Abstract
The circadian clock is a master regulator in coordinating daily oscillations of physiology and behaviors. Nevertheless, how the circadian rhythm affects endochondral ossification is poorly understood. Here we showed that endochondral bone formation exhibits circadian rhythms, manifested as fast DNA replication in the daytime, active cell mitosis, and matrix synthesis at night. Circadian rhythm disruption led to endochondral ossification deformities. The mechanistic dissection revealed that melatonin receptor 1 (MTR1) periodically activates the AMPKβ1 phosphorylation, which then orchestrates the rhythms of cell proliferation and matrix synthesis via destabilizing the clock component CRY1 and triggering BMAL1 expression. Accordingly, the AMPKβ1 agonist is capable of alleviating the abnormity of endochondral ossification caused by circadian dysrhythmias. Taken together, these findings indicated that the central circadian clock could control endochondral bone formation via the MTR1/AMPKβ1/BMAL1 signaling axis in chondrocytes. Also, our results suggested that the AMPKβ1 signaling activators are promising medications toward endochondral ossification deformities.
Subject terms: Development, Genetic interaction, Experimental models of disease
Introduction
Endochondral ossification is a developmental process by which many bones grow, including appendicular bones and craniofacial bones [1, 2]. Initially, mesenchyme condensation occurs, which is followed by chondrocyte differentiation. The chondrocytes then proliferate, undergo hypertrophy, and terminally differentiate into mineralized cartilage, subsequently replaced by bone [3]. Impaired endochondral ossification can lead to severe skeletal dysplasia, including short stature and craniofacial abnormalities [4–6]. However, the molecular mechanism that controls the sequential steps of endochondral ossification is not fully elucidated. Thus, exploring the mechanisms of endochondral bone formation could shed light on the pathophysiology of osteogenesis imperfecta and the therapeutic strategies.
The morphological and functional changes in proliferating and differentiating chondrocytes are not only regulated by a variety of local autocrine/paracrine factors, including parathyroid hormone-related peptide (PTHRP) and Indian hedgehog (IHH) but also under the control of complex transcriptional networks, such as ex-determining region Y (SRY)-box 9 (SOX9) and circadian clock transcription outputs [3, 7–12]. Accumulating evidence suggests that circadian rhythm core component aryl hydrocarbon receptor nuclear translocator-like protein 1 (BMAL1) plays pivotal roles in bone and cartilage homeostasis. Its deficiency leads to osteopenia and predisposes knee cartilage to osteoarthritis-like damage [10, 13, 14]. Our recent study observed that BMAL1 stringently controls osteogenesis of the mandibular condyle in a hedgehog signaling-dependent manner [15]. However, the mechanism of regulating BMAL1 expression in chondrocytes remains a mystery.
Peripheral clock genes expression is tightly controlled by the central clock in the suprachiasmatic nucleus (SCN) [16]. The SCN is principally entrained by the light–dark cycle and acts as a pacemaker that transmits circadian information to tissues and organs to synchronize the expression of downstream genes in a cell-type-specific manner [17]. Melatonin receptors (MTRs) are required for central clock mediated cell responses [18]. The diurnal expression changes of genes were orchestrated by the clock transcription-translation feedback loops (TTFLs), which are responsible for intracellular circadian time-keeping in mammals [19]. The TTFLs involve transcriptional activation by CLOCK/BMAL1 heterodimers, which drive the expressions of Period 1/2/3 (Pers) and Cryptochrome 1/2 (Crys) genes through E-box (CANNTG) regulatory sequences. Following dimerization and transport to the nucleus, PER/CRY complexes repress CLOCK/BMAL1 activities until progressive degradation of PER/CRY allows initiation of a new cycle [19]. The TTFLs are universally active across mammalian tissues, so how cellular clocks interact to achieve multi-cellular integration and ultimately direct circadian rhythms of cell behaviors are the matters of considerable interest. This study sought to explore the activity rhythms in the chondrification center, especially cell proliferation and matrix synthesis. We found that the central circadian clock and MTR1 play essential roles in cartilage metabolism and bone growth. The melatonin in serum is closely involved in circadian rhythm-mediated endochondral ossification. Furthermore, we demonstrated that the AMPKβ1 activator can effectively rescue the endochondral ossification deformities caused by circadian clock disruption. These findings provided new insight into the pathogenesis of endochondral ossification deformities and presented potential therapeutic strategies for osteogenesis abnormity.
Results
Chondrification center possesses a circadian rhythm of cell proliferation and matrix synthesis
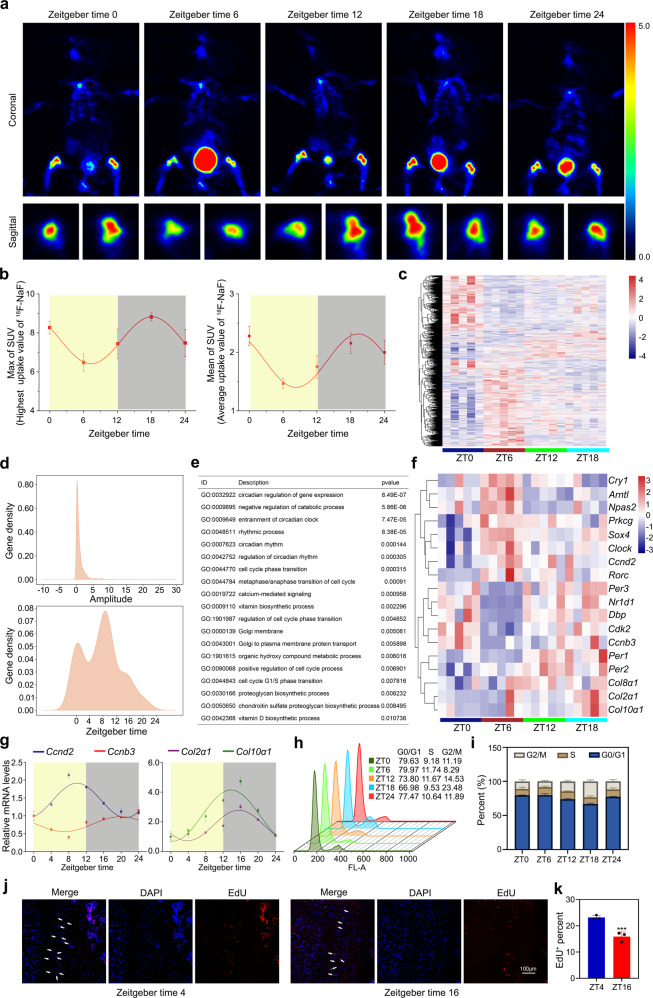
To identify the rhythm of bone formation in the chondrification center, we used a micro-positron emission tomography (PET) computerized tomography scanner to monitor 18F-labeled sodium fluoride (18F-NaF) uptake in C3H/HeN mice. During a 24-h dark–light cycle, we found a significant effect of time-of-day on 18F-NaF uptake, which was higher during the dark phase and reached the peak around ZT (Zeitgeber time) 18 or ZT0 (Fig. 1a). Although the individual errors resulted in the fluctuation of the amplitudes detected at ZT0 and ZT24, a cosinor curve was fitted through individual values, suggesting that the osteogenesis in the chondrification center has a circadian variation. The area under the ZT12-ZT24 curve was greater than that under the ZT0-ZT12 curve, which meant that the bone formation metabolism is relatively active at night (Fig. 1b).
Fig. 1. Chondrification center possesses a circadian rhythm of cell proliferation and matrix synthesis.
a Positron emission tomography (PET) scans of C57BL/6J mice after 18F-labeled sodium fluoride (18F-NaF) tail vein injection at indicated times (n = 3 per group). b PET analysis of max and mean SUV. c Heatmaps representing expression levels of circadian genes in the indicated genotypes. d The gene density of amplitude (upper) or phase (under) distribution of all circadian oscillation genes in wild-type (WT) mice. e Associated Gene Ontology (GO) across all circadian genes in WT. f Heatmap showed some circadian genes that were related to circadian rhythm, cell cycle, and collagen synthesis. g qRT-PCR analysis of the diurnal rhythms of Ccnd2, Ccnb3, Col2α1, and Col10α1 in the growth plate cartilages of distal femora at indicated times (n = 3 for each bar). h, i Cell-cycle distribution of the growth plate cartilages tissue determined by the flow cytometry (n = 3 for each bar). j, k EdU staining of proliferating cells in proliferation zone (PZ) from the growth plate cartilages at ZT4 and ZT16. Arrowheads indicate EdU+ cells, ***P < 0.001 (n = 3 for each bar). Scale bar, 100 μm.
To further delineate endochondral bone formation rhythm features, we performed time-course genome-wide RNA-sequence in the cartilaginous osteogenesis tissues (Fig. 1c). The data showed that ~5.4% of the transcriptome (1342 transcripts) appeared circadian in the chondrification center (Fig. 1d). The batch effect of total RNA content did not exist among the samples, which excluded its impact on the observed rhythms (Supplementary Fig. 1). As expected, many circadian transcripts were involved in the endochondral bone formation rhythm, including Bmal1 (brain and muscle arnt-like 1), Clock (circadian locomotor output cycles kaput), Per1/2 (period 1/2), Cry1 (cryptochromes 1), and Sox4 (sex-determining region Y-box 4) (Fig. 1e, f). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) validated that Col2α1 (type II collagen) and Col10α1 (type X collagen) were mainly expressed during the dark phase (Fig. 1g), which was consistent with the results of the PET scanner, suggesting that the matrix synthesis was active in the night.
