Abstract
The two main glucan polymers cellulose and callose in plant cell wall are synthesized at the plasma membrane by cellulose or callose synthase complexes. Cellulose is the prevalent glucan in cell wall and provides strength to the walls to support directed cell expansion. By contrast, callose is mainly produced in special cell wall and exercises important functions during development and stress responses. However, the structure and precise regulatory mechanism of callose synthase complex is not very clear. This review therefore compares and analyzes the regulation of callose and cellulose synthesis, and further emphasize the future research direction of callose synthesis.
Keywords: Callose, Callose synthase, Higher plants
Callose, Callose synthase, Higher plants.
1. Introduction
The cell wall is essential for plant growth and development. All plant cells are wrapped in cell walls, which protect plants from environmental hazards, define the cell expansion supporting plant morphology, and allow the transport of substances between cells and from roots to stems [1, 2].
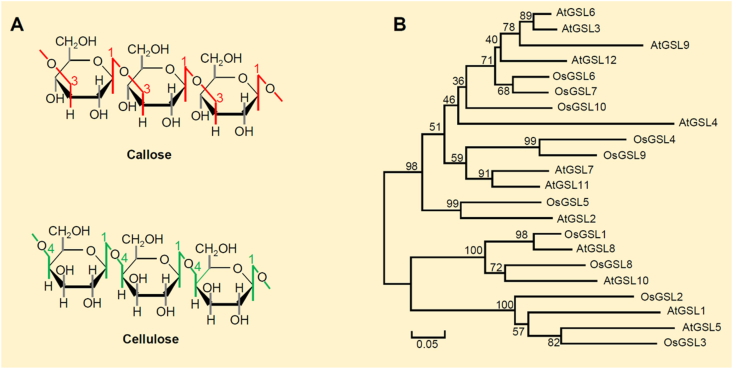
Although the composition and structure of the cell wall varied in response to environmental and developmental factors, the polysaccharides (including callose, cellulose, hemicellulose and pectin) provide the basic structure for the cell wall [2, 3, 4]. Callose and cellulose have a similar primary structure (Figure 1A). Callose mainly consists of repeating glucose units linked by β-1,3 glycosidic bonds, while cellulose is a polysaccharide of glucose mainly linked by β-1,4-glycosidic bonds [5, 6]. Different from hemicellulose and pectin that are synthesized and assembled in the Golgi apparatus, secreted into the apoplast, and modified and integrated into the cell wall, cellulose and callose are synthesized by plasma membrane-located glucan synthase complex, cellulose synthase (CesAs) and callose synthase (CalS also known as GSL for GLUCAN SYNTHASE-LIKE) [7, 8]. The two enzymes share UDP-glucose as the initial substrate [5]. Therefore, callose and cellulose may have similar synthesis mechanisms.
Figure 1.
Callose structure, and phylogenetic tree analysis of callose synthases. (A) Comparison of the structure of callose and cellulose. (B) Phylogenetic tree analysis of callose synthases (GSLs) in rice and Arabidopsis.
In this review, we summarized and analyzed the regulatory mechanism of callose biosynthesis and function in comparison to the cellulose. In addition, we especially focus on highlighting the unexplored areas and identifying new biological questions that need to be addressed to advance the understanding of callose synthesis regulation.
2. Callose synthase in higher plants
GSLs located on the plasma membrane directly catalyzes the synthesis of callose from UDP-glucose [5]. GSLs are widespread among various plant species and are present in special cell walls. Callose is synthesized by GSLs to respond to a wide range of environmental and physiological signals [9]. Like CesAs, the GSLs in plants often contains multiple members, forming a moderately sized gene family. In Arabidopsis, there are 12 GSL genes which were named as GSL1-GSL12 [9], while there are 10 predicted GSL genes in rice. Phylogenetic analysis revealed that the proteins are divided into two groups (Figure 1B). 4 rice GSLs (OsGSL1, OsGSL2, OsGSL3 and OsGSL8) and 4 Arabidopsis GSLs (AtGSL1, AtGSL5, AtGSL8 and AtGSL10) are grouped in a small clade. Other GSLs formed the neighboring subgroup. According to the functional characteristics of GSL genes, members of different subfamilies play partially redundant roles in regulation of multiple processes including plant growth, development and stress responses. A single GSL gene could be involved in multiple biological processes.
3. The function of callose in higher plants
The rigidity of cellulose supports the complete structure of plant cells, while also protecting plant cells from the influence of external stress [7, 10]. Callose also plays a role of support and barrier. However, callose is only distributed in specific cell wall, such as cell plates, outer wall of pollen wall and plasmodesmata [11, 12, 13, 14]. These all indicate a special function of callose in plants.
