Abstract
The gut microbiota is considered a key ‘metabolic organ’. Its metabolic activities play essential roles complementary to the host metabolic functions. The interplays between gut microbes and commonly used non-antibiotic drugs have garnered substantial attention over the years. Drugs can reshape the gut microorganism communities and, vice versa, the diverse gut microbes can affect drug efficacy by altering the bioavailability and bioactivity of drugs. The metabolism of drugs by gut microbial action or by microbiota–host cometabolism can transform the drugs into various metabolites. Secondary metabolites produced from the gut microbial metabolism of drugs contribute to both the therapeutic benefits and the side effects. In view of the significant effect of the gut microbiota on drug efficiency and clinical outcomes, it is pivotal to explore the interactions between drugs and gut microbiota underlying medical treatments. In this review, we describe and summarize the complex bidirectional interplays between gut microbes and drugs. We also illustrate the gut-microbiota profile altered by non-antibiotic drugs, the impacts and consequences of microbial alteration, and the biochemical mechanism of microbes impacting drug effectiveness. Understanding how the gut microbes interact with drugs and influence the therapeutic efficacy will help in discovering diverse novel avenues of regulating the gut microbes to improve the therapeutic effects and clinical outcomes of a drug in precision.
Keywords: gut microbiota, microbiome, drug, metabolism, efficacy
Introduction
A gigantic amount of microorganisms, including bacteria, viruses, fungi, and archaea, are co-residing in the human gastrointestinal (GI) tract, living in a commensal relationship with humans [1]. They are involved in the regulation of a multitude of host metabolic aspects, contributing to the digestion of foods, signaling transmission, and immunity development [2–5].
Intestinal microbiota can affect host physiology and disease pathogenesis through its structural component lipopolysaccharide or secreted metabolites that transmit via blood circulation [6]. Alterations of the gut microbiome are associated with several physical conditions, including gastrointestinal dysfunctions, cardiovascular diseases, metabolic disorders, and even diseases associated with psychiatric abnormalities [7–10]. In addition, the interest in associations between gut microbes and non-antibiotic drugs usage has been growing in recent years. Importantly, oral administration of medication is a convenient and widely used medication administration route by which drug digestion and absorption are mostly carried out in the GI tract. Therefore, gut microbes are considered a key participator in drug metabolism. Research has shown that the gut microbiota was able to influence the effect of >30 approved drugs [11] and that >200 approved compounds can inhibit the growth of at least one bacterium [12].
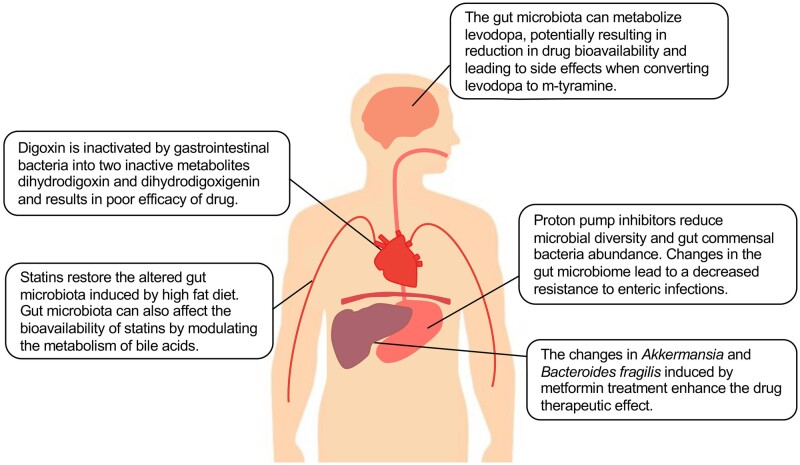
The clinical outcomes are various after the administration of the same medication in patients. Some individuals experience significant improvements whereas others see less of a benefit or even no improvements at all [13]. Considering that the human gut microbiota is highly individualized, it leads one to speculate that gut microbiota is one of the primary variates determining the effectiveness of the drugs. Knowledge of how human gut microbiota affects drug pharmacokinetics and pharmacodynamics over the last decades has made big strides and further advance our understanding of inter-individual variations in drug efficacy and adverse effects. In this review, we discuss the complex bidirectional interactions between commonly used non-antibiotic drugs and the gut microbiome, and describe the microbial impact on drug efficacy and safety underlying biochemical mechanisms. Elegant examples of drug–microbiome interactions are provided, including the drugs metformin and proton-pump inhibitors (PPIs) that modulate microbiome composition and functions [14, 15] (Figure 1). Meanwhile, the gut microbiota can influence the treatment effectiveness of a specific drug by impacting its concentration and bioactivity, such as in the cases of statin, levodopa, and digoxin (Figure 1). Understanding the relationship between the gut microbiota and non-antibiotic drugs could provide us with useful and valuable instructions for assessing current drug-administration routes and the development of precision medicine.
Figure 1.
Overview of interactions between non-antibiotic drugs and the gut microbes.