Furthermore, we noticed that a cohort of proliferation-related genes was rhythmically expressed, including Ccnb3 (Cyclin B3), Ccnd2 (CyclinD2), Cdk2 (Cyclin-dependent kinase 2) (Fig. 1e–g). To better understand the diurnal patterns of cell proliferation, we added flow cytometry to analyze the cell cycle of chondrocytes from the growth plate tissues at ZT0, 6, 12, 18, 24. The data showed that the S-phase cell number was much more at ZT6, and the G2–M phase cell number was concentrated at ZT18 (Fig. 1h, i). The EDU+ cells in the chondrification center during the light phase were more than those at night (Fig. 1j, k). Collectively, these findings demonstrated that endochondral bone formation exhibits a circadian rhythm, accompanying the fast DNA replication during the day, active cell mitosis, and matrix synthesis at night.
Disruption of circadian rhythm suppresses endochondral bone formation
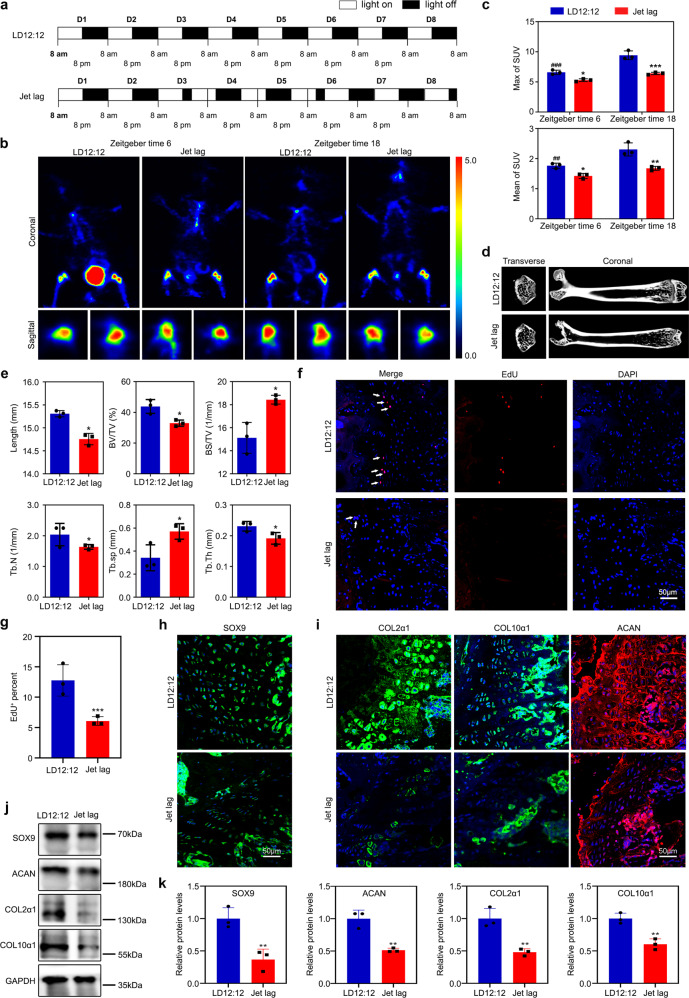
To clarify the role of circadian rhythm in endochondral ossification, we established a jet-lagged mouse model to assay the effect of circadian rhythm disruption. C3H/HeN mice were placed at a jet lag schedule with 8-h light advanced every 2–3 days for a total of 4 weeks, mimicking the circadian disturbance that humans undergo during shift work [20] (Fig. 2a). The PET results showed that the rate of 18F-NaF uptake was reduced, and its physiological variation was absent (Fig. 2b, c). By measuring the expression of the clock genes and osteogenesis biomarkers in the growth plate cartilages of the distal femora (Supplementary Fig. 2a, b), we found that the jet lag schedule reduced the expression of these genes. Notably, we noticed that femora length was decreased in the jet-lagged mice (Fig. 2d, e). Conventional morphological staining, western blot, and in vivo EdU assay showed that endochondral osteogenesis in the jet-lagged mice was inhibited (Fig. 2f–k and Supplementary Fig. 2c, d), suggesting that circadian rhythm disruption induced defects in cell proliferation, sequential differentiation, and matrix synthesis, which leads to short individuals and developmental defects.
Fig. 2. Disruption of circadian rhythm suppresses endochondral bone formation.
a The schematic shows experiment schedule. Zeitgeber time 0 is defined at the time the light was turned on (8 AM). b, c Regional uptake of 18F-NaF in femora and tibia as determined from PET image analysis in LD12:12 and jet-lagged mice after 18F-NaF tail vein injection at ZT6 and ZT18 (n = 3 per group). ##P < 0.01, ###P < 0.001, ZT6 compared with ZT18 in the control (LD12:12) mice. *P < 0.05, **P < 0.01, ***P < 0.001, LD12:12 compared with the Jet lag. d Representative images of micro-CT reconstruction of femora (n = 3 per group). e Micro-CT analysis of the total length, bone volume/total volume (BV/TV), bone surface area/bone volume (BS/BV), trabecular number (Tb.N), trabecular separation (Tb.Sp), and trabecular thickness (Tb.Th) of femora (n = 3 for each bar), *P < 0.05. f, g EdU staining analysis of proliferating cells in PZ from the growth plate cartilages. Arrowheads indicate EdU+ cells (n = 3 for each bar), ***P < 0.001. Scale bar, 50 μm. h, i Immunofluorescence of SOX9, COL2α1, COL10α1, and ACAN in the growth plate cartilages at ZT10. Scale bars, 50 μm. j, k Western blot analysis of the levels of SOX9, ACAN, COL2α1, and COL10α1 in the femoral metaphysis tissue from LD12:12 or jet-lagged mice.
MTR1 mediates the circadian rhythm in the chondrification center
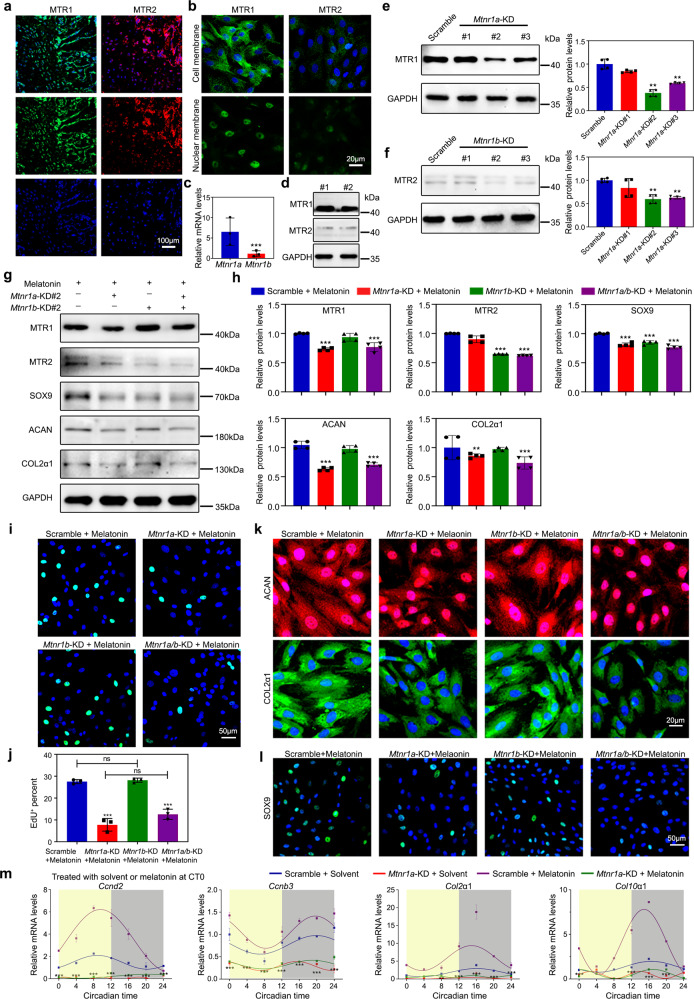
The MTR subfamily has been reported to be vital for mediating circadian rhythm [21]. Thus, we sought to test whether MTR mediates the circadian rhythm during endochondral bone formation. Interestingly, we noticed that the expression level of MTR1 was higher than that of MTR2 in chondrocytes (Fig. 3a–d). By knocking down Mtnr1a (the gene of MTR1) or/and Mtnr1b (the gene of MTR2), we observed whether melatonin was added at circadian time 0 (CT0) or CT12, its endochondral ossification enhancement was abolished in Mtnr1a-knockdown or Mtnr1a/b double-knockdown chondrocytes, including cell proliferation and matrix synthesis (Fig. 3e–m and Supplementary Fig. 3a). In Fig. 3g–l, the expressions of SOX9, ACAN, and COL2a1 were no substantial difference between Mtnr1a-knockdown chondrocytes and Mtnr1a/b double-knockdown chondrocytes, suggesting that the scanty MTR2 does not complement the effects of MTR1 very well, and their additive effect was not prominent. A similar effect on endochondral ossification was noted by antagonizing MTR1/2 with 100 μmol/L luzindole in melatonin-treated chondrocytes. In contrast, MTR2 antagonist 4P-PDOT (100 μmol/L) had little effect (Supplementary Fig. 3b–g). We also verified that melatonin could not effectively induce the rhythms of cell proliferation and matrix synthesis in Mtnr1a-knockdown chondrocytes (Fig. 3m and Supplementary Fig. 3a). In addition, we found that the efficacy of melatonin was in a dose-dependent manner, and moderate MTR1 overexpression (about 1.5-fold) could not create a synergistic effect in endochondral bone formation, suggesting that the amount of melatonin is the key (Supplementary Fig. 4a–h). Different from the effective concentration of melatonin reported in the literature [22], we showed that 100 μmol/L melatonin was responsible for promoting the proliferation of chondrocytes, which might be due to the differences in cell source, etc. Together these data suggested that MTR1 orchestrates the circadian rhythms of endochondral bone formation, which is different from bone marrow mesenchymal stem cells (BMSCs)-derived osteoblasts involving MTR2 [23].
