Summary
Pregnancy represents a unique tolerogenic immune state which may alter susceptibility to infection and vaccine response. Here, we characterized humoral immunity to seasonal influenza vaccine strains in pregnant and non-pregnant women. Although serological responses to influenza remained largely intact during late pregnancy, distinct modifications were observed. Pregnant women had reduced hemagglutinin subtype-1 (H1)- IgG, IgG1, IgG2, and IgG3, hemagglutination inhibition, and group 1 and 2 stem IgG titers. Intriguingly, H1-specific avidity and FcγR1 binding increased, and influenza antibodies had distinct Fc and Fab glycans characterized by increased di-galactosylation and di-sialylation. H1-specific Fc-functionality (i.e. monocyte phagocytosis and complement deposition) was moderately reduced in pregnancy. Multivariate antibody analysis revealed two distinct populations (pregnant vs. non-pregnant) segregated by H1 FcγR1 binding, H1-IgG levels, and Fab and Fc glycosylation. Our results demonstrated a structural and functional modulation of influenza humoral immunity during pregnancy that was antigen-specific and consistent with reduced inflammation and efficient placental transport
Subject areas: Virology, Pregnancy
Graphical abstract

Highlights
-
•
Pregnancy resulted in structural and functional modulation of influenza antibodies.
-
•
Antibodies had differential binding capacity, Fc/Fab glycosylation, and function.
-
•
Antibody glycans directed toward low inflammation and efficient placental transfer.
-
•
Multivariate analysis of immune markers segregated pregnant and non-pregnant women.
Virology; Pregnancy
Introduction
The immune system adapts in a precise and unique way during pregnancy to support fetal development and facilitate full-term delivery (Abu-Raya et al., 2020). These adaptations involve changes in cell phenotypes and frequencies (Th2 polarization, reduction of circulating NK and T cells, and reduction in the number of B cells) as well as modulation of cell function (i.e., cytokine production) (Kraus et al., 2010, 2012). In addition, antibodies produced during pregnancy display unique glycosylation patterns (Bondt et al., 2013, 2014; Einarsdottir et al., 2013). Collectively, these changes favor a non-inflammatory and tolerogenic environment (Alter et al., 2018; Lagattuta and Nigrovic, 2021). As a result, autoimmune conditions, such as rheumatoid arthritis, may transiently resolve during pregnancy (Förger and Förger, 2020). It has been proposed that a corollary of these alterations is increased maternal susceptibility to certain pathogens, which would also affect fetal health and development. For example, pregnant women are known to experience more severe influenza virus infection and are at higher risk of death, as evidenced during the 2009 H1N1 pandemic (Memoli et al., 2013; Kourtis et al., 2014). In the unfolding COVID-19 pandemic, symptomatic maternal infection posed a higher risk for unfavorable outcomes requiring hospitalization and invasive ventilation (Zambrano et al., 2020) and for fetal complications, including preterm birth, growth restriction, and miscarriage (Yee et al., 2020; Barrero-Castillero et al., 2020). These life-threatening health risks can be mitigated by interventions that strengthen prenatal immunity.
There is a long history of safe and effective immunization of pregnant women against tetanus and diphtheria (Rasmussen et al., 2014). In the past decade, the introduction of maternal acellular pertussis vaccine (ACIP, 2013) propitiously reduced infant hospitalizations and respiratory infections (Steinhoff et al., 2017; Amirthalingam et al., 2016). Seasonal influenza vaccination during pregnancy has been recommended to reduce the risk of infection and severity of disease of mothers and young infants (ACIP, 2013; Fiore et al., 2010; WHO, 2014). Several studies have demonstrated efficacy of this approach in reducing both maternal and infant hospitalization and illness (Thompson et al., 2019; Ohfuji et al., 2018; Regan et al., 2016)
Recognizing the value of this strategy to improve public health, a growing number of vaccine candidates intended to strengthen maternal-infant immunity through vaccination during pregnancy are advancing in the clinical pathway (Saso and Kampmann, 2020; Vojtek et al., 2018).
Conceivably, the profound physiological changes required to sustain pregnancy may also influence vaccine-induced immunity. Clinical studies of vaccination in pregnancy have focused primarily on the safety of these new interventions and their immunogenicity, through the analysis of serum antibody levels in mothers and their infants (Sperling et al., 2012; Schlaudecker et al., 2012; Ohfuji et al., 2018; Munoz et al., 2020). A few of these studies have examined maternal T cell responses (Huygen et al., 2015) and/or functional attributes of vaccine-induced antibodies (Jennewein et al., 2019). Some attention has been given to other factors influencing vaccine responses such as optimal gestational age to maximize infant seroprotection (Blanchard-Rohner et al., 2013; Eberhardt et al., 2016), and the effect of prior vaccination on maternal antibody levels (Christian et al., 2017).
The extent to which pregnancy per se influences vaccine-induced adaptive immunity and the implications of such changes for both maternal and infant health remain largely undefined. The wake of COVID-19 pandemic and rapid deployment of SARS-CoV-2 vaccines, created an opportunity to interrogate aspects in which pregnancy may impact vaccine-induced immunity (Shook et al., 2021). While knowledge is still evolving, some studies have found responses to be similar (Gray et al., 2021), while others observed reduced SARS-CoV-2 serum IgG and delayed antibody binding and effector functions in pregnant as compared to non-pregnant women (Bookstein Peretz et al., 2021; Atyeo et al., 2021). Recent results from a blood transcriptional profile analysis in pregnant women post influenza vaccination support the notion of pregnancy-intrinsic changes in interferon-stimulated and plasma cell gene expression that were associated with production of antibody (Giacomelli Cao et al., 2021).
Considering that antibodies are essential for protection against influenza, the goal of the present study was to examine the biophysical and functional properties of influenza-specific antibodies in pregnant and non-pregnant women, most of whom had received seasonal influenza vaccination. We tested the hypothesis that modifications of maternal antibody profiles during pregnancy would likely affect both canonical Fab- and extra-neutralizing Fc-mediated antibody functions, which are known to be key contributors to protective immunity (Boudreau and Alter, 2019). To this end, we conducted an in-depth characterization of antibodies to matched seasonal influenza vaccine strains in pregnant and non-pregnant women that included 1) serum antibody levels determined by hemagglutinin (HA) globular head and stem-specific IgG ELISA, hemagglutination inhibition (HAI), and microneutralization (MN); 2) HA-specific IgG avidity; 3) HA-specific IgG subclasses and Fc receptor binding by ELISA; 4) glycan profile of total and HA-specific IgG Fc and Fab molecules; and 5) Fc-mediated innate cell functions: antibody-dependent neutrophil phagocytosis (ADNP), antibody-dependent monocyte phagocytosis (ADCP), antibody-dependent complement deposition (ADCD), and antibody-dependent NK cell degranulation. Immunological outcomes in the two groups were compared using multivariate analysis. Distinct profiles of influenza-specific IgG Fab and Fc glycan profiles and function were identified between pregnant and non-pregnant women, with subtype-specific antibodies being differentially affected. Two well-defined segregated clusters were identified by aggregate multivariate data analysis. While influenza humoral immunity appeared to be largely unaffected during pregnancy, a deeper interrogation revealed distinct biophysical and functional antibody features. These results confirm previous reports of pregnancy-associated modulation of influenza immunity and provide evidence that support future-controlled clinical studies to inform the development of effective vaccines and guide immunization practices in this important population.
Results
Pregnancy-associated differences in influenza-specific serum antibody levels
To understand pregnancy-associated modulation of humoral immunity, we conducted a comparative analysis of the magnitude, structural features, and functionality of antibodies to seasonal vaccine strains in pregnant and non-pregnant women, the majority of whom had received seasonal influenza immunization (Table S1). Women’s age and time of blood collection post vaccination were similar among participants. An un-biased multivariate principal components analysis (PCA) of the study population confirmed that there were no differences in antibody profiles based on vaccination status in either group (Figure S1). Immunity in the non-vaccinated participants is attributed to exposure to vaccine-matching strains circulating during the influenza season. Likewise, no differences were observed when comparing all antibody profiles in black vs. non-black women (Figure S1).
The breadth of antibody responses in the two groups was determined through analysis of full-length HA- and HA-stem-specific serum IgG titers (Figures 1A and 1B) and canonical function of influenza antibodies: hemagglutinin inhibition (HAI) and microneutralization (MN) (Figures 1C and 1D). Antibody reactivity was tested against two of the subtypes included in the vaccine 2017–18 northern hemisphere flu vaccine (WHO, 2017): H1 MI (drifted strain of the 2009 H1N1 pandemic lineage which was included since the 2016–17 Northern Hemisphere vaccine--A/Michigan/45/2015, H1N1) and H3 HK (A/Hong Kong/480½014, H3N2), as well as against the original 2009 H1N1 pandemic strain H1 CA (A/California/07/2009, H1N1) that disproportionally and severely affected pregnant women during the 2009 pandemic (Jamieson et al., 2009; Louie et al., 2010).
Figure 1.
Pregnancy-associated differences in influenza-specific antibody reactivity, canonical functions, and binding strength
(A–E) HA-specific IgG (A), group 1- and group 2-stem specific IgG (B), hemagglutination inhibition (HAI) (C), microneutralization (MN) (D), and HA-specific IgG avidity (E) in pregnant and non-pregnant women. Data represent log transformed individual datapoints and geometric mean titers (bars) ±SD. ELISA, HAI, and MN titers were compared using one-tailed Mann-Whitney test. Difference in antibody avidity was evaluated using a two-tailed Mann-Whitney test ∗p < 0.05, ∗∗p < 0.01.
Pregnant women had significantly lower IgG titers against H1 MI HA, as compared to the non-pregnant, while titers against H1 CA were unaffected (Figure 1A). Serum IgG directed to the conserved group 1 and group 2 HA 2009 stem regions were both substantially reduced in the pregnant women (Figure 1B). Although both groups had similar HAI and MN titers for the contemporary vaccine strains H1 MI and H3 HK (Figures 1C and 1D), pregnant women did exhibit significantly reduced HAI titers against the original pandemic strain H1 CA (Figure 1C).
Intriguingly, the avidity of IgG against H1 CA HA was higher in the pregnant women as compared to the non-pregnant (Figure 1E). Likewise, a trend of higher IgG avidity against H1 MI HA was observed. Together, these results indicate that while serological responses to seasonal influenza remained largely unaffected, there were clear pregnancy-associated differences in magnitude, canonical functions, and strength of binding of influenza antibodies, which appeared to be antigen-specific.
