Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a major pathogen associated with severe morbidity and mortality and poses a significant threat to public health worldwide. The genetic diversity based on sequence types of MRSA strains was illustrated in previous studies; meanwhile, the diversity along with the predominant sequence type, especially in Egypt, remains unknown. The purpose of the current study was to determine the diversity of the predominant MRSA clone ST239-MRSA (n = 50) isolated from different hosts and clinical samples and to illustrate the correlation between the resistance patterns, toxin genes, and the genetic background in Port-said and El-Sharkia Governorates, Egypt. The ST239-MRSA clone was analyzed by phenotypic antibiotyping and various genotypic assays comprising SCCmec, agr, spa, coa, and coa-RFLP in addition to toxin gene profiles. Most of the analyzed strains (40/50, 80%) were multidrug resistant (MDR), belonged to SCCmec-III, agr-I, and coa genotype I, and harbored sea and pvl genes. A negative correlation between the toxin gene profiles and antimicrobial resistance was recorded. Meanwhile, the correlation between the toxin gene profiles and the genetic background was not observed in this study. Although ST239-MRSA strains belonged to a single sequence type, they exhibited a high degree of phenotypic and genotypic diversity, indicating weak clonality and adaptability. With such diversity, it is assumed that these strains may have undergone different evolutionary processes during transmission events among and/or within a single host or tissue niche.
Keywords: MRSA, ST239, MDR, genetic background, evolution
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has traditionally been considered a global pathogen (Gallagher et al., 2017). Indeed, most of the available studies have described the genetic diversity of MRSA strains belonging to multiple clones; meanwhile, few reports concerning a single clone have been reported. The proportions of both hospital and community-associated (CA) infections caused by single clones like ST239-MRSA strains have been increasing steadily in the past decades since the 1970s (Monecke et al., 2018). The ST239 is one of the dominant MRSA clones overtime worldwide, notably in Egypt (Abd El-Hamid et al., 2019), and it has been widely reported in Asia, Europe, the Middle East, e.g., Iran (Fatholahzadeh et al., 2009; Goudarzi et al., 2016; Sedaghat et al., 2018), and South and North America (Monecke et al., 2011). In terms of genetic structure, ST239-MRSA is an interesting clone as it harbors six of the seven multilocus sequence typing (MLST) housekeeping genes identical to ST8. However, ST239 and its clone relatives ST240 and ST241 differ from ST8 clone in arcC allele (arcC-2 in the former instead of arcC-3 in the latter) (Robinson and Enright, 2004).
Genetic typing is warranted to enable a better understanding of the infection dynamics. The typing methods used to determine the epidemiology of MRSA are sequence‐based methods such as MLST and staphylococcal protein A (spa) typing and non‐sequence‐based typing methods, which are based on staphylococcal cassette chromosome mec (SCCmec) elements, accessory gene regulator, agr (Magray et al., 2011) alleles, and restriction fragment length polymorphism (RFLP) patterns of coagulase (Coad et al., 2016) gene (coa-RFLP). Moreover, pulsed-field gel electrophoresis has been proposed as the gold standard for molecular epidemiological typing of MRSA bacterial strains (Ostojić, 2008). These methods have been recommended for determining the origin and the clonal relations and are used for enhanced discrimination of the closely related ST239-MRSA strains (Kong et al., 2017).
Similar to other MRSA clones, the ability of the ST239-MRSA clone to produce toxins and to resist certain antimicrobials affords a survival advantage and fitness under adverse conditions. Various sequence type (ST) clones of MRSA often display different antibiotic resistance and toxin patterns. Indeed, ST239-MRSA has been reported to be multidrug resistant (MDR), posing serious public health concerns (Kong et al., 2017). Another fitness feature of ST239-MRSA is the ability to produce several toxins, mainly staphylococcal enterotoxins (SE)A, K, and Q (sea, sek, and seq) and, to a lesser extent, toxic shock syndrome toxin (TSST), exfoliative toxins A and B (ETA and ETB), and Panton–Valentine leucocidin (PVL) leading to harmful toxic effects to the host (Xie et al., 2011; Boswihi et al., 2016; Alfouzan et al., 2019). However, many questions remain unanswered with regard to the correlation between the existence of toxin genes and antimicrobial resistance traits and many other factors such as the bacterial species, virulence, antimicrobial resistance, the ecological niche, and the host (Beceiro et al., 2013). Previous studies have indicated that some toxigenic strains belonged to a particular molecular type of MRSA (Baba et al., 2002; Chen et al., 2018). This implies that the molecular characteristics of MRSA strains affect their toxin gene profiles (Baba et al., 2002).
Although ST239-MRSA has been observed in several African countries (Kirchhelle, 2018); unfortunately, the molecular characteristics and virulence profiles of these strains are less clear in previous studies, and there are insufficient data on their epidemiological trends. Molecular epidemiological studies of clinical ST239-MRSA strains often depend on the application of different typing methods that produce various type assignments. Therefore, the present study aimed to investigate the molecular epidemiology and diversity of the ST239-MRSA clone belonging to different infected hosts and clinical sample types using a combination of typing approaches. Given the important consideration for planning and implementing the epidemiologic studies and outbreak investigations, we also aimed to evaluate the discriminatory abilities of the different typing methods and the strength of the correlation between them. The knowledge of the molecular epidemiology gained from this study would enable a better understanding of the characteristics of the ST239-MRSA clone and thus will be useful for the development of effective prevention and control measures.
Materials and Methods
Ethical Statement on Bacterial Strains
This is a case series study carried out to characterize some ST239-MRSA isolates during the period from June 2016 to August 2019. In this study, we investigated 105 S. aureus strains from both infected human (n = 60) and animal (n = 45) sources in Port-said and El-Sharkia Governorates, Egypt, including 30 previously identified strains as ST239-MRSA in one previous study for some of the co-authors (Abd El-Hamid et al., 2019). All human S. aureus strains associated with infections were kindly provided from different units in microbiology laboratories in Zagazig and Port-said University hospitals, which had acquired signed informed consents of the participating patients in this research. The human S. aureus strains were recovered from clinical samples including sputum (8), wound swabs (12), urine (14), pus (11), blood (9), cerebrospinal fluid (CSF) (4), and pericardial fluid (2). Meanwhile, S. aureus originating from milk samples of cows with mastitis were kindly supplied by the Department of Microbiology, Faculty of Veterinary Medicine, Zagazig University.
Identification of Staphylococcus aureus Strains
The identification of all S. aureus strains was based phenotypically on standard bacteriological procedures including morphological, cultural, and biochemical characterization (Becker et al., 2015) in addition to using API 20S identification kit (bioMerieux, Marcy l’Etoile, France). Moreover, the investigated strains were confirmed using PCR analyses of 16S rRNA and a species‐specific nuc genes (Abd El-Hamid and Bendary, 2015). The phenotypic identification of S. aureus strains carried out using standard bacteriological methods was based on mannitol fermentation on mannitol salt agar, β hemolysis on blood agar, the appearance of characteristic golden-yellow colonies, formation of Gram-positive grape-like clusters, and biochemical reaction results comprising the ability to coagulate rabbit plasma and the positive catalase reaction. For methicillin-resistance analysis, oxacillin and cefoxitin antibiotic susceptibility testing and PCR detection of mecA gene were performed (Abd El-Hamid and Bendary, 2015). The ST was characterized by MLST, which was based on seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpiA, and yqiL). Each sequence was then compared to the S. aureus MLST database (https://pubmlst.org/bigsdb?db=pubmlst_S.aureus_seqdef&page=profiles&scheme_id=1) to obtain an allele number as previously described (Enright et al., 2000).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility of all ST239-MRSA strains was tested by disk diffusion method using the standard antimicrobial discs (Oxoid, Hampshire, UK) and according to the guidelines of Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute, 2019). Strain susceptibility was examined against 13 types of antimicrobials including oxacillin (OX; 1 μg), cefoxitin (FOX; 30 μg), ceftriaxone (CRO; 30 μg), imipenem (IPM; 10 μg), vancomycin (VA; 30 μg), clindamycin (DA; 2 μg), rifamycin SV (RF; 30 μg), trimethoprim‐sulfamethoxazole (SXT; 1.25/23.75 μg), erythromycin (E; 15 μg), ciprofloxacin (CIP; 5 μg), tetracycline (TE; 30 μg), gentamicin (CN; 10 μg), and chloramphenicol (C; 30 μg). The oxacillin minimum inhibitory concentration (MIC) values for all ST239-MRSA strains were determined by the broth microdilution method, and the results were interpreted according to the CLSI guidelines (Clinical and Laboratory Standards Institute, 2019). Moreover, according to the recommendations included in the CLSI document (Clinical and Laboratory Standards Institute, 2019), disk testing is not reliable for assessing vancomycin resistance. Therefore, to determine the susceptibility of ST239-MRSA strains to such antimicrobial agent confirming them as vancomycin resistant, the MIC of vancomycin (Sigma Aldrich, Darmstadt, Germany) was determined phenotypically using the broth microdilution method (Franklin et al., 2012) with twofold increasing concentrations of vancomycin from 0.0625 to 1024 µg/mL. Moreover, the vancomycin resistance among all investigated strains was confirmed by a multiplex PCR amplification of vanA and vanB genes using the previously reported primer sets and under the same thermal conditions reported previously (Abd El-Aziz et al., 2018).