Metformin alters the gut microbiome of type 2 diabetes patients, contributing to its therapeutic effects
Type 2 diabetes is a disorder of blood glucose regulation (hyperglycemia) primarily arising from insulin resistance [16]. Treatment involves drug therapy and lifestyle intervention, where the blood-glucose-lowering compound metformin is the most viable drug option for type 2 diabetes [17]. The primary metabolic effect of metformin is inhibiting gluconeogenesis in the liver [18]. Compared with oral dosing, intravenously administered metformin does not enhance blood glucose regulation [19], underscoring a potentially critical role of the GI tract and its inhabiting microbes in this process. The fact that microbial mediation has beneficial effects on glucose metabolism under metformin treatment was revealed in both animals and humans [20, 21]. In humans, the changes in the abundance of the phyla Firmicutes and Bacteroidetes were significantly correlated with changes in serum cholic acid and its conjugates, which were elevated after metformin withdrawal [21]. It suggests that such microbiome changes may contribute to the therapeutic effect of metformin. In a murine study, higher abundance of the mucin-degrading bacteria Akkermansia and more mucin-producing goblet cells were observed in metformin-treated, high-fat diet (HFD)-fed mice than in HFD-fed mice without metformin treatment [20]. Increased Akkermansia muciniphila significantly enhanced glucose tolerance and attenuated adipose tissue inflammation in HFD-fed mice [20]. Meanwhile, another murine study revealed that the reduction in Bacteroides fragilis abundance and its selective bile salt hydrolase (BSH) activities both contributed to the improvement in glucose tolerance induced by metformin [22]. These data indicate that A. muciniphila and B. fragilis may enhance the therapeutic effect of metformin through immune-modulation and microbial metabolic processes.
Alterations in the gut microbiome composition and functions induced by metformin treatment were demonstrated in both mice [20] and humans [22, 23]. Moreover, metformin-induced changes in the gut microbiota are also diet-dependent [20]. HFD induced an increase in the abundance of Firmicutes and a decrease in the abundance of Bacteroidetes and Verrucomicrobia in mice compared with those fed with a normal chow diet (NCD) [20]. Metformin administration led to a profound shift in the fecal microbial profile of HFD-fed mice, where the abundances of Firmicutes, Bacteroidetes, and Verrucomicrobia were largely changed [20]. In contrast, there were no significant differences in these phyla between NCD-fed mice with and without metformin treatment. The abundances of 29 bacterial genera were differentiated between mice treated with and without metformin, which suggests that metformin is associated with changes in these taxa [20]. The relative abundance of Akkermansia, Parabacteroides, Odoribacter, Alistipes, Blautia, and Lactonifactor was altered by HFD, but metformin restored the abundances of these taxa to levels comparable with that in NCD-fed mice [20]. In particular, Akkermansia was considered a dominant contributor to the gut-microbiome difference between HFD-fed mice with and without metformin treatment [20].
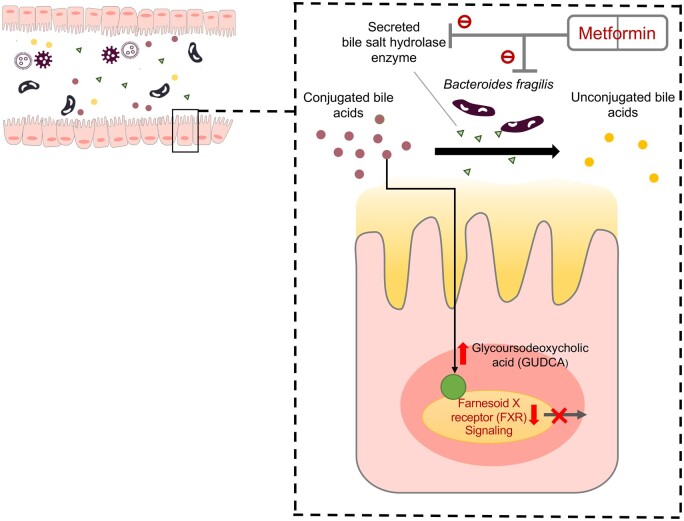
This finding was verified in humans, further supporting that A. muciniphila was the most increased taxon and the mere species that was increased in abundance in response to metformin [23]. Associations of metformin usage and the abundance of Bifidobacterium adolescentis [24], Escherichia coli [25] and A. muciniphila [20] were also observed. In vitro, the growth of B. adolescentis and A. muciniphila was promoted by metformin in pure cultures yet no promotion effect was seen for E. coli [23]. To further investigate the functionality changes in the gut microbiome in response to metformin exposure, fecal samples from humans were cultured in a gut-simulator system with the presence of metformin, followed by whole-genome shotgun sequencing at both the DNA and RNA levels [23]. The results showed that microbiome pathways involved in butyrate and pyruvate metabolism were enriched in the metformin-treated group compared to the control group [23]. When gut microbes obtained from metformin-treated subjects (before and post-treatment) were transferred to germ-free mice fed a HFD, glucose tolerance was enhanced in mice who received fecal microbiota from post-treatment samples as compared with those who received fecal microbiota from baseline (before-treatment) samples [23]. A greater abundance of Akkermansia induced by metformin was demonstrated to improve the glucose tolerance in mice fed a HFD, the beneficial effect of which was similar to that of metformin-administrated mice fed a HFD [20]. Akkermansia had an anti-diabetic effect by reducing stromal vascular fraction inflammation in visceral adipose tissue, which was involved in the pathophysiology of insulin resistance, and by restoring the levels of Treg proportion comparable with that in metformin-treated mice fed a HFD [20]. Akkermansia could increase insulin signaling in HFD-fed mice through regulating immune responses [26]. In addition, metformin improved