3.1. The role of callose in plant growth and development
During the process of cell division in higher plants, the cell plate determines correct assembly of cell wall layer. Callose forms a coat-like structure covering the surface of cell Plates [14, 15]. The changing process from the tubular network to a porous cell plate to the mature cell wall may be driven mainly by the synthesis of callose. Mutation of callose synthase gene GSL8/MASSUE delays callose deposition in the cell plates [16]. Moreover, gsl8/massue mutant displays a typical cytokinesis defect phenotype, such as bi- or multi-nucleate, with cell-wall stubs [16]. Therefore, callose regulates cell division through cell plates in plants.
In the process of plant reproduction development, callose is deposited between the primary cell wall and the plasma membrane, which can be used to maintain the morphology of the microspore mother cell and prevent the fusion and aggregation of the tetrad cells [17, 18]. AtGSL1 and AtGSL5 are responsible for forming the callose wall that separates the tetrad cells, and atgsl1 and atgsl5 mutants can cause abnormal microspore development [19]. In addition, callose seem to participate in the formation of primexine by providing mold for the construction of the outer wall of pollen during microspore formation. Mutation of AtGSL2 affects the deposition of callose in the primary cell wall of microspores and cannot form a proper sculpted outer wall, leading to the production of abortive pollen grains and male sterility [17, 20]. OsGSL5, homologous gene of AtGSL2 in rice, played a similar function [21]. Thus, callose is essential to plant reproductive development.
In higher plants, plasmodesmata between cells regulates the transport of nutrients and signal molecules between cells [22]. The callose is often found to be deposited in the plasmodesmata. The deposition of callose determines the size exclusion limit (SEL) of the plasmodesmata, which affects their permeability [22]. The change in permeability of plasmodesmata affects the development of stomata and sieve, the phototropism, and the transportation of phloem [23, 24, 25, 26]. Arabidopsis gsl8/chor mutant also showed reduced callose deposition in plasmodesmata, and increased SEL in leaf epidermal cells. Increased SEL affected stomatal development and phototropic response through regulating the distribution of stomatal's developmental regulator SPCH and auxin, respectively [24]. CANNOT REACH THE ROOF 1 (CRR1) in rice encodes a homologous protein of Arabidopsis GSL8 [23]. The crr1 mutant shows delayed ovary expansion and defective vascular cell pattern. It indicates that the increase in plasmodesmata permeability may induce the diffusion of cell fate determinants from the sieve element precursor cells to the adjacent dedifferentiated parenchyma cells, leading to the formation of clusters of sieve elements [23]. The Arabidopsis GSL7 gene regulates the deposition of callose at plasmodesmata in early-stage sieve plates, which results in sieve elements with fewer PD pores and affects phloem transport [25, 26]. In short, by controlling the developmental signals and the symplast transport mechanism of different solutes, callose in plasmodesmata is essential for plant development.
3.2. Callose deposition in response to stress
It is generally believed that callose positively regulates the stress response of plants [27, 28, 29, 30]. Under abiotic and biotic stress, plants often trigger the accumulation of callose. This process happens in a few seconds [29]. Callose deposition in cell wall, plasmodesmata, and sieve pore controls the cell wall permeability, which prevents the further penetration of the pathogen and deleterious metals into the tissue, and the loss of cellular water and solute to maintain the internal stabilities of cells [31].
Among the various abiotic stresses, chilling stress [32], water stress [33], heat [34], and heavy metals [35] are the major abiotic stresses challenging overall plant growth. These abiotic stresses could induce callose deposition. Action potentials arose in the phloem of maize leaf tips after chilling, and phloem transport of photoassimilates is inhibited [32]. This process is closely associated with callose deposition in leaf, which may be caused by the occlusion of plasmodesmata and phloem sieve pores. Tocopherol prevents abnormal callose deposition in phloem tissue, maintaining the transport of photoassimilates in cold conditions [36, 37]. Knockdown or knockdown of tocopherol cyclase in maize and potato resulted in vascular-specific callose deposition in leaves and defective sucrose export. Heavy metal ions such as Al3+ and Cd2+ are also involved in regulating callose deposition in plants [38]. Depolarization of the plasma membrane caused by aluminum induces callose deposition in tobacco cells [39]. In addition, increase of calcium concentration in the cytoplasm is required for aluminum-induced callose deposition [40]. These studies suggest that callose is involved in the regulation of plant resistance to abiotic stress.