Pregnancy influences HA IgG subclass and Fc receptor binding
To better understand the profile of influenza-specific antibodies elicited during pregnancy, we conducted a detailed class (IgG, IgA, and IgM) and subclass (IgG 1–4 and IgA 1–2) analysis of serum antibodies specific for full-length HA from H1 MI and H3 HK and their binding capacity to different Fc receptors using a multiplex Luminex assay (Figure 2). Consistent with the ELISA data described above (Figure 1A), pregnant women had significantly reduced levels of H1 MI HA-specific total IgG, IgG1, IgG2, and IgG3 as compared to non-pregnant women (Figures 2A and 2B). A trend of lower H3 HA-specific IgG and IgG1 was observed, while no differences were seen in either H1 MI or H3 HA-specific IgA1, IgA2, or IgM levels between the two groups (Figures 2A and 2B). Because of the different biophysical attributes and capacity of IgG subclasses to engage effector cells in extra-neutralizing effector function (Takai, 2002), the proportion (ratios) of IgG1 and IgG3 relative to IgG4 were calculated. Pregnant women had significantly lower ratios of IgG1:IgG4 and IgG3:IgG4 for both antigens, indicative of a higher proportion of IgG4 over the non-pregnant group (Figure 2C).
Figure 2.
Pregnancy influences HA-specific IgG subclass and Fc receptor binding
IgG, IgA, IgM, and IgG subclass and Fc receptor binding were assessed by Luminex. Dot plots depict individual levels for non-pregnant (NP, gray circles) and pregnant (P, blue circles) women.
(A–G) (A) Isotypes. (B) IgG subclasses. (C) IgG1/IgG4 and IgG3/IgG4 ratios. (D) Class 1 Fc receptor. (E) Class 2 Fc receptors. (F) Class 3 Fc receptors. (G) Neonatal Fc receptor. Lines at mean ± SD. Groups were compared by Mann-Whitney test, and asterisks represent differences between pregnant and non-pregnant individuals. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Clear differences were also seen in terms of HA-specific antibody binding to Fc receptors between the two groups (Figures 2D–2G). In contrast to the reduced HA-specific antibody binding patterns revealed by ELISA or Luminex assays, both H1 MI HA- and H3 HK HA-specific antibodies from pregnant women exhibited increased binding to FcγR1 as compared to the non-pregnant (Figure 2D). No differences in antibody binding capacity to other FcγR molecules were detected between two groups (Figures 2E–2G), except for reduced H1 HA-specific IgG binding to FcγR3 allotypic variant V in the pregnant group (Figure 2F).
Pregnancy alters antibody Fc and Fab N-glycosylation
To identify pregnancy-associated changes in influenza-specific antibody glycosylation, which can have a direct impact on antibody function (Alter et al., 2018; Jennewein et al., 2019; Boudreau and Alter, 2019), we examined the Fc- and Fab-glycan composition of total and H1 MI- and H3 HK-specific serum IgG in both pregnant and non-pregnant women. For each of the samples, a total of 24 distinct glycan peaks were captured by capillary electrophoresis, and overall changes in the four differentially added sugars: galactose, sialic acid, bisecting n-acetyl glucosamine (GlcNAc), and fucose, were calculated for bulk (Figures 3A and 3B) and for HA-specific (Figures 3C–3F and 3G–3V) IgG for each group.
Figure 3.
Pregnancy alters antibody Fc and Fab glycosylation. Antibody glycosylation for HA-specific and bulk antibodies was analyzed by capillary electrophoresis
(A–F) Radar plots depicting different glycosylation profiles of bulk IgG Fc (A) and Fab (B), H1-specific IgG Fc (C) and Fab (E) and H3-specific IgG Fc (D) and Fab (F) in non-pregnant and pregnant groups. G = galactose (0-2), F = fucose, B=Bisecting GlcNAc, S = sialic acid (1-2).
(G–V) These dot-line and stacked-bar plots depict the differences in HA1-specific antibody Fc (G–J) and Fab (O–R) and HA3-specific antibody Fc (K–N) and Fab (S–V) glycosylation between non-pregnant (NP) and pregnant (P) women. Bars at mean with whiskers at SD.
(W and X) Heatmaps showing the Spearman’s correlation between HA-specific Fab and Fc glycosylation for pregnant (W) and non-pregnant (X) women.
Significance for radar plots and glycosylation determined by Mann-Whitney test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Compared to bulk IgG of non-pregnant women, the bulk Fc glycosylation profile of pregnant women revealed shifts toward increased fucosylation (F) and sialylation (S1 and S2) consistent with an anti-inflammatory phenotype (Figure 3A) (Anthony et al., 2008; Kaneko et al., 2006; Washburn et al., 2015, Shields et al., 2002, Shinkawa et al., 2003). On the other hand, pregnant women’s bulk Fab had fully fucosylated (F), agalactosylated (G0 and G1), and bisected (B) glycans (Figures 3B and 3O–3V).
In contrast to the bulk molecules, Fc and Fab glycans of HA-specific antibodies were largely concordant and included digalactosylated (G2) structures with increased sialylation and reduced bisection (Figures 3C–3F and 3G–3V) in the pregnant group, whereas agalactosylated, monosialylated, and bisected structures were predominant in the non-pregnant group. Taken together, these results point to a distinct Fab and Fc glycan profile of antigen-specific antibodies produced during pregnancy consistent with distinct capacity for antigen-recognition (Fab) and Fc-mediated extra-neutralizing antibody function and finely modulated inflammatory properties.
To explore possible associations between Fab and Fc glycosylation, we performed correlation analyses between glycans of HA-specific antibodies in both groups (Figures 3W and 3X). Significant associations between specific glycan types were detected (Figures 3W and 3X), which suggest coordinated glycosylation of both Fab and Fc molecules. This was particularly evident in the non-pregnant group.
Antigen-specific extra-neutralizing antibody functions are selectively affected by pregnancy
Given the observed differences in magnitude and glycan composition of HA-specific antibodies during pregnancy, we next investigated their Fc-mediated effector functions. Responses were measured independently for the two vaccine strains (H1 MI and H3 HK). Monocyte phagocytosis (ADCP) and complement deposition (ADCD) activity of H1 HA-specific antibodies were significantly lower in pregnant women as compared to the non-pregnant women (Figures 4B and 4C). Intriguingly, no differences were seen in neutrophil phagocytosis (ADNP) and NK degranulation (Figures 4A, 4D, and 4E). Both H1 HA- and H3 HA-specific antibodies in the pregnant group exhibited reduced cytotoxicity (ADCC) although it did not reach statistical significance (Figure 4F). Overall, a consistent trend toward reduced antibody functionality across FcR-dependent functions was observed in the pregnant women. The predominant changes on H1 HA-specific antibody function further support pregnancy-associated impact on antigen-specific humoral immunity.
Figure 4.
Antigen-specific extra-neutralizing antibody functions are affected by pregnancy
(A–F) (A). Influenza H1- and H3-specific antibody-dependent neutrophil phagocytosis (ADNP), (B) monocyte phagocytosis (ADCP), (C) complement deposition (ADCD), (D and E) NK degranulation measured as CD107a, MIP-1β, and IFNγ and (F) cellular cytotoxicity (ADCC). Bars represent mean ± SD. Groups were compared using two-tailed Mann-Whitney test. ∗p < 0.05, ∗∗p < 0.01.
Multivariate signatures of antibodies during pregnancy
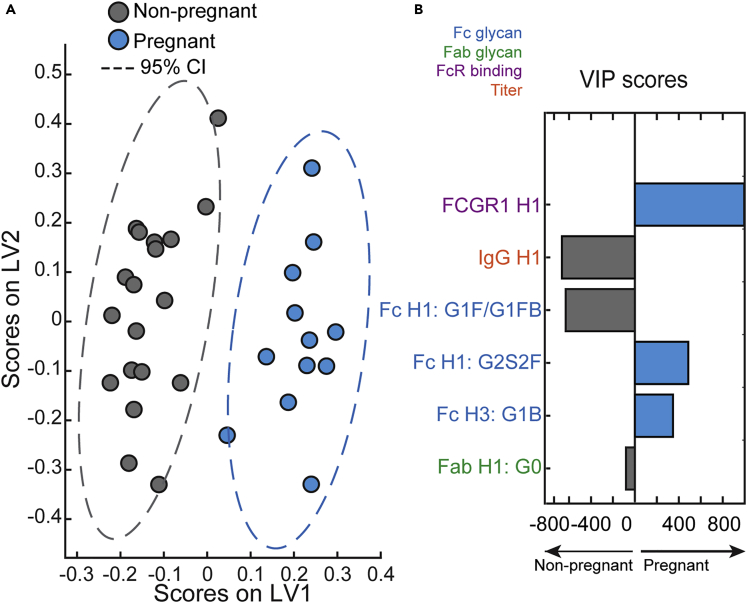
To determine whether pregnant and non-pregnant could be distinguished based on distinct features of influenza humoral immunity, we first examined the data using PCA as un-biased multivariate method (Figure S1). Pregnant and non-pregnant antibody features segregated within the PCA along PC1, with a 12% variance between the two populations. Given the differences seen in the PCA, and to further dissect the differences between antibodies in the two groups, a least absolute shrinkage and selection operator and LASSO Partial Least Squares Discriminant Analysis (LASSO-PLSDAs) was conducted. This multivariate computational modeling technique allowed the determination of features that contributed most strongly to variance between the two groups, and these predictors were then visualized on the PLSDA plot. This analysis included all the influenza-specific variables assessed (serum IgG, HAI and MN titers, antibody subtypes, Fc receptor binding, glycosylation, and Fc-effector functions), 79 in total. Readouts from pregnant and non-pregnant groups were separated and orthogonalized along LV1 (Figure 5A). The antibody features that contributed to a separation between groups were then ranked by their importance (Figure 5B). The top predictor was H1 HA-specific antibody binding to FcγR1, which was greatly enriched in pregnant women. Another predictor was the lower H1 HA-specific IgG titer in the pregnant group. The remaining predictors were mainly Fc and Fab glycan residues. The groups segregated in antigen-specific antibody galactosylation profiles; anti-inflammatory H3 HA Fc G1B and H1 HA Fc G2S2F glycans were preeminent in pregnant women whereas inflammatory H1 HA Fc-G1F/G1FB and H1 HA Fab-G0 dominated in the non-pregnant group. Thus, Fc glycosylation and Fc receptor binding were the primary driving forces in determining the model. Most of the parameters downselected with VIP scores were H1MI HA-specific even though H1 MI, H1 CA, and H3 HK HA-specific readouts were included in the model, reinforcing the notion of antigen dependency of pregnancy-induced antibody changes.
Figure 5.
