Abstract
The aryl hydrocarbon receptor (AhR) plays an important role in intestinal homeostasis, and some microbial metabolites of tryptophan are known AhR agonists. In this study, we assessed the impact of tryptophan supplementation on the formation of tryptophan metabolites, AhR activation, and microbiota composition in the simulator of the human intestinal microbial ecosystem (SHIME). AhR activation, microbial composition, and tryptophan metabolites were compared during high tryptophan supplementation (4 g/L tryptophan), control, and wash-out periods. During tryptophan supplementation, the concentration of several tryptophan metabolites was increased compared to the control and wash-out period, but AhR activation by fermenter supernatant was significantly decreased. This was due to the higher levels of tryptophan, which was found to be an antagonist of AhR signaling. Tryptophan supplementation induced most microbial changes in the transverse colon including increased relative abundance of lactobacillus. We conclude that tryptophan supplementation leads to increased formation of AhR agonists in the colon.
Keywords: aryl hydrocarbon receptor, SHIME, microbiota, tryptophan, tryptophan metabolites
Introduction
Species of the human gut microbiota produce numerous metabolites, including those generated by the metabolism of dietary compounds.1,2 Metabolite signaling through the aryl hydrocarbon receptor (AhR) is an example of an interaction that can have strong effects on the immune system and intestinal homeostasis.3−5 AhR is expressed in many different cell types in the body and is involved in many complex physiological processes, in particular, the regulation of xenobiotic metabolizing enzymes.4,6−8
AhR agonists can be found as such in the diet (e.g., certain polyphenols), they can be generated during digestion in the small intestine from dietary precursors (e.g., 3,3′-diindolylmethane from glucobrassicin) or they can be generated by microbial metabolism of tryptophan by specific species.9−11 Tryptophan (Trp) is abundant in protein-rich foods like beans and nuts, cheese, meat, fish, and eggs.12 The daily intake of Trp in the average “Western” diet is approximately 600–900 mg,13 of which approximately 30% is used for protein synthesis.13,14 Trp supplementation is used to treat sleep disorders and other neurological disorders linked to reduced secretion of serotonin because melatonin is synthesized in the pineal gland from Trp that is involved in the regulation of circadian.15 Trp is also metabolized by endogenous enzymes indoleamine 2,3-dioxygenase (IDO) and l-tryptophan 2,3-dioxygenase (TDO) in the kynurenine pathway of the host or metabolized by the gut microbiota9,16−21 to produce agonists of AhR. The metabolism of Trp to kynurenine and other AhR agonists or antagonists by microbiota is therefore of considerable interest in relation to host immune regulation. Kynurenine metabolites promote immunological tolerance through their activity on dendritic cells, T-cell anergy and apoptosis, and proliferation of Treg and Th17 cells.22 Microenvironmental factors regulate the kynurenine pathway to maintain immune homeostasis.
AhR agonists produced by the microbiota have been shown to be crucial for antifungal immunity in mouse infection models.9 IBD patients with polymorphisms in CARD9, leading to loss of function, have an altered microbiota that is deficient in producing AhR agonists, contributing to dysfunctional intestinal homeostasis.10 In turn, microbiota composition and metabolism are affected by Trp availability in the diet. It leads to an increase of lactobacilli, mainly L. reuteri, which subsequently affects the local immune response through the production of indole-3-aldehyde from Trp.9 Given the mounting evidence connecting gut Trp metabolism to health, there is interest in supporting human health through dietary supplementation with Trp.23 However, the effects of such supplementation on Trp metabolism and the production of different AhR agonists in the human intestine are currently unknown.
The aims of this study were to assess the impact of Trp supplementation on the formation of Trp metabolites by the human microbiota using the simulator of the human intestinal microbial ecosystem (SHIME) and to assess the activity of the individual and pooled metabolites on the AhR signaling pathway.
Materials and Methods
Chemicals
All chemicals were purchased from Sigma-Aldrich (USA) unless stated otherwise. Trp metabolites screened for AhR activation were tryptamine, indole-3-aldehyde, anthranilic acid, kynurenic acid, indole-3-propionic acid, 3-hydroxy-anthranilic acid, oxindole, indole, l-kynurenine and l-Trp in concentrations ranging from 0.1–100 μM.
AhR Activation
To measure the AhR activation, the dioxin responsive element (DR)-chemical activated luciferase gene expression (CALUX) rat hepatoma-derived reporter cells (BioDetection Systems, The Netherlands, mycoplasma free) were used as previously described.24 Following incubation with compounds binding to the aryl-hydrocarbon receptor (AhR), a ligand–receptor complex binds the DR in the promoter which is coupled to the firefly luciferase gene. This leads to expression of luciferase which is detected by addition of the substrate luciferin. In brief, cells were grown in α-MEM growth medium (Gibco, USA) with 10% heat-inactivated fetal calf serum (Gibco) and 1% penicillin/streptomycin (Gibco). The cells were grown in 96-wells white clear-bottom plates (Corning, USA) at a concentration of 7.5 × 104 cells per well for 24 h before stimulation. The cells were stimulated for 24 h in triplicate with Trp, and its derivatives dissolved in DMSO (Merck KGaA, Germany) or with 20% final volume of microbial supernatant. After stimulation, the cells were lysed with 20 μL lysis buffer (Promega, USA) per well and 100 μL luciferase assay buffer was added (Promega). The luminescence was measured using a Spectramax M5 (Molecular Devices, USA) immediately after the addition of the assay buffer. The results were expressed as a percentage of the positive control, 5 μM β-naphthoflavone.
In Vitro Human Fermentation
The SHIME was used to simulate the human intestinal tract, as previously described.25 The TWINSHIME setup was used where two faecal samples, each from an independent donor, were used to inoculate two sets of 4 vessels in parallel. Each set of vessels simulated a combined stomach and small intestine, followed by an ascending (AC, pH 5.6–5.9), transverse (TC, pH 6.15–6.4) and descending (DC, pH 6.6–6.9) colon part for each donor. The donors of the faecal sample were healthy and did not take antibiotics or prebiotics for 6 and 3 months before donation, respectively. Every 8 h, a new feeding cycle was started with a stomach phase, where a 140 mL feed with pH 2 (1.2 g/L arabinogalactan, 2 g/L pectin, 0.5 g/L xylan, 0.4 g/L glucose, 3 g/L yeast extract, 1 g/L special peptone, 3 g/L mucin, 0.5 g/L l-cysteine-HCl, and 4 g/L starch; pH 1.8–2.2) was incubated for 1.5 h, after which 60 mL of pancreatic juice was added (12.5 g/L NaHCO3, 6 g/L Oxgall, 0.9 g/L pancreatin; pH 7). After a further 1.5 h, the feed was transferred to the AC, TC, and DC connected in series. The experimental design consisted of 3 weeks, including a 1-week control period with the standard feed, a 1-week Trp supplementation period where 4 g/L l-tryptophan was added to the feed, and a 1-week wash-out period where the standard feed was provided again. Fermented samples were taken every day and immediately centrifuged at 9000g at 4 °C, after which the supernatant was filtered using a 0.2 μm RC filter (Phenomenex, Torrance, CA). The samples were stored at −20 °C until further analysis.