Diverse biotic stresses also cause callose deposition in plants. Viruses and the blast fungus infection triggers plasmodesmata callose deposition that limit cell-to-cell spread of viruses and the blast fungus [41]. Different Chrysanthemum species exhibited the different abilities to tolerate the chrysanthemum stunt viroid (CSVd) infection [42]. The level of plasmodesmata callose was much higher in the tolerant cultivars than the sensitive ones. Callose deposition may be an early defense mechanism in plants against pathogen attack. In contrast, studies have reported that callose also negatively regulates the disease resistance of plants. The loss-of-function mutants of GSL5/PMR4/CalS12 cannot synthesize callose in papillae, a physical barrier to slow pathogen invasion [43]. Unexpectedly, the reduction of callose in the gsl5 mutant makes plants more resistant to powdery mildew [44]. Further study found that the lack of callose in pmr4 mutants can enhance salicylic acid signaling, leading to increased resistance to pathogens [44]. These studies have shown that the response of plant pathogens involves multiple mechanisms in addition to callose. How to balance these two effects of callose in the stress response process, and the regulatory relationship between callose and other anti-stress factors remains to be studied.
4. Regulation of callose biosynthesis
4.1. Transcriptional regulation of callose synthase gene
Unlike cellulose, the production of callose in plants is relatively rare, ranging from about 0.3% (total cell wall content) in Arabidopsis to about 5% in the energy crop Miscanthus x giganteus [45]. In fact, callose induced by diverse development and stress responses is only deposited in a special cell wall, which may explain the specific expression of GSL gene family members cell or tissue types [9]. For instance, OsGSL5 specifically highly expressed in the anther has been proved to participate in callose deposition during rice anther development [21]. However, precise regulation mechanism of specific expression of callose synthase gene has not been demonstrated unequivocally. Plant hormones are believed to play a key role in this process. The plant hormones auxin, abscisic acid (ABA), gibberellin (GA), and salicylic acid (SA) regulate the expression of GSLs [24, 46, 47, 48, 49]. Auxin induces AtGSL8 expression through the auxin-responsive transcription factor ARF7 [24]. Then the asymmetric deposition of callose at plasmodesmata cause the auxin gradient in the Arabidopsis hypocotyl, which forms a feedback regulation loop and affects the phototropic of Arabidopsis [24]. High levels of ABA in poplar buds induces the expression of callose synthase 1 under short-day conditions, resulting in a large amount of callose accumulated at plasmodesmata [49]. Jasmonic acid (JA) and salicylic acid (SA) play crucial roles in the defense signaling pathways. During the flg22-triggered innate immune response, SA induces callose deposition in plasmodesmata, but JA plays an opposite role [33, 50]. Interestingly, JA was reported to be required for proper callose deposition during necrotrophic pathogens-triggered basal defense response [51]. SA enhances callose deposition in plasmodesmata through the action of GSL6/CalS1 and PDLP5 (plasmodesmal protein 5). PDLP5-mediated callose deposition are dependent on the EDS1/ICS/NPR1-associated SA signaling pathway. Future research might uncover details of how transcription factors regulate the expression pattern of GSLs in response to plant hormones.
4.2. Post-transcriptional regulation of callose synthesis
Cellulose is synthesized from CesA complexes with a diameter of 40–60 nm [52], which usually consists of a heterotrimeric CesAs and related proteins on the plasma membrane [7]. It is generally believed that GSLs will also form a complex similar to CesAs [9]. In Arabidopsis, GSL6 can be purified together with the cell-wall associated proteins, phragmoplastin, and UGT1 [53], so it is speculated that the GSL complexes may contain monomeric GTPase, UDP-glucose transferase (UGT), annexin and sucrose synthase. The proteins of these complexes may regulate the activity of GSLs to a certain extent.
The CesA complexes localized to the plasma membrane are assembled in the Golgi apparatus [7], and then transported to the plasma membrane through the trans-Golgi network (TGN) and small vesicle compartments [54, 55]. GSLs do not contain the signal peptides required for conventional secretory pathways, and their subcellular localization seems to require EXO70 family-mediated exocysts [56]. Whether the transport of GSL complexes is mediated by TGN still needs further corroboration.