Multivariate signatures of antibodies produced during pregnancy
Computational analysis was used to distinguish features enriched in pregnancy.
(A) LASSO Partial Least Squares Discriminant Analysis (LASSO-PLSDA) of influenza-specific antibody features between pregnant and non-pregnant women orthogonalized along latent variable 1 (LV1). LV1 explains 42.5% of the variance along the X axis while LV2 explained 17.1% of the variance.
(B) The Variable Importance in Projection (VIP) scores for the PLSDA indicate the prime factors driving the differences between pregnant and non-pregnant women. Factors pointing toward the left are enriched in non-pregnant samples, while those pointing right are enriched in pregnant samples. Bar color and length corresponds to relative importance. Antibody features are colored by category.
Discussion
Seasonal influenza vaccination during pregnancy is recommended by the Advisory Committee on Immunization Practices (Grohskopf et al., 2020) and the ACOG, (2018). The heightened susceptibility of pregnant women to severe influenza infection during the 2009 H1N1 pandemic (Siston et al., 2010; Jamieson et al., 2009; Louie et al., 2010) brought attention to the importance of vaccination for protection of both mothers and infants. The COVID-19 pandemic confirmed the urgent need for strategies and public health preparedness to protect pregnant women and their newborns (Shook et al., 2021). There has been renewed interest, in recent years, in expanding routine maternal immunization against other pathogens (e.g., respiratory syncytial virus, cytomegalovirus, group B streptococcus, etc.) that are important causes of neonatal and infant death. This approach, however, faces unique challenges. Fine-tuned regulatory mechanisms are deployed during pregnancy to balance the tolerogenic environment necessary to avoid unhealthy reactions to paternal antigens with the need to engage robust adaptive immunity against microbial threats (Mor and Cardenas, 2010; Schumacher et al., 2014). Profound changes in humoral immunity occur during gestation, which include decreased number of peripheral B cells (Watanabe et al., 1997; Faucette et al., 2015), and increased IL-10-secreting regulatory B cells (Rolle et al., 2013). These changes are also expected to impact vaccine-induced immunity.
In this study, we report, for the first time, distinct binding and functional features of antibodies specific for seasonal influenza vaccine strains, produced during pregnancy. Although overall humoral influenza immunity appeared to be largely intact, there were clear differences between the pregnant and non-pregnant groups. Among the most notable is the overt reduction in total IgG and IgG subclasses specific for the H1 MI vaccine strain. HAI titers against H1 CA—the original 2009 H1N1 pandemic strain—were also reduced below seroprotective levels. In contrast, HAI and MN titers to seasonal vaccine strains were largely unaffected, which suggests that canonical antibody activity (i.e. virus neutralization and cell-adhesion inhibition) was preserved during pregnancy, even though Fc-mediated extra-neutralizing function was greatly compromised, as discussed below.
Changes in antibody levels affected primarily those against H1 HA but not those against H3 HA. This could be due to immune imprinting, because H1 CA-like viruses have continued to circulate globally since the 2009 H1N1 pandemic (H1 MI is an antigenically drifted H1 CA-like virus) while the subclade of predominant H3 virus has changed almost every other year (WHO, 2022; Allen and Ross, 2018). In response to the antigenically drifted H3 virus, immune memory is recalled, favoring the production of antibodies that recognize common epitopes from previous infections or vaccinations, over de novo antibody responses to unseen epitopes. The history of prior influenza exposure of these women and preexisting immunity are not known and could have influenced the responses detected (Kosikova et al., 2018). Others have reported reduced HA-specific IgG and IgG subclass responses, as well as HAI titers to seasonal influenza vaccines during pregnancy (Schlaudecker et al., 2012, 2018; Bischoff et al., 2013). These studies examined pre-vaccination samples and yet found similar reductions in post-vaccine immunity during pregnancy regardless of basal antibody titers. Additionally, Schlaudecker et al. (2018) reported a decline in response to influenza vaccination as pregnancy progress, which suggests that the antibody modulations we and others have observed are intrinsic to the gestational period.
Pregnant women had significantly lower IgG1/IgG4 and IgG3/IgG4 ratios in response to both H1 and H3 vaccine strains, which is consistent with a Th2-type shift of humoral immunity during gestation. IgG1 and IgG3 are the main subclasses produced in response to viral antigens (Hjelholt et al., 2013), while IgG4 appears to be less functional, with lower affinity for Fc receptors (Jung et al., 2006; Bruhns et al., 2009; Vidarsson et al., 2014), and usually associated with allergic responses (Vidarsson et al., 2014). The increased proportion of IgG4 during pregnancy is consistent with impaired Fc-mediated cellular function. It has been proposed that antibody-dependent cellular function is less affected by antigenic drift and potentially provides influenza heterosubtypic protection (Von Holle and Moody, 2019). Thus, suboptimal cell engagement during pregnancy could reduce vaccine effectiveness and cross protective immunity. Further studies are warranted to understand influenza humoral immunity beyond traditional markers and in particular determinants of extra-neutralizing antibody function and its contribution to protection particularly in high-risk populations.
In addition to lower IgG response to the H1 vaccine strain, pregnant women had reduced IgG titers against highly conserved group 1 and group 2 HA stem regions. This observation is new and suggests that production of stem-specific antibodies during pregnancy might be impaired and that these antibodies could be limited in their capacity to recognize emerging virus variants. Additional investigations to understand HA stem-specific immunity in this group is important as stem antibodies are hypothesized to confer broad cross-protection (Wu and Wilson, 2017) and are candidate antigens for sought after universal flu vaccines (Sautto et al., 2018).
Intriguingly, we found that H1 HA-specific IgG in pregnant women had higher avidity compared to those from non-pregnant women. In addition, HA-specific antibodies produced during pregnancy had increased capacity to bind FcγR1. FcγR1 is an activating receptor expressed in monocyte/macrophages and dendritic cells, with high affinity for human monomeric IgG, particularly IgG1 (Stewart et al., 2014; Bruhns et al., 2009). On the other hand, H1 HA-specific IgG from pregnant women had reduced binding capacity to FcγRIIIa-V, a high affinity allotype receptor expressed in monocytes/macrophages and NK cells. The enhanced capacity of pregnancy-distinct antibodies to bind cognate antigens and to selectively engage in innate cell function via Fc-interaction may represent compensatory mechanisms to maximize response yet avoid harmful effects.
Another innovative aspect of our study was the analysis of extra-neutralizing FcR-mediated antibody functions in pregnant women. H1-specific ADCP, ADCD, and ADCC were reduced in this group, and this observation is consistent with reduced levels of H1 IgG, and in particular IgG1, which triggers these processes efficiently by engaging FcγRI (Gunn and Alter, 2016). The reduced capacity of H1 HA antibodies to bind to FcγRIII-V might have also contributed to this outcome. IgG1 has been implicated in activation of complement in the context of influenza infection; hence, reduced IgG1 in this group is consistent with lower C3 deposition.
Unlike FcR-mediated monocyte/macrophage uptake, neutrophil phagocytic activity and NK degranulation both appeared largely unaffected during pregnancy. Neutrophil phagocytosis can be activated by different and additional signals such as IgG2 interaction with FcγRIIa and IgA with FcαRI (Gunn and Alter, 2016). NK degranulation was maintained in the pregnant group despite their antibodies exhibiting lower binding affinity for FcγRIIIa (the cellular receptor that mediates NK degranulation). These observations further support the idea of compensatory mechanisms of the humoral immune system to offset pregnancy-intrinsic changes and maintain a healthy state.
Vastly different glycosylation of total and influenza-specific antibodies was observed in pregnant women compared to non-pregnant controls. The total Fc glycan profile in pregnant women, which included fucosylation and sialylation, was oriented toward reduced inflammation (Alter et al., 2018; Lagattuta and Nigrovic, 2021). In contrast, the total Fab glycans, which involved increased fucosylation, agalactosylation, and bisection, had been associated with increased inflammation (Alter et al., 2018; Lagattuta and Nigrovic, 2021). Non-inflammatory Fc galactosylation and sialylation as well as Fab N-acetylglucosamine bisection of bulk antibodies during pregnancy have been reported elsewhere (Bondt et al., 2014, 2016; Jansen et al., 2016; Ruhaak et al., 2014; Van De Geijn et al., 2009; Selman et al., 2012). Surprisingly, the Fc and Fab glycan profile of HA-specific antibodies were largely concordant within each group and included digalactosylated (G2) structures with increased sialylation and reduced bisection in the pregnant group, whereas agalactosylated, monosialylated, and bisected structures were predominant in the non-pregnant group. The association patterns remained broadly unchanged in the two groups, which indicate a coordinated global regulation of Fc and Fab glycosylation. However, other factors governing production of Fab and Fc glycans may reduce the overall level of coordination to reflect the putative greater level of Fab glycosylation during pregnancy (Van De Bovenkamp et al., 2016).
Two distinct populations emerged in the PLSDA analysis, with Fc and Fab glycosylation being primary discriminatory elements; the clear separation of the two groups confirms the robustness of the data. The similarity in Fc and Fab glycosylation for the influenza-specific antibodies suggests some level of coordination of post-translational modifications that differs according to the B cell population. This is the first demonstration of Fab and Fc glycosylation of vaccine-specific antibodies consistent with non-inflammatory activity in pregnant women and different from the more inflammatory pattern in the non-pregnant group. Despite the disparity in antibody glycosylation, all the relevant Fc-mediated influenza-specific cellular functions were detected in the pregnant group.
We have previously shown that digalactosylated antibodies involved in NK degranulation were enriched in cord blood; these digalactosylated antibodies preferentially bind to the neonatal Fc receptor (FcRn), which transits antibodies across the placenta (Jennewein et al., 2019; Martinez et al., 2019). Digalactosylated antibodies are reportedly more effective at facilitating NK degranulation, a function the neonatal immune system is better equipped to deploy as opposed to phagocytosis or other extra-neutralizing functions (Jennewein et al., 2019). For the mother, digalactosylation of antibodies during gestation may reflect another compensatory mechanism that maintains ADNP unaltered when other cellular functions may be reduced. Likewise, sialylation has been exploited in immunoglobulin therapy to endow anti-inflammatory benefits (Anthony et al., 2008; Pagan et al., 2018; Bruckner et al., 2017). Here, enhanced Fc-sialylation during pregnancy may contribute to dampened antibody-mediated inflammation consistent with the tolerizing state of pregnancy (Pagan et al., 2018).
Together, the shifts observed in influenza-specific antibodies during pregnancy are consistent with reduced inflammation, more efficient Fc receptor engagement to maintain antiviral function (as the magnitude of antibody available might be reduced), enhancement of placental transfer, reduction in antibody catabolism, and enrichment of antibodies that are more efficient for the newborn. The origin, regulation, evolution, and duration of these pregnancy-associated antibody changes remain to be investigated.