The multiple antibiotic resistance (MAR) index for each strain was calculated as previously described (Krumperman, 1983) by dividing the number of antimicrobials to which the strain was resistant by the total number of antimicrobials to which the strain was exposed in the current study. Calculated values of MAR indices greater than 0.2 indicate high-risk sources of contamination, where several antibiotics are often used. Of note, the strains were defined as MDR if they were resistant to at least one agent in three or more antimicrobial categories used in this study (Magiorakos et al., 2012).
Characterization of ST239-MRSA Genetic Background and Toxin Gene Profiles
All ST239-MRSA strains were analyzed for the detection of their genomic variations using uniplex PCR assays of coa and spa genes as stated previously (Hookey et al., 1998; Larsen et al., 2008). Digestion of coa PCR products with AluI restriction endonuclease (Sigma, St. Louis, MO, USA) was then performed according to the manufacturer’s recommended protocol to monitor the variations in MRSA populations on the basis of the sequence variation within the 3′ end coding region of coa gene. Moreover, multiplex PCRs were performed for agr and SCCmec typing as described previously (Gilot et al., 2002; Zhang et al., 2005). Characterization of toxin gene profiles by detecting genes encoding nine enterotoxins (sea, seb, sec, sed, see, seg, seh, sei, and sej), exfoliative toxins (eta and etb), TSST (tst), and PVL (lukSF-PV) was performed using the previously reported PCR conditions (Mehrotra et al., 2000). All PCR analyses were carried out in triplicate. For the quality control of the toxin genes detection including PVL, S. aureus ATCC25923 and Escherichia coli ATCC25922 were included with all PCR runs as positive and negative controls, respectively. MRSA strains NCTC10442, N315, 85/2082, JCSC4744, and D12 were utilized as reference strains for detecting SCCmec types I, II, III, IV, and V, respectively. For each investigated gene, all PCR runs were performed with relevant PCR positive controls (DNAs from MRSA isolates previously confirmed to harbor sequences for any of the target genes by gel electrophoresis of PCR products). These positive controls were provided from the National Laboratory for Veterinary Quality Control on Poultry Production (NLQP), Animal Health Research Institute, Giza, Egypt. Additionally, sequencing of the amplicons of SCCmec and lukSF-PV genes was employed to confirm the genetic diversity of the ST239-MRSA clone according to the manufacturer’s protocol (Elim Biopharmaceuticals, Inc., Hayward, CA, USA). The resulting partial (433 bp) lukSF-PV gene sequences of ST239-MRSA strains were aligned with the fragments corresponding to a region measuring 2041.196–2041.58 kb in the genome of a selected S. aureus reference strain 14505 (GenBank accession number CP053640.1). The lukSF-PV gene segments had high nucleotide sequence identity with the reference sequence (100%). The sequencing reactions were carried out with single-stranded DNA templates. Prior to the sequencing of the SCCmec elements, the amplification products for SCCmec types were gel extracted and purified using the QIAquick Gel Extraction kit (Qiagen, Germany), and their nucleotide sequences were then determined. By utilizing the SCCmec Finder software found at https://bitbucket.org/genomicepidemiology/sccmecfinder.git, SCCmec element types were then identified. Of note, we followed the guidelines of the PCR unidirectional workflow.
Bioinformatics and Statistical Analyses
Simpson’s index (Simpson, 1949) applied on the whole ST239-MRSA strains as well as on those per each host was used to reveal the diversity of the analyzed strains. Shapiro’s test (Royston, 1982) and Q-Q plot (Katayama et al., 2000) were used to assess and visualize the distribution of all variables, respectively. The correlation analyses were used to provide an estimate for the association among various variables, and then the correlation coefficient was visualized. The correlation analyses and visualization were done using R packages corrplot, heatmaply, hmisc, and ggpubr (Friendly, 2002; Galili et al., 2018; Jr, 2020). A heatmap supported by a hierarchical clustering dendrogram was generated to visualize the phenotype/genotype profiles of the examined strains. The analyses were conducted using the R environment (v. 3.6.2) (Kolde, 2018; Team, 2018). Fisher’s exact test was used to test whether the occurrence of resistance to certain antimicrobial differed significantly among studied hosts.
To assess the discriminatory power of the typing methods used in our study, the D-value of each typing method was assessed using Simpson’s index of diversity (Hunter and Gaston, 1988) calculating the probability that two unrelated strains sampled from the test population will be placed into different typing groups as follows:
where D is the index of discriminatory power, N is the number of unrelated strains tested, S is the number of different types, and xj is the number of strains belonging to the jth type assuming that strains will be classified into mutually exclusive categories. A D-value of 1.0 indicates that a typing method was able to distinguish each member of a strain population from all other members of that population; a D-value of 0.0 indicates that all members of a strain population were of an identical type. An index of 0.50 would mean that if one strain was chosen at random from a strain population, then there would be a 50% probability that the next strain chosen at random would be indistinguishable from the first.
Results
Multilocus Sequence Typing of Staphylococcus aureus Strains
According to the MLST results of 105 S. aureus strains, ST239 was proven for 50 strains. The detailed characteristics of all 105 strains are shown in Table S1 . The 50 ST239 strains were recovered from 18 milk samples of cows with mastitis (40%) and 32 human patients (53.3%), including 6 sputum (75%), 6 wound swabs (50%), 6 urine (42.9%), 7 pus (63.6%), 4 blood (44.4%), 2 CSF (50%), and 1 pericardial fluid (50%). The different allelic sequences of the newly identified 20 ST239-S. aureus strains defined using the MLST website on the basis of the sequences of the internal fragments of seven housekeeping genes have been deposited in the GenBank with accession numbers MN880894:MN881033.
Antimicrobial Resistance Profile of ST239-MRSA Clone
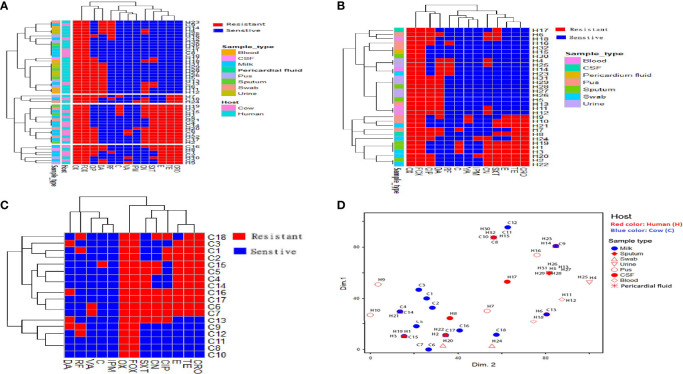
Analysis of the antimicrobial resistance patterns of all ST239 S. aureus strains (n = 50) revealed that all of them (100%) were uniformly resistant to oxacillin and cefoxitin. All strains exhibited high-level resistance to oxacillin with MIC values >128 µg/mL. The existence of mecA gene in all examined ST239 strains proved that they were identified as MRSA. Irrespective of the host, the highest resistance of ST239-MRSA strains was against ciprofloxacin (54%). Meanwhile, relatively low resistance rates were observed against vancomycin (8%) and imipenem (4%) ( Table S2 ). All strains derived from wound swabs were completely resistant to tetracycline, ceftriaxone, and gentamicin and sensitive only to vancomycin. Furthermore, all strains recovered from urine samples showed resistance to clindamycin and sensitivity to tetracycline, ceftriaxone, erythromycin, chloramphenicol, vancomycin, trimethoprim‐sulfamethoxazole, and imipenem ( Table 1 ). With the use of Fisher’s exact test, it was found that only the occurrence of clindamycin resistance was considered different among the studied hosts. Four ST239-MRSA strains (8%) had vancomycin MIC of >16 μg/mL, and therefore, they were initially named vancomycin-resistant S. aureus (VRSA). Screening for vanA and vanB resistance genes revealed that 2 human (6.25%) and 2 animal (11.1%) strains harbored vanA or vanB genes confirming that they were all VRSA ( Table S3 ). Interestingly, ST239-MRSA strains were resistant to at least 2–9 of the 13 antimicrobials generating MAR indices ranged from 0.15 to 0.69 (D-value = 0.86) with 24% of the strains showing a MAR index value of 0.30. However, only one isolate showed a MAR index of 0.23. High percentages of MDR were observed among our human (90.6%, 29/32) and cow (77.8% 14/18) strains. Antibiotyping of the examined strains revealed 26 different antimicrobial resistance patterns with D-value equal to 0.96 ( Figure 1A and Table S4 ). The antibiotyping showed low discriminatory power for the human isolates (D-value = 0.937, Figure 1B ) as compared to the animal ones (D-value = 0.967, Figure 1C ).