SCFA Analysis
Gas chromatography coupled with a flame-ionization detector (GC-FID, Shimadzu, Kyoto, Japan) was used to determine short-chain fatty acid (SCFA) composition in each sample. The samples and calibration standards were mixed in a ratio of 2:1 with an internal standard containing 0.45 mg/mL 2-ethylbutyric acid in 0.3 M HCl and 0.9 M oxalic acid. Subsequently, the solutions were centrifuged for 4 min at 20000g. One microliter of the supernatant was injected spitless in a Restek Stabilwax-DA column (30 m × 0.32 mm × 1 μm, Tmax = 240 °C, Restek, USA). Nitrogen was used as a carrier gas, with a flow rate of 2.51 mL/min. The makeup gases were nitrogen, hydrogen, and air with respective flow rates of 40, 30, and 400 mL/min. The temperature was initially held at 100 °C. After injection, the temperature was increased first to 180 °C and then to 240 °C and both temperatures were held for 2 min. The samples were compared to 6 calibration standard solutions containing acetic acid, propionic acid, butyric acid, valeric acid, isovaleric acid, and isobutyric acid. The results were processed using Chromeleon Edition 7 (Thermo Scientific, San Jose, CA).
Tryptophan Metabolites
Tryptophan metabolites were analyzed according to Koper et al.25 Samples were centrifuged (21700g, 10 min, 4 °C) and diluted in 0.1% formic acid, followed by filtration using a 0.22 μm cellulose filter (Phenomenex) and high-resolution mass spectrometry (HRMS) analysis. A silica modified Luna Polar C18 column (50 × 2.1 mm, 1.6 μm, Phenomenex) was used for the chromatographic separation of Trp and tryptophan metabolites. The mobile phases consisted of water (A) and acetonitrile (B) both with 0.1% v/v of formic acid, and the following gradient (min/%B) was used: (0/2), (0.50/2), (9.5/70), and (12/70). The flow rate was 200 μL/min, the column temperature was 40 °C, and 5 μL was injected. The U-HPLC system (Accela 1250, Thermo Fisher, Bremen, Germany) was interfaced to an Exactive Orbitrap HRMS (Thermo), and the analytes were detected through a heated electrospray interface (HESI-II) in positive mode by scanning the ions listed in Table S1 in the m/z range of 50–400. Analytical performances, mass spectrometry optimization, and linearity range were monitored according to Koper et al.25 Each sample was analyzed in duplicate, and the concentrations are given in micromolars.
Microbial Analysis
DNA extraction of the microbial pellets was performed using the QIAmp PowerFecal DNA Kit (Qiagen, USA). 16S rRNA sequencing of the V3–V4 region was performed by Novogene (Hong Kong). The results were analyzed using the CLC bio genomics workbench (Qiagen, The Netherlands), Microbial Genomics Toolbox with the SILVA 16S v132 99% as reference database. Further analysis and statistics were performed using the online MicrobiomeAnalyst tool (www.microbiomeanalyst.ca), with filtering steps: minimal count 4, prevalence 10%, and removal of 2% standard deviation. The Ward clustering analysis was shown as a heat map using Euclidean distance measurement at the family level.
Statistical Analysis
The statistical analyses were performed using GraphPad Prism 5 (La Jolla, USA). Results are shown as mean ± SEM, where *p < 0.05, **p < 0.01, and ***p < 0.001 were considered statistical differences. AhR activation was tested using one-way ANOVA followed by a Tukey post hoc analysis for the tryptophan derivatives and a repeated-measures ANOVA followed by a Tukey post hoc analysis for the fermentation samples. Different letters above bars represent statistically significant differences between responses to each concentration.
Results and Discussion
AhR Activation during Colonic Fermentation
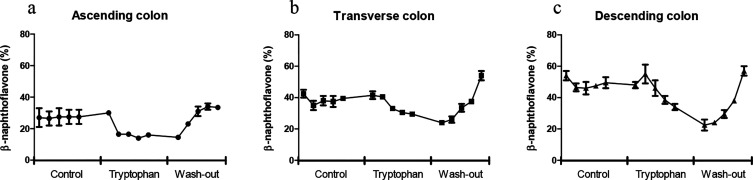
To study the effects of a high Trp diet in SHIME, 4 g/L l-tryptophan, corresponding to 560 mg per feeding, was added to the standard SHIME feed for 1 week in all feeding cycles. The amount of Trp was based on the highest reported dietary supplementation dose in human intervention studies, taking into account absorption in the small intestine.26 Each day, one sample was collected from each SHIME vessel just prior to the new feeding cycle. In the control period (i.e., one week before the Trp supplementation period), AhR activity of the samples remained constant in each part of the colon (Figure 1). The highest AhR activation was measured in the DC, followed by the TC and AC, which is in line with previous findings.25 Surprisingly, AhR activation decreased in all parts of the colon during the one week of Trp supplementation, with the largest decrease in the DC (Figure 1c). Finally, after the wash-out period, the AhR activation returned to the level measured in the control period in the AC and DC samples. However, on the last day of the wash-out period, AhR activity in the TC was significantly higher (p < 0.001) than in the control period (Figure 1b).
Figure 1.
AhR activation of (a) ascending, (b) transverse, and (c) descending colon, where each data point represents the AhR activity at each day of the control, tryptophan supplementation, and wash-out period. Measured with the CALUX reporter assay, expressed as percent of the positive control (β-naphthoflavone, 5 μM) using luciferase production as readout. Data are expressed as the mean of 2 donors ± SEM.