Rather than broad immune suppression during pregnancy (reviewed in Memoli et al., 2013), our data suggest that humoral immunity is selectively modulated by changes in antibody levels, avidity, FcR binding capacity, glycosylation patterns, and innate cellular functions. Our results also argue against universally compromised humoral immunity and in favor of discrete, antigen-dependent modifications. Whether these changes are transient (restricted to pregnancy) or more durable remain to be determined. Clinical studies to investigate modulation of influenza immunity in pregnant women longitudinally and beyond delivery are needed to corroborate these findings, and to deepen our understanding of this physiological process and the mechanisms involved.
The overall antibody modulations we have observed may not be restricted to influenza but could impact other vaccine antigens. Lower pertussis toxin (PT) serum IgG responses have been reported post vaccination in pregnant women that received the licensed tetanus-diphtheria-pertussis (TdaP) vaccine in the third trimester as compared to non-pregnant women (Peer et al., 2021). Similarly, a reduction of PT-specific IgG in TdaP-vaccinated pregnant women compared with non-pregnant had been reported earlier (Fortner et al., 2018)
Limitations of the study
Our study is limited by a small sample size, race/ethnicity distribution, and the retrospective analysis of immune responses to influenza seasonal vaccine strains. Nonetheless, it still provides a valuable snapshot of vaccine-specific immunity in the pregnant population.
Maternal infections place women and their infants at high risk. Understanding the nature and extent of pregnancy-associated changes in antibody production, post-translational modifications and function, as well as how these processes affect vaccine responses will inform vaccine design, immunization strategies, and the development of much needed preventive and therapeutic strategies, particularly those for use in emergency situations, to improve maternal and infant health.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| horseradish peroxidase-labeled goat-anti-human IgG | Invitrogen | # A18805 RRID:AB_2535582 |
| FITC Goat IgG anti-C3 | MP Biomedicals | CAT # 855385 |
| CD107a PE-Cy5 Mouse Anti-Human | BD Biosciences | CAT#555802; clone: H4A3; RRID:AB_396136 |
| Alexa Fluor® 700 Mouse Anti-Human CD3 | BD Biosciences | CAT#557943; clone: UCHT;1 RRID:AB_396952 |
| CD56 PE-Cy7 Mouse Anti-Human CD56 | BD Biosciences | CAT#557747; clone: B159 |
| APC-Cy™7 Mouse Anti-Human CD16 | BD Biosciences | CAT#557758; clone: 3G8; RRID:AB_396853 |
| APC Mouse Anti-Human IFN-g | BD Biosciences | CAT#554702; clone: B27; RRID:AB_398580 |
| PE MIP-1b Mouse anti-Human | BD Biosciences | CAT#550078; clone: D21-1351; RRID:AB_393549 |
| influenza A nucleoprotein-specific monoclonal antibody, clone A3 | Millipore | CAT# MAB8258 RRID:AB_95232 |
| influenza A nucleoprotein-specific monoclonal antibody , clone A1 | Millipore | Cat# MAB8257F-5, RRID:AB_570527 |
| anti-CD66b-Pacific blue | BioLegend | CAT#305112; clone: G10F5; RRID:AB_2563294) |
| Mouse Anti-Human IgG1-Fc PE | Southern Biotech | CAT # 9054-09; RRID:AB_2796628 |
| Mouse Anti-Human IgG2-Fc PE | Southern Biotech | CAT # 9060-09; RRID:AB_2796635 |
| Mouse Anti-Human IgG3-Hinge PE | Southern Biotech | CAT # 9210-09; RRID:AB_2796701 |
| Mouse Anti-Human IgG4-Fc PE | Southern Biotech | CAT # 9200-09; RRID:AB_2796693 |
| Mouse Anti-Human IgA1-Fc PE | Southern Biotech | CAT # 9130-09; RRID:AB_2796656 |
| Mouse Anti-Human IgA2-Fc PE | Southern Biotech | CAT # 9140-09; RRID:AB_2796664 |
| Mouse Anti-Human IgM-Fc PE | Southern Biotech | CAT # 9020-09; RRID:AB_2796577 |
| Bacterial and virus strains | ||
| H1N1 A/California/07/2009 | Produced in house | N.A. |
| H1N1 A/Michigan/45/2015 | Produced in house | N.A. |
| H3N2 A/Hong Kong/4801/2014 | Produced in house | N.A. |
| Biological samples | ||
| Cohort of pregnant/non-pregnant human serum samples | N.A. | N.A. |
| Chemicals, peptides, and recombinant proteins | ||
| G418 | ThermoFisher Scientific | CAT#10131027 |
| H3 HK HA | GeneScript | Custom made |
| chimeric HA bearing H1 A/Puerto Rico/8/1934 stalk (group 1) with mismatched H16 HA head | GeneScript | Custom made |
| chimeric HA bearing H3 HK stalk (group 2) with mismatched H4 HA head | Genescript | Custom made |
| H1 MI (HAΔM H1Ν1 Α/Michigan/45/2015) | Immune Technologies Corp. | CAT#: IT-003-00105ΔMp |
| H1 CA (HAΔTM A/California/07/2009) | Immune Technologies Corp. | CAT#: IT-003-00106ΔMp |
| H3 HK proteins (H3ΔTM H3N2 A/Hong Kong/4801/2014) | Immune Technologies Corp. | Cat: IT-003-004M24 |
| 1-Step Ultra TMB-ELISA substrate solution | ThermoFisher Scientific | CAT#34028 |
| receptor-destroying enzyme | Denka-Seiken | CAT#: 370013 |
| oseltamivir | selleckchem.com | CAT#: S2597 |
| veronal buffer supplemented with 0.1% fish skin gelatin | Boston Bio Products | CAT#: IBB-290X |
| Histopaque | ||
| Human IL-15 Recombinant Protein, eBioscience | ThermoFisher Scientific | CAT#: BMS319 |
| Brefeldin A Solution | Biolegend | CAT#420601 |
| Golgistop, Protein transport inhibitor (Containing Monensin) | BD Biosciences | CAT#554724 |
| FIX&Perm Cell Permeabilization Kit | ThermoFisher Scientific | CAT#GAS004 |
| EZ-Link NHS-LC-biotin | ThermoFisher Scientific | CAT#21343 |
| Bio-Glo luciferase assay reagent | Promega | CAT#: G7940 |
| BirA-500: BirA biotin-protein ligase standard reaction kit | Avidity | CAT#BirA500 |
| Streptavidin-R-Phycoerythrin | Prozyme | CAT#PJ31S |
| IDEZ-Protease | New England Biolabs | CAT#P0770S |
| GlycanAssure APTS kit | ThermoFisher Scientific | CAT#A28676 |
| RosetteSep™ Human NK Cell Enrichment Cocktail | Stem Cell Technologies | CAT#15065 |
| Human Fc receptors | Produced at the Duke Human Vaccine Institute, Boesch et al. 2014 | N/A |
| Experimental models: Cell lines | ||
| THP-1 Cells | ATCC | CAT#TIB-202 RRID: CVCL_0006 |
| Madin-Darby canine kidney-SIAT1 cells | Sigma Aldrich | CAT#05071502 |
| Jurkat effector cells | Promega | CAT#: G7015 |
| Recombinant DNA | ||
| pFastBac gp67 vector | GenScript | GenScript propriety |
| Software and algorithms | ||
| ForeCyt software | ||
| GlycanAssure Software | ThermoFisher Scientific | https://www.thermofisher.com/us/en/home/life-science/bioproduction/contaminant-and-impurity-testing/glycanAssure-Glycan-analysis.html |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| Other | ||
| MaxiSorp microtiter plates | Thermofisher | CAT#: 50-112-3685 |
| Victor 3V multilabel reader | PerkinElmer | 1420 multilabel counter |
| FluoSpheres NeutrAvidin-Labeled Microspheres, 1.0 μm, red fluorescent (505/515), 1% solids | Invitrogen | CAT # F8775 |
| guinea pig complement | Cedarlane | CAT#: CL4051 |
| Zeba Spin Desalting Column, 7 MWCO | ThermoFisher Scientific | CAT#: 89877 |
| FluoSpheres™ Carboxylate-Modified Microspheres, 1.0 μm, yellow-green fluorescent (505/515), 2% solids | ThermoFisher Scientific | CAT#F8823 |
| MagPlex microspheres | Luminex corporation | CAT#s:MC10015-YY, MC10026-YY, MC10042-YY, MC10062-YY, MC10068-YY, MC10072-YY |
| Streptavidin Magnetic Beads | New England Biolabs | CAT#: S1420S |
| protein G beads | New England Biolabs | CAT#: S1430S |
| Glyko® APTS Biantennary & High Mannose Partitioned Library | Prozyme | CAT#GKSP-520 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marcela Pasetti mpasetti@som.umaryland.edu
Materials availability
This study did not generate new unique reagents
Experimental modela and subject details
Serum samples were obtained from pregnant (end of third trimester) and non-pregnant women, the majority of whom had received inactivated influenza vaccine during the 2017-2018 flu season; serum samples were obtained within 4 months of vaccination (Table S1, Figure S1). Pregnant women also received TdaP vaccine during pregnancy. TdaP and influenza vaccination occurred within 3-4 months interval for all participants except for three who had record of TdaP and influenza vaccine given the same day. The vaccine components for the northern hemisphere 2017-2018 season influenza vaccine included H1N1 A/Michigan/45/2015, H3N2 A/Hong Kong/4801/2014, B/Phuket/3073/2013, and B/Brisbane/60/2008-like viruses((WHO), 2017). All subjects were HIV, HBV, and HCV negative and provided written informed consent before enrollment. Human subject research was approved by the University of Maryland, Baltimore Institutional Review Board (HP-00065842 and HP-00040025) and by Partners Human Research Committee (Approval Number 2018P001060).
Cell lines
Madin-Darby canine kidney-SIAT1 cells (SIAT1-MDCK) were grown in Minimal Essential Media (MEM) plus 50 μg/ml of G418, 5% fetal bovine serum, 1X GlutaMAX™, and penicillin/streptomycin. Jurkat cells were cultured in 90% RPMI 1640 with l-glutamine and 10% FBS supplemented with 100 μg/ml hygromycin, 250 μg/ml Antibiotic G-418, 1 mM Sulfate Solution, 0.1 m M sodium pyruvate, MEM and nonessential amino acids. THP-1 cells (ATCC, Manassas, VA, USA) were grown in R10 (RPMI plus 10% FBS L-glutamine and penicillin/streptomycin) supplemented with 0.01% β-mercaptoethanol.
