Abstract
Acinetobacter baumannii RYC 52763/97, a clinical isolate involved in a prolonged nosocomial outbreak at our hospital, was resistant to all β-lactams tested, including imipenem and meropenem, which had MICs of 128 and 256 μg/ml, respectively. This strain synthesized three β-lactamases: a plasmid-mediated TEM-1 β-lactamase (pI 5.4), an AmpC-type chromosomal cephalosporinase (pI 9.4), and a novel, presumptively chromosomally mediated OXA-related enzyme (pI 9.0) named OXA-24. After cloning and sequencing, the deduced amino acid sequence of the OXA-24 β-lactamase showed 40% homology with the OXA-10 (PSE-2) and OXA-7 β-lactamases, 39% homology with the OXA-11 and OXA-5 enzymes, and 33% homology with the LCR-1 β-lactamase. The amino acid sequence of the OXA-24 β-lactamase contained the STFK motif found in serine β-lactamases, but the typical class D triad KTG was replaced by KSG and the motif YGN was replaced by FGN. The OXA-24 β-lactamase hydrolyzed benzylpenicillin and cephaloridine but lacked activity against oxacillin, cloxacillin, and methicillin. The enzymatic activity was inhibited by chloride ions and by tazobactam (50% inhibitory concentration [IC50], 0.5 μM), sulbactam (IC50, 40 μM), and clavulanic acid (IC50, 50 μM). Carbapenem MICs for an Escherichia coli transformant (pBMB-1) expressing the cloned OXA-24 enzyme had a fourfold increase. Relative Vmax/Km values of 13 and 6 were obtained with imipenem and meropenem, respectively, and a positive microbiological assay result with imipenem was obtained with a purified enzymatic extract of this transformant strain. Therefore, we consider this new β-lactamase to be involved in the carbapenem resistance of A. baumannii RYC 52763/97.
Acinetobacter spp. are opportunistic pathogens with increasing relevance in nosocomial infections (5). They cause a wide range of clinical complications, such as pneumonia, septicemia, urinary tract infection, wound infection, and meningitis, especially in immunocompromised patients (21). Antimicrobial treatment of these clinical infections, particularly those caused by Acinetobacter baumannii clinical strains, may be compromised by multiple-drug resistance to β-lactams, aminoglycosides, and fluoroquinolones (4, 22). Regarding β-lactam antibiotics, different mechanisms are involved in the resistance of A. baumannii clinical strains, although as with other gram-negative rods, the main mechanism of resistance is the production of β-lactamases encoded either by the chromosome or by plasmids (3). The plasmid-mediated β-lactamases TEM-1, TEM-2, and CARB-5 have frequently been found in Acinetobacter spp. (19, 36). Moreover, the presence of different extended-spectrum β-lactamases, such as PER-1, ARI-1, ARI-2, and another as-yet-unnamed class D β-lactamase in A. baumannii clinical strains has recently been reported (7, 18, 27, 34, 37). On the other hand, the presence of different chromosomal cephalosporinases has also been reported (3, 28), and recently we reported the cloning, sequencing, and analysis of an ampC gene from an A. baumannii epidemic strain (6). In addition, the low permeability of the outer membrane of A. baumannii, resulting from small outer membrane pore size and/or limited porin production, has been involved in β-lactam resistance (3, 32). In the last few years, carbapenem-resistant A. baumannii isolates have been reported worldwide (1, 14, 20), and the loss of porins, penicillin-binding protein with reduced affinity, the ARI-1 and ARI-2 β-lactamases, and an oxacillin-hydrolyzing β-lactamase have been associated with resistance to carbapenems in A. baumannii clinical strains (7, 9, 16, 18, 27).
During the year 1997, a 10-month-long outbreak at our institution involving 29 patients, 23 of them hospitalized in five intensive care units, was caused by an imipenem- and meropenem-resistant A. baumannii strain (G. Bou, G. Cerveró, D. Malpica, M. Pérez-Vázquez, L. De Rafael, and J. Martínez-Beltrán, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. K-120, 1998). By isoelectric focusing, the sonicated extract of this epidemic strain showed, apart from TEM-1 and an AmpC-like β-lactamase, an unknown β-lactamase that focused at pI 9.0. The main purpose of the present work was to clone and sequence the gene encoding this enzyme.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A. baumannii RYC 52763/97 is a carbapenem-resistant clinical strain isolated in February 1997 from a bronchial aspirate of a patient admitted to the Medical Intensive Care Unit in the Ramón y Cajal Hospital, Madrid, Spain. It was the initial isolate obtained from the first patient involved in the carbapenem-resistant A. baumannii nosocomial outbreak occurring in 1997 at our hospital (Bou et al., 38th ICAAC). A. baumannii RYC 30222/97, a carbapenem-susceptible strain isolated in April 1997 from blood cultures of another patient treated in the hospital, was used for comparison. The two strains differed widely in their antimicrobial susceptibility patterns and were shown to be epidemiologically unrelated (Bou et al. 38th ICAAC). Escherichia coli BM21 [F− gyrA (λ+), nalidixic acid resistant] and Acinetobacter junii MA RYC95 (ampicillin susceptible) were used as recipients in conjugation experiments. E. coli TG1 [Δ(lac-pro) hsdD5 supE thi] was used as a host for plasmids. Plasmid pBGS18, carrying a kanamycin resistance marker (33), was used for cloning the OXA gene. Plasmid pUC18, carrying an ampicillin resistance marker (38), was used for nucleotide sequence reactions.