Considering relatively slow callose deposition by transcriptionally regulated GSLs, it seems likely that rapid accumulation of callose in plants might require modulation by faster post-translational modification regulation. GSLs are predicted to have multiple phosphorylation sites. Various abiotic and biotic stresses may induce the phosphorylation of multiple GSLs in Arabidopsis [57, 58, 59, 60, 61, 62, 63, 64]. The additional study from yeast indicated that phosphorylation status of GSLs regulates their activity [65, 66, 67]. For CesAs, phosphorylation is an important regulatory player [68]. A subset of phosphorylation sites mutation reveals different functions of phosphorylation for CesAs [69].
Secondary messengers such as Ca2+ and ROS are involved in callose deposition induced by stress [70]. the plasma membrane NADPH oxidases (RBOHs) are activated by stress to produce apoplastic ROS [71]. However, the mechanism by which stress-induced ROS controls callose deposition remains to be identified. CalS8/GSL4 has been identified as the key enzyme synthesizing plasmodesmal callose in response to ROS [33]. As the expression of GSL4 was not affected by H2O2 treatment, ROS-induced GSL4 activation may be subjected to post-transcriptional regulation. How GSL4 activity is controlled remains to be elucidated. Signaling players such as receptor-like proteins (RLPs) or receptor-like kinases (RLKs) possessing a ROS sensor ectodomain may link the signaling between ROS and GSL4 activation. Ca2+ sensed by CML (CALMODULIN-LIKE) proteins may act as the downstream of ROS [72]. One possibility is that CML member can bind to GSLs, which may directly activate GSLs. GSL5 might be a targeted regulation enzyme of CML41 during flg22-triggered immune response [29, 73]. The relationship between post-translational modification and GSL complex assembly and enzyme activity, as well as other post-translational protein modifications remains to be studied.
5. Future perspectives
In the context of climate change and environmental pollution, we are facing drought, high temperature, and heavy metal pollution, which are bringing about negative effects on many agricultural ecosystems around the world [74]. As an important plant glucan polymer, callose is involved in not only plant development but also response to various stresses. Genetic manipulation of enzyme activity and gene expression of GSLs therefore may contribute to improving the ecosystem function and agricultural productivity.
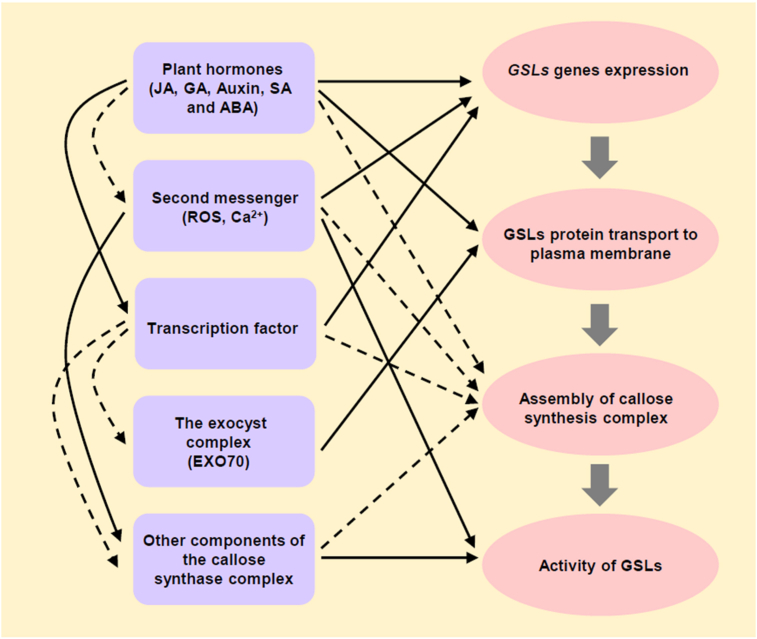
Like cellulose, callose is an important part of cell walls in plants. Both have similar primary structures, synthases, and common substrates. However, the difference is that callose only exists in special cell walls, and callose plays a different role from cellulose in the process of plant growth and development. Therefore, we give a general overview and brief summarization of the current understandings of function and the regulatory mechanism of callose biosynthesis (see Figure 2) in higher plants. Additionally, several critical questions remain unanswered: (i) What are the components of the callose synthase complex? (ii) how the callose synthase complex is assembled? (iii)How plants precisely regulate the function of the callose synthase complex? Although the assembly and regulation of callose synthase complex lack detailed potential molecular mechanisms, the comparative study of callose and cellulose should provide profound enlightenment and valuable insights for understanding their special functions and regulatory mechanisms in plants.
Figure 2.
Regulation of callose biosynthesis.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This work was supported by Zhejiang Provincial Natural Science Foundation of China (LGN22C130014), the National Natural Science Foundation of China (31600242), and the Young Talent Training Project of Zhejiang Academy of Agricultural Sciences (2019R24R08E01).
