the cytosol to the plasma membrane in MDA231 cells. As CXCL12 promotes membrane ruffle formation, we hypothesize that CXCL12‐induced membrane ruffling causes an increase in CCR7‐dependent signaling platform with CCR7 homodimer in plasma membrane.
In our study, in vitro migration, invasion, and in vivo metastasis in MDA231 cells are CCR7 dependent. This is consistent with previous reports showing that CCR7 signaling positively regulates cell migration and invasion, 20 , 41 and that in vitro invasiveness is well correlated with the in vivo metastatic potential of MDA231 cells. 42 In contrast to the finding of others, in which CCR7‐expressing tumors grew significantly greater than control cells, 20 our data showed that CCR7 overexpression did not significantly promote tumor growth in metastatic variants. Conversely, a decrease in the endogenous CCR7 expression reduced both tumor growth and lymph node metastasis, suggesting that tumor proliferative potential of cells potentially reflects lymph node metastasis. We speculate that this discrepancy results from the degree of changes in CCR7 expression in each experimental setting.
We demonstrated that CXCL12 promotes cell migration toward CCL21‐reconstituted lymphatic networks. The analysis of MDA231‐derived primary tumor tissues revealed that CXCL12 was detected in the peritumoral cell compartment, and no colocalization with cancer cells occurred throughout the tumor, suggesting that MDA231 cells are unlikely the source of CXCL12 in the tumor tissue. The CXCL12 expression level in cultured MDA231 cells was below the detection sensitivity of RT‐PCR, and that in the tumor tissue is predominantly derived from mouse cells, suggesting that MDA231 cells hardly express CXCL12 both in vitro and in vivo. These results are in agreement with several studies showing CXCL12 expression in stromal cells, 14 , 35 although there are conflicting reports whether CXCL12 cells expressed in MDA231 express 35 or not. 43 , 44 Tumor stromal cells are composed of heterogenous cell subsets, including tumor fibroblasts that could be divided into two subgroups, myofibroblastic CAFs (myCAFs) and inflammatory CAFs (iCAFs). 45 In line with our immunohistochemical analysis demonstrating that myCAFs are not colocalized with CXCL12, a recent work of single‐cell sequencing of bladder urothelial carcinoma demonstrated that iCAF is the major source of CXCL12. 46 Additional experiments are needed to directly assess the contribution of iCAF to CXCL12 expression in MDA231‐derived tumor. On the other hand, CCL21 expression has been reported to be produced by highly invasive cancer cells themselves and by lymphatic endothelial cells around the tumor mass. 22 The CCL21 transcripts in MDA231‐derived tumor are of mouse origin, and CCL21 was expressed primarily around LYVE‐1–positive lymph ducts, suggesting that CCL21 acts in a paracrine manner to induce CCR7‐dependent signaling in MDA231 cells. These results support the hypothesis that CXCR4 signaling reduces the sensitivity threshold of the cells to CCL21, thereby promoting CCR7‐dependent cancer cell migration toward CCL21‐expressing lymph vessels. Accordingly, in the tumor microenvironment, CXCL12 may indirectly regulate LN metastasis by promoting CCR7 signaling in cancer cells.
In growing tumors, many factors including inflammatory cytokines, hormones, and lipids regulate the chemokine production by cancer cells and tumor‐infiltrating leukocytes. 47 Our study demonstrated a possible functional collaboration between CXCL12 and CCR7 ligands in CXCR4‐ and CCR7‐positive breast cancer cell metastasis to LNs. CXCL12‐mediated CCR7 homodimerization and membrane ruffle formation may be the mechanism through which breast cancer cells metastasize to LNs in a CXCL12‐rich tumor microenvironment. In addition to these lymphoid chemokines, CCL1 and CXCL10 were reported to be involved in tumor metastasis to LNs. 48 , 49 To understand the contribution of chemokine signaling in LN metastasis, future studies should focus on the interplay between multiple chemokines in the tumor microenvironment.