Tryptophan Metabolites in Control and Supplemented Diet
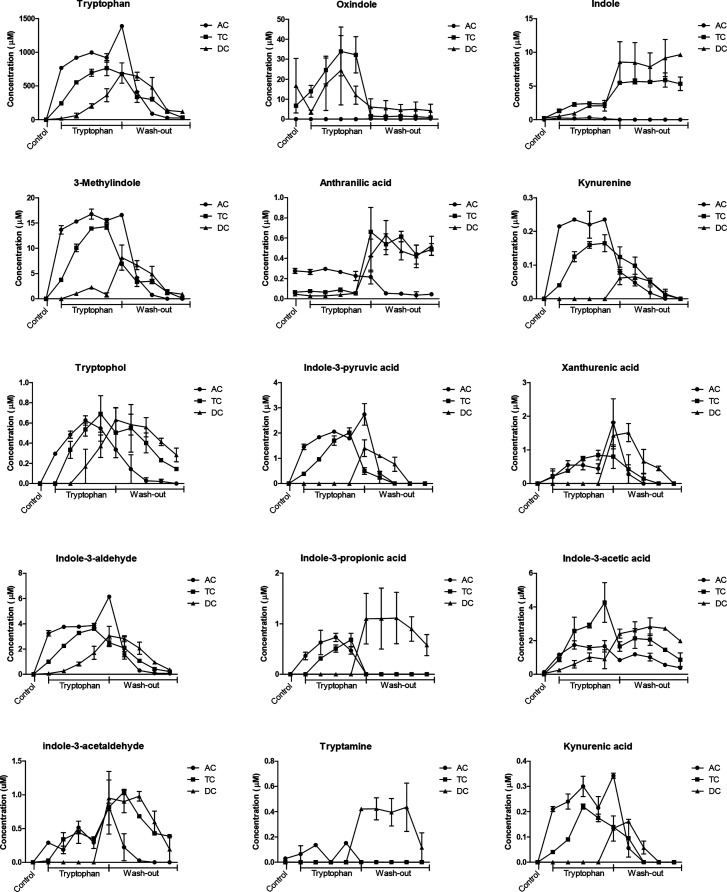
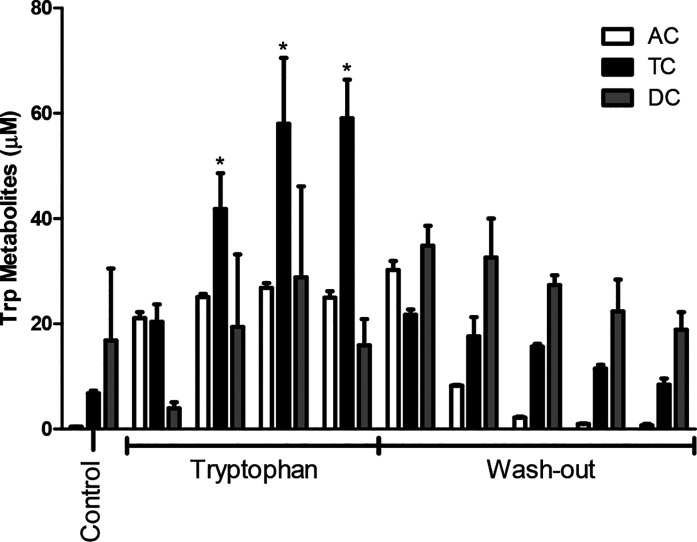
Trp and its metabolites were quantified in all sample supernatants, and as expected, Trp and all its metabolites increased during the Trp supplementation period (Figure 2). Trp reached a concentration of 995, 765, and 363 μM in the AC, TC, and DC, respectively. Trp supplementation increased the combined amount of Trp metabolites in AC, TC, and DC compared to the control (Figure 3). From Figure 3, it is also evident that Trp supplementation increased the conversion of Trp into its metabolites especially in TC but to a lesser extent in the AC and DC. The largest increase of Trp metabolites was measured in the TC (p < 0.0001), indicating the importance of the TC for Trp metabolism. After the wash out period, the total amount of Trp metabolites measured in the AC decreased to 0.7 μM, which is similar to the concentration present in the control period before Trp supplementation (0.5 μM). The same holds true for the concentration of Trp metabolites formed in the TC after the wash-out period indicating that the production of Trp metabolites by the microbiota is directly related to the amount of available Trp.
Figure 2.
Evolution of tryptophan and tryptophan metabolites concentration during control, tryptophan supplementation, and wash-out period, in the ascending (AC), transverse (TC), and descending colon (DC) during in vitro SHIME fermentation. n = 2 donors. Each data point represents the concentration at each day of the Trp supplementation and wash-out period. Only the last day of the control period is reported (first data point in the graph).
Figure 3.
Cumulative concentration of tryptophan metabolites quantified using Orbitrap during control, tryptophan supplementation, and wash-out period, in the ascending (AC), transverse (TC), and descending colon (DC) during in vitro SHIME fermentation. n = 2 donors. Each data point represents the concentration at each day of the Trp supplementation and wash-out period. Only the last day of the control period is reported (first data point in the graph). *p < 0.05.
The main metabolite formed during the period of Trp supplementation was the AhR ligand oxindole, which was at highest concentration (34 μM) in the TC, had fluctuating concentrations (3.5 to 25 μM) in the DC, and was absent in the AC (Figure 2). The second most abundant metabolite formed was 3-methylindole, with highest concentrations of 17 μM in the AC, 14 μM in the TC, and 2 μM in the DC. At the end of the wash-out period, Trp and most of its metabolites decreased to similar concentrations measured before Trp supplementation (control period in Figure 2).
The concentrations of some metabolites, namely tryptophol, indole-3-acetic acid, indole-3-acetaldehyde, and tryptamine were higher on the last day of the wash-out period than in the control period. Indole and anthranilic acid had higher concentrations during and after the wash-out in both the TC and DC compared to the beginning of the wash-out period.
As can be seen in Figure 2, we measured a larger increase of 3-methylindole, oxindole, and indole than the other Trp metabolites during Trp supplementation. This may be due to a higher rate of conversion for the reactions catalyzed by tryptophanase and pyruvate amino transferase compared to those catalyzed by arylformidase and transglutaminase, which are required for formation of kynurenine and xanthurenic acid.27,28 Alternatively, this may be due to the presence of bacterial species with different pathways for metabolism of Trp.
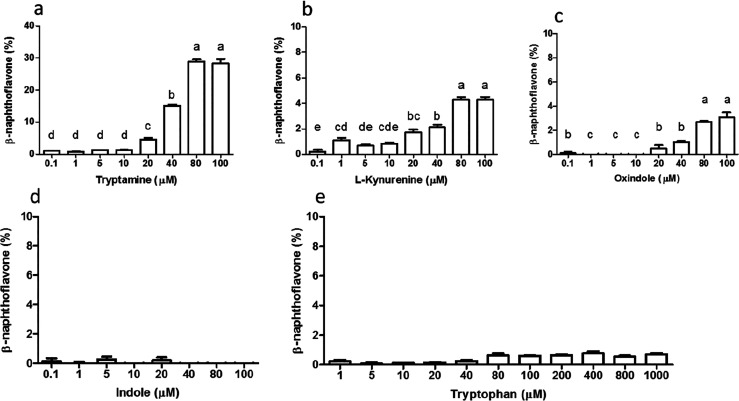
Of the metabolites formed (Figure 2), tryptamine, kynurenine, indole-3-acetaldehyde, indole-3-acetic acid and indole-3-aldehyde, indole, 3-methylindole, and oxindole have been reported as AhR ligands.9,29 Several Trp metabolites produced during colonic fermentation in the SHIME system (tryptamine, indole-3-aldehyde, anthranilic acid, kynurenic acid, indole-3-propionic acid, 3-hydroxy-anthranilic acid, oxindole, indole, l-kynurenine, and l-tryptophan) were screened for their capacity to activate AhR, based on their previously reported AhR-activating properties and their commercial availability as purified compounds. Of these, only tryptamine induced a relatively high concentration-dependent AhR activation (approximately 28% of the positive control) after stimulation with 80 and 100 μM (Figure 4a). l-kynurenine, oxindole, and indole induced significant but low levels of AhR activation (Figure 4b–d). Trp itself did not activate AhR in the range between 1 and 1000 μM. This is in line with other reports of AhR activation by Trp and its metabolites.24,30,31
Figure 4.