Fig. 3. MTR1 reconciles the circadian rhythm in the chondrification center.
a Immunofluorescence showed the expression levels of Melatonin receptor 1A (MTR1) and 1B (MTR2) in the growth plate cartilages from WT mice. Scale bar, 100 μm. b, c Immunofluorescence and qRT-PCR showed the expression levels of MTR1 and MTR2 in the primary chondrocytes, ***P < 0.001 (n = 3 for each bar). Scale bar, 20 μm. d Western blot showed the expression levels of MTR1 and MTR2 in the growth plate cartilages from WT rats. e, f MTR1 and MTR2 knockdown were assessed by western blot, **P < 0.01 (n = 4 for each bar). g, h Western blot analysis of the changes of related proteins in MTR1 or MTR2 knockdown chondrocytes with the addition of melatonin, **P < 0.01, ***P < 0.001 (n = 4 for each bar). i, j EdU staining analysis of proliferating cells after melatonin supplement in MTR1 or MTR2 knockdown chondrocytes. Scale bar, 50 μm. ***P < 0.001. k, l Immunofluorescence of ACAN, COL2α1, and SOX9 in MTR1 or MTR2 knockdown chondrocytes with melatonin or equal solvent. Scale bars, 20 and 50 μm. m qRT-PCR analysis of the mRNA levels of Ccnd2, Ccnb3, Col2α1, and Col10α1 at indicated time in Scramble or MTR1 knockdown chondrocytes treated with equal solvent or melatonin at CT0, **P < 0.01, ***P < 0.001, LD12:12 compared with the Jet lag (n = 3 per group). e, f, h, j Data were analyzed using one-way ANOVA with Tukey multiple comparisons test.
To characterize the role of MTR1 in mediating the circadian rhythms of the chondrification center, we constructed an in vitro cell model in which melatonin was supplemented into the chondrocytes at the time points of 0 and 24 h. We noticed that the expression peaks of the matrix synthesis biomarkers Col2α1 and Col10α1 came earlier than those of the cell cycle-related genes Ccnb3 and Ccnd2, and the phase difference was about 12 h (Supplementary Fig. 4i). This rhythm difference suggested that the mechanisms of MTR1-mediated cell proliferation and matrix synthesis are likely to be different.
Serum melatonin confers MTR1-mediated circadian rhythm
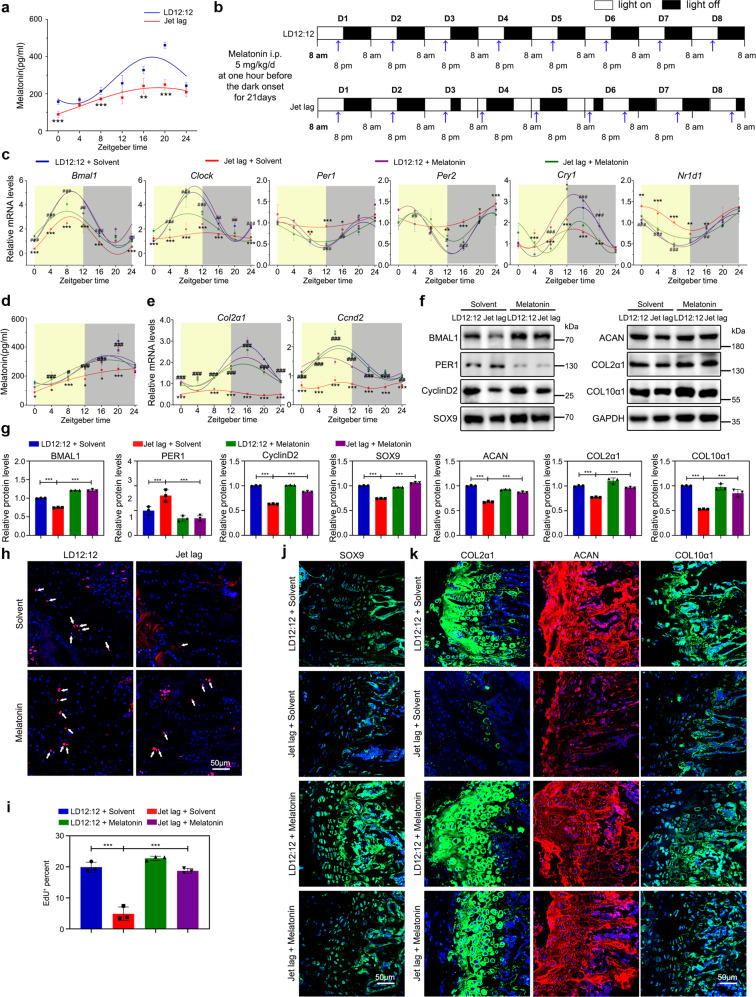
Our results suggested that MTR1 acts as an essential circadian rhythm transmitter in the chondrification center. To further establish the functional significance of MTR1, we tested whether the melatonin in serum holds a similar circadian rhythm. We noticed that the level of serum melatonin had prominent circadian rhythms in wild-type mice. Melatonin is significantly higher during the dark phase, reaching the peak around ZT18 (Fig. 4a). This daily change was consistent with the uptake rate of 18F-NaF in the chondrification center. In contrast, the melatonin level was reduced in the mice with circadian clock disruption-induced osteogenesis abnormity, accompanied by the loss of circadian pattern (Supplementary Fig. 5a, b). To corroborate the implication of melatonin in endochondral bone formation, we applied melatonin 1 h before the light-off in the jet-lagged mouse model (Fig. 4b). The data showed that the application reestablished the expression rhythms of clock genes in the chondrification center, including Bmal1 and Clock, and restored the circadian rhythm of melatonin (Fig. 4c, d). Likewise, the rhythms of cell proliferation and matrix synthesis in the growth plate cartilages were restored (Fig. 4e). Also, the chondrogenic defects and short individuals were effectively rescued (Fig. 4f–k and Supplementary Fig. 5a–f). It should be noted that the effect of melatonin was ambiguous in animals treated 1 h before the light on, reflecting the importance of obeying the physiological circadian rhythms (Supplementary Fig. 6). Taken together, our results indicated that serum melatonin is critical for orchestrating the circadian rhythms in endochondral bone formation.
Fig. 4. Melatonin in serum confers MTR1-mediated circadian rhythm.
a Detect the level of melatonin in serum at different time points by ELISA, **P < 0.01, ***P < 0.001 (n = 3 per group). b The schematic shows the experiment schedule. Blue arrowheads indicate the time of injection at one hour before the dark onset. c qRT-PCR analysis of the mRNA levels of Bmal1, Clock, Per1, Per2, Cry1, and Nr1d1 in the growth plate cartilages from LD12:12 or jet-lagged mice treated with equal solvent or melatonin (Sigma, M5250, 5 mg/kg/d, injected before the dark onset for 21 days, jet-lagged mice were pretreated with jet lag for 2 weeks before the melatonin injection) at the indicated time (n = 3 per group). *P < 0.05, **P < 0.01, ***P < 0.001, LD12:12 compared with the Jet lag. #P < 0.05, ##P < 0.01, ###P < 0.001, melatonin compared with equal solvent in jet-lagged mice. d ELISA analysis of the level of melatonin in serum at different time points after melatonin injection. e qRT-PCR analysis of the mRNA levels of Col2α1 and Ccnd2 in the growth plate cartilages of LD12:12 and jet-lagged mice treated with equal solvent or melatonin at indicated time (n = 3 per group). f, g Western blot analysis of the levels of BMAL1, PER1, CyclinD2, SOX9, ACAN, COL2α1, and COL10α1 in the femoral metaphysis tissue from LD12:12 or jet-lagged mice treated with equal solvent or melatonin, ***P < 0.001 (n = 3 per group). h, i EdU staining analysis of proliferating cells in PZ from the growth plate cartilages in LD12:12 and jet-lagged mice with melatonin or equal solvent, Arrowheads indicate EdU+ cells (n = 3 for each bar). Scale bar, 50 μm. j, k Immunofluorescence of SOX9, COL2α1, ACAN, and COL10α1 in the growth plate cartilages from LD12:12 and jet-lagged mice with melatonin or equal solvent at ZT10. Scale bars, 50 μm. g, i Data were analyzed using one-way ANOVA with Tukey multiple comparisons test.