Table 1.
Percentages of toxin genes and antimicrobials resistances of ST239-MRSA among different sample types.
| Sample type (no.) | Toxin genes % | Antimicrobial resistances % | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pvl | sea | see | sec | tst | eta | etb | CIP | RF | TE | CRO | E | C | VA | DA | SXT | CN | IPM | |
| Milk (18) | 39 | 44 | 6 | 0 | 6 | 33 | 11 | 39 | 22 | 61 | 61 | 61 | 6 | 11 | 22 | 50 | 39 | 5.6 |
| Sputum (6) | 67 | 33 | 17 | 0 | 17 | 0 | 0 | 50 | 0 | 50 | 50 | 50 | 17 | 0 | 33 | 50 | 33 | 0 |
| Wound swabs (6) | 100 | 50 | 0 | 0 | 0 | 0 | 0 | 67 | 17 | 100 | 100 | 67 | 33 | 0 | 17 | 67 | 100 | 17 |
| Urine (6) | 33 | 83 | 17 | 0 | 17 | 17 | 0 | 67 | 33 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 33 | 0 |
| Pus (7) | 57 | 29 | 29 | 14 | 0 | 14 | 0 | 57 | 14 | 43 | 43 | 29 | 14 | 29 | 57 | 29 | 14 | 0 |
| Blood (4) | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 75 | 25 | 0 | 0 | 0 | 0 | 0 | 75 | 25 | 75 | 0 |
| CSF (2) | 0 | 0 | 0 | 50 | 50 | 0 | 50 | 100 | 0 | 50 | 50 | 50 | 0 | 0 | 50 | 100 | 0 | 0 |
| Pericardial fluid (1) | 0 | 0 | 100 | 0 | 100 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
CSF, cerebrospinal fluid; pvl, Panton–Valentine leucocidin; sea, see, and sec, staphylococcal enterotoxins A, B, and C; tst, toxic shock syndrome toxin; eta and etb, exfoliative toxins A and B; CIP, ciprofloxacin; RF, rifamycin SV; TE, tetracycline; CRO, ceftriaxone; E, erythromycin; C, chloramphenicol; VA, vancomycin; DA, clindamycin; SXT, trimethoprim‐sulfamethoxazole; CN, gentamicin; IPM, imipenem.
Figure 1.
Distribution and hierarchical clustering of ST239 methicillin-resistant Staphylococcus aureus (MRSA) strains and the antimicrobial resistance variables based on the antimicrobial resistance profiles. (A–C) Heatmap showing the occurrence of antimicrobial resistance in the studied strains; all, human, and animal ST239 MRSA, respectively. Red and blue colors refer to resistance and sensitivity to a particular antimicrobial, respectively. The dendrogram represents the hierarchical clustering of the strains and antimicrobial resistance variables. The vertical lines dropped from the dendrogram branches show the 10 clusters that contained identical isolates. Different categories of hosts and sample types are color-coded on the right of the heatmap. (D) Non-metric multidimensional scaling analysis showing the overlap of ST239-MRSA strains belonging to different hosts and sample types based on antimicrobial resistance profile. CIP, ciprofloxacin; RF, rifamycin SV; TE, tetracycline; CRO, ceftriaxone; E, erythromycin; C, chloramphenicol; VA, vancomycin; DA, clindamycin; SXT, trimethoprim‐sulfamethoxazole; CN, gentamicin; IPM, imipenem; OX, oxacillin; FOX, cefoxitin.
Analysis of antimicrobial resistance patterns demonstrated a high variability among ST239-MRSA strains belonging to different hosts and from different sample types as evidenced by their large overlap revealed by the non-metric multidimensional scaling (nMDS) analyses ( Figure 1D ) with averaged binary distances equating to 0.4–0.5 among and within hosts ( Figure 2A ) and 0.2–0.5 among and within samples ( Figure 2B ), and averaged correlation coefficient, r(48) = 0.1–0.2 among hosts. It was evident that the antimicrobial resistance profiles of the strains colonizing the same host, particularly those within cows, were slightly more similar than those from the two hosts (i.e., considering cow and human strains together).
Figure 2.
Network depicting the differences among ST239-MRSA strains based on the antimicrobial resistance profile. (A) The average distance among strains from humans and cows. (B) The average distance among strains from various samples.
Correlation Among Resistances to Different Antimicrobials
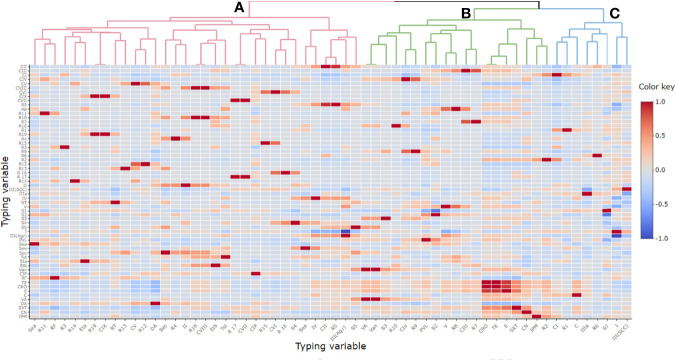
The correlation among individual antimicrobial resistance phenotypes was variable and showed an overall weak positive correlation, r(48) = 0.1. Ceftriaxone and tetracycline showed the highest positive correlation, r(48) = 1, while the clindamycin correlated negatively with tetracycline, r(48) = 0.4 ( Figure 3 ).
Figure 3.
Pairwise correlation (r) among different variables of typing methods. Red and blue colors indicate positive and negative correlation, respectively. The color key refers to correlation coefficient (r). The darker colors imply stronger positive or negative correlations. Hierarchical clustering of the variables is shown as a dendrogram illustrating different clusters with different colors and letters (e.g., (A–C). Variables that are identical among all strains are excluded and thus not shown in this figure. CIP, ciprofloxacin; RF, rifamycin SV; TE, tetracycline; CRO, ceftriaxone; E, erythromycin; C, chloramphenicol; VA, vancomycin; DA, clindamycin; SXT, trimethoprim‐sulfamethoxazole; CN, gentamicin; IPM, imipenem; CI-CIX, coagulase genotypes; R1–R19, coa-RFLP patterns; SCCmec (II, III, IV, and V), staphylococcal cassette chromosome mec; NT, non-typeable; agr (I and III), accessory gene regulator; S1–S5, spa PCR products; van, vancomycin resistance gene; pvl, Panton–Valentine leucocidin; sea, see, and sec; staphylococcal enterotoxins A, B, and C; tst, toxic shock syndrome toxin; eta and etb, exfoliative toxins A and B.
Molecular Typing of ST239-MRSA
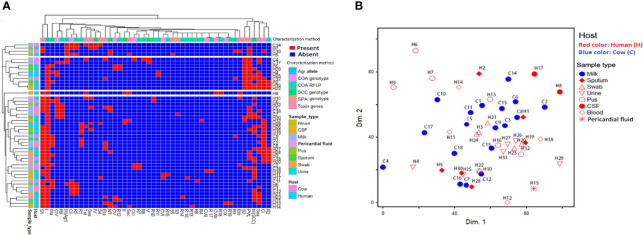
Coagulase genotyping revealed that ST239-MRSA strains presented diverse patterns with D-value equal to 0.79. Nine different coagulase genotypes (CI–CIX) were detected ( Figure 4A ). The CI (band size = 750 bp) was the predominant coagulase type being identified in 38% of the strains (19/50). Moreover, coa-RFLP was the best discriminatory method for investigating the ST239-MRSA strains, as it gave rise to 19 coa-RFLP patterns with a high D-value (0.92) ( Figure 4A , Table S5 ).
Figure 4.
Distribution and hierarchical clustering of ST239-MRSA strains and the different genotypes based on different molecular typing methods. (A) Heatmap showing the occurrence of particular genotype/toxin gene in the studied ST239-MRSA strains. Red and blue colors refer to presence and absence of a particular feature, respectively. The dendrogram represents the hierarchical clustering of the strains and the genotype variables. Different categories of genotyping methods, sample types, and hosts are color-coded on the right of the heatmap. (B) Non-metric multidimensional scaling analysis indicating the presence of each subject in the 2-dimensional space and thus showing how the subjects could cluster or be separated from each other. CI–CIX, coagulase genotypes; R1–R19, coa-RFLP patterns; SCCmec (II, III, IV, and V), staphylococcal cassette chromosome mec; NT, non-typeable; agr (I and III), accessory gene regulator; S1–S5, spa PCR products; van, vancomycin resistance gene; pvl, Panton–Valentine leucocidin; sea, see, and sec, staphylococcal enterotoxins A, B, and C; tst, toxic shock syndrome toxin; eta and etb, exfoliative toxins A and B.