Antimicrobial agents and susceptibility tests.
The antimicrobial agents used in this study were kindly supplied in the form of standard laboratory powders of known potency by the indicated sources: ampicillin, clavulanic acid, ticarcillin, methicillin, oxacillin, and cloxacillin by SmithKline Beecham, Madrid, Spain; piperacillin and tazobactam by Wyeth Lederle, Madrid, Spain; sulbactam by Pfizer, Madrid, Spain; cephaloridine, cefazolin, and tobramycin by Lilly, Madrid, Spain; cefuroxime and ceftazidime by Glaxo-Wellcome, Madrid, Spain; cefotaxime and cefpirome by Hoechst Marion-Roussel, Barcelona, Spain; aztreonam and cefepime by Bristol-Myers Squibb, Madrid, Spain; cefoxitin and imipenem by Merck Sharp & Dohme, Madrid, Spain; meropenem by Zeneca Farma, Madrid, Spain; benzylpenicillin by Sigma, Madrid, Spain; and ciprofloxacin by Bayer, Barcelona, Spain. All antibiotic solutions were prepared immediately before use.
Susceptibility testing was performed by agar dilution in accordance with the guidelines of the National Committee for Clinical Laboratory Standards (26). MICs were determined with serial twofold dilutions of antibiotics in Mueller-Hinton agar (Oxoid, Basingstoke, United Kingdom) with an inoculum of about 104 CFU per spot. MICs of ampicillin, ceftazidime, and imipenem were determined alone and in combination with a fixed concentration of 4 μg of clavulanic acid, sulbactam, and tazobactam/ml. E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 (reference strains) were used as controls for MIC determinations.
Analytical isoelectric focusing.
β-Lactamases were characterized by isoelectric focusing of ultrasonic bacterial extracts (24). Bacteria growing exponentially at 37°C in Luria-Bertani (LB) medium were harvested, and cell-free lysates were prepared by sonication (35). β-Lactamases were analyzed by isoelectric focusing of cell extracts on polyacrylamide gels containing ampholytes with a pH range of 3.5 to 9.5 (Ampholine PAGplate; Pharmacia Biotech) in a Multiphor II system (Pharmacia-LKB). The focused β-lactamases were detected by overlaying the gel with nitrocefin (0.5 mg/ml) in phosphate buffer (100 mM, pH 7.0). pI values were determined by comparison with those of β-lactamases with known pI: TEM-1 (5.4), TEM-3 (6.3), SHV-1 (7.6), MIR-1 (8.4), and A. baumannii RYC 52763/97 AmpC (9.4).
Conjugation experiments.
Transfer of resistance by conjugation was attempted using strains E. coli BM21 and A. junii MA RYC95 as recipients. Overnight filter mating experiments were performed at 30 and 37°C, and the transconjugants were selected on MacConkey agar plates supplemented with ampicillin (25 μg/ml) and nalidixic acid (50 μg/ml) for E. coli and on Columbia agar plates supplemented with d-glucose (2%, wt/vol), neutral red, and ampicillin (25 μg/ml) for A. junii.
DNA extraction.
The A. baumannii RYC 52763/97 strain was grown overnight on MacConkey agar plates at 37°C, and growth from approximately one-quarter of a plate was resuspended in 180 μl of distilled water. A total of 200 μl of buffer solution (0.01 M Tris-Cl [pH 7.8], 0.005 M EDTA, 0.5% sodium dodecyl sulfate) and 20 μl of proteinase K (1 mg/ml) were added. The mixture was incubated at 55°C for 2 h, and then 400 μl of a phenol-chloroform solution was added, mixed with gentle agitation, and centrifuged at 11,000 × g for 5 min. The supernatant was collected and DNA was precipitated after the addition of 0.5 volume of 7.5 M ammonium acetate and 2 volumes of ethanol. DNA was washed with 70% ethanol, dried, and resuspended with 100 μl of the Tris-EDTA buffer.
Cloning experiments and DNA sequencing.
Plasmid purifications by the alkaline lysis method and cloning procedures were performed as described by Sambrook et al. (29). Restriction enzymes were purchased from Boehringer (Mannheim, Germany) and were used according to the manufacturer's directions. The pI 5.4 bla gene carried on the 22-kb plasmid pAB1 was cloned into plasmid pBGS18 after amplification by PCR using blaTEM-specific primers C1 (5′-GGGAATTCTCGGGGAAATGTGCGCGGAAC) and C2 (5′-GGGATCCGAGTAAACTTGGTCTGACAG) (TEM-1 type). For cloning the pI 9.0 bla gene, chromosomal DNA from A. baumannii RYC 52763/97 was digested with restriction enzyme BglII. The resulting fragments were ligated into the pBGS18 plasmid digested with the restriction enzyme BamHI, and the mixture was transformed into E. coli TG1 made competent by the calcium chloride method. After transformation, a few clones grew on kanamycin (10 μg/ml) and ampicillin (25 μg/ml) LB plates. They harbored an identical plasmid with an insert of about 4.5 kb. The bla gene of this plasmid was subcloned into pBGS18 with XbaI, yielding plasmid pBMB-1 with an insert of 1.5 kb.