AhR activation of (a) tryptamine, (b) l-kynurenine, (c) oxindole, (d) indole, and (e) tryptophan, measured with the CALUX reporter assay, expressed as percent of the positive control (β-naphthoflavone, 5 μM) using luciferase production as the readout. Data are expressed as mean of 3 replicates ± SEM, and different letters above the bar represent statistically significant different responses compared to each concentration.
The AhR ligand indole-3-acetaldehyde32 had a higher concentration in the TC after the wash-out period than in the control and Trp supplementation period (Figure 2). This change may partly explain the higher AhR activity in the TC after the wash-out period (Figure 1b). This may involve the interconversion between tryptophol (which is not an AhR agonist) and indole-3-acetaldehyde.30 Again, this implies a relevant role of the TC regarding the production of AhR ligands that are formed further down the Trp metabolism pathway.
Liang et al. studied supplementation of Trp in pigs and although they did not discriminate between different colon parts, they found different levels of Trp metabolites in colonic content of pigs with different Trp treatments (control, 0.2% Trp, and 0.4% Trp).33 Indole-3-acetic acid was increased the most in the cecal content after Trp supplementation, but no increase was measured in the colonic content. In our study, Trp supplementation elevated the amount of Trp available to microbiota in the AC, TC, and DC by 46, 31, and 21 times, respectively, even though the total amount of Trp metabolites was not proportionally higher compared to the control (Figure 3).
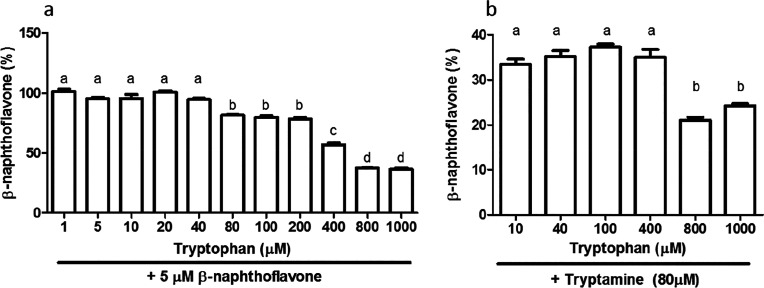
To explain the fact that AhR activity was lower than the control during Trp supplementation (Figure 4), we hypothesized that Trp itself might have an antagonistic effect on AhR signaling. Indeed, we found that above concentrations of 200 μM, Trp inhibits AhR activation by β-naphthoflavone (Figure 5a) and tryptamine (Figure 5b), one of the most potent microbial-derived AhR agonists. In the AC, Trp supplementation increased concentrations to above 200 μM which would account for the decreased AhR activity in AC compared to the control. This is, as far as we know, the first time Trp has been reported to antagonize AhR signaling by other ligands. In vivo, the plasma concentration of tryptophan is around 40 μΜ in healthy subjects.34 In this same study, tryptophan concentrations were 87 pmol/g of intestine, 26.8 pmol/g liver, and 42.2 pg/g of spleen although metabolites of kynurenine (i.e., anthranilic acid and kyneuric acid) with reported AhR activity were between 78 and 161 pmol/g in all tissues. Thus, the inhibition we see in the SHIME samples is due to the relatively high dose of tryptophan (15g/day26) and the lack of metabolite absorption in SHIME resulting in a high Trp to/Trp-metabolite ratio. However, this is an intrinsic limitation of any in vitro model of colon fermentation. The reviewer might also consider that the trp:metabolites ratio in our experiment is expected to be way higher than that produced by a typical diet which provides a lower amount of tryptophan to gut microbiota.
Figure 5.
AhR activation measured with the CALUX reporter assay, expressed as percent of the positive control (β-naphthoflavone, 5 μM) using luciferase production as readout, with (a) different concentrations of tryptophan combined with 5 μM β-naphthoflavone and (b) different concentrations of tryptophan combined with 80 μM tryptamine. n = 3. Data are expressed as mean ± SEM, and different letters above the bar represent statistically significant different responses compared to each concentration.
Effects of Trp Supplementation on Microbial and Composition SCFA Production
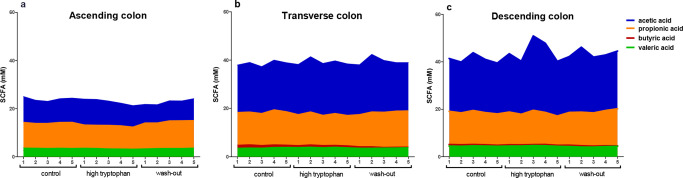
SCFAs and the microbial composition were analyzed in order to determine whether there was a change in microbial composition and fermentation leading to an altered production of SCFA. Figure 6 shows that the total production of SCFA was highest in the DC and lowest in the AC, which is in accordance with Van den Abbeele et al.35 Overall, the SCFAs concentrations in all parts of the colon remained at similar levels independently of Trp supplementation. This is in accordance with the findings of Liang et al., who did not find any differences in SCFA production with different amounts of Trp supplementation in vivo.33 Also, Van den Abbeele et al., showed that the SHIME microbiota is stable over time without changed fermentation conditions like SCFAs.35 Recently, it was shown that butyrate can activate the AhR pathway in vitro, in a dose-dependent manner at concentrations above 1 mM.36−38 In previous studies using different cell lines AhR activity was not altered by 10 to 20 mM acetate, whereas propionate induced AhR-dependent gene expression at concentrations of 5 mM or 10 mM depending on the cell line.37,38 It is known that results may differ between cell lines and species due to assay sensitivity or transport of AhR agonists.39,40 In our AhR activation assay butyrate concentrations were less than 1 mM, but SCFAs might be important for AhR activation in vivo. However, butyrate is found in scarce amounts in the blood, due to metabolism by epithelial cells, which makes its effect largely restricted to cells in the intestinal epithelium.41,42
Figure 6.
SCFA profile (mM) for the (a) ascending colon, (b) transverse colon and (c) descending colon, after a control, tryptophan supplementation and wash-out period of 5 days. Each data point represents the SCFA profile at each day of the control, Trp supplementation and wash-out period. n = 2 donors.