MTR1 modulates rhythmic phosphorylation of AMPKβ1 in chondrocytes
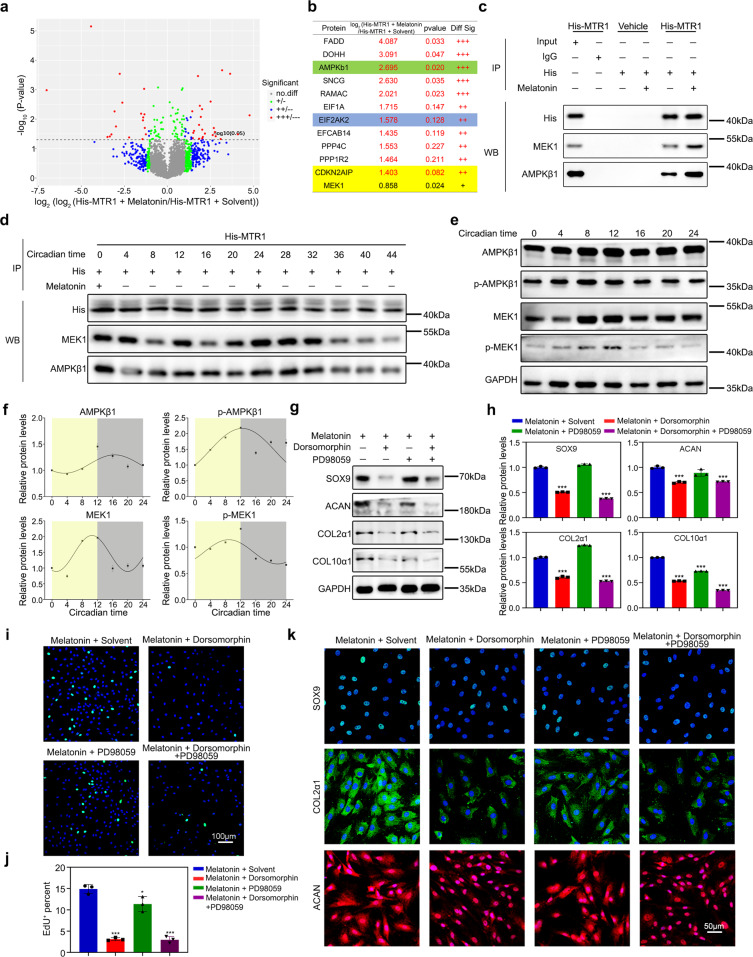
To explore the intracellular mechanism of circadian signal transduction in chondrocytes, we performed co-immunoprecipitation and mass spectrometry (Co-IP/MS) assays to screen the candidate proteins binding to MTR1 intracellular segments in His-MTR1 expressing chondrocytes. In contrast to the solvent group, a total of 544 proteins in the melatonin-treated chondrocytes were pulled down by the anti-6X-His antibody (Fig. 5a). Among the identified interactive proteins, two protein kinases, AMP-activated protein kinase subunit beta-1 (AMPKβ1) and mitogen-activated protein kinase kinase 1 (MEK1), were significantly upregulated in chondrocytes treated with 100 μmol/L melatonin (Fig. 5b). The interactions of AMPKβ1 and MEK1 with MTR1 were validated by a Co-IP assay (Fig. 5c). Next, we examined the interactions of MTR1 with AMPKβ1 or MEK1 at an interval of 4 h over the 48-h cycle, in which melatonin was supplemented into the chondrocytes at time points of 0 and 24 h (Fig. 5d). We found that the binding of MTR1 with AMPKβ1 or MEK1 was enhanced and presented a circadian rhythm upon the melatonin supplementation (Fig. 5d). Importantly, the levels of phosphorylated AMPKβ1 and MEK1 also showed a circadian fashion (Fig. 5e, f), suggesting that MTR1 can mediate the rhythmic phosphorylation of AMPKβ1 and MEK1.
Fig. 5. MTR1 promotes AMPKβ1 phosphorylation in chondrocytes.
a Scatter plot of Co-IP/MS analysis of the differentially expressed protein in chondrocytes treated with equal solvent or melatonin (n = 5 pairs per group). b Significantly increased proteins in Co-IP/MS analysis. c Co-immunoprecipitation (Co‐IP) of the chondrocytes expressing AMPKβ1 and MEK1 in melatonin at 100 μmol/L concentrations. 6X-His antibody was used for immunoprecipitation. d Co‐immunoprecipitation (Co‐IP) of the chondrocytes expressing AMPKβ1 and MEK1 in 100 μmol/L concentrations melatonin at CT0, 4, 8, 12, 16, 20, 24, 28, 32, 36, 40, 44. 6X-His antibody was used for immunoprecipitation. e, f Western blot analysis of the total and phosphorylation levels of AMPKβ1 and MEK1 protein at the indicated time. g, h Western blot analysis of the changes of relative protein after melatonin supplementation with Dorsomorphin or PD98059 or Dorsomorphin and PD98059. ***P < 0.001 (n = 3 for each bar). i, j EdU staining analysis of proliferating cells after melatonin supplement with Dorsomorphin or PD98059. Scale bar, 100 μm. ***P < 0.001. k Immunofluorescence of SOX9, COL2α1, and ACAN in chondrocytes after melatonin supplementation with Dorsomorphin or PD98059 (n = 3 for each bar). Scale bar, 50 μm. h, j Data were analyzed using one-way ANOVA with Tukey multiple comparisons test.
To determine whether AMPKβ1 and/or MEK1 regulate MTR1-mediated cell proliferation and matrix synthesis, we used Dorsomorphin and PD89059 to inhibit the activation of AMPKβ1 and MEK1, respectively. The data of EdU staining, immunofluorescent staining and western blotting assays showed that Dorsomorphin inhibited the cell proliferation and matrix synthesis of melatonin-treated chondrocytes (Fig. 5g–k). MEK inhibitor PD98059 could only reverse the overexpression of COL10α1, so there was no reason to think it is the effective mediator of melatonin-MTR1 in matrix synthesis (Fig. 5g, h). Taken together, these findings suggested that MTR1 acts as a critical regulator of circadian bone formation via modulating rhythmic phosphorylation of AMPKβ1.
AMPKβ1 can activate BMAL1 expression via phosphorylating CRY1
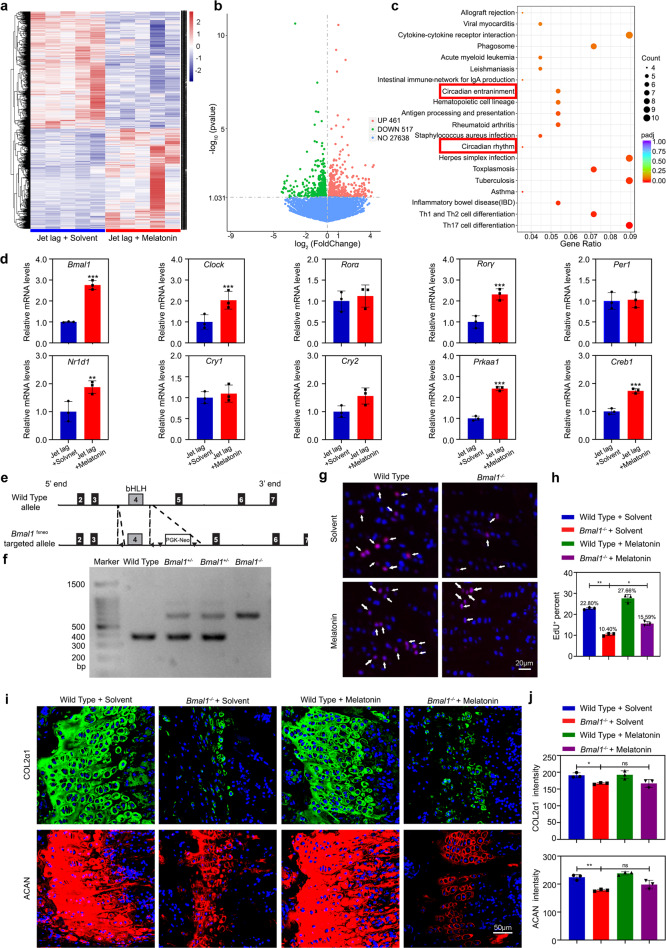
To delineate the mechanisms by which AMPKβ1 signaling confers the circadian rhythms, we executed a genome-wide RNA-sequencing to acquire the transcriptional profile of distal femur cartilage tissues. A total of 978 genes were found to be differentially expressed in melatonin-treated jet-lagged mice (Fig. 6a, b). Gene Ontology analysis revealed that circadian clock genes were enriched (Fig. 6c), and the core clock gene Bmal1 was upregulated (Fig. 6d). In vitro, expressional knockdown of Mtnr1a suppressed BMAL1 expression, suggesting that BMAL1 is affiliated with MTR1/AMPKβ1 signaling (Supplementary Fig. 7a, b).
Fig. 6. BMAL1 acts as a principal molecule in circadian cell proliferation and matrix synthesis.
a Heatmap of differentially expressed genes (DEGs) in jet-lagged mice treated with equal solvent or melatonin (n = 5 pairs per group). b Volcano plot of jet-lagged mice treated with equal solvent or melatonin. c Associated Gene Ontology (GO) across jet-lagged mice treated with equal solvent and melatonin. d Confirmation of the DEGs in the growth plate tissue from jet-lagged mice treated with equal solvent or melatonin by qRT-PCR (n = 3 for each bar). Data represented as mean ± SD, **P < 0.01, ***P < 0.001. e, f The genotypes of WT, Bmal1+/-, and Bmal1-/- mice analysis by PCR. g, h EdU staining of proliferating cells in PZ from the growth plate cartilages in wild-type and Bmal1-/- mice with melatonin or equal solvent. Arrowheads indicate EdU+ cells (n = 3 for each bar). Scale bar, 20 μm. i, j COL2α1 and ACAN immunofluorescence images and quantitative analysis of labeling intensity in the growth plate cartilages from wild-type and Bmal1-/- mice with melatonin or equal solvent at ZT10 (n = 3 for each bar). *P < 0.05, **P < 0.01, ns, nonsignificant. Data were analyzed using one-way ANOVA with Tukey multiple comparisons test. Scale bar, 50 μm. (Sigma, M5250, 5 mg/kg/d, Bmal1-/- mice were injected from 3 weeks of age, injected before the dark onset for 28 days).