To determine the nucleotide sequence, the insert was subcloned into pUC18, yielding plasmid pBMB-2. Templates were sequenced on both strands by the method of Sanger et al. (30). Sequencing was carried out with the Taq DyeDeoxiTerminator cycle sequencing kit using primers specific to the coding sequence, and the sequence was analyzed in an automatic DNA sequencer (377 ABI Prism; Perkin-Elmer).
Determination of kinetic and inhibition parameters.
For kinetic studies, a cell-free lysate was obtained by sonication of the sediment from a 1-liter exponentially growing culture of E. coli TG1 harboring the OXA-24 enzyme (pBMB-1 plasmid) at 37°C in LB broth containing 50 μg of ampicillin per ml. The sonicated extract was dialyzed overnight at 4°C in 0.05 M phosphate buffer (pH 7.4) and then loaded into a 300-ml (75- by 2.5-cm) Sephadex G100 column (Pharmacia Fine Chemicals AB, Uppsala, Sweden) previously equilibrated with the same buffer. The β-lactamase was eluted with 0.05 M phosphate buffer (pH 7.4), and its activity was tested with the nitrocefin method. Fractions containing β-lactamase activity were collected, concentrated with Centricon (Amicon B15; W. R. Grace and Co., Danvers, Mass.), stored for a maximum of 1 week at −70°C, and used for the determination of kinetic constants.
Initial hydrolysis rates were monitored spectrophotometrically (UVIKON-930) at 25°C in 0.05 M phosphate buffer (pH 7.4). β-Lactamase activities were determined by measuring the change in absorbance for the following antibiotics at the indicated wavelengths: benzylpenicillin, 235 nm; cephaloridine, 295 nm; oxacillin, cloxacillin, and methicillin, 263 nm; cefotaxime and ceftazidime, 257 nm; cefepime, 260 nm; and imipenem and meropenem, 299 nm. Kinetic parameters were determined in duplicate experiments by making linear plots of the initial steady-state rates at different substrate concentrations (Lineweaver-Burk transformation). Hydrolysis rates were calculated using saturation concentrations of the substrate, and apparent Km and Vmax values were calculated for comparison of the enzyme activities by using the program Excel 5.0.
Inhibition assays were carried out by preincubation of different concentrations of clavulanic acid, sulbactam, and tazobactam with the OXA-24 β-lactamase extract for 10 min at 37°C before testing the rate of nitrocefin (25 μg/ml) hydrolysis. The concentration required to inhibit 50% of enzyme activity (IC50) was determined by spectrophotometric assay at 37°C. Moreover, inhibition of enzymatic activity by NaCl and EDTA was assayed using nitrocefin as substrate by measuring the residual β-lactamase activity after incubation of the OXA-24 extract for 10 min at 37°C in the presence of a 1 mM concentration of both compounds.
Microbiological assay of β-lactamase activity.
In order to study the inactivation of imipenem by the A. baumannii OXA-24 β-lactamase, a microbiological disk assay was performed with a modification of the method of Masuda et al. (23). An imipenem disk (10 μg) was placed in the center of a Mueller-Hinton agar plate seeded with the E. coli ATCC 25922 strain. Four filter paper disks, one each containing 20, 10, or 5 μl of the enzyme preparation or 20 μl of sodium phosphate buffer (pH 7.0), were applied 15 mm from the imipenem disk. Plates were incubated at 37°C overnight, and inactivation of imipenem was shown by growth of the indicator strain within the expected inhibition zone.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been assigned EMBL accession number AJ239129.
RESULTS
Antibiotic susceptibility pattern.
The antimicrobial susceptibility profile of each strain included in this study is shown in Table 1. A. baumannii RYC 52763/97 showed a high level of resistance to all β-lactam antibiotics tested, with MICs of imipenem and meropenem of 128 and 256 μg/ml, respectively. This high level of β-lactam resistance was not restored with clavulanic acid, sulbactam, or tazobactam. With the exception of tobramycin (MIC, 4 μg/ml) and colistin (MIC, 4 μg/ml), the multiresistance pattern of this strain included also resistance to gentamicin, amikacin, and ciprofloxacin. In contrast, A. baumannii RYC 30222/97, the other clinical strain studied for comparison, was susceptible to ticarcillin, extended-spectrum cephalosporins, and carbapenems. All attempts to transfer the β-lactam resistance by conjugation from A. baumannii RYC 52763/97 to E. coli and A. junii recipient strains were unsuccessful. On the other hand, MICs of β-lactams for E. coli TG1 harboring recombinant plasmid pBMB-1 carrying the blaOXA-24 gene indicated resistance to penicillins; susceptibility to extended-spectrum cephalosporins, albeit a slight increase of ceftazidime, cefepime, cefpirome, and aztreonam MICs; and a moderate decrease in sensitivity to carbapenems, with MICs of imipenem (1 μg/ml) and meropenem (0.125 μg/ml) fourfold higher than that for the E. coli TG1 host strain.