Concerning the SCCmec typing, ST239-MRSA strains were discriminated into five types (D-value = 0.46) with SCCmec-III being the most common one (72%) ( Table S6 ). The SCCmec-II, SCCmec-IV, and SCCmec-V types were detected among 3 human, 2 human, and 2 (one human and one animal) isolates, respectively. The carriage of these uncommon SCCmec types by these ST239 strains was also validated by DNA sequencing of their PCR products with accession numbers of MW563684:MW563719, MW563720:MW563722, MW563723:MW563724, and MW563725:MW563726 for SCCmec-III, SCCmec-II, SCCmec-IV, and SCCmec-V, respectively.
Grouping of agr alleles revealed only agr types I and III with D-value equal to 0.22 ( Table S6 ). The agr type I was the most prevalent [88%, (44/50)]. All SCCmec type III strains (72%; 36/50) were of agr type I.
According to PCR amplification of spa-X region, the ST239-MRSA strains were grouped into 5 spa types with D-value equal to 0.53. The majority of the strains (64%, 32/50) were typed as S1 (400 bp). The S1 type was associated mainly with SCCmec-III and agr-I types being present in 71.9% of the strains (23/32) ( Figure 4A and Table S6 ).
Toxin Gene Profiles
Toxin gene profiles of ST239-MRSA strains revealed the presence of one or more toxin genes in 80% (40/50) of the tested strains. The most prevalent genes among the toxigenic strains were enterotoxins (75%) and lukSF-PV (57.5%) ( Figure 4A ). All strains derived from wound swabs possessed lukSF-PV gene (100%) ( Table 1 ). The high prevalence of lukSF-PV gene among our isolates was confirmed by sequencing of the PCR products with accession numbers of MW570780:MW570802. Our study indicated that only one et allele (eta or etb) was detected in all ST239-MRSA strains. Of the nine enterotoxin genes examined, only sea, sec, and see were identified, and all the toxigenic strains possessed only one type of these genes. The most prevalent enterotoxin gene among the examined strains was sea (40%). The results also revealed that lukSF-PV, see, sec, and tst genes were more prevalent among the human strains as compared to the animal ones, but the reverse occurred with sea, eta, and etb genes. The combinative data of toxin gene profiles revealed 19 different toxin gene profiles with D-value equal to 0.91 ( Figure 4A ).
Distribution of Different Methicillin-Resistant Staphylococcus aureus Genotypes Among Different Hosts and Sample Types
Depending on all molecular typing methods (coa, SCCmec, agr, and spa), the analyzed ST239-MRSA strains showed a considerable overlap ( Figure 4B ) with no segregation into defined clusters and with high averaged binary distance reaching 0.8 and low average positive correlation coefficient, r(48) = 0.2–0.3 among cow and human strains and between cow and human strains. The dissimilarity among strains of the same host was almost similar to that among different hosts (average binary distance = 0.78) ( Figure 5A ). Of note, the strains from the same sample were highly diverse with the range of averaged binary distance between 0.6 and 0.8. The CSF strains showed the highest heterogeneity (average binary distance = 0.8) and a very weak positive correlation (r(48) = 0.03), and the swab strains constituted the least diverse pool (average binary distance = 0.6) ( Figure 5B ). The homogeneity of the ST239-MRSA strains within each sample type and among different samples was quite similar, which was consistent for each of the molecular typing approaches. Moreover, the majority of ST239-MRSA strains have different genetic backgrounds when analyzed by combined molecular typing methods ( Figure 4A ). The coa-RFLP typing demonstrated the highest discriminatory power for strains within and among hosts in opposition to the agr typing.
Figure 5.
Network depicting the differences among ST239-MRSA strains based on both genotypes and toxin gene profile. (A) The average distance among strains from humans and cows. (B) The average distance among strains from various samples.
Correlation Among Antimicrobial Resistance Patterns, Molecular Typing Variables, and Toxin Gene Profiles
Considering all ST239-MRSA strains, the correlation analyses segregated all typing features into three main clusters (A, B, and C) distinguished by different colors as seen in the top of the dendrogram in Figure 3 and each cluster contained a pool of typing parameters, each belonging to different typing methods. The majority of the typing methods (76.2%, 16/21) correlated to each other positively, and only five of them correlated negatively.
Correlation Between the Existence of Antimicrobial Resistances and Toxin Genes
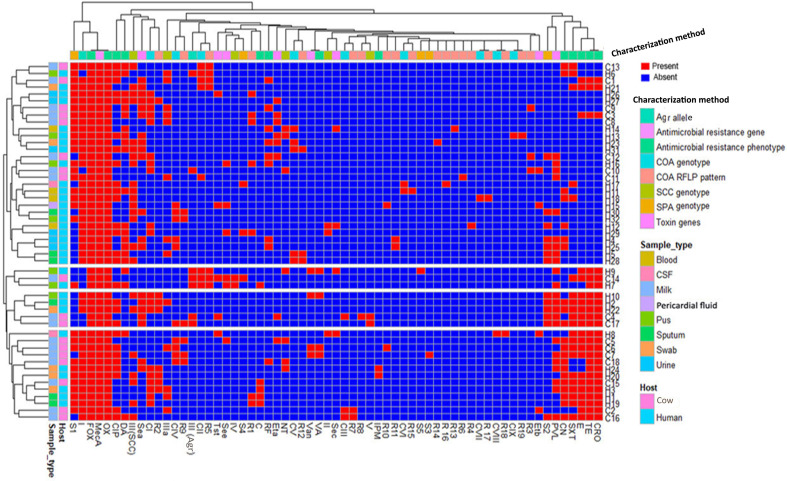
Our study also revealed a negative correlation between the existence of toxin genes and antimicrobial resistances. Notably, MDR strains with maximum antimicrobial resistance patterns were associated with the least toxin gene profiles. Overall, 50% of toxigenic strains showed low MAR indices (≤0.38), but higher MAR indices (≥0.38) were observed in 80% of non-toxigenic strains ( Figure 6 ).
Figure 6.
Heatmap showing the hierarchical clustering and overall distribution of ST239-MRSA strains based on their genotypes and antimicrobial resistance and toxin gene profiles. Red and blue colors refer to presence and absence of a particular feature, respectively. The dendrogram represents the hierarchical clustering of the strains and various features. Different categories of characterization methods, sample types, and hosts are color-coded on the right of the heatmap. CIP, ciprofloxacin; RF, rifamycin SV; TE, tetracycline; CRO, ceftriaxone; E, erythromycin; C, chloramphenicol; VA, vancomycin; DA, clindamycin; SXT, trimethoprim‐sulfamethoxazole; CN, gentamicin; IPM, imipenem; CI–CIX, coagulase genotypes; R1–R19, coa-RFLP patterns; SCCmec (II, III, IV, and V), staphylococcal cassette chromosome mec; NT, non-typeable; agr (I and III), accessory gene regulator; S1–S5, spa PCR products; van, vancomycin resistance gene; pvl, Panton–Valentine leucocidin; sea, see, and sec, staphylococcal enterotoxins A, B, and C; tst, toxic shock syndrome toxin; eta and etb, exfoliative toxins A and B.
Association Between Genetic Background and Toxin Genes
The majority of PVL (78.3%, 18/23) and enterotoxins (83.3%, 25/30) positive strains belonged to agr-I genotype. Moreover, the majority of the strains harboring enterotoxin genes (56.7%, 17/30) were found to belong to SCCmec-III, and 38.9% (14/36) of ST239-MRSA-SCCmec-III strains possessed sea. Meanwhile, sec was found among ST239-MRSA SCCmec-II strains. The ST239-MRSA SCCmec-IV and SCCmec-V were found to carry see and sea genes, respectively. Furthermore, the majority of lukSF-PV positive strains (65.2%) belonged to SCCmec-III. All ST239-MRSA SCCmec-IV and SCCmec-V and the majority of non-typeable strains had more than one toxin gene. Moreover, the majority of toxin-producing ST239-MRSA strains belonged to spa type 1 (57.5%, 23/40) and coa type I (40%, 16/40) ( Figure 4A ).
Discussion
The increase in the incidence of MDR bacterial and fungal infections has become an emerging threat to public health (Ghaith et al., 2020; Ghaly et al., 2020). The ST239-MRSA lineage is one of the most successful and persistent hybrid that showed high levels of drug resistance (Feil et al., 2008). Antimicrobial susceptibility testing of ST239-MRSA strains revealed the increasing prevalence of resistance to tetracycline, erythromycin, and ciprofloxacin as previously reported in China (Kong et al., 2017; Sedaghat et al., 2018). The haphazard use of antimicrobials to treat bacterial infections in animals often increases the resistance to these antimicrobials (Kirchhelle, 2018). The adapted ability of ST239-MRSA clone to enhanced antimicrobial resistance is extremely serious. Interestingly, more than 80% of our strains, especially of human origin, were MDR. This is consistent with a previous study conducted in India (Woodyard, 2011). Meanwhile, the MDR rate among human strains in our study (90.6%) is slightly lower than that previously reported in China (100%) (Kong et al., 2017). The indiscriminate use of antimicrobials in Egypt has rendered the commonly used antimicrobials completely ineffective in the treatment of this clone (Singh et al., 2016).