hyperglycemia via the B. fragilis–intestinal farnesoid X receptor (FXR) axis [22]. FXR is involved in multiple metabolic disorders by influencing glucose and lipid homeostasis [27, 28]. FXR deficiency was associated with reduced adipose tissue mass and was accompanied by glucose homeostasis and improved adipose tissue insulin sensitivity [29]. Activation of FXR induced the expression of fibroblast growth factor 15 (in mice) and 19 (in humans and other species), while metformin could suppress its activation by acting on its signaling processes [22]. Interestingly, the level of FXR expression was comparable between the mice treated with metformin plus antibiotics and mice treated with antibiotics only [22]. Hence, gut microbiota is an essential factor in intestinal FXR signaling suppression during metformin treatment. As bile acids are significant messengers in the FXR signaling, mice were gavaged with a FXR agonist taurocholic acid along with a mixture of bile acids including chenodeoxycholic acid (CDCA), glycoursodeoxycholic acid (GUDCA), and tauroursodeoxycholic acid (TUDCA) to probe their roles in the metformin–microbiome–the host axis [22]. CDCA promoted taurocholic acid-induced intestinal FXR signaling while GUDCA and TUDCA alleviated this activated signaling, suggesting that GUDCA and TUDCA are potential FXR antagonists [22]. Further, the abundance of B. fragilis was found to be positively correlated with FXR-activated signaling and negatively correlated with the level of potential FXR antagonists, including GUDCA and TUDCA, in stool and serum samples [22]. Interestingly, metformin could inhibit the growth of B. fragilis and downregulated BSH gene expression in B. fragilis to increase GUDCA levels to inhibit FXR signaling [22] (Figure 2). Collectively, metformin has an anti-diabetic effect through mediating the level and activities of B. fragilis and influencing the FXR signaling in the gut. Bacteroidesfragilis administration markedly abrogated the effect of metformin, characterized by impaired glucose tolerance and insulin sensitivity [22]. All these studies in rodents and humans suggest that gut microbial changes induced by metformin might enhance its therapeutic effect. The B. fragilis–GUDCA–FXR axis represents a critical inner working mechanism for the effect of metformin–microbiome cooperation on the host.
Figure 2.
Regulation of gut microbiota–bile acid–farnesoid X receptor (FXR) axis to improve type 2 diabetes. Metformin reduces the abundance of B. fragilis and inhibits bile salt hydrolase (BSH) activity. These changes can further increase levels of GUDCA (endogenous FXR antagonists) and suppress the FXR signaling. It shows beneficial effects on metabolic diseases dependent on intestinal FXR inhibition.
Alterations in gut microbiota by cholesterol-lowering drugs and its possible beneficial roles in drug metabolism
The relationship between rosuvastatin (a commonly used cholesterol-lowering drug of the statin class) and the gut microbiota was extrapolated from a clinical trial [30]. Blood lipid levels of 64 patients with hyperlipidemia showed a significant reduction after the rosuvastatin treatment [30]. Interestingly, blood lipid levels of half of patients slumped to normal levels, presented by a reduction in the low-density lipoprotein cholesterol (LDL-C) levels (58.5%) and total cholesterol levels (26.6%), while blood lipid levels of the other patients remained high after the rosuvastatin therapy [30]. On the consideration that the various gut-microbiome compositions across humans might account for the observed variations in rosuvastatin treatment efficacy, the authors found that bacteria from Firmicutes and Fusobacteria had a negative correlation with LDL-C levels while Cyanobacteria and Lentisphaerae had a positive correlation with LDL-C levels [30]. In a separate study conducted in mice, blood levels of triglycerides, total cholesterol, and LDL-C were reduced after a 2-week statin treatment [31]. However, the level of LDL-C from the statin-plus-antibiotics-treated group (microbes-depleted) was remarkably higher than the LDL-C level from the statin-treated group [31]. These data together suggest that the intestinal microbiome contributed to the effectiveness of the statin [31].
The genera Bacteroides, Butyricimonas, and Mucispirillum were found to be enriched in atorvastatin- and rosuvastatin-treated mice, where rosuvastatin was more effective than atorvastatin in restoring the altered gut microbiota induced by HFD [32]. Moreover, the abundances of these bacteria were closely correlated with host inflammation markers [32]. In addition, fecal microbiota from rosuvastatin-treated mice improved serum glucose and glucose tolerance in HFD-fed mice [32]. In humans, Blautia and Anaerostipes were positively associated with butyric acid production, whereas these bacteria were depleted in patients with acute coronary syndrome. However, acute coronary syndrome patients treated with statins had a comparable level of Blautia and Anaerostipes to that of healthy individuals [33]. These results imply that statins can restore the gut-microbiota profile that are altered in a disease setting, whereas the microbiome may work in synergism with the drug on the host.