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We acknowledge with apologies the many excellent studies and topics that space did not permit us to discuss or cite.
References
- 1.Somerville C., et al. Toward a systems approach to understanding plant cell walls. Science. 2004;306:2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- 2.Vaahtera L., Schulz J., Hamann T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants. 2019;5:924–932. doi: 10.1038/s41477-019-0502-0. [DOI] [PubMed] [Google Scholar]
- 3.Mohnen D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Scheller H.V., Ulvskov P. Hemicelluloses. Annu. Rev. Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 5.Galatis B., Apostolakos P. A new callose function: involvement in differentiation and function of fern stomatal complexes. Plant Signal. Behav. 2010;5:1359–1364. doi: 10.4161/psb.5.11.12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barghahn S., et al. Mixed linkage β-1, 3/1, 4-glucan oligosaccharides induce defense responses in Hordeum vulgare and Arabidopsis thaliana. Front. Plant Sci. 2021;12:1201. doi: 10.3389/fpls.2021.682439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McFarlane H.E., Döring A., Persson S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014;65:69–94. doi: 10.1146/annurev-arplant-050213-040240. [DOI] [PubMed] [Google Scholar]
- 8.Ellinger D., Voigt C.A. Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade. Ann. Bot. 2014;114:1349–1358. doi: 10.1093/aob/mcu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verma D.P.S., Hong Z. Plant callose synthase complexes. Plant Mol. Biol. 2001;47:693–701. doi: 10.1023/a:1013679111111. [DOI] [PubMed] [Google Scholar]
- 10.Polko J.K., Kieber J.J. The regulation of cellulose biosynthesis in plants. Plant Cell. 2019;31:282–296. doi: 10.1105/tpc.18.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy A., Erlanger M., Rosenthal M., Epel B.L. A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J. 2007;49:669–682. doi: 10.1111/j.1365-313X.2006.02986.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen X.Y., et al. The Arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol. 2009;150:105–113. doi: 10.1104/pp.108.133918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radja A., Horsley E.M., Lavrentovich M.O., Sweeney A.M. Pollen cell wall patterns form from modulated phases. Cell. 2019;176:856–868.810. doi: 10.1016/j.cell.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Han X., et al. Plasmodesmata-related structural and functional proteins: the long sought-after secrets of a cytoplasmic channel in plant cell walls. Int. J. Mol. Sci. 2019;20:2946. doi: 10.3390/ijms20122946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samuels A.L., Giddings T.H., Jr., Staehelin L.A. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. JCB (J. Cell Biol.) 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiele K., et al. The timely deposition of callose is essential for cytokinesis in Arabidopsis. Plant J. 2009;58:13–26. doi: 10.1111/j.1365-313X.2008.03760.x. [DOI] [PubMed] [Google Scholar]
- 17.Dong X., Hong Z., Sivaramakrishnan M., Mahfouz M., Verma D.P. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 2005;42:315–328. doi: 10.1111/j.1365-313X.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang B., et al. Rice LecRK5 phosphorylates a UGPase to regulate callose biosynthesis during pollen development. J. Exp. Bot. 2020;71:4033–4041. doi: 10.1093/jxb/eraa180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enns L.C., et al. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol. Biol. 2005;58:333–349. doi: 10.1007/s11103-005-4526-7. [DOI] [PubMed] [Google Scholar]
- 20.Nishikawa S., Zinkl G.M., Swanson R.J., Maruyama D., Preuss D. Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol. 2005;5:22. doi: 10.1186/1471-2229-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi X., et al. GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol. 2015;56:497–509. doi: 10.1093/pcp/pcu193. [DOI] [PubMed] [Google Scholar]
- 22.Wu S.W., Kumar R., Iswanto A.B.B., Kim J.Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018;69:5325–5339. doi: 10.1093/jxb/ery317. [DOI] [PubMed] [Google Scholar]
- 23.Song L., Wang R., Zhang L., Wang Y., Yao S. CRR1 encoding callose synthase functions in ovary expansion by affecting vascular cell patterning in rice. Plant J. 2016;88:620–632. doi: 10.1111/tpj.13287. [DOI] [PubMed] [Google Scholar]