The role of AMPK in phosphorylating and thus destabilizing the clock component CRY1 has been proposed [24]. To validate this notion, we examined the variation of CRY1 phosphorylation. We found that CRY1 phosphorylation was significantly promoted in the melatonin-treated chondrocytes or chondrocytes treated with AMPKβ1 signal activator, while its protein level was decreased (Supplementary Fig. 7c, d). We then applied the CRY1 stabilizer KL001 into the melatonin-treated chondrocytes and recorded the expression of BMAL1, COL2α1, and COL10α1. Of note, the melatonin-induced expression rhythms of Bmal1, Col2α1, Ccnb3, and Ccnd2 were lost, and significant downregulation of levels was detected in the KL001-treated chondrocytes (Supplementary Fig. 7e–g). Together these results suggested that CRY1 acts as a negative regulator in MTR1/AMPKβ1-mediated BMAL1 activation.
BMAL1 acts as a principal rhythm mediator during endochondral ossification
To consolidate the role of BMAL1 in circadian endochondral ossification, we constructed BMAL1-deficiency (Bmal1-/-) mice (Fig. 6e, f). We found that the efficacy of melatonin in encouraging cell proliferation and matrix synthesis was abolished in Bmal1-/- mice (Fig. 6g–j). Also, we extracted primary BMAL1-deficient chondrocytes and found that melatonin could not induce the circadian rhythms of cell proliferation and cartilage matrix production (Supplementary Fig. 8). These results indicated that BMAL1 is essential for responding to the AMPKβ1 signal initiated by the central circadian clock and conferring the rhythms of cell proliferation and matrix synthesis.
AMPKβ1 agonist ameliorates osteogenesis imperfecta caused by circadian irregularities
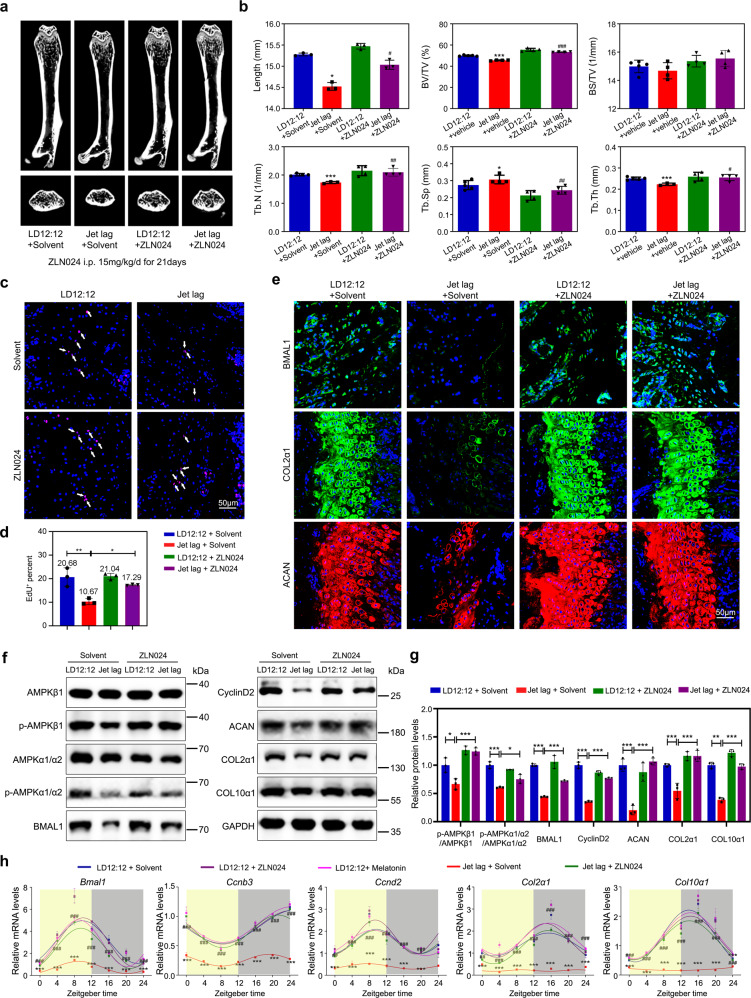
Thus far, our results suggested that the MTR1/AMPKβ1/BMAL1 signaling axis is essential for endochondral bone formation. To explore the strategy of alleviating the low bone mass phenotype, we injected AMPKβ1 activator ZLN024 into jet-lagged mice (15 mg/kg/d, a total of 21 days), according to the literature [25]. Micro-CT results showed that skeletal development was substantially improved (Fig. 7a, b). The EdU staining assay revealed a prominent enhancement in chondrocyte proliferation (Fig. 7c, d). Moreover, the expression of BMAL1 and cartilage matrix components (COL2α1 and ACAN) increased in ZLN024-treated jet-lagged mice (Fig. 7e). Accordingly, western blot showed that the expressions of CyclinD2, COL2α1, ACAN, and COL10α1 were restored in ZLN024-treated jet lag mice, as the same with the microscopy shown (Fig. 7f, g). The 24-h rhythms of these proteins were restored in ZLN024-treated jet-lagged mice that was similar to the melatonin treatment, neither was effective in the normal LD12:12 mice (Fig. 7h). Together, our results suggested that the AMPKβ1 activator could be a potential strategy for the clinical treatment of endochondral ossification deformities.
Fig. 7. AMPKβ1 agonist ameliorates osteogenesis imperfecta caused by circadian irregularities.
a Representative images of micro-CT reconstruction of femora from LD12:12 and jet-lagged mice injected AMPK activator (ZLN024 hydrochloride, MCE, 15 mg/kg/d, injected before the dark onset for 21 days, jet-lagged mice were pretreated with jet lag for 2 weeks before the ZLN024 injection experiment) or equal solvent (1% DMSO + 99% Normal saline). b Micro-CT analysis of the total femora length, BV/TV, BS/TV, Tb.N, Tb.Sp, and Tb.Th of LD12:12 and jet-lagged mice injected ZLN024 or equal solvent (n = 3–5 per group). #P < 0.05, ##P < 0.01, ###P < 0.001, ZLN024 compared with solvent in jet-lagged mice. *P < 0.05, **P < 0.01, ***P < 0.001, LD12:12 compared with the Jet lag. c, d EdU staining of proliferating cells in PZ from the growth plate cartilages in LD12:12 and jet-lagged mice injected AMPK activator ZLN024 hydrochloride or equal solvent. Arrowheads indicate EdU+ cells (n = 3 for each bar). Scale bar, 50 μm. e Immunofluorescence of BMAL1, COL2α1, and ACAN in the growth plate cartilages from LD12:12 and jet-lagged mice injected AMPK activator ZLN024 hydrochloride or equal solvent (n = 3 per group), Scale bar, 50 μm. f, g Western blot analysis of the levels of AMPKβ1, p-AMPKβ1, AMPKα1/α2, p-AMPKα1/α2, BMAL1, CyclinD2, ACAN, COL2α1, and COL10α1 in the femoral metaphysis tissue from LD12:12 and jet-lagged mice treated with equal solvent or ZLN024 (n = 3 per group). h qRT-PCR analysis of the circadian rhythms of Bmal1, Ccnb3, Ccnd2, Col2α1, and Col10α1 in LD12:12 and jet-lagged mice treated with equal solvent or ZLN024 or melatonin supplementation at the indicated time (n = 3 per group). b, g Data were analyzed using one-way ANOVA with Tukey multiple comparisons test.
Discussion
Traditional hereditary and epigenetic factors, such as PTHRP, IHH, SOX9 [3, 7–12], failed to fully account for the risk of endochondral ossification abnormity, suggesting the existence of additional mechanisms. Here, we showed that circadian rhythm is essential for the maintenance of cartilage homeostasis. MTR1 relays the central rhythm via an MTR1/AMPKβ1/ BMAL1 axis. Notably, the AMPKβ1 phosphorylation activator can be used to alleviate the osteogenesis abnormity caused by circadian dysrhythmia. Taken together, our results proposed a new strategy to treat the endochondral ossification deformities.
Circadian rhythms in the growth plate have been observed in mitotic activity and mineralization [26, 27]. Our studies showed that the peripheral rhythms in the chondrification center could be orchestrated by SCN through melatonin in serum, which is essential for bone growth. Although a large sample of epidemiological studies is required, circadian dysrhythmia should be considered a pathological syndrome that significantly impacts heteroplasia. In addition, the prevalence of circadian rhythm disruption is rising as more people adopted nocturnal lifestyles. Therefore, circadian rhythm disruption should be recognized as a significant public health issue.