Viruses
H1N1 A/California/07/2009 (H1 CA), H1N1 A/Michigan/45/2015 (H1 MI), and H3N2 A/Hong Kong/4801/2014 (H3 HK) were grown in 9-10 day old embryonated eggs. Infectious viral titer, i.e., 50% tissue culture infectious dose (TCID50), was determined using a nucleoprotein-based enzyme-linked immunosorbent assay (ELISA) (Kosikova et al., 2018).
Recombinant proteins
For titer and avidity determination, genes coding H3 HK HA, chimeric HA bearing H1 A/Puerto Rico/8/1934 stalk (group 1) with mismatched H16 HA head and chimeric HA bearing H3 HK stalk (group 2) with mismatched H4 HA head were synthesized as previously described (Stevens et al., 2004; Pica et al., 2012; Radvak et al., 2021) and were subcloned into the pFastBac gp67 vector. Recombinant H3 HK HA, chimeric group 1 or group 2 stalk proteins expressed in Sf9 cells were column purified. For functional assays and Luminex Fc receptor binding and titer, recombinant H1 MI (HAΔM H1Ν1 Α/Michigan/45/2015), H1 CA (HAΔTM A/California/07/2009) and H3 HK proteins (H3ΔTM H3N2 A/Hong Kong/4801/2014) expressed in 293 cells were used.
Method details
Anti-HA ELISA and avidity
HA-specific ELISA and avidity assays were performed as described (Kosikova et al., 2018). Briefly, serum samples were serially diluted and added to MaxiSorp microtiter plates coated with 0.2 μg/mL of recombinant protein. Plates were incubated at room temperature for 60 min. For avidity determination, antigen-antibody complexes were dissociated by incubation with 100 μl of 4 M urea for 15 min and plates blocked again for an additional hour (Kosikova et al., 2018). Bound antibodies were detected by addition of horseradish peroxidase-labeled goat anti-human IgG followed by 1-Step Ultra TMB substrate. Absorbance at 450 nm was measured in a Victor V multilabel reader. Antibody titers were determined by interpolation in a Sigmoidal four parameter logistic regression curve using Prism 9 (GraphPad, San Diego, CA), and avidity index was calculated based on the area under the curve (Kosikova et al., 2018).
Hemagglutination inhibition (HAI)
Sera were pre-treated with receptor-destroying enzyme (RDE), serially diluted two-fold (starting at 1:10) and incubated with 4 HA units per 25 μl of viruses at room temperature for 30 min. Hemagglutination was determined using 0.5% turkey red blood cells for H1N1 viruses or 0.75% guinea pig red blood cells in the presence of 20 nM oseltamivir for H3N2 viruses, respectively (Xie et al., 2011, 2015). HAI titers were reported as the reciprocal of the highest serum dilution that resulted in complete HAI. A titer of 5 was assigned if no inhibition was observed at the starting dilution.
Complement deposition
Antibody-dependent complement deposition (ADCD) was performed as previously described (Fischinger et al., 2019). Biotinylated recombinant HAΔM H1Ν1 Α/Michigan/45/2015 and H3ΔTM H3N2 A/Hong Kong/4801/2014 were coupled to red fluorescent NeutrAvidin microspheres, incubated with sera and added to single wells as described above. Lyophilized guinea pig complement was resuspended in ice-cold water and diluted 1:100 in cold veronal buffer supplemented with 0.1% fish skin gelatin. 200 μl of diluted complement was added to each well, and plates were incubated for 20 min at 37°C, 5% CO2. Plates were then washed twice with ice-cold 15 mM EDTA-PBS and stained with anti-guinea pig C3-FITC. After a final wash, beads were re-suspended in 50 μl of PBS and analyzed on the iQue Screener PLUS. Red-fluorescent beads were gated based on size. Complement deposition was determined as the geometric mean fluorescent intensity divided by 1,000. Data presented is area under the curve of the complement score averaged for two replicates.
NK cell activation
ELISA plates were coated with recombinant HAΔM H1Ν1 Α/Michigan/45/2015 and H3ΔTM H3N2 A/Hong Kong/4801/2014 at 3 μg/mL of PBS for 2 h at 37°C, 5% CO2. Plates were then washed three times with PBS and blocked overnight with 200 μl of 5% BSA in PBS. The next day, plates were washed, and samples were added in three serial dilutions (1:10, 1:50, 1:100) in a 50 μl volume and incubated 2 h at 37°C, 5% CO2. NK cells were isolated from healthy adult PBMC using a RosetteSep NK Cell Enrichment Kit, according to manufacturer’s instructions, followed by density gradient centrifugation in Histopaque. A suspension of 1.5×106 cells/ml in R10 was prepared, supplemented with 1 ng/ml rhIL15 and incubated overnight at 37°C, 5% CO2. Prior to the assay, a suspension of 2.5×105 cells/ml was prepared and 2.5 μg/ml of Brefeldin A and Golgistop and 2.5 μl of anti-CD107a were added. NK cells (5 × 104 in 200 μl) were added to each well containing Ag-Ab, and plates were incubated for 5 h at 37°C, 5% CO2. Following the incubation and washing, cells were stained with anti-CD3, anti-CD56, and anti-CD16, washed, fixed, permeabilized with FIX&Perm kit, and stained intracellularly with anti-IFNγ and MIP-1β. Cells were resuspended in PBS and analyzed on the iQue Screener as described above. Influenza-specific IgG and PBS (diluent) were included as positive and negative controls. Negative control wells were used to set gates. Data were reported as the percentage of CD16+CD56+CD3- NK cells positive for CD107a, IFNγ, or MIP-1β; reported values are the area under the curve of the mean of three replicates divided by 100,000.
Microneutralization (MN)
H1N1 and H3N2-specific MN titers were determined using a nucleoprotein-based ELISA as previously described (Kosikova et al., 2018). Briefly, RDE-treated sera were incubated with 100 50% tissue culture infectious dose (TCID50) of virus for 1 h at room temperature, 5% CO2, and then overlaid onto SIAT1-MDCK cells overnight. Infected cells were detected using influenza A nucleoprotein-specific monoclonal antibodies. MN titers were reported as the reciprocal of the highest serum dilution with ≥50% neutralization.
Phagocytosis
The antibody-dependent monocyte phagocytosis assay (ADCP) used was adapted from Ackerman et al. (2011) and the antibody-dependent neutrophil phagocytosis assay (ADNP) was performed as described (Karsten et al., 2019). Briefly, influenza antigens, recombinant HAΔM H1Ν1 Α/Michigan/45/2015 and HAΔTM H3N2 A/Hong Kong/4801/2014 were biotinylated with a 20-fold excess of 10 mM EZ-Link NHS-LC-biotin and unbound biotin was removed using a Zeba Spin Desalting Column. Antigens were then mixed with yellow-green fluorescent NeutrAvidin microspheres at a ratio of 10 μl of beads per 10 μg of biotinylated protein and incubated at 37°C, 5% CO2 for 2 h. Following incubation, beads were washed twice with PBS-0.01% BSA and resuspended in 1 mL of PBS-0.01% BSA. Serum samples were diluted 1:10, 1:50, and 1:100 in PBS, and 10 μl of each dilution was mixed with 10 μl of antigen-coupled fluorescent beads in a 96-well round bottom plate and incubated at 37°C, 5% CO2, for 2 h. The beads were then washed with 200 μl of PBS. For the monocyte phagocytosis, THP-1 cells (2.5×104 cells in 200 μl per well) were added to wells containing the beads, and plates were incubated for 16 h at 37°C, 5% CO2. For the neutrophil phagocytosis, fresh neutrophils were isolated from whole blood of healthy adults and added (5×105 cells in 200 μl) to bead-containing wells. Plates were incubated for 1h at 37°C, 5% CO2. Following incubation, cells were spun down and stained with anti-CD66b for 15 min. For both assays, cells were fixed with 100 μl of 4% paraformaldehyde (PFA) for 10 min, resuspended in 50 μl of PBS and analyzed by Flow cytometry using an iQue Screener PLUS and ForeCyt software. Influenza-specific IgG and PBS were included as positive and negative controls, respectively. Phagocytosis scores were calculated as the percentage of bead positive cells, multiplied by the geometric mean fluorescent intensity of the beads for bead positive cells, divided by 1×107. All reported values are the area under the curve of the mean of two replicates.
Cellular cytotoxicity
Antibody-dependent cellular cytotoxicity (ADCC) assay was performed as previously described (Jacobsen et al., 2017) with modifications. Briefly, SIAT1-MDCK cells were seeded onto 96-well white flat-bottom plates (PerkinElmer), infected with H1 MI or H3 HK viruses, and incubated for 6 h at 37°C. RDE-treated sera (25 μl/well) and Jurkat effector cells (25 μl/well) were added to infected cells and incubated overnight at 37°C. Bio-Glo luciferase assay reagent was then added, and luminescence was measured using a Victor V multilabel reader.
Antibody subclass and FcR binding
IgG, IgA, IgM, and Fc receptor binding analyses were performed using a multiplex luminex assay as previously described (Brown et al., 2012). Briefly, recombinant HAΔM H1Ν1 Α/Michigan/45/2015 and H3ΔTM H3N2 A/Hong Kong/4801/2014 were coupled to MagPlex microspheres at a ratio of 25 μg of protein to 400 μl of beads. Microspheres were activated with 100 mM monobasic sodium phosphate pH 6.2 in the presence of 50 mg/mL EDC and 50 mg/ml sulfo-NHS, washed with 0.05 M 2[N-Morpholino] ethanesulfonic acid (MES) pH 5.0, incubated with antigen for 2 h at 37°C, 5% CO2, and blocked with PBS-TBN (0.1% BSA, 0.02% tween-20, 0.05% Azide in PBS, pH 7.4) for 30 min. The beads were washed in PBS-0.05% tween-20, resuspended in 250 μl of PBS. Beads were added to 384-well plates and incubated with sera at a final 1:500 dilution in luminex wash buffer (PBS-0.05% BSA-0.001% tween-20) for 2 h at room temperature and with shaking (800 rpm).
To detect antibody subclasses, following incubation, beads were washed in luminex buffer and incubated with PE-labeled anti-IgG, -IgG1, -IgG2, - IgG3, -IgG4, -IgA1, -IgA2, and -IgM for 1 h at room temperature with shaking. After washing, beads were resuspended in Qsol buffer and plates were read on an iQue Screener PLUS. H1N1 and H3N2 coupled beads were distinguished based on size and granularity, and data were analyzed as median fluorescent intensity of PE for each bead population.