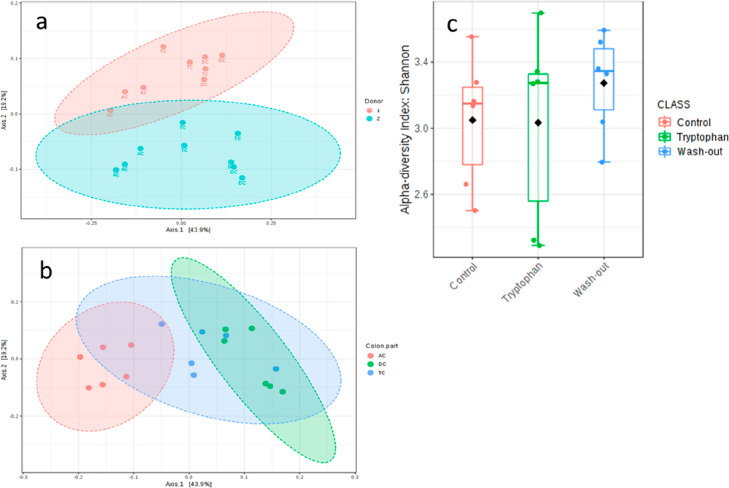
The microbiota was compared between the donors and in the different colon compartments over the period of supplementation and wash-out. The microbiota of the two human donors was stable but differed in composition during the control period (Figure 7a, p < 0.019). As anticipated, the physiological conditions in each simulated colon compartment altered the composition, independently of the donor (Figure 7b, p < 0.001). No significant differences were measured in the alpha diversity during the Trp supplementation compared to the control and wash-out period (Figure 7c).
Figure 7.
(a,b) Beta diversity at the OTU level, using the Bray–Curtis PCoA index, showing the effects of donor (a) and colon part (b) on the overall microbial composition. (c) Alpha-diversity of the control vs tryptophan supplementation vs wash-out period at OTU level using the Shannon index.
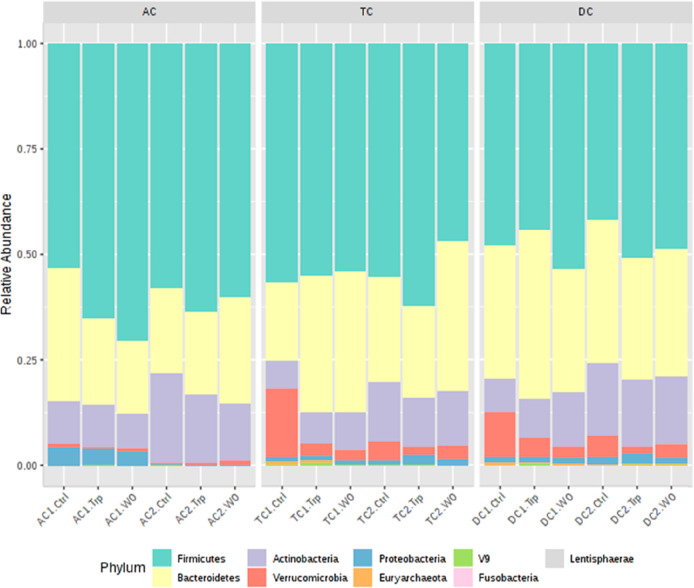
The effect of Trp supplementation on the phylum level composition was analyzed for both donors in each simulated colon compartment. In the AC of donor 1 (AC1), the relative abundance of Firmicutes phylum increased from 53% in the control period to 65% after the high Trp and 70% after the wash-out period (Figure 8). This coincided with a decreased relative abundance of the Bacteroidetes phylum, from 31% to 20% and 17% during the control, Trp supplementation, and wash-out periods, respectively. However, an increase in Firmicutes was not observed in AC of donor 2 (AC2) during the Trp supplementation and wash-out period. Instead, the AC2 showed a small but significant decrease in abundance of Actinobacteria phylum during the Trp (16%) and wash-out period (14%) compared to the control (21%). Trp supplementation induced similar changes in the microbiota composition of the TC and DC compartments of both donors. For donor 1, the Verrucomicrobia (mainly Akkermansia) decreased during and after the Trp period (from 16% to 2 and 3% in TC1 and from 11% to 5 and 3% in DC1). The decrease in Verrucomicrobia coincided with an increase in Bacteroidetes (from 18% to 33%) in TC1 and in Actinobacteria in DC1 (from 7 to 13%). In donor 2, Verrucomicrobia also decreased in the TC and DC compartments although to a lesser extent. Akkermansia muciniphila, the only species of the phylum Verrucomicrobia is indicated to be beneficial in maintaining intestinal integrity.43 For this reason the big decrease observed in the TC and DC of both donors (Table S2) might indicate intestinal imbalance, although our in vitro system does not represent the full intestinal complexity. Particularly, it lacks the mucous layer and A. muciniphila is a mucin-degrading species. The Bacteroidetes in DC1 were highest in abundance after the Trp period (40%) compared to the control (32%) and wash-out period (29%). In donor 2, a small increase in Firmicutes was observed during the Trp supplementation in both the TC and DC (TC2 from 55 to 62% and DC2 from 41 to 51%).
Figure 8.
Relative abundance of 16S rRNA sequencing at the phylum level for donor 1 and donor 2, in the ascending (AC1/2), transverse (TC1/2), and descending (DC1/2) colon at the control period (Ctrl), tryptophan supplementation period (Trp), and wash-out period (WO).
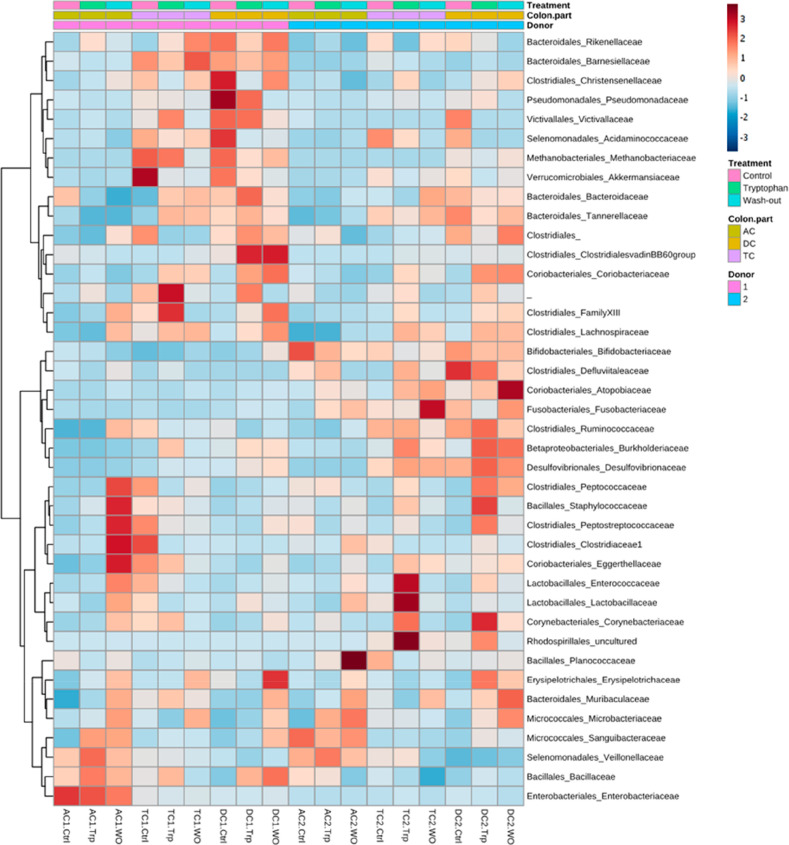
The microbiota differences between donors and colon parts are shown in more detail in a heat map cluster analysis at the family level of taxonomy for the two donors in all colon compartments after the control, Trp supplementation, and wash-out period (Figure 9, Table S2).