TABLE 1.
MICs for A. baumannii RYC 52763/97, recipient strain E. coli TG1, clone E. coli TG1 (OXA-24) harboring the plasmid pBMB-1, and the A. baumannii RYC 30222/97 clinical strain
| Antibiotic(s) | MIC (μg/ml) for:
|
|||
|---|---|---|---|---|
| A. baumannii RYC 52763/97 (OXA-24, AmpC, TEM-1) | E. coli TG1 | E. coli TG1(pBMB-1) (OXA-24) | A. baumannii RYC 30222/97 | |
| Ampicillin | >1,024 | 4 | 128 | 32 |
| Ampicillin + clavulanic acida | >1,024 | 4 | 64 | 16 |
| Ampicillin + sulbactama | >1,024 | 2 | 64 | 4 |
| Ampicillin + tazobactama | >1,024 | 2 | 32 | 16 |
| Ticarcillin | >1,024 | 4 | 256 | 8 |
| Cefazolin | >256 | 8 | 32 | >256 |
| Cefuroxime | >256 | 4 | 4 | 64 |
| Cefoxitin | >256 | 4 | 4 | 128 |
| Cefotaxime | >256 | 0.06 | 0.06 | 8 |
| Ceftazidime | >256 | 0.12 | 0.25 | 2 |
| Ceftazidime + clavulanic acid | >256 | 0.12 | 0.25 | 2 |
| Ceftazidime + sulbactam | >256 | 0.12 | 0.25 | 0.5 |
| Ceftazidime + tazobactam | >256 | 0.12 | 0.12 | 0.5 |
| Cefepime | 256 | 0.06 | 0.12 | 4 |
| Cefpirome | >256 | 0.06 | 0.12 | 8 |
| Aztreonam | >256 | 0.06 | 0.12 | 16 |
| Imipenem | 128 | 0.12 | 1 | 0.25 |
| Imipenem + clavulanic acid | 128 | 0.12 | 1 | 0.25 |
| Imipenem + sulbactam | 128 | 0.12 | 1 | 0.06 |
| Imipenem + tazobactam | 64 | 0.12 | 0.5 | 0.06 |
| Meropenem | 256 | 0.03 | 0.125 | 0.5 |
| Tobramycin | 4 | 0.12 | 0.12 | 0.5 |
| Ciprofloxacin | 128 | 0.03 | 0.03 | 0.25 |
Concentration of β-lactamase inhibitor was 4 μg/ml.
β-Lactamase and plasmids of A. baumannii RYC 52763/97.
By isoelectric focusing, the sonicated extract of A. baumannii RYC 52763/97 showed three β-lactamase activity bands at pIs 5.4, 9.0, and 9.4. The β-lactamase of pI 5.4 (TEM-1 type) was identified in the only plasmid (pAB1) revealed by electrophoresis in the strain. The blaTEM-1 gene was cloned by PCR with blaTEM-specific primers described in Materials and Methods. The β-lactamase focusing at pI 9.4 was chromosomally mediated, and after cloning, nucleotide sequencing revealed homology with AmpC β-lactamases (6). The third β-lactamase, focused at pI 9.0, failed to be transferred by conjugation experiments but was detected as a single band in E. coli TG1 harboring recombinant plasmid pBMB-1. This previously unknown enzyme was genetically and biochemically characterized in this study.
Cloning and sequencing of the OXA-24 gene.
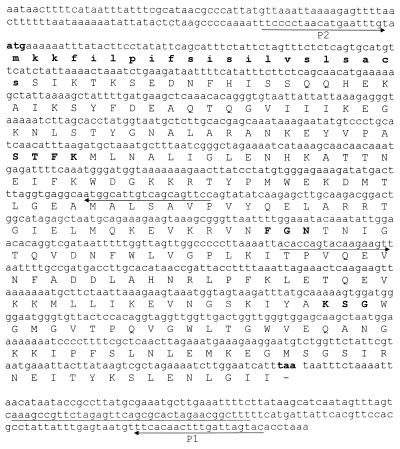
Chromosomal DNA from the A. baumannii RYC 52763/97 strain was digested with the restriction enzyme BglII. The resulting fragments were ligated into the pBGS18 plasmid digested with the restriction enzyme BamHI, and the reaction mixture was transformed into E. coli TG1 competent cells. All transformants harbored an identical plasmid with an insert of about 4.5 kb. The bla gene was subcloned by enzymatic restriction with XbaI, yielding the plasmid pBMB-1 with an insert of 1.5 kb. In order to determine the nucleotide sequences, the insert was subcloned into the pUC18 plasmid, yielding the plasmid pBMB-2. This cloned DNA fragment was entirely sequenced on both strands, and analysis of this insert for coding regions revealed one 825-bp open reading frame encoding a 274-amino-acid protein. The nucleotide sequence of the open reading frame and the deduced amino acid sequence are shown in Fig. 1. The amino acid sequence of the OXA-24 β-lactamase contained the motif found in serine β-lactamases: the active site of the enzyme contained a serine-threonine-phenyalanine-lysine tetrad (STFK). However, the typical motifs tyrosine-glycine-asparagine (YGN) and lysine-threonine-arginine (KTG), which are characteristic of other class D β-lactamases, were replaced in the OXA-24 enzyme by FGN and KSG, respectively. The nucleotide sequence of the flanking regions of the OXA-24 gene (about 400 bp on each side) did not show inverted repeated sequences suggestive of the presence of a transposable element. The OXA-24 gene was probably not inserted into an integron, since the 59-base element (specific to gene cassettes inserted in integron structures) was not observed on the flanking regions. How the OXA-24 gene was inserted into the chromosomal DNA remains to be elucidated.