To investigate Fc receptor binding, recombinant Fc receptors with an AviTag were biotinylated using a Bir500 kit according to manufacturer’s instructions and purified using a Zeba Spin Desalting Column. Biotin-labeled FcRs were then incubated with streptavidin-PE for 10 min and excess streptavidin was quenched with 20 μΜ biotin for another 10 min. Following bead and serum incubation, PE-Fc receptors were then added and incubated for a further 1 h at room temperature with shaking. Plates were then washed and analyzed on an iQue Screener as described above. Data reported for each sample represents the mean of two replicates.
Glycosylation analysis
Antigen-specific glycosylation analysis was performed as described in Jennewein et al., (2019). Briefly, recombinant HAΔM H1Ν1 Α/Michigan/45/2015 and H3ΔTM H3N2 A/Hong Kong/4801/2014 were biotinylated and coupled to NeutrAvidin magnetic beads at a ratio of 2.5 μg of protein to 25 μl of beads. Sera (200 μl) were first incubated with non-coated NeutrAvidin beads to remove non-specific binding and then added to the antigen-coated beads and incubated for 1 h at 37°C. Antibodies were eluted by incubation in 50 μl of pH 2.9 citrate buffer for 30 min at 37C. Samples were then spun down and the eluted antibodies, contained in the supernatant, were neutralized with 30 μl pH 8.9 potassium phosphate buffer. Antibodies were then coupled to protein G beads. After beads were washed, IDEZ was used to cleave the Fab (in the supernatant) from the Fc (remained on the magnetic beads) for 1 h at 37°C and collected. The two fragments were deglycosylated and fluorescently labeled using a GlycanAssure APTS kit according to manufacturer’s instructions. For the Fc fragment, which remained bound to the protein G beads, an additional magnetic separation after PNGase glycan cleavage separated the glycans from remaining magnetic beads and the protocol then proceeded per manufacturer’s instructions. Glycans were analyzed on a 3500xL genetic analyzer. Glycan fucosyl and afucosyl libraries were used to assign 24 discrete glycan peaks using GlycanAssure software. Data are reported as percentages of total glycans for each of the glycan peaks.
Quantification and statistical analysis
Mann-Whitney test (univariate analysis) was used to examine differences in readouts between pregnant and non-pregnant groups. p values are two-sided, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Data on radar plots were Z scorez-score normalized individually along each radar to an average of 0 with a standard deviation of 1. Multivariate Principal Component
Principal Component Analysis (PCA) and Least Absolute Shrinkage and Selection Operator (LASSO) and Partial Least Squares Discriminant Analysis (PLSDA, LASSO-PLSDA) were used to distinguish pregnant vs non-pregnant immune profiles. PCA was used to visualize whether pregnant and non-pregnant samples separate into discrete groups within the two-dimensional principal component (PC) space. LASSO-PLSDA was used to identify the features that were most important in separating the pregnant and non-pregnant samples (Lau et al., 2011; Arnold et al., 2016). LASSO is a regression analysis that selects a minimal set of variables that contribute to the differences between samples arranged along an ordinal dependent variable. It selects features that are required to create a maximum separation along a linear axis. These variables are then plotted and visualized on the PLSDA plot. Missing values were imputed using the k-nearest neighbor. Variables were centered and scaled to a standard deviation of 1. The LASSO-PLSDA model had a calibration success of 100% with a leave-1-out CV success of 97.0588%. Univariate and PCA analysis were performed with Prism 9 (GraphPad, San Diego, CA). MATLAB was used to perform PLSDA multivariate analysis.
Acknowledgments
H.X. was supported by the FDA Office of Women’s Health, award #20-01-0010. This work was supported in part by NIH/NIAID U19AI145825 to H.X. and M.F.P. (Project 1) and G.A. (Core 1).
Author contributions
Conceptualization, M.F.J., H.X., G.A., and M.F.P.; Clinical samples: M.F.P, W.H.C., and J.D.C. Investigation; M.F.J., M.K., F.J.N., P.R., and C.M.B.; Resources, H.X., G.A., and M.F.P.; Writing-Original draft, M.F.J, H.X., G.A., and M.F.P.; Writing-review and editing, J.D.C and W.H.C.; Supervision, H.X., G.A., and M.F.P.
Declaration of interests
C.B. is an employee and equity holder of Leyden Labs, a company developing pandemic prevention therapeutics. G.A. is a founder and equity holder of Seromyx Systems, a company developing a platform technology that describes the antibody immune response. G.A. is an employee and equity holder of Leyden Labs, a company developing pandemic prevention therapeutics. G.A.'s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ + community.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104088.
Contributor Information
Hang Xie, Email: hang.xie@fda.hhs.gov.
Galit Alter, Email: galter@mgh.harvard.edu.
Marcela F. Pasetti, Email: mpasetti@som.umaryland.edu.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ackerman M.E., Moldt B., Wyatt R.T., Dugast A.S., McAndrew E., Tsoukas S., Jost S., Berger C.T., Sciaranghella G., Liu Q., Irvine D.J., Burton D.R., Alter G. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods. 2011;366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACOG Committee opinion no. 732: Influenza Vaccination During Pregnancy. Obstet. Gynecol. 2018;131(4):e109–e114. doi: 10.1097/AOG.0000000000002588. [DOI] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2014. Safety of immunization during pregnancy: a review of the evidence: Global Advisory Committee on Vaccine Safety.https://apps.who.int/iris/handle/10665/340577 [Google Scholar]
- WHO . World Health Organization; 2017. Recommended Composition of Influenza Virus Vaccines for Use in the 2017-2018 Northern Hemisphere Influenza Season.https://www.who.int/publications/m/item/recommended-composition-of-influenza-virus-vaccines-for-use-in-the-2017-2018-northern-hemisphere-influenza-season [Google Scholar]
- Abu-Raya B., Michalski C., Sadarangani M., Lavoie P.M. Maternal Immunological Adaptation During Normal Pregnancy. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.575197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACIP Updated recommendations for use of tetanus toxoid, reduced diphtheria toxoid, and acellular pertussis vaccine (Tdap) in pregnant women. 07 ed. MMWR Morb. Mortal. Wkly Rep. 2013;60:1424–1426. [PubMed] [Google Scholar]
- Allen J.D., Ross T.M. H3N2 influenza viruses in humans: viral mechanisms, evolution, and evaluation. Hum. Vaccin. Immunother. 2018;14:1840–1847. doi: 10.1080/21645515.2018.1462639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G., Ottenhoff T.H.M., Joosten S.A. Antibody glycosylation in inflammation, disease and vaccination. Semin. Immunol. 2018;39:102–110. doi: 10.1016/j.smim.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirthalingam G., Campbell H., Ribeiro S., Fry N.K., Ramsay M., Miller E., Andrews N. Sustained Effectiveness of the Maternal Pertussis Immunization Program in England 3 Years Following Introduction. Clin. Infect. Dis. 2016;63(4):S236–S243. doi: 10.1093/cid/ciw559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony R.M., Nimmerjahn F., Ashline D.J., Reinhold V.N., Paulson J.C., Ravetch J.V. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008;320:373–376. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K.B., Burgener A., Birse K., Romas L., Dunphy L.J., Shahabi K., Abou M., Westmacott G.R., Mccorrister S., Kwatampora J., et al. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal Immunol. 2016;9:194–205. doi: 10.1038/mi.2015.51. [DOI] [PubMed] [Google Scholar]
- Atyeo C., Deriso E.A., Davis C., Bordt E.A., De Guzman R.M., Shook L.L., Yonker L.M., Fasano A., Akinwunmi B., Lauffenburger D.A., et al. COVID-19 mRNA vaccines drive differential antibody Fc-functional profiles in pregnant, lactating, and nonpregnant women. Sci. Transl. Med. 2021;13:eabi8631. doi: 10.1126/scitranslmed.abi8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Castillero A., Beam K.S., Bernardini L.B., Ramos E.G.C., Davenport P.E., Duncan A.R., Fraiman Y.S., Frazer L.C., Healy H., Herzberg E.M., et al. COVID-19: neonatal-perinatal perspectives. J. Perinatol. 2020;41:940–951. doi: 10.1038/s41372-020-00874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff A.L., Folsgaard N.V., Carson C.G., Stokholm J., Pedersen L., Holmberg M., Bisgaard A., Birch S., Tsai T.F., Bisgaard H. Altered response to A(H1N1)pnd09 vaccination in pregnant women: a single blinded randomized controlled trial. PLoS One. 2013;8:e56700. doi: 10.1371/journal.pone.0056700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard-Rohner G., Meier S., Bel M., Combescure C., Othenin-Girard V., Swali R.A., Martinez De Tejada B., Siegrist C.A. Influenza vaccination given at least 2 weeks before delivery to pregnant women facilitates transmission of seroprotective influenza-specific antibodies to the newborn. Pediatr. Infect. Dis. J. 2013;32:1374–1380. doi: 10.1097/01.inf.0000437066.40840.c4. [DOI] [PubMed] [Google Scholar]
- Boesch A.W., Brown E.P., Cheng H.D., Ofori M.O., Normandin E., Nigrovic P.A., Alter G., Ackerman M.E. Highly parallel characterization of IgG Fc binding interactions. MAbs. 2014;6:915–927. doi: 10.4161/mabs.28808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondt A., Rombouts Y., Selman M.H., Hensbergen P.J., Reiding K.R., Hazes J.M., Dolhain R.J., Wuhrer M. Immunoglobulin G (IgG) Fab glycosylation analysis using a new mass spectrometric high-throughput profiling method reveals pregnancy-associated changes. Mol. Cell Proteomics. 2014;13:3029–3039. doi: 10.1074/mcp.M114.039537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondt A., Selman M.H., Deelder A.M., Hazes J.M., Willemsen S.P., Wuhrer M., Dolhain R.J. Association between galactosylation of immunoglobulin G and improvement of rheumatoid arthritis during pregnancy is independent of sialylation. J. Proteome Res. 2013;12:4522–4531. doi: 10.1021/pr400589m. [DOI] [PubMed] [Google Scholar]