In terms of the susceptibility patterns of our MRSA isolates to the tested β-lactam antibiotics, all our investigated strains appeared resistant to oxacillin and cefoxitin; meanwhile, 48% and 4% of the isolates were resistant to ceftriaxone and imipenem, respectively. Consistent with a report from China, all ST239-MRSA strains were resistant to oxacillin and cefoxitin (Li et al., 2021), confirming the isolates as MRSA. Moreover, a previous research paper carried out in Israel revealed that 97.9% of MRSA isolates were sensitive to imipenem (Samba and Gadba, 1993). The highly mentioned susceptibility patterns to imipenem are reflective of its use as an effective antistaphylococcal agent against MRSA isolates as previously proven (Fan et al., 1986). However, a striking difference was detected in the susceptibility patterns of MRSA isolates to ceftriaxone. In Beijing, China, all isolates belonging to ST398-MRSA were resistant to ceftriaxone (Zhang et al., 2011). Additionally, Mushtaq et al. reported that only 16.7% of MRSA strains showed susceptibility to ceftriaxone (Mushtaq et al., 2017), confirming the misuse of ceftriaxone in these areas. Notably, the changing in the resistance profiles of MRSA clones to this antibiotic among various studies indicated that the traditional ceftriaxone-resistant MRSA isolates became less frequent with the emergence and spread of MRSA clones that were susceptible to this drug.
Currently, the emergence and spread of VRSA strains remain a challenging global health crisis. The clinical significance of VRSA is attributed to its potential to cause worldwide noticeable mortality in the absence of effective control and treatment options (Cong et al., 2020). Therefore, it crucially important to diagnose VRSA cases to facilitate its rapid isolation and recognition that help in implementing effective infection prevention and control practices. Interestingly, the current study revealed high resistance of human and animal isolates to vancomycin. In accordance with our study, some ST239-MRSA strains in Australia showed reduced vancomycin susceptibility (Holmes et al., 2014). Indeed, vancomycin resistance in ST-239 isolates has been rarely reported around the world and has not been reported previously in Egypt. Previous studies from Iran had indicated that a variant of ST-239 (named as ST1283 strain) was resistant to vancomycin (MIC of 512 μg/mL) (Azimian et al., 2012), and ST239-SCCmec-III/t037 from a university hospital showed resistance to vancomycin (Shekarabi et al., 2017). The latter clone was also reported to be resistant to vancomycin from hospitalized patients in Iran (Kouhsari et al., 2020). Similar to other bacteria, it is evidenced that the emergence of ST239 VRSA is a multifactorial phenomenon that could result from factors such as the intensive use of vancomycin in treating life-threatening infections (McDonald et al., 1997), bacterial genetic background, and yet-unknown evolutionary mechanisms. Therefore, there is a great challenge associated with the treatment of this clone, and this study further demonstrates the need to organize standard infection-control precautions to control the spread of vancomycin resistance in Egypt. Finally, these findings support the necessity for future surveillance studies on ST239-VRSA strains to keep their transmission to a minimum.
Concerning ciprofloxacin, clindamycin, and gentamicin, human strains showed higher resistance rates than animal ones, while the reverse occurred with tetracycline, ceftriaxone, and erythromycin. The continuous prescription of several Egyptian hospitals of ciprofloxacin, clindamycin, and gentamicin for chest infections (Eldeeb, 2006) and the large applications of tetracycline and erythromycin in veterinary fields (Zwjuang, 2005) lead to the variations in the resistance patterns to those antimicrobials among human and animal strains. The co-administration of erythromycin and tetracycline in the clinical field reflects the positive correlation between their resistances among our strains. Meanwhile, the differences in the mechanisms of resistance and genetic elements associated with the resistance to tetracycline and clindamycin (Fletcher and Macrina, 1991) may illustrate the negative correlation between these antimicrobials. The significant differences in the clindamycin resistance observed among our human and animal strains may be attributed to the absence of injection dosage form or the high cost, which limits the veterinary use of this antimicrobial in Egypt in comparison with other antimicrobials. Therefore, we confirm that the resistance pattern of the ST239-MRSA clone is well correlated with the extensive usage of antibiotics as announced in several reports (Havaei et al., 2017; Sedaghat et al., 2018)
Our results regarding the high prevalence of ST239-MRSA SCCmec-III agree with a previous report from India, which indicated that the majority (66.6%) of the ST-239 MRSA isolates belonged to SCCmec-III (Jain et al., 2019). Indeed, ST239-MRSA-SCCmec type III is considered to be an epidemic strain of hospital-associated MRSA in Asia (Chongtrakool et al., 2006). Moreover, ST-239 MRSA of the SCCmec-III is responsible for many outbreaks in healthcare settings all over the world and it is able to cause serious disseminated infections (Botelho et al., 2019). Moreover, a lower proportion of ST239-SCCmec type II (2%) was observed in a previous study conducted in Korea (Chong et al., 2013). Obviously, being the lineage with the highest prevalence in our isolates does not imply a general view on Egyptian ST-239 MRSA clone, as we did these analyses on only 50 isolates, and additional wide-scale analyses are needed to generalize these data.
Regarding the analysis of ST239-MRSA toxin gene profiles, our study indicated that nearly half of the strains were positive for lukSF-PV gene. In China, five identified ST239-MRSA isolates were all positive for lukSF-PV gene (Yu et al., 2008). Meanwhile, lukSF-PV-positive ST239-MRSA isolates were distinctly rare in many previous studies from various geographic areas (Alfouzan et al., 2013; Boswihi et al., 2016), suggesting that the carriage of lukSF-PV gene may be an unreliable marker for classifying the ST239-MRSA strains. Usually, the ST239-MRSA lineage lacks tst gene, while few studies have reported the presence of this gene in ST239 strains (Iwao et al., 2012; Kong et al., 2016) as was presented in our finding, where only few strains carried tst gene. Although previous reports (Xie et al., 2011; Jain et al., 2019) indicated the lack of eta gene in ST239 strains, it was determined to be the major exfoliative toxin type among our strains. The variable distribution of this virulent determinant was possible because it is located on a prophage that is integrated into S. aureus chromosome phages. To date, few ETA phages have been induced from bacterial cells and were found to be capable of transferring eta gene into eta-negative S. aureus strains, converting them into eta producers (Yoshizawa et al., 2000). Notably, sea was the most common enterotoxin gene detected among ST239-MRSA strains in previous studies (Tekeli et al., 2016; Jain et al., 2019). Similarly, our study showed that sea is the most prevalent enterotoxin gene among our ST239-MRSA strains.
Frequently, the MRSA strains with SCCmec-IV carry lukSF-PV gene encoding Panton–Valentine leucocidin (Bendary et al., 2016). Nevertheless, our results confirmed the association of lukSF-PV positive ST239 strains to SCCmec-III. This finding collaborated with an earlier report conducted in China (Yu et al., 2008), where five ST239 SCCmec-III isolates were PVL-positive. Taken together, the discrepancies between publications support that there is no absolute correlation between toxin profiles and the genetic background of this clone worldwide.
In the current analyses, it was expected that the ST239-MRSA strains belonging to different hosts (i.e., humans and cows) would exhibit more divergence compared to that among strains of the same host, and this was accepted phenotypically. Meanwhile, the diversity of both human and animal strains was almost similar to that among the same host depending on the genetic background. These results also suggest that the host is not the driving factor for the diversity of this clone, and possibly ST239-MRSA strains within the same origins may have undergone different evolution processes. The great diversity of these strains provides a wide array of defense strategies and challenges to both prevention methods and therapies (Yamamoto et al., 2012).
Despite the evolutions in the genetic background of the ST239-MRSA clone, the oldest truly pandemic MRSA strain, it is still the common and widespread clone. Of note, the mutation or the alteration in the housekeeping genes may lead to the emergence of new ST-MRSA clones as it was previously observed in arcC-allele leading to the emergence of ST239 from canonical ST8-MRSA clone. Unexpectedly, the ST239-MRSA clone can have genetic divergence without the emergence of a new ST-clone due to the conservation of the housekeeping gene sequences. It was previously reported that genetic divergence in this clone was related to the diverse geographic origins (Castillo-Ramírez et al., 2012). Therefore, the ST239-MRSA clone was clustered into 4 major clades including “European”, “Latin American”, “Turkish”, and “Asian” (Harris et al., 2010; Monecke et al., 2018). Surprisingly, in this study and in accordance with previous reports (Harris et al., 2010; Chen et al., 2014), the emerging ST239-MRSA subclones of one geographic origin and/or one host were announced. Therefore, the geographic origin and the host specificity were not the only driving factors for such genetic diversity. Possibly, the horizontal acquisition of the mobile genetic elements harbored antibiotic resistance genes, and virulence genes (Chen et al., 2014), clinical practice, prophage, and transgenerational adaptations were the major drivers alongside the geographic origin and host specificity for the genetic diversity within this clone, which may evolve and emerge as novel ST-MRSA clones in the near future. Crucially, ST239-MRSA clone has a selective advantage of the resistance provided by SCCmec-III; meanwhile, the prevalence of ST239 has recently declined, supporting the hypothesis that the recombination events, especially in the type of SCCmec, create a low fitness (Baltrus, 2013; San Millan and Maclean, 2017).