Figure 9.
Heat map of the 16S rRNA microbial analysis at family level of donor 1 and donor 2, ascending (AC1/2), transverse (TC1/2), and descending (DC1/2) colon at control period (Ctrl), tryptophan supplementation period (Trp), and wash-out period (WO). Red indicates a higher abundance and blue a lower abundance.
There was a higher abundance of Peptococcacea, Peptostreptococcaceae, Staphylococcaceae, Clostridiacaea, Eggerthellaceae, Enterococcaceae, and Lactobacillacae in the AC of donor 1 in the wash-out period. This was also observed in the AC of donor 2, but the increase in abundance of Lactobacillacae was less. Some donor-specific changes in the composition of the microbiota were also evident, for example, the large increase of Planococcaceae in the AC of donor 1 in the wash-out period. Some of these families are symbiotic and produce bioactives with anti-inflammatory effects, for example Eggerthellacacae, which are well-known to metabolize ellagitannins into urolithins.44 A major increase in Lactobacillacae and Enterococcaceae was found in TC2 after Trp supplementation. Besides, Trp supplementation increased relative abundance of Corynebacteriaceae (uncultured species) and Rhodospirillales (uncultured) in DC2. In donor 1, the abundance of these species was not significantly altered, but Trp supplementation led to an increase in several families of the Clostridiales order in both the TC and DC. There was also a decrease in Akkermansiaceae in TC1 after the Trp supplementation period compared to the control period (from 16 to 2%), which is consistent with changes at the phylum level (Figure 8). In the TC and DC of both donors, there was a major increase in Lachnospiraceae after the Trp supplementation, which includes butyrate-producing species45 that have beneficial roles in the intestine.46,47 At the end of the wash-out period, the Lachnospiraceae were still increased in abundance in the TC and DC and also increased in the AC of both donors compared to the control period. The increase in Lachnospiraceae in both donors indicates that the Trp supplementation had an effect on butyrate producers and can play a role in anti-inflammatory properties after supplementation.
Lactobacillacae and Peptococcacea are reported to be able to produce AhR ligands. Besides, an increase in Lactobacillacae in a high Trp diet is confirmed by several studies.9,48,49 Further analysis of Lactobacillacae and Enterococcaceae on species level, as found in TC2 after Trp supplementation, appear to be mainly an increased abundance of Enterococcus faecalis (from 0.2 to 0.4 to 0.1%), Lactobacillus reuteri and Lactobacillus murinus (0.02 to 0.09 to 0.02%), which are all known to be AhR ligand producers.49 The differences in microbiota after the Trp supplementation found between the two donors, combined with having a similar AhR activity, suggest that probably not only the Lactobacillacae and Peptococcacea are important when converting Trp in AhR ligands. Besides, as only relative abundance was measured, there might be differences in absolute amounts of, e.g., Lactobacillacae, and a different amount of AhR ligands produced. Although the microbiota composition differs between donors, the AhR activation and the SCFA production were not significantly different. It can be hypothesized that different microbial species can exert similar metabolism, thereby maintaining a balanced microbial ecosystem. A limiting factor in this study design was that only 2 donors could be utilized, given the technical characteristics of the TWINSHIME model. To generalize our findings and to study interindividual differences in responses to trp supplementation, more donors should be investigated in the future.
This is the first time that an intestinal model of the human microbiota ecosystem has been used to investigate the detailed profile of Trp metabolites generated in different parts of the colon and the effect of Trp supplementation on the microbial composition. Even though Trp supplementation increased the amount of Trp metabolites in the fermenter supernatants, the overall AhR activity was lower than in the control period. This was due to an antagonistic effect of Trp on AhR activation. However, little is known on the relative transport rate of tryptophan and its microbial metabolites across the colon epithelium. This means that the relative concentrations of tryptophan and AhR ligands measured in our in vitro fermenter may not fully represent the concentration ratios occurring within the colon cells.
Most Trp metabolites were produced in the TC where the largest effect of Trp on the microbiota composition was observed. In the TC, the AhR ligand producers Enterococcus faecalis, Lactobacillus reuteri, and Lactobacillus murinus were increased as a result of Trp supplementation. Overall, these findings indicate that Trp supplementation can increase Trp metabolism and the production of AhR agonists.
Acknowledgments
Erik Meulenbroeks and Wolf Tettelaar are gratefully acknowledged for their help operating the SHIME.
Glossary
Abbreviations Used
- AC
ascending colon
- AhR
aryl hydrocarbon receptor
- CALUX
chemical activated luciferase gene expression
- DC
descending colon
- SCFA
short-chain fatty acid
- SHIME
simulator of the human intestinal microbial ecosystem
- TC
transverse colon
- Trp
tryptophan
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jafc.1c04145.