The bioavailability of cholesterol-lowering drugs could be affected by gut microbiota via modulating the metabolism of bile acids. In humans, bile acids and statins share the same three transporters in the intestine: multidrug resistance gene 1 P-glycoprotein, multidrug resistance-associated protein 2, and organic anion-transporting polypeptide 1B1 [34, 35]. Therefore, the bile acids pool can compete with cholesterol-lowering drugs for transporters, consequently affecting the bioavailability and therapeutic efficacy of these drugs. In this process, the gut microbiota may play a role by regulating the bile acid profile. Study has revealed that increased plasma concentrations of simvastatin are positively correlated with the levels of several secondary bile acids [36]. One hypothesis is that the gut microbiota calibrates the bile acid profile impacting the competition between bile acids and statins in contending for transporters on host cells, leading to bioavailability differences in statins and its therapeutic effectiveness across different individuals due to variations in gut-microbiome configurations.
Gut microbes reduce the bioavailability of levodopa through decarboxylation
Accumulating evidence suggests that a low diversity of gut microbiota is associated with mental-health disorders such as attention deficit hyperactivity disorder [37, 38]. When the mice got a fecal transplant from depressed patients, recipient mice developed depression-like behaviors, indicating a crucial role for microbiome in such a disease [39, 40]. Similarly, the gut microbiome can influence the effect of psychotropic drugs in modulating host mental health [41]. Parkinson’s disease (PD) is a neurological movement disorder that leads to shaking and difficulty in walking and balance, affecting >1% of the population aged >60 years [42]. The most potent medication for PD is levodopa (L-dopa), which is prescribed to alleviate motor symptoms [43]. L-dopa is absorbed into the intestine and must enter the brain so as to be converted by the aromatic amino acid decarboxylase to the neurotransmitter dopamine for it to be functional in coordinating signaling from the brain to muscles [44]. However, the GI tract is one of the major sites for dopa decarboxylation, rendering that dopamine synthesized in the periphery hardly crosses the blood–brain barrier, resulting in ineffective medication of dopa [45, 46]. One major pathway contributing to this ineffectiveness is the consecutive gut microbial dihydroxylation that converts L-dopa into non-therapeutic m-tyramine [47, 48]. Thereby, the bioavailability of L-dopa to the brain at the site of active gut microbial metabolism is one key factor determining drug efficacy. Eradication of specific bacterial clusters via antibiotics was found to improve the L-dopa therapy in both humans and mice, suggesting that drug efficacy is counteracted by certain gut bacteria [49, 50]. Moreover, peripheral dopamine may result in side effects in the GI tract as well as orthostatic hypotension and cardiac arrhythmias [51]. Overall, the gut-microbiota-mediated L-dopa metabolism may lead to poor clinical outcomes and side effects [52].
The impact of L-dopa on the gut microbiota of PD patients has also been revealed in a longitudinal study to examine the gut microbiota composition in PD patients before and after L-dopa administration. A lower relative abundance of Clostridium group IV was observed in PD patients who experienced an obvious or moderate improvement in motor impairment in response to L-dopa compared with those with a small response [53]. Alterations in the microbiome functions were also observed after treatment. A strong positive correlation between bacterial tyrosine decarboxylase (tdc) gene relative abundance and L-dopa treatment dose as well as the duration of disease was observed [54]. Moreover, the tdc gene in the fecal microbiota was significantly correlated with L-dopa dosage, suggesting bacterial tdc may play a role in L-dopa efficacy.
Rekdal and his colleagues [55] identified a candidate L-dopa decarboxylating enzyme, PLP-dependent tyrosine decarboxylase (pyridoxal 5′-phosphate TyrDC) that was encoded by the bacterium Enterococcus faecalis. TyrDC was involved in drug metabolism by catalysizing the decarboxylation of L-dopa to dopamine [55]. Another dopamine dehydroxylating strain from the species Eggerthella lenta was also isolated, where expression association was found between PLP-dependent decarboxylase and molybdenum cofactor-dependent dopamine dehydroxylase (Dadh) enzyme, corroborating that L-dopa could be sequentially metabolized into m-tyramine [55]. The abundance of E. faecalis, TyrDC, dadh, L-dopa, and dopamine metabolism in the gut microbiota from PD patients were all inter-connected, suggesting that these microorganisms and their metabolism are relevant in L-dopa conversion [55]. Hence, to make L-dopa more effective in PD patients harboring such L-dopa-utilizing bacteria, inhibitors of gut bacterial L-dopa decarboxylation are proposed to be co-administered. Considering tyrosine is a substrate preference by TyrDCs, a mimic (S)-α-fluoromethyltyrosine (AFMT) was considered and used to reduce L-dopa decarboxylation, which improved the therapeutic effect in PD mice [55]. When a mixture of L-dopa and AFMT was administered to E. faecalis-colonized mice, it led to an elevation in the level of L-dopa in serum [55].