Our study revealed an overall negative correlation between the existence of antimicrobial resistance and the toxin genes. The toxigenic ST239-MRSA clone had a non-MDR profile, and they were susceptible to most antimicrobial agents, while MDR ST239-MRSA strains were less toxicogenic. This indicates the possibility of the acquisition of toxin genes on mobile elements at the expense of extended antibiotic resistance and vice versa. The consequence of these practices among ST239-MRSA strains may provide a fortuitous opportunity to correlate phenotypic antibiotic resistance patterns with the occurrence of toxin genes (Bendary et al., 2016).
The present study has some limitations. The lack of funds during the time of investigation precluded us from doing DNA sequencing for spa using the well-established spa typing method or whole-genome sequencing on the isolates, which are definitely the best validation approaches when it comes to validate their genetic diversity. However, we compensated for this via using PCR including positive and negative controls in addition to using sequencing techniques for unexpected results such as the high prevalence of lukSF-PV gene and the occurrence of different SCCmec genotypes. Additionally, the small number of strains used in this study hinders us to reach solid conclusions regarding the genetic characterization of the ST-239 isolates. Obviously, large isolate numbers should be used in future investigations to provide the information needed for infection control plans. Characterizing the current isolates and taking into consideration their origin [healthcare-associated (HA) versus CA] would be more interesting, yet the logistic difficulties of obtaining full data about the history of the patients, from whom the samples were taken, made this difficult and limited the scope of this study. In future investigations, applying whole-genome sequencing on the isolates would be appropriate to unravel strain diversity.
Conclusions
The current study showed a high diversity of the ST239-MRSA clone possibly due to their recombination or evolutionary dynamism. Most strains were MDR and belonged to SCCmec-III, agr-I, coa-I (750 bp), and spa-I (400 bp) genotypes and harbored sea and lukSF-PV genes. These results have important implications, especially with regard to understanding the complex epidemiology of this clone, which is ultimately linked to strategies of its prevention and control. Therefore, there is an urgent need for more specific recommendations in terms of the control and prevention of such pathogens as well as their resistance such as applying more restricted isolation guidelines within each host/hospital unit and also avoiding the misuse of antibiotics, especially in veterinary fields. Future studies featuring a large sample size are needed to reach solid conclusions regarding more impacted infection control decisions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Materials .
Ethics Statement
Ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements. The patients/participants provided their written informed consents to participate in this study.
Author Contributions
MB* and MA designed the study, carried out the antibiotyping and molecular analyses, and participated in the data analyses. MS performed the bioinformatics and statistical analyses and prepared the drafts and final version of the figures. MA-S, EK, MA, MB*, RM, and AS revised and discussed the figures, tables, and results. AS, HA, and WH wrote the primary version of the manuscript and participated in the design, antibiotyping, and molecular analyses. DG, HA, WA, and RM conceived the study and participated in the analysis. HR, MB and MG reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
MA-S extends his gratitude to the deputyship for research and innovation at the Ministry of Education in Saudi Arabia for supporting this work through project number 375213500.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Materials for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.782045/full#supplementary-material
References
- Abd El-Aziz N. K., Abd El-Hamid M. I., Bendary M. M., El-Azazy A. A., Ammar A. M. (2018). Existence of Vancomycin Resistance Among Methicillin Resistant S. aureus Recovered From Animal and Human Sources in Egypt. Slov. Vet. Res. 55, 221–230. doi: 10.26873/SVR-649-2018 [DOI] [Google Scholar]
- Abd El-Hamid M. I., Bendary M. (2015). Comparative Phenotypic and Genotypic Discrimination of Methicillin Resistant and Susceptible Staphylococcus aureus in Egypt. Cell. Mol. Biol. 61, 101–112. doi: 10.14715/cmb/2015.61.4.18 [DOI] [PubMed] [Google Scholar]
- Abd El-Hamid M. I., Bendary M. M., Merwad A. M. A., Elsohaby I., Mohammad Ghaith D., Alshareef W. A. (2019). What Is Behind Phylogenetic Analysis of Hospital-, Community- and Livestock-Associated Methicillin-Resistant Staphylococcus Aureus ? Transbound. Emerg. Dis. 66, 1506–1517. doi: 10.1111/tbed.13170 [DOI] [PubMed] [Google Scholar]
- Alfouzan W., Al-Haddad A., Udo E., Mathew B., Dhar R. (2013). Frequency and Clinical Association of Panton-Valentine Leukocidin-Positive Staphylococcus aureus Isolates: A Study From Kuwait. Med. Princ. Pract. 22, 245–249. doi: 10.1159/000343906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfouzan W., Udo E. E., Modhaffer A. (2019). Molecular Characterization of Methicillin-Resistant Staphylococcus aureus in a Tertiary Care Hospital in Kuwait. Sci. Rep. 9, 1–8. doi: 10.1038/s41598-019-54794-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimian A., Havaei S. A., Fazeli H., Naderi M., Ghazvini K., Samiee S. M., et al. (2012). Genetic Characterization of a Vancomycin-Resistant Staphylococcus aureus Isolate From the Respiratory Tract of a Patient in a University Hospital in Northeastern Iran. J. Clin. Microbiol. 50, 3581–3585. doi: 10.1128/JCM.01727-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T. F., Kuroda M., Yuzawa H., Aoki K. (2002). Genome and Virulence Determinants of High Virulence Community-Acquired MRSA. Lancet 359, 1819–1827. doi: 10.1016/S0140-6736(02)08713-5 [DOI] [PubMed] [Google Scholar]
- Baltrus D. A. (2013). Exploring the Costs of Horizontal Gene Transfer. Trends Ecol. Evol. 28, 489–495. doi: 10.1016/j.tree.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Beceiro A., Tomás M., Bou G. (2013). Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 26, 185–230. doi: 10.1128/CMR.00059-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K., Skov R. L., Von Eiff C. (2015). Staphylococcus, Micrococcus, and Other Catalase-Positive Cocci. Manual Clin. Microbiol. 1, 354–382. doi: 10.1128/9781555817381.ch21 [DOI] [Google Scholar]
- Bendary M., Solyman S., Azab M., Mahmoud N., Hanora A. (2016). Genetic Diversity of Multidrug Resistant Staphylococcus aureus Isolated From Clinical and Non Clinical Samples in Egypt. Cell. Mol. Biol. 62, 55–61. doi: 10.14715/cmb/2016.62.10.9 [DOI] [PubMed] [Google Scholar]
- Boswihi S. S., Udo E. E., Al-Sweih N. (2016). Shifts in the Clonal Distribution of Methicillin-Resistant Staphylococcus aureus in Kuwait Hospitals: 1992-2010. PloS One 11, e0162744. doi: 10.1371/journal.pone.0162744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botelho A. M. N., Cerqueira E Costa M. O., Moustafa A. M., Beltrame C. O., Ferreira F. A., Cortes M. F., et al. (2019). Local Diversification of Methicillin-Resistant Staphylococcus aureus ST239 in South America After Its Rapid Worldwide Dissemination. Front. Microbiol. 10, 82. doi: 10.3389/fmicb.2019.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Ramírez S., Corander J., Marttinen P., Aldeljawi M., Hanage W. P., Westh H., et al. (2012). Phylogeographic Variation in Recombination Rates Within a Global Clone of Methicillin-Resistant Staphylococcus Aureus . Genome Biol. 13, 1–13. doi: 10.1186/gb-2012-13-12-r126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Han C., Huang X., Liu Y., Guo D., Ye X. (2018). A Molecular Epidemiological Study of Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus Contamination in the Airport Environment. Infect. Drug Resist. 11,2363. doi: 10.2147/IDR.S178584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Yang X., Wang Q., Zhao C., Li H., He W., et al. (2014). Insights on Evolution of Virulence and Resistance From the Whole-Genome Analysis of a Predominant Methicillin-Resistant Staphylococcus aureus Clone Sequence Type 239 in China. Chin. Sci. Bull. 59, 1104–1112. doi: 10.1007/s11434-014-0149-1 [DOI] [Google Scholar]