The SCN governs the release of melatonin from the pineal gland via sympathetic neurons of the superior cervical ganglion [28]. Being a circadian hormone, melatonin regulates peripheral circadian clocks [29]. It has been reported that melatonin can upregulate the expression of Col2α1, Sox9, and Acan in porcine articular chondrocytes at 1~10 ng/mL, suggesting that melatonin has chondroprotective activities [30]. This notion was corroborated by Hong et al., who showed that 1 μmol/L melatonin improved extracellular matrix production in chondrocytes [31]. Here, we showed that MTR1 in chondrocytes transmits rhythm signals by phosphorylating AMPKβ1, promoting endochondral bone formation via phosphorylating and destabilizing the clock component CRY1 and activating the expression of BMAL1. E-box elements are found in the promoters of collagens, suggesting that their expressions can be directly regulated by BMAL1/CLOCK dimers consistent with other E-box genes, including Per1/2, Nr1d1, Dbp, showed an increase around ZT18 in chondrocytes. Bmal1 and Clock were both regulated by REV-ERB/ROR through binding the RORE elements, so their expression phases were opposite to those of E-box genes such as Per1/2. The expression peak of Sox4 was consistent with Clock, suggesting that it might also be regulated by REV-ERB/ROR. Of note, in bone tissues, the matured osteoblasts and osteocytes mainly express MTR2, which triggers Gαi-dependent cAMP and ERK signaling pathways and promotes the osteogenic differentiation of BMSCs [32]. It is also shown that melatonin can inhibit H2O2-induced chondrocyte death and promote chondrocyte proliferation independently of BMAL1 [33], indicating that its antioxidant effect is nonnegligible during osteogenesis. In addition, our study showed that melatonin treatment decreases CRY1 level and increases BMAL1 expression, which concurs with the previous study [22]. Melatonin-induced BMAL1 expression can modulate double targets of Hedgehog signaling, which is indispensable for endochondral ossification [10, 15]. Specifically, Hedgehog signaling indirectly regulates the chondrocyte differentiation via a negative feedback loop with PTHRP, whereas it acts on chondrocyte proliferation in a direct manner, which may be the underlying cause of the phase difference in MTR1-mediated cell proliferation and matrix synthesis [34]. Together, the data indicated that MTR1/AMPKβ1/BMAL1 signaling axis is required explicitly for endochondral ossification.
Plastic operation is a clinical strategy to remedy the endochondral ossification deformities [35]. But the operation is complicated, and the side-effects are serious. Several in vitro studies have reported the efficacy of melatonin on chondrocyte metabolism [30, 36]. For example, Pei et al. showed that the expression of COL2α1, SOX9, and ACAN was enhanced by adding 1~10 ng/mL melatonin to porcine articular chondrocytes [30]. Our results are consistent with these reports, showing that melatonin enhanced chondrocyte proliferation and the expression of cartilage-associated genes. However, a previous study using cultured chondrocytes from rat vertebral body growth plates showed that a relatively high concentration of melatonin could cause inhibition of cell proliferation and mRNA expression of Col2α1, Sox9, Acan, and Smad4 [37], suggesting that the effects of melatonin may vary among cartilage types, even among hyaline cartilages and permanent cartilages. Moreover, the dose and administration time of melatonin, cell concentration, etc., were closely related to the effects of melatonin [38–40].
In addition, melatonin is an indoleamine with potent multifunctional pharmacological effects, both receptor-dependent and receptor-independent, including antioxidant, anticancer, antitumor, anti-inflammatory, anti-aging, anti-diabetic, antiviral, neuroprotective activities [41]. Thus, melatonin is not a promising drug to effectively modulate osteogenesis. Our results revealed that AMPK signaling activator ZLN024 improved the bone formation rhythm and relieved bone mass reduction caused by circadian rhythm disruption. Being a small molecule, ZLN024 can be a potential candidate for the clinical treatment of endochondral ossification deformities.
Materials and methods
Animals
A total of 410 three-week-old male C3H/HeN mice were obtained from the Beijing Charles River Bioscience (Beijing, China) and randomized placed into LD12:12 or jet lag (set 8:00 am as zeitgeber time 0, ZT0). For jet lag, mice were randomly placed in altered light-cycle conditions with 8-h light advance every 2–3 days. Four weeks later, animals were sacrificed to obtain femora at the indicated time points. For BMAL1-deficiency mice (Bmal1-/-, B6.129-Arntltm1Bra/J mice), as previous [15], homozygous mice were produced by breeding heterozygous BMAL1-deficiency mating pairs (Bmal1+/–) that were kindly provided by Dr Ying Xu (Soochow University, China) and obtained originally from the Jackson Laboratory. Investigators were blinded at the time of group allocation before and after jet lag model establishment. Mice in poor health were excluded from the study. All animal studies were approved by the Institutional Animal Care and Use Committee of Tongji Medical College (IACUC number: S739).
Micro-CT
Femora were harvested and fixed from the mice without soft tissues. Then the bones were scanned at a resolution of 9 μm using the Micro-CT (SkyScan 1176, Broker). The images were used to reconstruct and quantify by InstaRecon/NRecon Research Workplace software. Bone volume/total volume, trabecular thickness, bone surface area/bone volume, trabecular number, trabecular separation, and total length of femora were indexes to show difference among mice.
PET imaging
Mice were kept under 12 h of light, 12 h of dark (LD) cycle for 4 weeks. Half of the mice were maintained on an LD cycle 12 h out of the phase to scan at two opposite time points simultaneously. Measurements were made on three separate occasions, first measuring time points concentrated on ZT6 and ZT18, second centered on ZT0 and ZT12, and finally, a third day during which images were acquired for all four different time points. PET imaging was performed using a Trans-PET BioCalibure 700 system (Raycan Technology Co., Ltd, Suzhou, China). A total of 4.5 MBq sodium fluoride labeled with fluorine-18 (18F-NaF) was intravenously injected into the animals 30 min before imaging. Quantification of bone formation metabolism was obtained using the standardized uptake value (SUV) normalized to body weight. The SUV was defined as the ratio of the radioactivity of 18F-NaF ingested by the chondrification center of femora and tibia refer to the average radioactivity of the whole body. The maximum standardized uptake value (SUV max) and the mean standardized uptake value (SUV mean) of the chondrification center were analyzed using PMOD software. SUV = CT/DInj/Ws. CT is the maximum/mean 18F-NaF radioactivity per unit volume in the chondrification center, DInj is the injection dose, and WS is the mice’s weight.
RNA-sequencing
Total RNA from the tissue of LD12:12 mice at ZT0, ZT6, ZT12, ZT18, Jet lag mice with solvent or melatonin was prepared with RNAiso Plus (Invitrogen, USA) at ZT6. The samples were sent to Novogene (Wuhan, China) for quantification, RNA-Seq library construction, and sequencing. In brief, purification of mRNA using poly-T oligo-attached magnetic beads, fragmentation, first- and second-strand cDNA synthesis and ligation of adaptors, cDNA fragments of 250~300 bp were selected and purified. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer, followed by purification of PCR products (AMPure XP system). RNA integrity of the sample libraries was assessed by Bioanalyzer 2100 system (Agilent Technologies, CA, USA). RNA sequencing was performed using the Novaseq platform (Illumina, San Diego, CA, USA), and 150 bp paired-end reads were generated. Fastq files were mapped to the mouse genome (NCBI37/mm9) using Hisat2 (version 2.0.5) [42]. The read numbers mapped to each gene were counted by featureCounts. Differential expression analysis of two conditions/groups (five biological replicates per condition) was performed using the DESeq2 R package (version 3.6.1). Pathway enrichment analyses of the Kyoto Encyclopedia of Genes and Genomes were performed to categorize the considerably enriched circadian rhythm. According to the literature [43], we used the JTK_Cycle analysis conducted by the MetaCycle package to obtain the circadian oscillation genes (https://openwetware.org/wiki/HughesLab:JTK_Cycle), including their amplitudes and phases. The gene density was the probability that the amplitude (upper) or phase (under) of a particular rhythm falls within the “unit width” at the x-axis, which was calculated using KernelDensity from “sklearn.”
Chondrocyte isolation, viral infection, and treatments
The growth plate cartilage slices of distal femora were isolated from the distal femora of 3-week-old rats. The cartilage slices were washed in sterile PBS containing 1% penicillin (100 U/mL)/streptomycin (100 μg/mL) (Beyotime), and then digested for 30 min with 0.25% pancreatin, followed by incubated with 0.2% type II collagenase in DMEM/F12 medium (Hyclone) overnight. After incubation, the cells were resuspended in DMEM/F12 medium containing 10% fetal bovine serum (Gibco), 1% penicillin (100 U/mL)/streptomycin (100 μg/mL). Then the cells were cultured in a humidified incubator at 37 °C, 5% CO2. The primary chondrocytes were transfected with lentivirus gene expression vector (hU6-MCS-CMV-Puromycin, GV112, GENECHEM Biosciences, China) containing rMtnr1a-RNAi (NM_053676) or lentivirus gene expression vector (hU6-MCS-CMV-Neomycin, GV152, GENECHEM Biosciences, China) containing rMtnr1b-RNAi (NM_001100641) sequence. For Bmal1 knockdown, the CRISPR/CAS9 system (pLV[Exp]-CBh>hCas9, VB160923-1033trt, REBIOSCI SCIENTIFIC, China) with Lentiviral Vector (pLV[2gRNA]-Puro-U6, VB180124-1097pnr, REBIOSCI SCIENTIFIC, China) was used. For IP-MS, the chondrocytes were transfected with lentivirus gene expression vector (Ubi-MCS-SV40-EGFP-IRES, GV367, GENECHEM Biosciences, China) containing rMtnr1a (NM_053676-6his) gene sequence. The details of the vectors target sequence are shown in Supplementary Table 2. Viral packaging was purchased from GENECHEM Biosciences Inc. (Shanghai, China). For viral inoculation, we incubated second-generation chondrocytes with the viral supernatants (MOI = 10) in DMEM/F12 supplemented with 10% FBS at 37 °C. For 12 h we replaced it with a fresh complete medium. For cell synchronization, the chondrocytes were grown to 70% confluency and synchronized by culturing in DMEM containing 1% serum for 24 h, followed by DMEM containing 50% serum for 2 h. Subsequently, the media was changed to DMEM containing 10% serum (normal condition, set as circadian time 0, CT0). Individual cells were harvested for total RNA and protein extraction every 4 h. For western blot, chondrocytes were incubated for 72 h in the presence of melatonin (1, 10, 50, 100, 200, 500, 1000 μmol/L), luzindole (100 μmol/L), 4P‐PDOT (100 μmol/L), Dorsomorphin (200 nmol/L), PD89059 (10 μmol/L), or a solvent control (Ethanol or DMSO or PBS ≤ 0.1%). The third-generation chondrocytes were used for EdU assay, and fifth-generation chondrocytes were used for matrix synthesis assay by immunofluorescence.