FIG. 1.
Nucleotide sequence of the blaOXA-24 gene. The boldface atg and taa represent the initiation and termination codons, respectively. The deduced amino acid sequence of OXA-24 is shown below the nucleotide sequence. Amino acids of the signal peptide, written in lowercase and bold letters, were identified using the Swiss-Prot annotated protein sequence database (http://www.expasy.ch/sprot/sprot-top.html/). The β-lactamase active site STFK, the typical motif FGN, and the triad KSG are indicated by bold letters. The locations of the primers used to sequence the gene are underlined.
To verify the chromosomal localization of the OXA-24 gene, cesium chloride gradients were used (29). Plasmid DNA and chromosomal DNA were separated, and each template was used for cloning experiments. By the PCR technique, only a TEM-1 gene was found in the plasmid preparation, while the OXA gene was exclusively located in the chromosomal DNA. To check for putative plasmid contamination, a PCR assay using TEM-specific primers was done with chromosomal DNA as a template, and again a negative result was obtained. Therefore, only the TEM-1 gene can be assigned to the plasmid. Moreover, the nucleotide sequence of the flanking regions of the OXA-24 gene did not show the presence of any known plasmid gene. In contrast, homology with several eucaryotic chromosomal genes (up to 53%) were obtained (EMBL accession numbers A1055840 and AC013607).
Nucleotide and peptide sequence alignment.
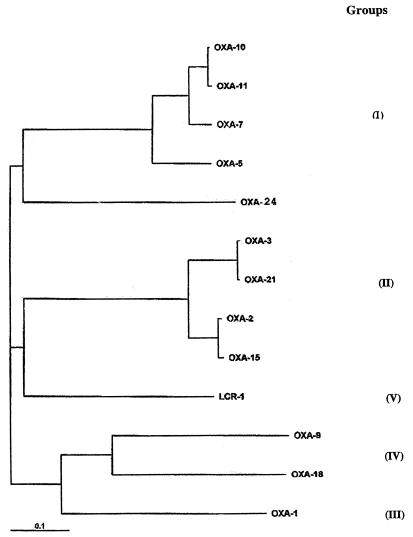
The nucleotide and peptide sequences of the OXA-24 β-lactamase were compared with those of known oxacillinases, and a dendrogram was constructed to relate OXA-24 to 12 other class D β-lactamases (Fig. 2). The highest similarity ratio for the OXA-24 β-lactamase in the Swiss-Prot database was obtained with the OXA-10 and OXA-7 (40%) and OXA-11 and OXA-5 (39%) β-lactamases. Considering the molecular homology, the OXA-24 β-lactamase may be assigned to oxacillinases belonging to group I (31).
FIG. 2.
Phylogram relating OXA-24 to 12 other class D β-lactamases. Analysis was performed using the CLUSTAL W multiple sequence alignment program. A gap penalty of three was applied, and 100 bootstrap replications were applied.
OXA-24 kinetic and inhibition profiles and imipenem hydrolysis.
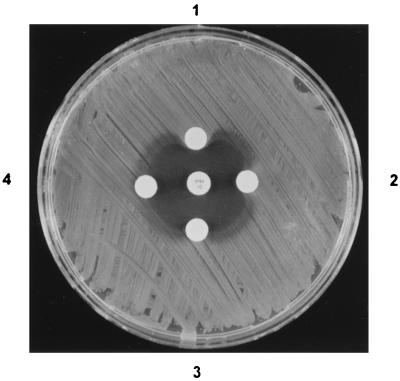
The kinetic and inhibition parameters of the OXA-24 β-lactamase, obtained with a partially purified extract of E. coli TG1 harboring the recombinant plasmid pBMB-1, are summarized in Table 2. The enzyme hydrolyzed benzylpenicillin and cephaloridine, but surprisingly, hydrolysis of oxacillin, cloxacillin, and methicillin was not detected, in contrast to other class D β-lactamases. With regard to carbapenems, the OXA-24 β-lactamase showed a moderate rate of hydrolysis, with relative Vmax/Km values of 13 and 6 for imipenem and meropenem, respectively. In addition, a positive Masuda test result was also obtained with imipenem, and this result confirmed imipenem inactivation by the OXA-24 β-lactamase (Fig. 3). These results correlated well with the increase in the carbapenem MICs obtained for the E. coli TG1 strain harboring the OXA-24 β-lactamase (pBMB-1).