(Table S1) High-resolution mass spectrometry performances, RT (retention time, min), EC (elemental composition), TM (theoretical mass, [M + H]+), EM (experimental mass, [M + H]+), Δppm (mass accuracy) and (Table S2) relative abundance as percentage of the 16S rRNA microbial analysis at the family level (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Selma M. V.; Espin J. C.; Tomas-Barberan F. A. Interaction between phenolics and gut microbiota: role in human health. J. Agric. Food Chem. 2009, 57 (15), 6485–6501. 10.1021/jf902107d. [DOI] [PubMed] [Google Scholar]
- Li M.; Wang B.; Zhang M.; Rantalainen M.; Wang S.; Zhou H.; Zhang Y.; Shen J.; Pang X.; Zhang M. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (6), 2117–2122. 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss E. A.; Vonarbourg C.; Kopfmann S.; Hobeika E.; Finke D.; Esser C.; Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 2011, 334 (6062), 1561–1565. 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Li Y.; Innocentin S.; Withers D. R.; Roberts N. A.; Gallagher A. R.; Grigorieva E. F.; Wilhelm C.; Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell 2011, 147 (3), 629–640. 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Murray I. A.; Nichols R. G.; Zhang L.; Patterson A. D.; Perdew G. H. Expression of the aryl hydrocarbon receptor contributes to the establishment of intestinal microbial community structure in mice. Sci. Rep. 2016, 6, 33969. 10.1038/srep33969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher T. H.; Maggirwar S. B.; Baglole C. J.; Lakatos H. F.; Gasiewicz T. A.; Phipps R. P.; Sime P. J. Aryl hydrocarbon receptor-deficient mice develop heightened inflammatory responses to cigarette smoke and endotoxin associated with rapid loss of the nuclear factor-κB component RelB. American journal of pathology 2007, 170 (3), 855–864. 10.2353/ajpath.2007.060391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi T.; Kato Y.; Ooneda O.; Mimura J.; Takada T.; Mochizuki H.; Yamamoto M.; Fujii-Kuriyama Y.; Furusako S. Effects of aryl hydrocarbon receptor signaling on the modulation of TH1/TH2 balance. J. Immunol. 2005, 175 (11), 7348–7356. 10.4049/jimmunol.175.11.7348. [DOI] [PubMed] [Google Scholar]
- Xu C.; Li C. Y.-T.; Kong A.-N. T. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Archives of pharmacal research 2005, 28 (3), 249. 10.1007/BF02977789. [DOI] [PubMed] [Google Scholar]
- Zelante T.; Iannitti R. G.; Cunha C.; De Luca A.; Giovannini G.; Pieraccini G.; Zecchi R.; D’Angelo C.; Massi-Benedetti C.; Fallarino F.; Carvalho A.; Puccetti P.; Romani L. Tryptophan Catabolites from Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via Interleukin-22. Immunity 2013, 39 (2), 372–385. 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Lamas B.; Richard M. L.; Leducq V.; Pham H.-P.; Michel M.-L.; Da Costa G.; Bridonneau C.; Jegou S.; Hoffmann T. W.; Natividad J. M.; Brot L.; Taleb S.; Couturier-Maillard A.; Nion-Larmurier I.; Merabtene F.; Seksik P.; Bourrier A.; Cosnes J.; Ryffel B.; Beaugerie L.; Launay J.-M.; Langella P.; Xavier R. J.; Sokol H. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22 (6), 598–605. 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.; Ma N.; He T.; Johnston L. J.; Ma X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit. Rev. Food Sci. Nutr. 2020, 60, 1760–1768. 10.1080/10408398.2019.1598334. [DOI] [PubMed] [Google Scholar]
- Roager H. M.; Licht T. R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9 (1), 3294. 10.1038/s41467-018-05470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegri G.; Costa C. V.; Bertazzo A.; Biasiolo M.; Ragazzi E. Enzyme activities of tryptophan metabolism along the kynurenine pathway in various species of animals. Il Farmaco 2003, 58 (9), 829–836. 10.1016/S0014-827X(03)00140-X. [DOI] [PubMed] [Google Scholar]
- Richard D. M.; Dawes M. A.; Mathias C. W.; Acheson A.; Hill-Kapturczak N.; Dougherty D. M. L-Tryptophan: Basic Metabolic Functions, Behavioral Research and Therapeutic Indications. International Journal of Tryptophan Research 2009, 2, S2129. 10.4137/IJTR.S2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuhkuri K.; Sihvola N.; Korpela R. Diet promotes sleep duration and quality. Nutrition research 2012, 32 (5), 309–319. 10.1016/j.nutres.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Opitz C. A.; Litzenburger U. M.; Sahm F.; Ott M.; Tritschler I.; Trump S.; Schumacher T.; Jestaedt L.; Schrenk D.; Weller M.; Jugold M.; Guillemin G. J.; Miller C. L.; Lutz C.; Radlwimmer B.; Lehmann I.; von Deimling A.; Wick W.; Platten M. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011, 478 (7368), 197. 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- Wikoff W. R.; Anfora A. T.; Liu J.; Schultz P. G.; Lesley S. A.; Peters E. C.; Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (10), 3698–3703. 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.Tryptophan nutrition and metabolism: an overview. In Kynurenine and serotonin pathways; Springer, 1991; pp 345–358. [DOI] [PubMed] [Google Scholar]
- Oxenkrug G. F. Genetic and hormonal regulation of tryptophan-kynurenine metabolism. Ann. N.Y. Acad. Sci. 2007, 1122 (1), 35–49. 10.1196/annals.1403.003. [DOI] [PubMed] [Google Scholar]
- Fujigaki S.; Saito K.; Takemura M.; Fujii H.; Wada H.; Noma A.; Seishima M. Species Differences inl-Tryptophan-Kynurenine Pathway Metabolism: Quantification of Anthranilic Acid and Its Related Enzymes. Archives of biochemistry and biophysics 1998, 358 (2), 329–335. 10.1006/abbi.1998.0861. [DOI] [PubMed] [Google Scholar]
- Hubbard T. D.; Murray I. A.; Perdew G. H. Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab. Dispos. 2015, 43 (10), 1522–35. 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Leek A. P.; Yanishevsky Y.; Kozyrskyj A. L. The kynurenine pathway as a novel link between allergy and the gut microbiome. Frontiers in immunology 2017, 8, 1374. 10.3389/fimmu.2017.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B.; Hernandez-Galan L.; Galipeau H. J.; Constante M.; Clarizio A.; Jury J.; Breyner N. M.; Caminero A.; Rueda G.; Hayes C. L.; McCarville J. L.; Bermudez Brito M.; Planchais J.; Rolhion N.; Murray J. A.; Langella P.; Loonen L. M. P.; Wells J. M.; Bercik P.; Sokol H.; Verdu E. F. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci. Transl Med. 2020, 12 (566), eaba0624. 10.1126/scitranslmed.aba0624. [DOI] [PubMed] [Google Scholar]
- Koper J. E. B.; Loonen L. M. P.; Wells J. M.; Troise A. D.; Capuano E.; Fogliano V. Polyphenols and Tryptophan Metabolites Activate the Aryl Hydrocarbon Receptor in an in vitro Model of Colonic Fermentation. Mol. Nutr Food Res. 2019, 63 (3), 1800722 10.1002/mnfr.201800722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koper J. E. B.; Loonen L. M. P.; Wells J. M.; Troise A. D.; Capuano E.; Fogliano V. Polyphenols and Tryptophan Metabolites Activate the Aryl Hydrocarbon Receptor in an in vitro Model of Colonic Fermentation. Mol. Nutr. Food Res. 2019, 63 (3), 1800722. 10.1002/mnfr.201800722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E.; Cravens J.; List S. Hypnotic effects of L-tryptophan. Arch Gen Psychiatry 1974, 31 (3), 394–7. 10.1001/archpsyc.1974.01760150098014. [DOI] [PubMed] [Google Scholar]
- Kanehisa Laboratories Tryptophan metabolism reference pathway (00380). Kyoto Enceclopedia of Genes and Genomes (KEGG); 25–04–2019 ed.; 2019.