DISCLOSURE
The authors declare no competing financial interests.
Supporting information
Figure S1‐S5
ACKNOWLEDGEMENTS
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (Grant‐in‐Aid for Scientific Research, 19K07278 to H.H.). We thank Drs. Ichio Shimada (The University of Tokyo), Takaaki Aoba (The Nippon Dental University), and Yasushi Hiraoka (Osaka University) for helpful suggestions. We thank Ms. Yuko Wakabayashi and Dr. Naoko Sasaki for technical assistance. The authors gratefully thank the Division of Joint Research Center, Kindai University for providing a flow cytometer. The authors would like to thank Enago (www.enago.jp) for the English language review.
Hayasaka H, Yoshida J, Kuroda Y, et al. CXCL12 promotes CCR7 ligand–mediated breast cancer cell invasion and migration toward lymphatic vessels. Cancer Sci. 2022;113:1338–1351. doi: 10.1111/cas.15293
REFERENCES
- 1. Miyasaka M, Tanaka T. Lymphocyte trafficking across high endothelial venules: dogmas and enigmas. Nat Rev Immunol. 2004;4:360‐370. [DOI] [PubMed] [Google Scholar]
- 2. Worbs T, Mempel TR, Bolter J, von Andrian UH, Forster R. CCR7 ligands stimulate the intranodal motility of T lymphocytes in vivo. J Exp Med. 2007;204:489‐495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ohl L, Mohaupt M, Czeloth N, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady‐state conditions. Immunity. 2004;21:279‐288. [DOI] [PubMed] [Google Scholar]
- 4. Müller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50‐56. [DOI] [PubMed] [Google Scholar]
- 5. Tanaka T, Bai Z, Srinoulprasert Y, Yang BG, Hayasaka H, Miyasaka M. Chemokines in tumor progression and metastasis. Cancer Sci. 2005;96:317‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farnsworth RH, Karnezis T, Maciburko SJ, Mueller SN, Stacker SA. The interplay between lymphatic vessels and chemokines. Front Immunol. 2019;10:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell‐derived factor 1 (SDF‐1). J Exp Med. 1996;184:1101‐1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol. 2004;14:171‐179. [DOI] [PubMed] [Google Scholar]
- 9. Saur D, Seidler B, Schneider G, et al. CXCR4 expression increases liver and lung metastasis in a mouse model of pancreatic cancer. Gastroenterology. 2005;129:1237‐1250. [DOI] [PubMed] [Google Scholar]
- 10. Darash‐Yahana M, Pikarsky E, Abramovitch R, et al. Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J. 2004;18:1240‐1242. [DOI] [PubMed] [Google Scholar]
- 11. Rubin JB, Kung AL, Klein RS, et al. A small‐molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci USA. 2003;100:13513‐13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kochetkova M, Kumar S, McColl SR. Chemokine receptors CXCR4 and CCR7 promote metastasis by preventing anoikis in cancer cells. Cell Death Differ. 2009;16:664‐673. [DOI] [PubMed] [Google Scholar]
- 13. Kryczek I, Lange A, Mottram P, et al. CXCL12 and vascular endothelial growth factor synergistically induce neoangiogenesis in human ovarian cancers. Can Res. 2005;65:465‐472. [PubMed] [Google Scholar]
- 14. Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF‐1/CXCL12 secretion. Cell. 2005;121:335‐348. [DOI] [PubMed] [Google Scholar]
- 15. Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ‐specific metastasis. Nat Rev Immunol. 2011;11:597‐606. [DOI] [PubMed] [Google Scholar]
- 16. Mashino K, Sadanaga N, Yamaguchi H, et al. Expression of chemokine receptor CCR7 is associated with lymph node metastasis of gastric carcinoma. Can Res. 2002;62:2937‐2941. [PubMed] [Google Scholar]