TABLE 2.
Kinetic and inhibition parameters of the OXA-24 β-lactamasea
| Antibiotic | Km (μM) | Vmax (μmol/min/μl) | Relative Vmax/Kmb | Relative hydrolysis rateb | IC50 (μM)c |
|---|---|---|---|---|---|
| Benzylpenicillin | 65 | 370 | 100 | 100 | |
| Cephaloridine | 395 | 200 | 9 | 64 | |
| Oxacillin | NDd | ND | ND | <1 | |
| Cloxacillin | ND | ND | ND | <1 | |
| Methicillin | ND | ND | ND | <1 | |
| Imipenem | 20 | 15 | 13 | 4 | |
| Meropenem | 775 | 280 | 6 | 2 | |
| Clavulanic acid | 50 | ||||
| Sulbactam | 40 | ||||
| Tazobactam | 0.5 |
Kinetic experiments were performed using a protein extract with a specific activity of 13.61 μmol of nitrocefin hydrolyzed/min/μg of protein.
Obtained by normalizing the value of each antibiotic to that for benzylpenicillin (taken as 100).
Nitrocefin was used as the substrate (25 μg/ml) following 10 min of preincubation of enzyme and inhibitor.
ND, hydrolysis not detected.
FIG. 3.
Microbiological assay plate showing inactivation of imipenem (central disk) by the A. baumannii OXA-24 enzyme. (1) Twenty microliters of semipurified OXA-24 protein (nitrocefin-specific activity, 251.5 μmol/min/μl); (2) 10 μl of the same extract; (3) 5 μl of the same extract; (4) 20 μl of phosphate-buffered saline.
The results of the IC50 determination of enzyme activity with nitrocefin as the substrate were as follows: clavulanic acid, 50 μM; sulbactam, 40 μM; and tazobactam, 0.5 μM. The activity of the enzyme was inhibited by NaCl but not by EDTA.
DISCUSSION
In this work we describe a novel oxacillinase, OXA-24, produced by a multiresistant A. baumannii clinical strain; this oxacillinase possesses structural elements which are characteristic of class D β-lactamases. Experimental data suggest that the OXA-24 enzyme is mediated chromosomally and has a moderate hydrolytic activity against carbapenems. This activity correlates with the moderate increase in the MICs of imipenem and meropenem observed for E. coli TG1 harboring the blaOXA-24 gene.
Oxacillinases are enzymes belonging to the molecular class D β-lactamases (2) and are included in group 2d of the classification of Bush et al. (8). From a biochemical point of view, these enzymes are characterized by their hydrolytic activity for isoxazolyl penicillins and methicillin. Twenty-three oxacillinases have been characterized so far, and several of them are derived from OXA-2 (11), OXA-3 (37), and OXA-10 (10, 12, 13, 17, 25) and are mainly produced by P. aeruginosa. They hydrolyze extended-spectrum cephalosporins and aztreonam and are considered class D extended-spectrum β-lactamases. The phylogenetic tree of the OXA β-lactamases shows at least five groups on the basis of their amino acid sequences (31).
Comparison of the OXA-24 protein with the oxacillinases belonging to group I reveals a homology (39 to 40%) of the OXA-24 β-lactamase with the OXA-10, OXA-7, OXA-11, and OXA-5 β-lactamases, thus allowing the inclusion of OXA-24 in group I. Despite these similarities, some interesting and differing features between previous oxacillinases and this new β-lactamase are worth mentioning. (i) In contrast with the other oxacillinases, OXA-24 lacks hydrolytic activity against oxacillin, cloxacillin, and methicillin. (ii) With the exception of the ARI-1 β-lactamase (OXA-23) (15, 27) and another, yet-unnamed oxacillinase defined only by its biochemical properties (18), both involved in the carbapenem resistance of A. baumannii, the antibiotic susceptibility pattern associated with OXA-24 in E. coli TG1 differs from that of previous oxacillinases, displaying a moderate level of resistance to carbapenems. (iii) In contrast with most oxacillinases, OXA-24, like ARI-1 (OXA-23) and the other, unnamed oxacillinase, showed by spectrophotometry and bioassay a moderate hydrolysis of imipenem and meropenem.
Whether these biochemical properties result from changes in the primary structure of the OXA enzyme is currently unknown. Certainly, in the OXA-24 β-lactamase, the typical class D triad KTG is replaced by the KSG domain, and in the typical motif YGN, tyrosine is replaced by phenylalanine (FGN). Although the relevance of these substitutions to the enzymatic properties of the protein remains to be elucidated, it is important to remark that the ARI-1 (OXA-23) enzyme also contains the FGN replacement, and this β-lactamase also produces a moderate carbapenem hydrolysis and increases the carbapenem resistance levels in A. baumannii (15, 27; H. M. Donald, S. G. B. Amyes, and H. K. Young, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1462, 1999). Therefore, this amino acid change may be involved in the structural requirement for carbapenem hydrolysis; however, mutagenesis experiments are necessary to confirm this hypothesis. On the other hand, the presence of aspartic acid at position 157 in different extended-spectrum oxacillinases has been associated with the high level of ceftazidime resistance in P. aeruginosa (10, 12, 17). The low level of ceftazidime resistance conferred by OXA-24 on E. coli TG1 could be associated with a lack of aspartic acid at that position in the amino acid sequence.