- Bondt A., Wuhrer M., Kuijper T.M., Hazes J.M., Dolhain R.J. Fab glycosylation of immunoglobulin G does not associate with improvement of rheumatoid arthritis during pregnancy. Arthritis Res. Ther. 2016;18:274. doi: 10.1186/s13075-016-1172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein Peretz S., Regev N., Novick L., Nachshol M., Goffer E., Ben-David A., Asraf K., Doolman R., Levin E.G., Regev Yochay G., Yinon Y. Short-term outcome of pregnant women vaccinated with BNT162b2 mRNA COVID-19 vaccine. Ultrasound Obstet. Gynecol. 2021;58:450–456. doi: 10.1002/uog.23729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau C.M., Alter G. Extra-Neutralizing FcR-Mediated Antibody Functions for a Universal Influenza Vaccine. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.P., Licht A.F., Dugast A.S., Choi I., Bailey-Kellogg C., Alter G., Ackerman M.E. High-throughput, multiplexed IgG subclassing of antigen-specific antibodies from clinical samples. J. Immunol. Methods. 2012;386:117–123. doi: 10.1016/j.jim.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner C., Lehmann C., Dudziak D., Nimmerjahn F. Sweet SIGNs: IgG glycosylation leads the way in IVIG-mediated resolution of inflammation. Int. Immunol. 2017;29:499–509. doi: 10.1093/intimm/dxx053. [DOI] [PubMed] [Google Scholar]
- Bruhns P., Iannascoli B., England P., Mancardi D.A., Fernandez N., Jorieux S., Daeron M. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- Christian L.M., Beverly C., Mitchell A.M., Karlsson E., Porter K., Schultz-Cherry S., Ramilo O. Effects of prior influenza virus vaccination on maternal antibody responses: implications for achieving protection in the newborns. Vaccine. 2017;35:5283–5290. doi: 10.1016/j.vaccine.2017.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt C.S., Blanchard-Rohner G., Lemaitre B., Boukrid M., Combescure C., Othenin-Girard V., Chilin A., Petre J., De Tejada B.M., Siegrist C.A. Maternal immunization earlier in pregnancy maximizes antibody transfer and expected infant seropositivity against pertussis. Clin. Infect. Dis. 2016;62:829–836. doi: 10.1093/cid/ciw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einarsdottir H.K., Selman M.H., Kapur R., Scherjon S., Koeleman C.A., Deelder A.M., Van Der Schoot C.E., Vidarsson G., Wuhrer M. Comparison of the Fc glycosylation of fetal and maternal immunoglobulin G. Glycoconj. J. 2013;30:147–157. doi: 10.1007/s10719-012-9381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucette A.N., Unger B.L., Gonik B., Chen K. Maternal vaccination: moving the science forward. Hum. Reprod. Update. 2015;21:119–135. doi: 10.1093/humupd/dmu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore A.E., Uyeki T.M., Broder K., Finelli L., Euler G.L., Singleton J.A., Iskander J.K., Wortley P.M., Shay D.K., Bresee J.S., et al. Prevention and control of influenza with vaccines: recommendations of the advisory committee on immunization practices (ACIP), 2010. MMWR Recomm. Rep. 2010;59:1–62. [PubMed] [Google Scholar]
- Fischinger S., Fallon J.K., Michell A.R., Broge T., Suscovich T.J., Streeck H., Alter G. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods. 2019;473:112630. doi: 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förger F., Förger P.M. Immunological adaptations in pregnancy that modulate rheumatoid arthritis disease activity. Nat Rev Rheumatol. 2020;16:113–122. doi: 10.1038/s41584-019-0351-2. [DOI] [PubMed] [Google Scholar]
- Fortner K.B., Swamy G.K., Broder K.R., Jimenez-Truque N., Zhu Y., Moro P.L., Liang J., Walter E.B., Heine R.P., Moody M.A., et al. Reactogenicity and immunogenicity of tetanus toxoid, reduced diphtheria toxoid, and acellular pertussis vaccine (Tdap) in pregnant and nonpregnant women. Vaccine. 2018;36:6354–6360. doi: 10.1016/j.vaccine.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli Cao R., Christian L., Xu Z., Jaramillo L., Smith B., Karlsson E.A., Schultz-Cherry S., Mejias A., Ramilo O. Early changes in interferon gene expression and antibody responses following influenza vaccination in pregnant women. J. Infect. Dis. 2021;225:341–351. doi: 10.1093/infdis/jiab345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K.J., Bordt E.A., Atyeo C., Deriso E., Akinwunmi B., Young N., Baez A.M., Shook L.L., Cvrk D., James K., et al. Coronavirus disease 2019 vaccine response in pregnant and lactating women: a cohort study. Am. J. Obstet. Gynecol. 2021;225:303.e1–303.e17. doi: 10.1016/j.ajog.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohskopf L.A., Alyanak E., Broder K.R., Blanton L.H., Fry A.M., Jernigan D.B., Atmar R.L. Prevention and control of seasonal influenza with vaccines: recommendations of the advisory committee on immunization practices - United States, 2020-21 influenza season. MMWR Recomm. Rep. 2020;69:1–24. doi: 10.15585/mmwr.rr6908a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn B.M., Alter G. Modulating antibody functionality in infectious disease and vaccination. Trends Mol. Med. 2016;22:969–982. doi: 10.1016/j.molmed.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelholt A., Christiansen G., Sorensen U.S., Birkelund S. IgG subclass profiles in normal human sera of antibodies specific to five kinds of microbial antigens. Pathog. Dis. 2013;67:206–213. doi: 10.1111/2049-632X.12034. [DOI] [PubMed] [Google Scholar]
- Huygen K., Cabore R.N., Maertens K., Van Damme P., Leuridan E. Humoral and cell mediated immune responses to a pertussis containing vaccine in pregnant and nonpregnant women. Vaccine. 2015;33:4117–4123. doi: 10.1016/j.vaccine.2015.06.108. [DOI] [PubMed] [Google Scholar]
- Jacobsen H., Rajendran M., Choi A., Sjursen H., Brokstad K.A., Cox R.J., Palese P., Krammer F., Nachbagauer R. Influenza virus hemagglutinin stalk-specific antibodies in human serum are a surrogate marker for in vivo protection in a serum transfer mouse challenge model. MBio. 2017;8 doi: 10.1128/mBio.01463-17. e01463–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson D.J., Honein M.A., Rasmussen S.A., Williams J.L., Swerdlow D.L., Biggerstaff M.S., Lindstrom S., Louie J.K., Christ C.M., Bohm S.R., et al. H1N1 2009 influenza virus infection during pregnancy in the USA. Lancet. 2009;374:451–458. doi: 10.1016/S0140-6736(09)61304-0. [DOI] [PubMed] [Google Scholar]
- Jansen B.C., Bondt A., Reiding K.R., Lonardi E., De Jong C.J., Falck D., Kammeijer G.S., Dolhain R.J., Rombouts Y., Wuhrer M. Pregnancy-associated serum N-glycome changes studied by high-throughput maldi-Tof-ms. Sci. Rep. 2016;6:23296. doi: 10.1038/srep23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein M.F., Goldfarb I., Dolatshahi S., Cosgrove C., Noelette F.J., Krykbaeva M., Das J., Sarkar A., Gorman M.J., Fischinger S., et al. Fc glycan-mediated regulation of placental antibody transfer. Cell. 2019;178:202–215. doi: 10.1016/j.cell.2019.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D., Giallourakis C., Mostoslavsky R., Alt F.W. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kaneko Y., Nimmerjahn F., Ravetch J.V. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science. 2006;313:670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- Karsten C.B., Mehta N., Shin S.A., Diefenbach T.J., Slein M.D., Karpinski W., Irvine E.B., Broge T., Suscovich T.J., Alter G. A versatile high-throughput assay to characterize antibody-mediated neutrophil phagocytosis. J. Immunol. Methods. 2019;471:46–56. doi: 10.1016/j.jim.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosikova M., Li L., Radvak P., Ye Z., Wan X.F., Xie H. Imprinting of repeated influenza A/H3 exposures on antibody quantity and antibody quality: implications for seasonal vaccine strain selection and vaccine performance. Clin. Infect. Dis. 2018;67:1523–1532. doi: 10.1093/cid/ciy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis A.P., Read J.S., Jamieson D.J. Pregnancy and infection. N. Engl. J. Med. 2014;370:2211–2218. doi: 10.1056/NEJMra1213566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus T.A., Engel S.M., Sperling R.S., Kellerman L., Lo Y., Wallenstein S., Escribese M.M., Garrido J.L., Singh T., Loubeau M., Moran T.M. Characterizing the pregnancy immune phenotype: results of the viral immunity and pregnancy (VIP) study. J. Clin. Immunol. 2012;32:300–311. doi: 10.1007/s10875-011-9627-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus T.A., Sperling R.S., Engel S.M., Lo Y., Kellerman L., Singh T., Loubeau M., Ge Y., Garrido J.L., Rodriguez-Garcia M., Moran T.M. Peripheral blood cytokine profiling during pregnancy and post-partum periods. Am. J. Reprod. Immunol. 2010;64:411–426. doi: 10.1111/j.1600-0897.2010.00889.x. [DOI] [PubMed] [Google Scholar]
- Lagattuta K.A., Nigrovic P.A. Estrogen-driven changes in immunoglobulin G Fc glycosylation. Exp. Suppl. 2021;112:341–361. doi: 10.1007/978-3-030-76912-3_11. [DOI] [PubMed] [Google Scholar]
- Lau K.S., Juchheim A.M., Cavaliere K.R., Philips S.R., Lauffenburger D.A., Haigis K.M. In vivo systems analysis identifies spatial and temporal aspects of the modulation of TNF-alpha-induced apoptosis and proliferation by MAPKs. Sci. Signal. 2011;4:ra16. doi: 10.1126/scisignal.2001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie J.K., Acosta M., Jamieson D.J., Honein M.A., California Pandemic Working, Group Severe 2009 H1N1 influenza in pregnant and postpartum women in California. N. Engl. J. Med. 2010;362:27–35. doi: 10.1056/NEJMoa0910444. [DOI] [PubMed] [Google Scholar]