- 17. Ding Y, Shimada Y, Maeda M, et al. Association of CC chemokine receptor 7 with lymph node metastasis of esophageal squamous cell carcinoma. Clin Cancer Res. 2003;9:3406‐3412. [PubMed] [Google Scholar]
- 18. Takanami I. Overexpression of CCR7 mRNA in nonsmall cell lung cancer: correlation with lymph node metastasis. Int J Cancer. 2003;105:186‐189. [DOI] [PubMed] [Google Scholar]
- 19. Wiley HE, Gonzalez EB, Maki W, Wu MT, Hwang ST. Expression of CC chemokine receptor‐7 and regional lymph node metastasis of B16 murine melanoma. J Natl Cancer Inst. 2001;93:1638‐1643. [DOI] [PubMed] [Google Scholar]
- 20. Cunningham HD, Shannon LA, Calloway PA, et al. Expression of the C‐C chemokine receptor 7 mediates metastasis of breast cancer to the lymph nodes in mice. Transl Oncol. 2010;3:354‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shields JD, Emmett MS, Dunn DB, et al. Chemokine‐mediated migration of melanoma cells towards lymphatics–a mechanism contributing to metastasis. Oncogene. 2007;26:2997‐3005. [DOI] [PubMed] [Google Scholar]
- 22. Shields JD, Fleury ME, Yong C, Tomei AA, Randolph GJ, Swartz MA. Autologous chemotaxis as a mechanism of tumor cell homing to lymphatics via interstitial flow and autocrine CCR7 signaling. Cancer Cell. 2007;11:526‐538. [DOI] [PubMed] [Google Scholar]
- 23. D'Agostino G, Cecchinato V, Uguccioni M. Chemokine heterocomplexes and cancer: A novel chapter to be written in tumor immunity. Front Immunol. 2018;9:2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bai Z, Hayasaka H, Kobayashi M, et al. CXC chemokine ligand 12 promotes CCR7‐dependent naive T cell trafficking to lymph nodes and Peyer's patches. J Immunol (Baltimore, MD: 1950). 2009;182(3):1287‐1295. [DOI] [PubMed] [Google Scholar]
- 25. Hayasaka H, Kobayashi D, Yoshimura H, Nakayama EE, Shioda T, Miyasaka M. The HIV‐1 Gp120/CXCR4 axis promotes CCR7 ligand‐dependent CD4 T cell migration: CCR7 homo‐ and CCR7/CXCR4 hetero‐oligomer formation as a possible mechanism for up‐regulation of functional CCR7. PLoS One. 2015;10:e0117454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kobayashi D, Endo M, Ochi H, Hojo H, Miyasaka M, Hayasaka H. Regulation of CCR7‐dependent cell migration through CCR7 homodimer formation. Sci Rep. 2017;7:8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hauser MA, Schaeuble K, Kindinger I, et al. Inflammation‐induced CCR7 oligomers form scaffolds to integrate distinct signaling pathways for efficient cell migration. Immunity. 2016;44:59‐72. [DOI] [PubMed] [Google Scholar]
- 28. Cabioglu N, Yazici MS, Arun B, et al. CCR7 and CXCR4 as novel biomarkers predicting axillary lymph node metastasis in T1 breast cancer. Clin Cancer Res. 2005;11:5686‐5693. [DOI] [PubMed] [Google Scholar]
- 29. Tucker KL, Beard C, Dausmann J, et al. Germ‐line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 1996;10:1008‐1020. [DOI] [PubMed] [Google Scholar]
- 30. Akagi T, Shishido T, Murata K, Hanafusa H. v‐Crk activates the phosphoinositide 3‐kinase/AKT pathway in transformation. Proc Natl Acad Sci USA. 2000;97:7290‐7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nishiguchi A, Yoshida H, Matsusaki M, Akashi M. Rapid construction of three‐dimensional multilayered tissues with endothelial tube networks by the cell‐accumulation technique. Adv Mater. 2011;23:3506‐3510. [DOI] [PubMed] [Google Scholar]
- 32. Asano Y, Shimoda H, Matsusaki M, Akashi M. Transplantation of artificial human lymphatic vascular tissues fabricated using a cell‐accumulation technique and their engraftment in mouse tissue with vascular remodelling. J Tissue Eng Regen Med. 2018;12:e1501‐e1510. [DOI] [PubMed] [Google Scholar]