In the last few years, there has been a growing concern about infections caused by carbapenem-resistant A. baumannii strains (1, 14, 20). The main mechanisms of resistance to β-lactams have been reported for A. baumannii strains with resistance to carbapenems. Thus, new β-lactamases (such as PER-1, ARI-1, ARI-2, and the one as-yet-unnamed oxacillinase), diminished permeability, and penicillin-binding protein changes have been associated with resistance to carbapenems in A. baumannii strains (3, 7, 9, 16, 18, 27). Experimental data obtained in this study reveal a putative role for the OXA-24 β-lactamase in carbapenem resistance: enzymatic imipenem hydrolysis and increased carbapenem MICs for E. coli TG1 transformants harboring the OXA-24 gene. However, carbapenem resistance conferred by the OXA-24 β-lactamase on the E. coli host strain did not reach the level of resistance observed in the original A. baumannii strain, thus revealing that other mechanisms are certainly involved in the resistance to carbapenems of the A. baumannii RYC 52763/97 strain. In gram-negative bacteria, diminished outer membrane permeability and multidrug efflux pumps make a major contribution to intrinsic resistance. In the A. baumannii RYC 52763/97 strain, a reduction in the expression of two porins of 22 and 33 kDa was observed; however, no differences in the β-lactam MICs were detected when reserpine (25 and 50 μg/ml) was added, suggesting that a putative efflux pump mechanism was not present in this strain (G. Bou and J. Martínez-Beltrán, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1461, 1999). Therefore, an interesting point will be to elucidate the level of carbapenem resistance conferred by the OXA-24 β-lactamase on A. baumannii. With this purpose, experiments are in progress to transfer the pBMB-2 plasmid into an imipenem-susceptible A. baumannii strain.
In summary, different oxacillinases with imipenem hydrolysis activity have previously been described for A. baumannii strains (15, 18, 27), but apart from the plasmid-mediated ARI-1 (OXA-23) β-lactamase (15), this is the first report describing the nucleotide and amino acid sequence of a new chromosomally mediated OXA-derived β-lactamase with imipenem hydrolysis activity in an A. baumannii strain. We propose the designation of OXA-24 for this new β-lactamase.
ACKNOWLEDGMENTS
We thank Luis de Rafael and Gonzalo Cerveró for their critical comments and Dolores Malpica for her excellent technical assistance.
REFERENCES
- 1.Afzal M S, Livermore D. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J Antimicrob Chemother. 1998;41:576–577. doi: 10.1093/jac/41.5.576. [DOI] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of β-lactamases. Philos Trans R Soc Lond Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Amyes S G B, Young H-K. Mechanism of antibiotic resistance in Acinetobacter spp.—genetics of resistance. In: Bergogne-Berezin E, Joly-Guillou M L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management, 1996. New York, N.Y: CRC Press; 1996. pp. 185–223. [Google Scholar]
- 4.Bergogne-Bérézin E. Resistance of Acinetobacter spp. to antimicrobials—overview of clinical resistance patterns and therapeutic problems. In: Bergogne-Berezin E, Joly-Guillou M L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management, 1996. New York, N.Y: CRC Press; 1996. pp. 133–183. [Google Scholar]
- 5.Bergogne-Bérézin E, Towner K J. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou G, Martínez-Beltrán J. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2000;44:428–432. doi: 10.1128/aac.44.2.428-432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown S, Bantar C, Young H K, Amyes S G B. Limitation of Acinetobacter baumannii treatment by plasmid-mediated carbapenemase ARI-2. Lancet. 1998;351:186–187. doi: 10.1016/S0140-6736(05)78210-6. [DOI] [PubMed] [Google Scholar]
- 8.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark R B. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33–36 KDa outer membrane protein. J Antimicrob Chemother. 1996;38:245–251. doi: 10.1093/jac/38.2.245. [DOI] [PubMed] [Google Scholar]
- 10.Danel F, Hall L M C, Gur D, Livermore D M. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1881–1884. doi: 10.1128/aac.39.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danel F, Hall L M C, Gur D, Livermore D M. OXA-15, an extended-spectrum variant of OXA-2 β-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob Agents Chemother. 1997;41:785–790. doi: 10.1128/aac.41.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danel F, Hall L M C, Gur D, Livermore D M. OXA-16, a further extended-spectrum variant of OXA-10 β-lactamase, from two Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother. 1988;42:3117–3122. doi: 10.1128/aac.42.12.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danel F, Hall L M C, Duke B, Gur D, Livermore D M. OXA-17, a further extended-spectrum variant of OXA-10 β-lactamase, isolated from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1362–1366. doi: 10.1128/aac.43.6.