- Martinez D.R., Fong Y., Li S.H., Yang F., Jennewein M.F., Weiner J.A., Harrell E.A., Mangold J.F., Goswami R., Seage G.R., 3rd, et al. Fc characteristics mediate selective placental transfer of IgG in HIV-Infected women. Cell. 2019;178:190–201. doi: 10.1016/j.cell.2019.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli M.J., Harvey H., Morens D.M., Taubenberger J.K. Influenza in pregnancy. Influenza Other Respir. Viruses. 2013;7:1033–1039. doi: 10.1111/irv.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor G., Cardenas I. The immune system in pregnancy: a unique complexity. Am. J. Reprod. Immunol. 2010;63:425–433. doi: 10.1111/j.1600-0897.2010.00836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz F.M., Patel S.M., Jackson L.A., Swamy G.K., Edwards K.M., Frey S.E., Petrie C.R., Sendra E.A., Keitel W.A. Safety and immunogenicity of three seasonal inactivated influenza vaccines among pregnant women and antibody persistence in their infants. Vaccine. 2020;38:5355–5363. doi: 10.1016/j.vaccine.2020.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohfuji S., Deguchi M., Tachibana D., Koyama M., Takagi T., Yoshioka T., Urae A., Ito K., Kase T., Maeda A., et al. Protective effect of maternal influenza vaccination on influenza in their infants: a prospective cohort study. J. Infect. Dis. 2018;217:878–886. doi: 10.1093/infdis/jix629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan J.D., Kitaoka M., Anthony R.M. Engineered sialylation of pathogenic antibodies in vivo attenuates autoimmune disease. Cell. 2018;172:564–577.e13. doi: 10.1016/j.cell.2017.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer V., Muhsen K., Betser M., Green M.S. Antibody response to pertussis vaccination in pregnant and non-pregnant women-the role of sex hormones. Vaccines (Basel) 2021;9:637. doi: 10.3390/vaccines9060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pica N., Hai R., Krammer F., Wang T.T., Maamary J., Eggink D., Tan G.S., Krause J.C., Moran T., Stein C.R., et al. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc. Natl. Acad. Sci. U S A. 2012;109:2573–2578. doi: 10.1073/pnas.1200039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radvak P., Kosikova M., Kuo Y.C., Li X., Garner R., Schmeisser F., Kosik I., Ye Z., Weir J.P., Yewdell J.W., Xie H. Highly pathogenic avian influenza A/Guangdong/17SF003/2016 is immunogenic and induces cross-protection against antigenically divergent H7N9 viruses. NPJ Vaccin. 2021;6:30. doi: 10.1038/s41541-021-00295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.A., Watson A.K., Kennedy E.D., Broder K.R., Jamieson D.J. Vaccines and pregnancy: past, present, and future. Semin Fetal Neonatal Med. 2014;19(3):161–169. doi: 10.1016/j.siny.2013.11.014. [DOI] [PubMed] [Google Scholar]
- Regan A.K., De Klerk N., Moore H.C., Omer S.B., Shellam G., Effler P.V. Effect of maternal influenza vaccination on hospitalization for respiratory infections in newborns: a retrospective cohort study. Pediatr. Infect. Dis. J. 2016;35:1097–1103. doi: 10.1097/INF.0000000000001258. [DOI] [PubMed] [Google Scholar]
- Rolle L., Memarzadeh Tehran M., Morell-Garcia A., Raeva Y., Schumacher A., Hartig R., Costa S.D., Jensen F., Zenclussen A.C. Cutting edge: IL-10-producing regulatory B cells in early human pregnancy. Am. J. Reprod. Immunol. 2013;70:448–453. doi: 10.1111/aji.12157. [DOI] [PubMed] [Google Scholar]
- Ruhaak L.R., Uh H.W., Deelder A.M., Dolhain R.E., Wuhrer M. Total plasma N-glycome changes during pregnancy. J. Proteome Res. 2014;13:1657–1668. doi: 10.1021/pr401128j. [DOI] [PubMed] [Google Scholar]
- Saso A., Kampmann B. Maternal immunization: nature meets nurture. Front. Microbiol. 2020;11:1499. doi: 10.3389/fmicb.2020.01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautto G.A., Kirchenbaum G.A., Ross T.M. Towards a universal influenza vaccine: different approaches for one goal. Virol. J. 2018;15:17. doi: 10.1186/s12985-017-0918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaudecker E.P., Ambroggio L., Mcneal M.M., Finkelman F.D., Way S.S. Declining responsiveness to influenza vaccination with progression of human pregnancy. Vaccine. 2018;36:4734–4741. doi: 10.1016/j.vaccine.2018.05.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaudecker E.P., Mcneal M.M., Dodd C.N., Ranz J.B., Steinhoff M.C. Pregnancy modifies the antibody response to trivalent influenza immunization. J. Infect. Dis. 2012;206:1670–1673. doi: 10.1093/infdis/jis592. [DOI] [PubMed] [Google Scholar]
- Schumacher A., Costa S.D., Zenclussen A.C. Endocrine factors modulating immune responses in pregnancy. Front. Immunol. 2014;5:196. doi: 10.3389/fimmu.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman M.H., De Jong S.E., Soonawala D., Kroon F.P., Adegnika A.A., Deelder A.M., Hokke C.H., Yazdanbakhsh M., Wuhrer M. Changes in antigen-specific IgG1 Fc N-glycosylation upon influenza and tetanus vaccination. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014563. M111.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R.L., Lai J., Keck R., O'connell L.Y., Hong K., Meng Y.G., Weikert S.H., Presta L.G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002;277:26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- Shinkawa T., Nakamura K., Yamane N., Shoji-Hosaka E., Kanda Y., Sakurada M., Uchida K., Anazawa H., Satoh M., Yamasaki M., et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003;278:3466–3473. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- Shook L.L., Fallah P.N., Silberman J.N., Edlow A.G. Covid-19 vaccination in pregnancy and lactation: current Research and gaps in understanding. Front. Cell Infect. Microbiol. 2021;11:735394. doi: 10.3389/fcimb.2021.735394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siston A.M., Rasmussen S.A., Honein M.A., Fry A.M., Seib K., Callaghan W.M., Louie J., Doyle T.J., Crockett M., Lynfield R., et al. Pandemic 2009 influenza A(H1N1) virus illness among pregnant women in the United States. JAMA. 2010;303:1517–1525. doi: 10.1001/jama.2010.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling R.S., Engel S.M., Wallenstein S., Kraus T.A., Garrido J., Singh T., Kellerman L., Moran T.M. Immunogenicity of trivalent inactivated influenza vaccination received during pregnancy or postpartum. Obstet. Gynecol. 2012;119:631–639. doi: 10.1097/AOG.0b013e318244ed20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff M.C., Katz J., Englund J.A., Khatry S.K., Shrestha L., Kuypers J., Stewart L., Mullany L.C., Chu H.Y., LeClerq S.C., Kozuki N., et al. Year-round influenza immunisation during pregnancy in Nepal: a phase 4, randomised, placebo-controlled trial. Lancet Infect Dis. 2017;17(9):981–989. doi: 10.1016/S1473-3099(17)30252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J., Corper A.L., Basler C.F., Taubenberger J.K., Palese P., Wilson I.A. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Stewart R., Hammond S.A., Oberst M., Wilkinson R.W. The role of Fc gamma receptors in the activity of immunomodulatory antibodies for cancer. J. Immunother. Cancer. 2014;2:29. [Google Scholar]
- Takai T. Roles of Fc receptors in autoimmunity. Nat. Rev. Immunol. 2002;2:580–592. doi: 10.1038/nri856. [DOI] [PubMed] [Google Scholar]
- Thompson M.G., Kwong J.C., Regan A.K., Katz M.A., Drews S.J., Azziz-Baumgartner E., Klein N.P., Chung H., Effler P.V., Feldman B.S., et al. Influenza vaccine effectiveness in preventing influenza-associated hospitalizations during pregnancy: a multi-country retrospective test negative design study, 2010-2016. Clin. Infect. Dis. 2019;68:1444–1453. doi: 10.1093/cid/ciy737. [DOI] [PubMed] [Google Scholar]
- Van De Bovenkamp F.S., Hafkenscheid L., Rispens T., Rombouts Y. The emerging importance of IgG Fab glycosylation in immunity. J. Immunol. 2016;196:1435–1441. doi: 10.4049/jimmunol.1502136. [DOI] [PubMed] [Google Scholar]
- Van De Geijn F.E., Wuhrer M., Selman M.H., Willemsen S.P., De Man Y.A., Deelder A.M., Hazes J.M., Dolhain R.J. Immunoglobulin G galactosylation and sialylation are associated with pregnancy-induced improvement of rheumatoid arthritis and the postpartum flare: results from a large prospective cohort study. Arthritis Res. Ther. 2009;11:R193. doi: 10.1186/ar2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek I., Dieussaert I., Doherty T.M., Franck V., Hanssens L., Miller J., Bekkat-Berkani R., Kandeil W., Prado-Cohrs D., Vyse A. Maternal immunization: where are we now and how to move forward? Ann. Med. 2018;50:193–208. doi: 10.1080/07853890.2017.1421320. [DOI] [PubMed] [Google Scholar]
- Von Holle T.A., Moody M.A. Influenza and antibody-dependent cellular cytotoxicity. Front. Immunol. 2019;10:1457. doi: 10.3389/fimmu.2019.01457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn N., Schwab I., Ortiz D., Bhatnagar N., Lansing J.C., Medeiros A., Tyler S., Mekala D., Cochran E., Sarvaiya H., et al. Controlled tetra-Fc sialylation of IVIg results in a drug candidate with consistent enhanced anti-inflammatory activity. Proc. Natl. Acad. Sci. U S A. 2015;112:E1297–E1306. doi: 10.1073/pnas.1422481112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Iwatani Y., Kaneda T., Hidaka Y., Mitsuda N., Morimoto Y., Amino N. Changes in T, B, and NK lymphocyte subsets during and after normal pregnancy. Am. J. Reprod. Immunol. 1997;37:368–377. doi: 10.1111/j.1600-0897.1997.tb00246.x. [DOI] [PubMed] [Google Scholar]
- WHO Global Influenza Programme. Recommendations for influenza vaccine composition. https://www.who.int/teams/global-influenza-programme/vaccines/who-recommendations.
- Wu N.C., Wilson I.A. A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J. Mol. Biol. 2017;429:2694–2709. doi: 10.1016/j.jmb.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Jing X., Li X., Lin Z., Plant E., Zoueva O., Yang H., Ye Z. Immunogenicity and cross-reactivity of 2009-2010 inactivated seasonal influenza vaccine in US adults and elderly. PLoS One. 2011;6:e16650. doi: 10.1371/journal.pone.0016650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Wan X.F., Ye Z., Plant E.P., Zhao Y., Xu Y., Li X., Finch C., Zhao N., Kawano T., et al. H3N2 mismatch of 2014-15 northern hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci. Rep. 2015;5:15279. doi: 10.1038/srep15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee J., Kim W., Han J.M., Yoon H.Y., Lee N., Lee K.E., Gwak H.S. Clinical manifestations and perinatal outcomes of pregnant women with COVID-19: a systematic review and meta-analysis. Sci. Rep. 2020;10:18126. doi: 10.1038/s41598-020-75096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano L.D., Ellington S., Strid P., Galang R.R., Oduyebo T., Tong V.T., Woodworth K.R., Nahabedian J.F., 3rd, Azziz-Baumgartner E., Gilboa S.M., et al. Update: characteristics of symptomatic women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status - United States, January 22-October 3, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:1641–1647. doi: 10.15585/mmwr.mm6944e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.