Co-IP/MS
6X-His-MTR1 overexpression, MTR1 wild-type along with the empty vector as negative control were applied to cross-linking Co-IP/MS. For all chondrocytes, 2 × 107 cells were fixed with 0.5% formaldehyde for 10 min at room temperature and terminated by adding 1.25 M glycine (10×) for 5 min. Cells were collected and washed three times in cold PBS. The pellets were suspended in 300 μL lysis buffer (50 mM HEPES/KOH, pH 7.5; 500 mM NaCl; 1% Triton X-100; 1 mM EDTA; 0.1% SDS and PI) with protein inhibitors (Complete Protease Inhibitor Cocktail, EDTA-free, Roche), and sonicated in Bioruptor Plus (Diagenode) at low power (2 s on, 1 s off) for 30 cycles and centrifuged at 12,000 rpm for 5 min. The supernatant was collected and precleared with 80 μL Protein A/G Magnetics Beads (MCE, HY-K0202, USA). Next, 2 μg 6X-His antibody was added to the lysate and incubated at 4 °C for 5 h. The beads were washed five times with lysis buffer and boiled in 40 μL SDS loading buffer for 15 min. The supernatant was separated by SDS–PAGE electrophoresis for one-third of the gel, which was minimally stained with Fast silver stain (Beyotime, P0017S, China). Liquid chromatography-tandem mass spectrometry analysis was performed with Orbitrap Fusion Lumos equipped with an online Easy-nLC 1000 nano-HPLC system (ThermoFisher) as previously described.
Western blot analysis
For western blot, chondrocytes were lysed in RIPA buffer containing protease and phosphatase inhibitors. Proteins were separated on the SDS–PAGE with 10% polyacrylamide gels, and transferred to PVDF membranes. The membrane was blocked in 5% nofat milk and then incubated with primary antibodies against BMAL1 (Abcam, ab93806, 1:1000), COL2α1 (Abcam, ab34712, 1:1000), SOX9 (Abcam, ab185966, 1:500), ACAN (ABclonal, 13880-1-AP, 1:800; ThermoFisher, MA3-16888, 1:500), COL10α1 (ABclonal, A11645, 1:1000), CRY1(Abcam, ab54649, 1:800), Phospho-CRY1-S506 (ABclonal, AP1236, 1:500), AMPKβ1(Abcam, ab32112, 1:1000), Phospho-AMPKβ1-S108 (ABclonal, AP0597, 1:1000), MEK1 (CST, 2352, 1:1000), Phospho-MEK1-S217 (ABclonal, AP00623, 1:1000), AMPKα1/α2 (Abcam, ab207442, 1:1000), Phospho-AMPKα1-T183/AMPKα2-T172 (ABclonal, AP0116, 1:500), CyclinD2 (ABclonal, A1773, 1:1000), 6X-His tag® (Abcam, ab18184, 1:1000), GAPDH (Proteintech, 10494-1-AP,1:10000) overnight at 4 °C. Next day, membranes were incubated with secondary anti-rabbit antibodies (Proteintech, SA00001-2, 1:5000) or anti-mouse antibodies (Proteintech, SA00001-1, 1:5000), followed by exposing in the appearance of the Western Blotting Detection Kit (GE Healthcare, cat#: RPN2106).
Histology and immunofluorescence
For Safranin O staining, femora were fixed by 4% paraformaldehyde and decalcified by 20% EDTA solution. The decalcified specimens were prepared for paraffin section and Safranin O staining. Safranin O staining was performed on the standard protocols. Immunofluorescence was performed using antibodies COL2α1 (Abcam, ab34712, 1:100; ThermoFisher, MA5-12789, 1:200), ACAN (ABclonal, 13880-1-AP, 1:50; ThermoFisher, MA3-16888, 1:100), COL10α1 (ABclonal, A6889, 1:100), SOX9 (Abcam, ab185966, 1:100), and fluorescent secondary antibody (1:200) was then added for 1 h. Digital images were taken with a laser scanning confocal microscope (Nikon, Japan) and NIS-Elements Viewer software.
Flow cytometry
To detecting cell-cycle alternation, dissociated growth plate cartilages tissue were obtained at ZT0, ZT6, ZT12, ZT18, and ZT24, single‐cell suspension (1 × 106 cells per milliliter) was prepared with 0.25% pancreatin for 30 min, 0.2% type II collagenase for 5 h, then fixed in cold 70% (vol/vol) ethanol and harvested on the second day. Single‐cell suspension was treated with 100 µg/mL RNase A (C1052, Beyotime, China), and stained with 50 µg/mL PI (C1052, Beyotime, China). Samples were then analyzed on a FACS Calibur (BD Biosciences). Cell-cycle distribution was analyzed using FlowJo (Tree Star Inc.) by fitting the histogram of DNA content to the Watson Pragmatic Model.
EdU staining assay
For in vitro EdU assay, the third-generation chondrocytes were cultured for 2 h in DMEM/F12 medium containing 20 μmol/L EdU (BeyoClick™ EdU-488, China). The cells were fixed and permeabilized by 0.5% Triton X-100 for 10 min and were stained by Azide Alexa Fluor 488. For in vivo EdU staining assay, mice were injected intraperitoneally with EdU (ST067-250mg, Beyotime, China) at a dose of 20 μg/g 4 h before harvest. According to the manufacturer’s instructions, paraffin sections were subjected to a click reaction (BeyoClick™ EdU-594, China).
Quantitative reverse transcription (qRT)-PCR analysis
Total RNA was isolated from the growth plate tissue of distal femora with RNAiso Plus (Invitrogen, USA). The cDNA was synthesized using the HiScript® II Q Select RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China). Quantitative analysis was performed with the ChamQTM SYBR® qPCR Master Mix (Vazyme, China). The specific primers used are listed in Supplementary Table 1. GAPDH was used for normalization, and the relative mRNA expression was calculated by the 2–△△Ct method.
Statistical analysis
Sample sizes were determined based on the amount of data required to give the statistical significance. The numbers of in vitro and in vivo experiments used for each analysis are indicated in each figure legend. All data were presented as mean ± standard deviation. The GraphPad Prism 8.0 software was used for statistical analysis. The Shapiro–Wilk test was used to analyze the normality and equal distribution of variance between the different groups. For data with normal distribution, statistical analyses were performed with Student’s t-test or ANOVA analyses of variance unless otherwise stated. P value <0.05 was considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Prof. Luoying Zhang for valuable advice.
Author contributions
SY and QT conceived and designed experiments; SY, GC, and XL performed experiments; SY, GC, XL, YY, MX, YL, WZ, and FG participated in the data acquisition; SY analyzed and interpreted the data; QT and SY wrote the paper; LS and AS gave crucial advice and technical support; LC provided funding, supervised all the experiments, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Natural Science Foundation of China for Excellent Young Scholars (31725011, to LC) and Key Program Projects (82030070, to LC), Hubei Provincial Natural Science Fund for Creative Research (2020CFA014, to LC), and Innovation Team Project by Health Commission of Hubei Province (WJ2019C001, to LC).
Data availability
All data supporting the results reported here are available in the article and Supplementary files or from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethical approval
All animal studies were approved by the Institutional Animal Care and Use Committee of Tongji Medical College (IACUC number: S739).
Footnotes
Edited by M Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shaoling Yu, Qingming Tang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00919-4.