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Da Silva G J, Leitão R, Peixe L. Emergence of carbapenem-hydrolyzing enzymes in Acinetobacter baumannii clinical isolates. J Clin Microbiol. 1999;37:2109–2110. doi: 10.1128/jcm.37.6.2109-2110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donald H M, Scaife W, Amyes S G B, Young H-K. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000;44:196–199. doi: 10.1128/aac.44.1.196-199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehrlein M, Leying H, Cullmann W, Wendt S, Opferkuch W. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin-binding proteins. Chemotherapy. 1991;37:405–412. doi: 10.1159/000238887. [DOI] [PubMed] [Google Scholar]
- 17.Hall L M C, Livermore D M, Gur D, Akova M, Akalin H E. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:1637–1644. doi: 10.1128/aac.37.8.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornstein M, Sautjeau-Rostoker C, Peduzzi J, Vessieres A, Hong L T, Barthelemy M, Scavizzi M, Labia R. Oxacillin-hydrolyzing beta-lactamase involved in resistance to imipenem in Acinetobacter baumannii. FEMS Microbiol Lett. 1997;153:333–339. doi: 10.1111/j.1574-6968.1997.tb12593.x. [DOI] [PubMed] [Google Scholar]
- 19.Joly-Guillou M L, Bergogne-Bérézin E, Philippon A. Distribution of β-lactamases and phenotypic analysis in clinical strains of Acinetobacter calcoaceticus. J Antimicrob Chemother. 1988;22:597–604. doi: 10.1093/jac/22.5.597. [DOI] [PubMed] [Google Scholar]
- 20.Jones M E, Thornsberry C, Livermore D M, Sahma D F. Prevalence of Acinetobacter spp. with reduced susceptibility to imipenem, as determined by a USA-wide electronic surveillance network. J Antimicrob Chemother. 1999;43:429–431. doi: 10.1093/jac/43.3.429. [DOI] [PubMed] [Google Scholar]
- 21.Levi I, Rubinstein E. Acinetobacter infections—overview of clinical features. In: Bergogne-Berezin E, Joly-Guillou M L, Towner K J, editors. Acinetobacter: microbiology, epidemiology, infections, management, 1996. New York, N.Y: CRC Press; 1996. pp. 101–115. [Google Scholar]
- 22.Martínez-Beltrán J, Alvarez M, Sierra M P. Acinetobacter baumannii en pacientes críticos. Madrid, Spain: Merck Sharp & Dohme de España S.A.; 1998. pp. 21–43. [Google Scholar]
- 23.Masuda G, Tomioka S, Hasegawa M. Detection of β-lactamase production by Gram-negative bacteria. J Antibiot (Tokyo) 1976;29:662–664. doi: 10.7164/antibiotics.29.662. [DOI] [PubMed] [Google Scholar]
- 24.Matthew M, Harris A M, Marshall M, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 25.Mugnier P, Casin I, Bouthors A T, Collatz E. Novel OXA-10-derived extended-spectrum β-lactamases selected in vivo or in vitro. Antimicrob Agents Chemother. 1998;42:3113–3116. doi: 10.1128/aac.42.12.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 27.Paton R, Miles R S, Hood J, Amyes S G B. ARI-1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2:81–88. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 28.Perilli M, Felici A, Oratore A, Cornaglia G, Bonfiglio G, Rossolini G M, Amicosante G. Characterization of the chromosomal cephalosporinases produced by Acinetobacter lwoffii and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother. 1996;40:715–719. doi: 10.1128/aac.40.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanschagrin F, Couture F, Levesque R C. Primary structure of OXA-3 and phylogeny of oxacillin-hydrolyzing class D β-lactamases. Antimicrob Agents Chemother. 1995;39:887–893. doi: 10.1128/aac.39.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato K, Nakae T. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J Antimicrob Chemother. 1991;28:33–45. doi: 10.1093/jac/28.1.35. [DOI] [PubMed] [Google Scholar]
- 33.Spratt B G, Hedge P J, Heesen T S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 34.Vahaboglu H, Öztürk R, Aygün G, Coşkunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vecoli C, Prevost F E, Ververis J J, Medeiros A A, O'Leary G P., Jr Comparison of polyacrylamide and agarose gel thin-layer isoelectric focusing for the characterization of β-lactamases. Antimicrob Agents Chemother. 1983;24:186–189. doi: 10.1128/aac.24.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vila J, Marcos A, Marco F, Abdalla S, Vergara Y, Reig R, Gomez-Lus R, Jiménez De Anta T. In vitro antimicrobial production of β-lactamases, aminoglycoside-modifying enzymes, and chloramphenicol acetyltransferase by and susceptibility of clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 1993;37:138–141. doi: 10.1128/aac.37.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vila J, Navia M, Ruiz J, Casals C. Cloning and nucleotide sequence analysis of a gene encoding an OXA-derived β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 1997;41:2757–2759. doi: 10.1128/aac.41.12.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]