PPIs alter the composition of the gut microbiota and increase the risk of enteric bacterial infection
PPIs are commonly used to reduce stomach acid in acid-related disorders and prevent gastroduodenopathy and bleeding [56]. Although few side effects are reported in PPI users, the absolute number of PPI users presenting adverse drug responses is still high [57]. 16S rDNA-based study revealed that patients with inflammatory bowel disease and patients with irritable bowel syndrome are associated with lower diversity and changes in 20% of the gut microbiota (with relative abundances decreased or increased) after PPIs treatment [58]. While the disease itself is a confounding factor when interrogating the effect of PPIs on the gut microbiome, it precludes us from dissecting the effects of disease vs PPIs usage on the gut microbiome. To tease apart the effect of PPIs usage vs disease in affecting the gut microbiome, there have been a handful of studies investigating the effect of PPIs intake on gut-microbiome composition in healthy individuals. Two separate trials on 12 healthy volunteers [14] and 1,827 healthy twins [59] who voluntarily took PPIs showed that PPIs induced considerable changes in taxonomic composition. The former study found that taxa associated with Clostridioides difficile infection were significantly changed after PPIs administration [14], whereas the latter study found that the gut microbiome of PPI users was characterized by a lower abundance of commensal bacteria and lower microbial diversity compared with non-users [59]. Overall, the gut microbiome of PPI users had a reduction in the abundance of Ruminococcaceae and Bifidobacteriaceae, and an elevation in the abundance of Enterobacteriaceae, Enterococcaceae, and Lactobacillaceae compared with non-PPI users [14]. Changes in the microbial taxon and functionality were positively correlated with a higher drug dosage [60]. A higher resolution of taxonomic and functional pathway interrogation was facilitated by metagenomic sequencing. PPIs accounted for most of the observed associations between drugs and gut-microbiome alterations amongst the associations between 42 commonly used drugs and the gut microbiome [60]. Beyond that, PPIs were the only drug category associated with gut-microbiome compositional changes across all cohorts [60]. PPI-induced changes in the predicted microbial functions include fatty acid, lipid and L-arginine biosyntheses [60].
It is vital to recognize that microbiome changed by PPIs may actually contribute to the pathogenesis or progression of some clinical diseases. Accumulating studies found that loss of specific bacterial taxa results in weakened resistance to enteric infections, including those caused by Clostridioidesdifficile and by Salmonella, which were frequently observed in PPI users [61, 62]. The odds ratios were estimated 1.5–1.8 for infection with C. difficile and 2.0–4.0 for infections with other pathogens after PPIs treatment [63]. It is known that antibiotics administration induces dysbiosis of gut microbial ecology that makes one vulnerable to C. difficile infection afterwards; this is also true for PPIs usage [64]. In addition, one study showed that PPI treatment was associated with the clinical course in decompensated liver cirrhosis, where the gut microbiome mediated this process [65]. PPI abuse in early childhood was associated with long-term changes in the gut-microbiome development and obesity in later life [66]. Although the efficacy and safety profile of PPIs are favorable, the medical community should embark on assessing the functional consequences and impacts of the changed gut microbiome by PPIs.
Gut flora causes inter-individual variations in the metabolism of digoxin
Orally administered cardiac glycoside drug digoxin is known to control heart problems, such as irregular heartbeats (arrhythmias) including atrial fibrillation, and it also helps to manage the symptoms of heart failure [67]. However, ∼10% of patients experienced a lower benefit of digoxin due to the substantial conversion of digoxin into relatively inactive metabolites, dihydrodigoxin and dihydrodigoxigenin [68]. One of the major sites of this conversion is again carried out in the GI tract, where metabolism of digoxin by gut microbes was well established [69, 70]. Moreover, antibiotic pretreatment reduced the secretion of dihydrodigoxin in urine and increased the level of digoxin in blood, hinting at an increase in digoxin bioavailability in the host [68]. More than 40% of ingested digoxin was converted into inactivate metabolites before its absorption in the gut; the role of gut microbiota in digoxin efficiency reveals an extra layer of information about the microbial contribution to host health that is independent of human metabolism [71].