- 33. Luker KE, Gupta M, Luker GD. Imaging CXCR4 signaling with firefly luciferase complementation. Anal Chem. 2008;80:5565‐5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wells CM, Walmsley M, Ooi S, Tybulewicz V, Ridley AJ. Rac1‐deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J Cell Sci. 2004;117:1259‐1268. [DOI] [PubMed] [Google Scholar]
- 35. Allinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17‐32. [DOI] [PubMed] [Google Scholar]
- 36. Kerjaschki D, Regele HM, Moosberger I, et al. Lymphatic neoangiogenesis in human kidney transplants is associated with immunologically active lymphocytic infiltrates. J Am Soc Nephrol. 2004;15:603‐612. [DOI] [PubMed] [Google Scholar]
- 37. Yang BG, Tanaka T, Jang MH, Bai Z, Hayasaka H, Miyasaka M. Binding of lymphoid chemokines to collagen IV that accumulates in the basal lamina of high endothelial venules: its implications in lymphocyte trafficking. J Immunol. 2007;179:4376‐4382. [DOI] [PubMed] [Google Scholar]
- 38. Laufer JM, Hauser MA, Kindinger I, Purvanov V, Pauli A, Legler DF. Chemokine receptor CCR7 triggers an endomembrane signaling complex for spatial Rac activation. Cell Rep. 2019;29:995‐1009 e1006. [DOI] [PubMed] [Google Scholar]
- 39. Thelen M, Legler DF. Membrane lipid environment: Potential modulation of chemokine receptor function. Cytokine. 2018;109:72‐75. [DOI] [PubMed] [Google Scholar]
- 40. Poltavets V, Faulkner JW, Dhatrak D, Whitfield RJ, McColl SR, Kochetkova M. CXCR4‐CCR7 heterodimerization is a driver of breast cancer progression. Life. 2021;11(10):1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu B, Zhou M, Qiu W, Ye J, Feng Q. CCR7 mediates human breast cancer cell invasion, migration by inducing epithelial‐mesenchymal transition and suppressing apoptosis through AKT pathway. Cancer Med. 2017;6:1062‐1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abdelkarim M, Vintonenko N, Starzec A, et al. Invading basement membrane matrix is sufficient for MDA‐MB‐231 breast cancer cells to develop a stable in vivo metastatic phenotype. PLoS One. 2011;6:e23334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wendt MK, Cooper AN, Dwinell MB. Epigenetic silencing of CXCL12 increases the metastatic potential of mammary carcinoma cells. Oncogene. 2008;27:1461‐1471. [DOI] [PubMed] [Google Scholar]
- 44. Bachelder RE, Wendt MA, Mercurio AM. Vascular endothelial growth factor promotes breast carcinoma invasion in an autocrine manner by regulating the chemokine receptor CXCR4. Can Res. 2002;62:7203‐7206. [PubMed] [Google Scholar]
- 45. Ohlund D, Handly‐Santana A, Biffi G, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Z, Zhou L, Liu L, et al. Single‐cell RNA sequencing highlights the role of inflammatory cancer‐associated fibroblasts in bladder urothelial carcinoma. Nat Commun. 2020;11:5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gorbachev AV, Fairchild RL. Regulation of chemokine expression in the tumor microenvironment. Crit Rev Immunol. 2014;34:103‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Das S, Sarrou E, Podgrabinska S, et al. Tumor cell entry into the lymph node is controlled by CCL1 chemokine expressed by lymph node lymphatic sinuses. J Exp Med. 2013;210:1509‐1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kawada K, Hosogi H, Sonoshita M, et al. Chemokine receptor CXCR3 promotes colon cancer metastasis to lymph nodes. Oncogene. 2007;26:4679‐4688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1‐S5