- Chong Y. P., Kim E. S., Park S.-J., Park K.-H., Kim T., Kim M.-N., et al. (2013). Accessory Gene Regulator (Agr) Dysfunction in Staphylococcus aureus Bloodstream Isolates From South Korean Patients. Antimicrob. Agents Chemother. 57, 1509–1512. doi: 10.1128/AAC.01260-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chongtrakool P., Ito T., Ma X. X., Kondo Y., Trakulsomboon S., Tiensasitorn C., et al. (2006). Staphylococcal Cassette Chromosome Mec (SCC Mec) Typing of Methicillin-Resistant Staphylococcus aureus Strains Isolated in 11 Asian Countries: A Proposal for a New Nomenclature for SCCmec Elements. Antimicrob. Agents Chemother. 50, 1001–1012. doi: 10.1128/AAC.50.3.1001-1012.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute . (2019). Performance Standards for Antimicrobial Susceptibility Testing. Wayne, Pa, USA: CLSI Document M100-S15, Clinical and Laboratory Standards Institute, 15th International Supplement. [Google Scholar]
- Coad B. R., Griesser H. J., Peleg A. Y., Traven A. (2016). Anti-Infective Surface Coatings: Design and Therapeutic Promise Against Device-Associated Infections. PloS Pathog. 12, e1005598. doi: 10.1371/journal.ppat.1005598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Yang S., Rao X. (2020). Vancomycin Resistant Staphylococcus aureus Infections: A Review of Case Updating and Clinical Features. J. Adv. Res. 21, 169–176. doi: 10.1016/j.jare.2019.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldeeb A. (2006). Microbiological Study on Respiratory Tract Infections in Libya. Egypt J. Hosp. Med. 24, 442–459. doi: 10.21608/ejhm.2006.17908 [DOI] [Google Scholar]
- Enright M. C., Day N. P., Davies C. E., Peacock S. J., Spratt B. G. (2000). Multilocus Sequence Typing for Characterization of Methicillin-Resistant and Methicillin-Susceptible Clones of Staphylococcus Aureus . J. Clin. Microbiol. 38, 1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W., Del Busto R., Love M., Markowitz N., Cendrowski C., Cardenas J., et al. (1986). Imipenem-Cilastatin in the Treatment of Methicillin-Sensitive and Methicillin-Resistant Staphylococcus aureus Infections. Antimicrob. Agents Chemother. 29, 26–29. doi: 10.1128/AAC.29.1.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatholahzadeh B., Emaneini M., Aligholi M., Gilbert G., Taherikalani M., Jonaidi N., et al. (2009). Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Clones From a Teaching Hospital in Tehran. Jpn. J. Infect. Dis. 62, 309–311. [PubMed] [Google Scholar]
- Feil E. J., Nickerson E. K., Chantratita N., Wuthiekanun V., Srisomang P., Cousins R., et al. (2008). Rapid Detection of the Pandemic Methicillin-Resistant Staphylococcus aureus Clone ST 239, a Dominant Strain in Asian Hospitals. J. Clin. Microbiol. 46, 1520–1522. doi: 10.1128/JCM.02238-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher H. M., Macrina F. L. (1991). Molecular Survey of Clindamycin and Tetracycline Resistance Determinants in Bacteroides Species. Antimicrob. Agents Chemother. 35, 2415–2418. doi: 10.1128/AAC.35.11.2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin R. C., Matthew A., Jeff A., Michael N., George M. (2012). “Performance Standards for Antimicrobial Susceptibility Testing,” in Twenty-Second Informational Supplement, M100-S22 (USA: Wayne; ). [Google Scholar]
- Friendly M. (2002). Corrgrams. Am. Stat. 56, 316–324. doi: 10.1198/000313002533 [DOI] [Google Scholar]
- Galili T., O’callaghan A., Sidi J., Sievert C. (2018). Heatmaply: An R Package for Creating Interactive Cluster Heatmaps for Online Publishing. Bioinformatics 34, 1600–1602. doi: 10.1093/bioinformatics/btx657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher R., Motohashi N., Vanam A. (2017). Global Methicillin-Resistant Staphylococcus aureus (MRSA) Infections and Current Research Trends. Arch. Gen. Intern. Med. 1 (2), 3–11. [Google Scholar]
- Ghaith D. M., Zafer M. M., Said H. M., Elanwary S., Elsaban S., Al-Agamy M. H., et al. (2020). Genetic Diversity of Carbapenem-Resistant Klebsiella Pneumoniae Causing Neonatal Sepsis in Intensive Care Unit, Cairo, Egypt. Eur. J. Clin. Microbiol. Infect. Dis. 39, 583–591. doi: 10.1007/s10096-019-03761-2 [DOI] [PubMed] [Google Scholar]
- Ghaly M., Shaheen A., Bouhy A., Bendary M. (2020). Alternative Therapy to Manage Otitis Media Caused by Multidrug-Resistant Fungi. Arch. Microbiol. 202, 1231–1240. doi: 10.1007/s00203-020-01832-z [DOI] [PubMed] [Google Scholar]
- Gilot P., Lina G., Cochard T., Poutrel B. (2002). Analysis of the Genetic Variability of Genes Encoding the RNA III-Activating Components Agr and TRAP in a Population of Staphylococcus aureus Strains Isolated From Cows With Mastitis. J. Clin. Microbiol. 40, 4060–4067. doi: 10.1128/JCM.40.11.4060-4067.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M., Goudarzi H., Sá Figueiredo A. M., Udo E. E., Fazeli M., Asadzadeh M., et al. (2016). Molecular Characterization of Methicillin Resistant Staphylococcus aureus Strains Isolated From Intensive Care Units in Iran: ST22-SCC Mec IV/t790 Emerges as the Major Clone. PloS One 11, e0155529. doi: 10.1371/journal.pone.0155529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. R., Feil E. J., Holden M. T., Quail M. A., Nickerson E. K., Chantratita N., et al. (2010). Evolution of MRSA During Hospital Transmission and Intercontinental Spread. Science 327, 469–474. doi: 10.1126/science.1182395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaei S. A., Halaji M., Vidovic S., Dillon J. A. R., Karbalaei M., Ghanbari F., et al. (2017). Prevalence and Genotyping of Methicillin-Resistant and-Susceptible Staphylococcus aureus Strains Isolated From Patients in a University Hospital, Isfahan, Iran. Jundishapur J. Microbiol. 10 (5), e13571. doi: 10.5812/jjm.13571 [DOI] [Google Scholar]
- Holmes N., Turnidge J., Munckhof W., Robinson J., Korman T., O’sullivan M., et al. (2014). Genetic and Molecular Predictors of High Vancomycin MIC in Staphylococcus aureus Bacteremia Isolates. J. Clin. Microbiol. 52, 3384–3393. doi: 10.1128/JCM.01320-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hookey J. V., Richardson J. F., Cookson B. D. (1998). Molecular Typing of Staphylococcus aureus Based on PCR Restriction Fragment Length Polymorphism and DNA Sequence Analysis of the Coagulase Gene. J. Clin. Microbiol. 36, 1083–1089. doi: 10.1128/JCM.36.4.1083-1089.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P. R., Gaston M. A. (1988). Numerical Index of the Discriminatory Ability of Typing Systems: An Application of Simpson’s Index of Diversity. J. Clin. Microbiol. 26, 2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwao Y., Khokhlova O., Takano T., Hung W.-C., Isobe H., Peryanova O., et al. (2012). Fatal Pneumonia in HIV-Infected Patients From a Novel ST239 Methicillin-Resistant Staphylococcus aureus Carrying the Toxic Shock Syndrome Toxin-1 Gene in Krasnoyarsk, Siberian Russia. Jpn. J. Infect. Dis. 65, 184–186. [PubMed] [Google Scholar]
- Jain S., Chowdhury R., Datta M., Chowdhury G., Mukhopadhyay A. K. (2019). Characterization of the Clonal Profile of Methicillin Resistant Staphylococcus aureus Isolated From Patients With Early Post-Operative Orthopedic Implant Based Infections. Ann. Clin. Microbiol. Antimicrob. 18, 1–7. doi: 10.1186/s12941-019-0307-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jr F. E. H. (2020) Package ‘Hmisc’. Available at: https://cran.r-project.org/web/packages/Hmisc/Hmisc.pdf.