References
- 1.Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 2.Pacifici M. Retinoid roles and action in skeletal development and growth provide the rationale for an ongoing heterotopic ossification prevention trial. Bone. 2018;109:267–75. doi: 10.1016/j.bone.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–6. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 4.Krakow D, Rimoin DL. The skeletal dysplasias. Genet Med. 2010;12:327–41. doi: 10.1097/GIM.0b013e3181daae9b. [DOI] [PubMed] [Google Scholar]
- 5.Stoll C, Dott B, Roth MP, Alembik Y. Birth prevalence rates of skeletal dysplasias. Clin Genet. 1989;35:88–92. doi: 10.1111/j.1399-0004.1989.tb02912.x. [DOI] [PubMed] [Google Scholar]
- 6.Voss LD, Sandberg DE. The psychological burden of short stature: evidence against. Eur J Endocrinol. 2004;151:S29–33. doi: 10.1530/eje.0.151s029. [DOI] [PubMed] [Google Scholar]
- 7.Long F, Ornitz DM. Development of the endochondral skeleton. Cold Spring Harb Perspect Biol. 2013;5:a008334. doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hata K, Takahata Y, Murakami T, Nishimura R. Transcriptional network controlling endochondral ossification. J Bone Metab. 2017;24:75–82. doi: 10.11005/jbm.2017.24.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams SL, Cohen AJ, Lassova L. Integration of signaling pathways regulating chondrocyte differentiation during endochondral bone formation. J Cell Physiol. 2007;213:635–41. doi: 10.1002/jcp.21262. [DOI] [PubMed] [Google Scholar]
- 10.Takarada T, Kodama A, Hotta S, Mieda M, Shimba S, Hinoi E, et al. Clock genes influence gene expression in growth plate and endochondral ossification in mice. J Biol Chem. 2012;287:36081–95. doi: 10.1074/jbc.M112.408963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akagi R, Akatsu Y, Fisch KM, Alvarez-Garcia O, Teramura T, Muramatsu Y, et al. Dysregulated circadian rhythm pathway in human osteoarthritis: NR1D1 and BMAL1 suppression alters TGF-beta signaling in chondrocytes. Osteoarthr Cartil. 2017;25:943–51. doi: 10.1016/j.joca.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maronde E, Schilling AF, Seitz S, Schinke T, Schmutz I, van der Horst G, et al. The clock genes Period 2 and Cryptochrome 2 differentially balance bone formation. PLoS One. 2010;5:e11527. doi: 10.1371/journal.pone.0011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudek M, Gossan N, Yang N, Im HJ, Ruckshanthi JP, Yoshitane H, et al. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J Clin Investig. 2016;126:365–76. doi: 10.1172/JCI82755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, et al. Circadian clock interaction with HIF1alpha mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab. 2017;25:86–92. doi: 10.1016/j.cmet.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu S, Tang Q, Xie M, Zhou X, Long Y, Xie Y, et al. Circadian BMAL1 regulates mandibular condyle development by hedgehog pathway. Cell Prolif. 2020;53:e12727. doi: 10.1111/cpr.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–62. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cermakian N, Sassone-Corsi P. Multilevel regulation of the circadian clock. Nat Rev Mol cell Biol. 2000;1:59–67. doi: 10.1038/35036078. [DOI] [PubMed] [Google Scholar]
- 18.Gengatharan A, Malvaut S, Marymonchyk A, Ghareghani M, Snapyan M, Fischer-Sternjak J, et al. Adult neural stem cell activation in mice is regulated by the day/night cycle and intracellular calcium dynamics. Cell. 2021;184:709–722. doi: 10.1016/j.cell.2020.12.026. [DOI] [PubMed] [Google Scholar]
- 19.Nobel A. Pursuit may not run like clockwork. Cell. 2017;171:1246–51. doi: 10.1016/j.cell.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Zhou X, Tang Q, Yu R, Yu S, Long Y, et al. BMAL1 deficiency contributes to mandibular dysplasia by upregulating MMP3. Stem Cell Rep. 2018;10:180–95. doi: 10.1016/j.stemcr.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jockers R, Delagrange P, Dubocovich ML, Markus RP, Renault N, Tosini G, et al. Update on melatonin receptors: IUPHAR Review 20. Br J Pharmacol. 2016;173:2702–25. doi: 10.1111/bph.13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu S, Kuwahara M, Uchida Y, Koudo S, Hayashi D, Shimomura Y, et al. Circadian production of melatonin in cartilage modifies rhythmic gene expression. J Endocrinol. 2019;241:161–73. [DOI] [PubMed]
- 23.Maria S, Samsonraj RM, Munmun F, Glas J, Silvestros M, Kotlarczyk MP, et al. Biological effects of melatonin on osteoblast/osteoclast cocultures, bone, and quality of life: Implications of a role for MT2 melatonin receptors, MEK1/2, and MEK5 in melatonin-mediated osteoblastogenesis. J Pineal Res. 2018;64:e12465. [DOI] [PMC free article] [PubMed]
- 24.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–40. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang LN, Xu L, Zhou HY, Wu LY, Li YY, Pang T, et al. Novel small-molecule AMP-activated protein kinase allosteric activator with beneficial effects in db/db mice. PLoS One. 2013;8:e72092. doi: 10.1371/journal.pone.0072092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons DJ. Diurnal periodicity in epiphyseal growth cartilage. Nature. 1962;195:82–83. doi: 10.1038/195082a0. [DOI] [PubMed] [Google Scholar]
- 27.Igarashi K, Saeki S, Shinoda H. Diurnal rhythms in the incorporation and secretion of 3H-proline and 3H-galactose by cartilage cells and osteoblasts in various bone-forming sites in growing rats. Orthodontic Waves. 2013;72:11–15. doi: 10.1016/j.odw.2012.09.001. [DOI] [Google Scholar]
- 28.Zimmerman NH, Menaker M. The pineal gland: a pacemaker within the circadian system of the house sparrow. Proc Natl Acad Sci USA. 1979;76:999–1003. doi: 10.1073/pnas.76.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandi-Perumal SR, Srinivasan V, Maestroni GJ, Cardinali DP, Poeggeler B, Hardeland R. Melatonin: nature’s most versatile biological signal? FEBS J. 2006;273:2813–38. doi: 10.1111/j.1742-4658.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- 30.Pei M, He F, Wei L, Rawson A. Melatonin enhances cartilage matrix synthesis by porcine articular chondrocytes. J Pineal Res. 2009;46:181–7. doi: 10.1111/j.1600-079X.2008.00646.x. [DOI] [PubMed] [Google Scholar]
- 31.Hong Y, Kim H, Lee Y, Lee S, Kim K, Jin Y, et al. Salutary effects of melatonin combined with treadmill exercise on cartilage damage. J Pineal Res. 2014;57:53–66. doi: 10.1111/jpi.12143. [DOI] [PubMed] [Google Scholar]
- 32.Cecon E, Oishi A, Jockers R. Melatonin receptors: molecular pharmacology and signalling in the context of system bias. Br J Pharmacol. 2018;175:3263–80. doi: 10.1111/bph.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim HD, Kim YS, Ko SH, Yoon IJ, Cho SG, Chun YH, et al. Cytoprotective and anti-inflammatory effects of melatonin in hydrogen peroxide-stimulated CHON-001 human chondrocyte cell line and rabbit model of osteoarthritis via the SIRT1 pathway. J Pineal Res. 2012;53:225–37. doi: 10.1111/j.1600-079X.2012.00991.x. [DOI] [PubMed] [Google Scholar]
- 34.Ohba S. Hedgehog signaling in skeletal development: roles of Indian hedgehog and the mode of its action. Int J Mol Sci. 2020;21:6665. [DOI] [PMC free article] [PubMed]
- 35.Chiu G, Chang C, Roberts WE. Interdisciplinary treatment for a compensated Class II partially edentulous malocclusion: orthodontic creation of a posterior implant site. Am J Orthod Dentofacial Orthop. 2018;153:422–35. doi: 10.1016/j.ajodo.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 36.Gao W, Lin M, Liang A, Zhang L, Chen C, Liang G, et al. Melatonin enhances chondrogenic differentiation of human mesenchymal stem cells. J Pineal Res. 2014;56:62–70. doi: 10.1111/jpi.12098. [DOI] [PubMed] [Google Scholar]
- 37.Zhong ZM, Li T, Xu ZX, Meng TT, Zeng JH, Zheng S, et al. Effect of melatonin on the proliferation and differentiation of chondrocytes from rat vertebral body growth plate in vitro. Int J Med Sci. 2013;10:1392–8. doi: 10.7150/ijms.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu L, Zhang L, Wang Z, Li C, Li S, Li L, et al. Melatonin suppresses estrogen deficiency-induced osteoporosis and promotes osteoblastogenesis by inactivating the NLRP3 inflammasome. Calcif Tissue Int. 2018;103:400–10. doi: 10.1007/s00223-018-0428-y. [DOI] [PubMed] [Google Scholar]
- 39.Mir AR, Siddiqui H, Alam P, Hayat S. Melatonin modulates photosynthesis, redox status, and elemental composition to promote growth of Brassica juncea—a dose-dependent effect. Protoplasma. 2020;257:1685–1700. doi: 10.1007/s00709-020-01537-6. [DOI] [PubMed] [Google Scholar]
- 40.Estrada-Reyes R, Valdés-Tovar M, Arrieta-Baez D. The Timing of Melatonin Administration Is Crucial for Its Antidepressant-Like Effect in Mice. 2018;19:2278. [DOI] [PMC free article] [PubMed]
- 41.Habtemariam S, Daglia M, Sureda A, Selamoglu Z, Gulhan MF, Nabavi SM. Melatonin and respiratory diseases: a review. Curr Top Med Chem. 2017;17:467–88. doi: 10.2174/1568026616666160824120338. [DOI] [PubMed] [Google Scholar]
- 42.Kim D, Paggi JM, Park C, Bennett C. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–15. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welz PS, Zinna VM, Symeonidi A, Koronowski KB, Kinouchi K, Smith JG, et al. BMAL1-driven tissue clocks respond independently to light to maintain homeostasis. Cell. 2019;177:1436–47. doi: 10.1016/j.cell.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the results reported here are available in the article and Supplementary files or from the corresponding author upon reasonable request.