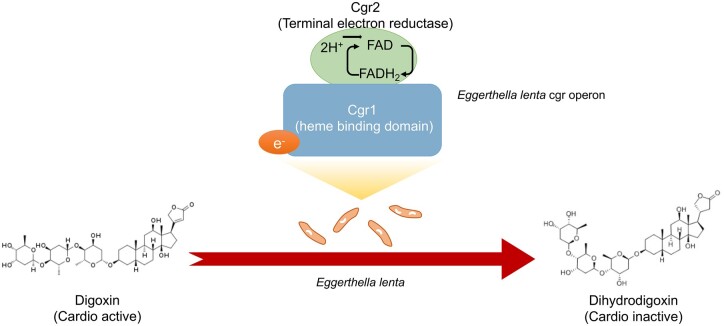
Research has identified some strains of digoxin-metabolizing gut bacteria, such as E. lenta, in individuals who can reduce the level of digoxin [72]. A two-gene cytochrome-encoding operon (namely the cardiac glycoside reductase, cgr) was also significantly upregulated in the presence of digoxin [73]. This cgr operon functions by producing a protein–Cgr1–Cgr2 complex that binds to digoxin and accounts for digoxin's consequent reduction due to the proteins that are homologous to bacterial cytochromes and are therefore potentially capable of using digoxin as an alternative electron acceptor (Figure 3). Two E. lenta strains that lack the operon were unable to inactivate digoxin [74]. Others found that amino acids, especially arginine, serve as the main source of nitrogen and carbon for E. lenta but it simultaneously inhibits digoxin inactivation [72, 73]. Therefore, a high-protein diet can help to improve the efficacy of digoxin in those patients who carry cgr + E. lenta.
Figure 3.
Schematic representation of digoxin-to-dihydrodigoxin conversion with the involvement of E. lenta. The heme binding domain of Cgr1 transfers electrons to the extra-cytoplasmic terminal electron reductase Cgr2 through heme, resulting in reduction of digoxin to dihydrodigoxin.
Conclusion and perspectives
We herein describe and summarize the relationship and interplays between non-antibiotic drugs and the gut microbiome. Clinicians and scientists should be aware that, beyond antibiotics, non-antibiotic drugs can also influence the gut-microbiome configuration and development, which may ultimately enhance or impair host health and clinical outcomes. Meanwhile, as the pharmaco-microbiomes field is coming to the surface and getting attention, a comprehensive understanding of how gut microbes metabolize/utilize/bio-transform drugs will open new potential avenues for regulating the gut microbiome to improve the efficacy of drugs and biologics. There are many clinical trials underway. For example, the clinical trial NCT04208958 (EudraCT number 2010–022394-34) is evaluating the safety and efficacy of VE800, a commensal bacterial strain formulation, in combination with Nivolumab in patients with several metastatic cancer. Another ongoing clinical trial (NCT03637803) aims to investigate the safety and efficacy of pembrolizumab in combination with a single bacterial strain Enterococcus gallinarum in patients with solid tumors. These trials and results would influence clinical practice in the foreseeable future.
Authors’ Contributions
Y.T.W. conceived of the study and drafted the manuscript. T.Z. provided significant intellectual contribution and constructive advice, and revised the manuscript.
Funding
This work was supported by the Municipal Key Research and Development Program of Guangzhou [grant number 202206010014], the National Natural Science Foundation of China [grant numbers 82172323 and 32100134], and a joint seed fund from the Sixth Affiliated Hospital of Sun Yat-sen University and Sun Yat-sen University, China.
Acknowledgements
None.
Conflicts of Interest
We declare that we have no conflict of interest.
References
- 1. Hooper LV, Gordon JI.. Commensal host-bacterial relationships in the gut. Science 2001;292:1115–8. [DOI] [PubMed] [Google Scholar]
- 2. Cox LM, Weiner HL.. Microbiota signaling pathways that influence neurologic disease. Neurotherapeutics 2018;15:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bengmark S. Gut microbiota, immune development and function. Pharmacol Res 2013;69:87–113. [DOI] [PubMed] [Google Scholar]
- 4. Holmes E, Li JV, Athanasiou T. et al. Understanding the role of gut microbiome–host metabolic signal disruption in health and disease. Trends Microbiol 2011;19:349–59. [DOI] [PubMed] [Google Scholar]
- 5. Vernocchi P, Del Chierico F, Putignani L.. Gut microbiota metabolism and interaction with food components. IJMS 2020;21:3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sherwin E, Bordenstein SR, Quinn JL. et al. Microbiota and the social brain. Science 2019;366(6465): eaar2016. [DOI] [PubMed] [Google Scholar]
- 7. Novakovic M, Rout A, Kingsley T. et al. Role of gut microbiota in cardiovascular diseases. World J Cardiol 2020;12:110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lloyd-Price J, Arze C, Ananthakrishnan AN. et al. ; IBDMDB Investigators. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou W, Sailani MR, Contrepois K. et al. Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 2019;569:663–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sampson TR, Debelius JW, Thron T. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 2016;167:1469–80.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun C, Chen L, Shen Z.. Mechanisms of gastrointestinal microflora on drug metabolism in clinical practice. Saudi Pharm J 2019;27:1146–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maier L, Pruteanu M, Kuhn M. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018;555:623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Savage N. The complex relationship between drugs and the microbiome. Nature 2020;577:S10–1. [DOI] [PubMed] [Google Scholar]