- Katayama Y., Ito T., Hiramatsu K. (2000). A New Class of Genetic Element, Staphylococcus Cassette Chromosome Mec, Encodes Methicillin Resistance in Staphylococcus Aureus . Antimicrob. Agents Chemother. 44, 1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhelle C. (2018). Pharming Animals: A Global History of Antibiotics in Food Productio –2017). Palgrave Commun. 4, 1–13. doi: 10.1057/s41599-018-0152-2 [DOI] [Google Scholar]
- Kolde R. (2018). Package ‘Pheatmap’: Pretty Heat Map. Available at: https://CRAN.R-project.org/package=pheatmap. Retrieved from: https://CRAN.R-project.org/package=pheatmap. [Google Scholar]
- Kong H., Yu F., Zhang W., Li X., Wang H. (2017). Molecular Epidemiology and Antibiotic Resistance Profiles of Methicillin-Resistant Staphylococcus aureus Strains in a Tertiary Hospital in China. Front. Microbiol. 8, 838. doi: 10.3389/fmicb.2017.00838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z., Zhao P., Liu H., Yu X., Qin Y., Su Z., et al. (2016). Whole-Genome Sequencing for the Investigation of a Hospital Outbreak of MRSA in China. PloS One 11, e0149844. doi: 10.1371/journal.pone.0149844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouhsari E., Hosseini M., Ahmadi A., Khadivar P., Rahimi S., Amini A. (2020). Molecular Characterizations, Virulence Determinants and Antimicrobial Resistance Profiles of Methicillin-Resistant Staphylococcus aureus (MRSA) in the North of Iran. doi: 10.21203/rs.3.rs-41456/v1 [DOI] [PubMed] [Google Scholar]
- Krumperman P. H. (1983). Multiple Antibiotic Resistance Indexing of Escherichia Coli to Identify High-Risk Sources of Fecal Contamination of Foods. Appl. Environ. Microbiol. 46, 165–170. doi: 10.1128/aem.46.1.165-170.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Stegger M., Sørum M. (2008). Spa Typing Directly From a Meca, Spa and Pvl Multiplex PCR Assay—A Cost-Effective Improvement for Methicillin-Resistant Staphylococcus aureus Surveillance. Clin. Microbiol. Infect. 14, 611–614. doi: 10.1111/j.1469-0691.2008.01995.x [DOI] [PubMed] [Google Scholar]
- Li X., Zhang J., Zhang Y., Zhou J., Li X., Feng R., et al. (2021). Methicillin-Resistant Staphylococcus aureus of the Clonal Lineage ST5-SCCmecII-T2460 was Associated With High Mortality in a Wuhan Hospital. Braz. J. Microbiol. 52, 1929–1936. doi: 10.1007/s42770-021-00557-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorakos A.-P., Srinivasan A., Carey R., Carmeli Y., Falagas M., Giske C., et al. (2012). Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Magray M. S., Kumar A., Rawat A. K., Srivastava S. (2011). Identification of Escherichia Coli Through Analysis of 16S rRNA and 16S-23S rRNA Internal Transcribed Spacer Region Sequences. Bioinformation 6, 370–371. doi: 10.6026/97320630006370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L. C., Kuehnert M. J., Tenover F. C., Jarvis W. R. (1997). Vancomycin-Resistant Enterococci Outside the Health-Care Setting: Prevalence, Sources, and Public Health Implications. Emerg. Infect. Dis. 3, 311–317. doi: 10.3201/eid0303.970307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra M., Wang G., Johnson W. M. (2000). Multiplex PCR for Detection of Genes for Staphylococcus aureus Enterotoxins, Exfoliative Toxins, Toxic Shock Syndrome Toxin 1, and Methicillin Resistance. J. Clin. Microbiol. 38, 1032–1035. doi: 10.1128/JCM.38.3.1032-1035.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S., Coombs G., Shore A. C., Coleman D. C., Akpaka P., Borg M., et al. (2011). A Field Guide to Pandemic, Epidemic and Sporadic Clones of Methicillin-Resistant Staphylococcus Aureus . PloS One 6, e17936–e17936. doi: 10.1371/journal.pone.0017936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke S., Slickers P., Gawlik D., Muller E., Reissig A., Ruppelt-Lorz A., et al. (2018). Molecular Typing of ST239-MRSA-III From Diverse Geographic Locations and the Evolution of the SCCmec III Element During its Intercontinental Spread. Front. Microbiol. 9, 1436. doi: 10.3389/fmicb.2018.01436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq S., Khan J. A., Rabbani F., Latif U., Arfan M., Yameen M. A. (2017). Biocompatible Biodegradable Polymeric Antibacterial Nanoparticles for Enhancing the Effects of a Third-Generation Cephalosporin Against Resistant Bacteria. J. Med. Microbiol. 66, 318–327. doi: 10.1099/jmm.0.000445 [DOI] [PubMed] [Google Scholar]
- Ostojić M. (2008). Epidemiologic Genotyping of Methicillin-Resistant Staphylococcus aureus (MRSA) by Pulsed-Field Gel Electrophoresis (PFGE). Bosnian J. Basic Med. Sci. 8, 259. doi: 10.17305/bjbms.2008.2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D. A., Enright M. C. (2004). Evolution of Staphylococcus aureus by Large Chromosomal Replacements. J. Bacteriol. 186, 1060. doi: 10.1128/JB.186.4.1060-1064.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royston P. (1982). An Extension of Shapiro and Wilk’s W Test for Normality to Large Samples. Appl. Stat. 31, 115–124. doi: 10.2307/2347973 [DOI] [Google Scholar]
- Samba Z., Gadba R. (1993). Antibiotic Susceptibility and Phage Typing of Methicillin-Resistant Staphylococcus aureus Clinical Isolates From Blood Cultures of 692 Patients in 15 Israeli Hospitals. Eur. J. Epidemiol. 9, 559–562. doi: 10.1007/BF00209536 [DOI] [PubMed] [Google Scholar]
- San Millan A., Maclean R. C. (2017). Fitness Costs of Plasmids: A Limit to Plasmid Transmission. Microbiol. Spectr. 5, MTBP-0016-2017. doi: 10.1128/microbiolspec.MTBP-0016-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghat H., Esfahani B. N., Halaji M., Jazi A. S., Mobasherizadeh S., Havaei S. R., et al. (2018). Genetic Diversity of Staphylococcus aureus Strains From a Teaching Hospital in Isfahan, Iran: The Emergence of MRSA ST639- SCCmec III and ST343- SCCmec III. Iran. J. Microbiol. 10 (2), 82–89. [PMC free article] [PubMed] [Google Scholar]
- Shekarabi M., Hajikhani B., Salimi Chirani A., Fazeli M., Goudarzi M. (2017). Molecular Characterization of Vancomycin-Resistant Staphylococcus aureus Strains Isolated From Clinical Samples: A Three Year Study in Tehran, Iran. PloS One 12, e0183607. doi: 10.1371/journal.pone.0183607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E. H. (1949). Measurement of Diversity. Nature 163, 688. doi: 10.1038/163688a0 [DOI] [Google Scholar]
- Singh V. K., Kumar A., Yadav S. K. (2016). Antimicrobial Susceptibility Profiling of Milk Samples From Bovine Clinical Mastitis. Int. J. Med. Microbiol. Trop. Dis. 2, 52–55. doi: 10.5958/2455-6807.2016.00004.0 [DOI] [Google Scholar]
- Team R. C. R. (2018). A Language and Environment for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing; ). Available at: https://www.R-project.org/. [Google Scholar]
- Tekeli A., Ocal D. N., Ozmen B. B., Karahan Z. C., Dolapci I. (2016). Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Bloodstream Isolates in a Turkish University Hospital Between 2002 and 2012. Microb. Drug Resist. 22, 564–569. doi: 10.1089/mdr.2015.0116 [DOI] [PubMed] [Google Scholar]
- Woodyard C. (2011). Exploring the Therapeutic Effects of Yoga and Its Ability to Increase Quality of Life. Int. J. Yoga 4, 49. doi: 10.4103/0973-6131.85485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., He Y., Gehring A., Hu Y., Li Q., Tu S.-I., et al. (2011). Genotypes and Toxin Gene Profiles of Staphylococcus aureus Clinical Isolates From China. PloS One 6, e28276. doi: 10.1371/journal.pone.0028276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Takano T., Higuchi W., Iwao Y., Singur O., Reva I., et al. (2012). Comparative Genomics and Drug Resistance of a Geographic Variant of ST239 Methicillin-Resistant Staphylococcus aureus Emerged in Russia. PloS One 7, e29187. doi: 10.1371/annotation/05a851e1-8627-4ea4-b12a-cfdba09f1e61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa Y., Sakurada J., Sakurai S., Machida K., Kondo I., Masuda S. (2000). An Exfoliative Toxin A-Converting Phage Isolated From Staphylococcus aureus Strain ZM. Microbiol. Immunol. 44, 189–191. doi: 10.1111/j.1348-0421.2000.tb02481.x [DOI] [PubMed] [Google Scholar]
- Yu F., Chen Z., Liu C., Zhang X., Lin X., Chi S., et al. (2008). Prevalence of Staphylococcus aureus Carrying Panton–Valentine Leukocidin Genes Among Isolates From Hospitalised Patients in China. Clin. Microbiol. Infect. 14, 381–384. doi: 10.1111/j.1469-0691.2007.01927.x [DOI] [PubMed] [Google Scholar]
- Zhang W., Hao Z., Wang Y., Cao X., Logue C. M., Wang B., et al. (2011). Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Strains From Pet Animals and Veterinary Staff in China. Vet. J. 190, e125–e129. doi: 10.1016/j.tvjl.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Zhang K., Mcclure J.-A., Elsayed S., Louie T., Conly J. M. (2005). Novel Multiplex PCR Assay for Characterization and Concomitant Subtyping of Staphylococcal Cassette Chromosome Mec Types I to V in Methicillin-Resistant Staphylococcus Aureus . J. Clin. Microbiol. 43, 5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwjuang (2005). Study of Antibiotic Resistance and Epidemiological Typing of Staphylococcus aureus Isolated From Bovine Mastitis in Taiwan (Taiwan: National Chung Hsing University, Department of Veterinary Medicine; ). Master Thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Materials .