- Kanehisa M.; Furumichi M.; Tanabe M.; Sato Y.; Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45 (D1), D353–D361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T. D.; Murray I. A.; Bisson W. H.; Lahoti T. S.; Gowda K.; Amin S. G.; Patterson A. D.; Perdew G. H. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep. 2015, 5, 12689. 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agus A.; Planchais J.; Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell host & microbe 2018, 23 (6), 716–724. 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Jin U.-H.; Lee S.-O.; Sridharan G.; Lee K.; Davidson L. A.; Jayaraman A.; Chapkin R. S.; Alaniz R.; Safe S. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol. Pharmacol. 2014, 85 (5), 777–788. 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeev E. E.; Lanis J. M.; Kao D. J.; Campbell E. L.; Kelly C. J.; Battista K. D.; Gerich M. E.; Jenkins B. R.; Walk S. T.; Kominsky D. J.; Colgan S. P. Microbiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor. American journal of pathology 2018, 188 (5), 1183–1194. 10.1016/j.ajpath.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H.; Dai Z.; Liu N.; Ji Y.; Chen J.; Zhang Y.; Yang Y.; Li J.; Wu Z.; Wu G. Dietary L-Tryptophan Modulates the Structural and Functional Composition of the Intestinal Microbiome in Weaned Piglets. Frontiers in Microbiology 2018, 9, 1736. 10.3389/fmicb.2018.01736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak D.; Tankiewicz A.; Matys T.; Buczko W. Peripheral distribution of kynurenine metabolites and activity of kynurenine pathway enzymes in renal failure. J. Physiol Pharmacol 2003, 54 (2), 175–189. [PubMed] [Google Scholar]
- Van den Abbeele P.; Grootaert C.; Marzorati M.; Possemiers S.; Verstraete W.; Gerard P.; Rabot S.; Bruneau A.; El Aidy S.; Derrien M.; Zoetendal E.; Kleerebezem M.; Smidt H.; Van de Wiele T. Microbial community development in a dynamic gut model is reproducible, colon region specific, and selective for Bacteroidetes and Clostridium cluster IX. Appl. Environ. Microbiol. 2010, 76 (15), 5237–5246. 10.1128/AEM.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli L.; Martin-Gallausiaux C.; Bourhis J.-M.; Béguet-Crespel F.; Blottière H. M.; Lapaque N. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci. Rep. 2019, 9 (1), 643. 10.1038/s41598-018-37019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korecka A.; Dona A.; Lahiri S.; Tett A. J.; Al-Asmakh M.; Braniste V.; D’Arienzo R.; Abbaspour A.; Reichardt N.; Fujii-Kuriyama Y.; Rafter J.; Narbad A.; Holmes E.; Nicholson J.; Arulampalam V.; Pettersson S. Bidirectional communication between the Aryl hydrocarbon Receptor (AhR) and the microbiome tunes host metabolism. NPJ. Biofilms Microbiomes 2016, 2, 16014. 10.1038/npjbiofilms.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.; Schoones T.; Wells J. M.; Fogliano V.; Capuano E. Substrate-Driven Differences in Tryptophan Catabolism by Gut Microbiota and Aryl Hydrocarbon Receptor Activation. Mol. Nutr Food Res. 2021, 65 (13), 2100092 10.1002/mnfr.202100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F.; Hao F.; Murray I. A.; Smith P. B.; Koo I.; Tindall A. M.; Kris-Etherton P. M.; Gowda K.; Amin S. G.; Patterson A. D.; Perdew G. H. Intestinal microbiota-derived tryptophan metabolites are predictive of Ah receptor activity. Gut Microbes 2020, 12 (1), 1788899. 10.1080/19490976.2020.1788899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Jin U. H.; Allred C. D.; Jayaraman A.; Chapkin R. S.; Safe S. Aryl Hydrocarbon Receptor Activity of Tryptophan Metabolites in Young Adult Mouse Colonocytes. Drug Metab. Dispos. 2015, 43 (10), 1536–43. 10.1124/dmd.115.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H.; Park J.; Kim M. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune network 2014, 14 (6), 277–288. 10.4110/in.2014.14.6.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H. Immune regulation by microbiome metabolites. Immunology 2018, 154 (2), 220–229. 10.1111/imm.12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerlings S.; Kostopoulos I.; de Vos W.; Belzer C. Akkermansia muciniphila in the human gastrointestinal tract: when, where, and how?. Microorganisms 2018, 6 (3), 75. 10.3390/microorganisms6030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selma M. V.; Beltrán D.; Luna M. C.; Romo-Vaquero M.; García-Villalba R.; Mira A.; Espín J. C.; Tomás-Barberán F. A. Isolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin a from ellagic acid. Frontiers in microbiology 2017, 8, 1521. 10.3389/fmicb.2017.01521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L.; Gao R.; Huang J.; Qin H. Supplementation of triple viable probiotics combined with dietary intervention is associated with gut microbial improvement in humans on a high-fat diet. Experimental and therapeutic medicine 2019, 18 (3), 2262–2270. 10.3892/etm.2019.7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaoul R.; Day A. S. Nutritional regulators of intestinal inflammation. Curr. Opin Gastroenterol 2019, 35, 486. 10.1097/MOG.0000000000000585. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Song L.; Wang Y.; Liu C.; Zhang L.; Zhu S.; Liu S.; Duan L. Beneficial effect of butyrate-producing Lachnospiraceae on stress-induced visceral hypersensitivity in rats. Journal of gastroenterology and hepatology 2019, 34, 1368. 10.1111/jgh.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M.; Luo C.; Kolde R.; d’Hennezel E.; Annand J. W.; Heim C. E.; Krastel P.; Schmitt E. K.; Omar A. S.; Creasey E. A.; Garner A. L.; Mohammadi S.; O’Connell D. J.; Abubucker S.; Arthur T. D.; Franzosa E. A.; Huttenhower C.; Murphy L. O.; Haiser H. J.; Vlamakis H.; Porter J. A.; Xavier R. J. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell host & microbe 2017, 22 (1), 25–37. 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamas B.; Natividad J. M.; Sokol H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunology 2018, 11 (4), 1024–1038. 10.1038/s41385-018-0019-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kanehisa Laboratories Tryptophan metabolism reference pathway (00380). Kyoto Enceclopedia of Genes and Genomes (KEGG); 25–04–2019 ed.; 2019.