- 14. Freedberg DE, Toussaint NC, Chen SP. et al. Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: a crossover trial. Gastroenterology 2015;149:883–5.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forslund K, Hildebrand F, Nielsen T. et al. ; MetaHIT consortium. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528:262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weyer C, Bogardus C, Mott DM. et al. The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J Clin Invest 1999;104:787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holman R. Metformin as first choice in oral diabetes treatment: the UKPDS experience. Journ Annu Diabetol Hotel Dieu 2007;13–20. [PubMed] [Google Scholar]
- 18. Hundal RS, Krssak M, Dufour S. et al. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes 2000;49:2063–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonora E, Cigolini M, Bosello O. et al. Lack of effect of intravenous metformin on plasma concentrations of glucose, insulin, C-peptide, glucagon and growth hormone in non-diabetic subjects. Curr Med Res Opin 1984;9:47–51. [DOI] [PubMed] [Google Scholar]
- 20. Shin N-R, Lee J-C, Lee H-Y. et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014;63:727–35. [DOI] [PubMed] [Google Scholar]
- 21. Napolitano A, Miller S, Nicholls AW. et al. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One 2014;9:e100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun L, Xie C, Wang G. et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med 2018;24:1919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu H, Esteve E, Tremaroli V. et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 2017;23:850–8. [DOI] [PubMed] [Google Scholar]
- 24. Bradley CA. Gut microbiota: trust your gut—metformin and diabetes. Nat Rev Endocrinol 2017;13:440. [DOI] [PubMed] [Google Scholar]
- 25. Walsh J, Griffin BT, Clarke G. et al. Drug–gut microbiota interactions: implications for neuropharmacology. Br J Pharmacol 2018;175:4415–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Greer RL, Dong X, Moraes ACF. et al. Akkermansia muciniphila mediates negative effects of IFNγ on glucose metabolism. Nat Commun 2016;7:13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsubara T, Li F, Gonzalez FJ.. FXR signaling in the enterohepatic system. Mol Cell Endocrinol 2013;368:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duran-Sandoval D, Mautino G, Martin G. et al. Glucose regulates the expression of the farnesoid X receptor in liver. Diabetes 2004;53:890–8. [DOI] [PubMed] [Google Scholar]
- 29. Prawitt J, Abdelkarim M, Stroeve JH. et al. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes 2011;60:1861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Song X, Zhou H. et al. Gut microbiome associates with lipid-lowering effect of rosuvastatin in vivo. Front Microbiol 2018;9:530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, Wang Y, Wang H. et al. The influence of the intestinal microflora to the efficacy of Rosuvastatin. Lipids Health Dis 2018;17:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim J, Lee H, An J. et al. Alterations in gut microbiota by statin therapy and possible intermediate effects on hyperglycemia and hyperlipidemia. Front Microbiol 2019;10:1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu X, Li H, Zhao X. et al. Multi-omics study reveals that statin therapy is associated with restoration of gut microbiota homeostasis and improvement in outcomes in patients with acute coronary syndrome. Theranostics 2021;11:5778–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holtzman CW, Wiggins BS, Spinler SA.. Role of P‐glycoprotein in statin drug interactions. Pharmacotherapy 2006;26:1601–7. [DOI] [PubMed] [Google Scholar]
- 35. Chen C, Mireles RJ, Campbell SD. et al. Differential interaction of 3-hydroxy-3-methylglutaryl-coa reductase inhibitors with ABCB1, ABCC2, and OATP1B1. Drug Metab Dispos 2005;33:537–46. [DOI] [PubMed] [Google Scholar]
- 36. Kaddurah-Daouk R, Baillie RA, Zhu H. et al. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS One 2011;6:e25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dawson S, Dash S, Jacka F.. The importance of diet and gut health to the treatment and prevention of mental disorders. Int Rev Neurobiol 2016;131:325–46. [DOI] [PubMed] [Google Scholar]
- 38. Prehn-Kristensen A, Zimmermann A, Tittmann L. et al. Reduced microbiome alpha diversity in young patients with ADHD. PLoS One 2018;13:e0200728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kelly JR, Borre Y, C OB. et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res 2016;82:109–18. [DOI] [PubMed] [Google Scholar]
- 40. Zheng P, Zeng B, Zhou C. et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol Psychiatry 2016;21:786–96. [DOI] [PubMed] [Google Scholar]
- 41. Cussotto S, Clarke G, Dinan TG. et al. Psychotropics and the microbiome: a chamber of secrets…. Psychopharmacology (Berl) 2019;236:1411–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reeve A, Simcox E, Turnbull D.. Ageing and Parkinson's disease: why is advancing age the biggest risk factor? Ageing Res Rev 2014;14:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Surmeier DJ. Determinants of dopaminergic neuron loss in Parkinson's disease. FEBS J 2018;285:3657–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagatsu T, Sawada M.. L-dopa therapy for Parkinson's disease: past, present, and future. Parkinsonism Relat Disord 2009;15:S3–8. [DOI] [PubMed] [Google Scholar]
- 45. Papavasiliou PS, Cotzias GC, Düby SE. et al. Levodopa in Parkinsonism: potentiation of central effects with a peripheral inhibitor. N Engl J Med 1972;286:8–14. [DOI] [PubMed] [Google Scholar]
- 46. Bergmann S, Curzon G, Friedel J. et al. The absorption and metabolism of a standard oral dose of levodopa in patients with Parkinsonism. Br J Clin Pharmacol 1974;1:417–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goldman P, Peppercorn MA, Goldin BR.. Metabolism of drugs by microorganisms in the intestine. Am J Clin Nutr 1974;27:1348–55. [DOI] [PubMed] [Google Scholar]
- 48. Goodwin B, Ruthven C, King G. et al. Metabolism of 3, 4-dihydroxyphenylalanine, its metabolites and analogues in vivo in the rat: urinary excretion pattern. Xenobiotica 1978;8:629–51. [DOI] [PubMed] [Google Scholar]
- 49. Hashim H, Azmin S, Razlan H. et al. Eradication of Helicobacter pylori infection improves levodopa action, clinical symptoms and quality of life in patients with Parkinson's disease. PLoS One 2014;9:e112330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fasano A, Bove F, Gabrielli M. et al. The role of small intestinal bacterial overgrowth in Parkinson's disease. Mov Disord 2013;28:1241–9. [DOI] [PubMed] [Google Scholar]
- 51. Noack C, Schroeder C, Heusser K. et al. Cardiovascular effects of levodopa in Parkinson's disease. Parkinsonism Relat Disord 2014;20:815–8. [DOI] [PubMed] [Google Scholar]
- 52. Sandler M, Goodwin B, Ruthven C.. Therapeutic implications in Parkinsonism of m-tyramine formation from L-dopa in man. Nature 1971;229:414–6. [DOI] [PubMed] [Google Scholar]
- 53. Palacios N, Hannoun A, Flahive J. et al. Effect of levodopa initiation on the gut microbiota in Parkinson's disease. Front Neurol 2021;12:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van Kessel SP, Frye AK, El-Gendy AO. et al. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat Commun 2019;10:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rekdal VM, Bess EN, Bisanz JE. et al. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 2019;364(6445): eaau6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wolfe MM, Proton Pump Inhibitors: Overview of Use and Adverse Effects in the Treatment of Acid Related Disorders. Waltham, MA: UpToDate, 2018. [Google Scholar]
- 57. Forgacs I, Loganayagam A.. Overprescribing proton pump inhibitors. BMJ 2008;336:2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Imhann F, Bonder MJ, Vila AV. et al. Proton pump inhibitors affect the gut microbiome. Gut 2016;65:740–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jackson MA, Goodrich JK, Maxan M-E. et al. Proton pump inhibitors alter the composition of the gut microbiota. Gut 2016;65:749–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vila AV, Collij V, Sanna S. et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun 2020;11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Buffie CG, Bucci V, Stein RR. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015;517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Buffie CG, Pamer EG.. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013;13:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Leonard J, Marshall JK, Moayyedi P.. Systematic review of the risk of enteric infection in patients taking acid suppression. Am J Gastroenterology 2007;102:2047–56. [DOI] [PubMed] [Google Scholar]
- 64. Baur D, Gladstone BP, Burkert F. et al. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis 2017;17:990–1001. [DOI] [PubMed] [Google Scholar]
- 65. Bajaj JS, Acharya C, Fagan A. et al. Proton pump inhibitor initiation and withdrawal affects gut microbiota and readmission risk in cirrhosis. Am J Gastroenterol 2018;113:1177–86. [DOI] [PubMed] [Google Scholar]
- 66. Stark CM, Susi A, Emerick J. et al. Antibiotic and acid-suppression medications during early childhood are associated with obesity. Gut 2019;68:62–9. [DOI] [PubMed] [Google Scholar]
- 67. Group DI. The effect of digoxin on mortality and morbidity in patients with heart failure. N Engl J Med 1997;336:525–33. [DOI] [PubMed] [Google Scholar]
- 68. Lindenbaum J, Rund DG, Butler VP Jr. et al. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N Engl J Med 1981;305:789–94. [DOI] [PubMed] [Google Scholar]
- 69. Vick RL, Kahn JB Jr, Acheson GH.. Effects of dihydro-ouabain, dihydrodigoxin and dihydrodigitoxin on the heart-lung preparation of the dog. J Pharmacol Exp Ther 1957;121:330–9. [PubMed] [Google Scholar]
- 70. Dobkin JF, Lindenbaum J.. Inactivation of digoxin by the gut flora and its reversal by antibiotics. Pediatr Cardiol 1986;1240–1. [Google Scholar]
- 71. Robertson L, Chandrasekaran A, Reuning R. et al. Reduction of digoxin to 20R-dihydrodigoxin by cultures of Eubacterium lentum. Appl Environ Microbiol 1986;51:1300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Saha JR, Butler VP Jr, Neu HC. et al. Digoxin-inactivating bacteria: identification in human gut flora. Science 1983;220:325–7. [DOI] [PubMed] [Google Scholar]
- 73. Haiser HJ, Gootenberg DB, Chatman K. et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 2013;341:295–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Haiser HJ, Seim KL, Balskus EP. et al. Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes 2014;5:233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]