- 24.Han X., et al. Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev. Cell. 2014;28:132–146. doi: 10.1016/j.devcel.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Barratt D.H., et al. Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol. 2011;155:328–341. doi: 10.1104/pp.110.166330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie B., Wang X., Zhu M., Zhang Z., Hong Z. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 2011;65:1–14. doi: 10.1111/j.1365-313X.2010.04399.x. [DOI] [PubMed] [Google Scholar]
- 27.Flors V., Ton J., Jakab G., Mauch-Mani B. Abscisic acid and callose: team players in defence against pathogens? J. Phytopathol. 2005;153:377–383. [Google Scholar]
- 28.Krzeslowska M. The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy. Acta Physiol. Plant. 2011;33:35–51. [Google Scholar]
- 29.Luna E., et al. Callose deposition: a multifaceted plant defense response. Mol. Plant Microbe Interact. 2011;24:183–193. doi: 10.1094/MPMI-07-10-0149. [DOI] [PubMed] [Google Scholar]
- 30.Hunter K., et al. CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 2019;180:2004–2021. doi: 10.1104/pp.19.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piršelová B., Matušíková I. Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol. Plant. 2013;35:635–644. [Google Scholar]
- 32.Fromm J., Hajirezaei M.-R., Becker V.K., Lautner S. Electrical signaling along the phloem and its physiological responses in the maize leaf. Front. Plant Sci. 2013;4:239. doi: 10.3389/fpls.2013.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui W., Lee J.-Y. Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants. 2016;2:1–9. doi: 10.1038/nplants.2016.34. [DOI] [PubMed] [Google Scholar]
- 34.Rinne P.L., et al. Tobacco plants respond to the constitutive expression of the Tospovirus movement protein NSM with a heat-reversible sealing of plasmodesmata that impairs development. Plant J. 2005;43:688–707. doi: 10.1111/j.1365-313X.2005.02489.x. [DOI] [PubMed] [Google Scholar]
- 35.Ueki S., Citovsky V. Vol. 102. 2005. Identification of an interactor of cadmium ion-induced glycine-rich protein involved in regulation of callose levels in plant vasculature; pp. 12089–12094. (Proceedings of the National Academy of Sciences). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofius D., et al. RNAi-mediated tocopherol deficiency impairs photoassimilate export in transgenic potato plants. Plant Physiol. 2004;135:1256–1268. doi: 10.1104/pp.104.043927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Botha C.E., Cross R.H., Van Bel A., Peter C.I. Phloem loading in the sucrose-export-defective (SXD-1) mutant maize is limited by callose deposition at plasmodesmata in bundle sheath-vascular parenchyma interface. Protoplasma. 2000;214:65–72. [Google Scholar]
- 38.O’Lexy R., et al. Exposure to heavy metal stress triggers changes in plasmodesmatal permeability via deposition and breakdown of callose. J. Exp. Bot. 2018;69:3715–3728. doi: 10.1093/jxb/ery171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivaguru M., Yamamoto Y., Rengel Z., Ahn S.J., Matsumoto H. Early events responsible for aluminum toxicity symptoms in suspension-cultured tobacco cells. New Phytol. 2005;165:99–109. doi: 10.1111/j.1469-8137.2004.01219.x. [DOI] [PubMed] [Google Scholar]
- 40.Bhuja P., McLachlan K., Stephens J., Taylor G. Accumulation of 1, 3-β-D-glucans, in response to aluminum and cytosolic calcium in Triticum aestivum. Plant Cell Physiol. 2004;45:543–549. doi: 10.1093/pcp/pch068. [DOI] [PubMed] [Google Scholar]
- 41.Beffa R.S., Hofer R.-M., Thomas M., Meins F., Jr. Decreased susceptibility to viral disease of [beta]-1, 3-glucanase-deficient plants generated by antisense transformation. Plant Cell. 1996;8:1001–1011. doi: 10.1105/tpc.8.6.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., et al. Invasion of shoot apical meristems by Chrysanthemum stunt viroid differs among Argyranthemum cultivars. Front. Plant Sci. 2015;6:53. doi: 10.3389/fpls.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobs A.K., et al. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell. 2003;15:2503–2513. doi: 10.1105/tpc.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura M.T., et al. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science. 2003;301:969–972. doi: 10.1126/science.1086716. [DOI] [PubMed] [Google Scholar]
- 45.Falter C., et al. Glucanocellulosic ethanol: the undiscovered biofuel potential in energy crops and marine biomass. Sci. Rep. 2015;5:13722. doi: 10.1038/srep13722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., et al. Mechanisms of callose deposition in rice regulated by exogenous abscisic acid and its involvement in rice resistance to Nilaparvata lugens Stal (Hemiptera: Delphacidae) Pest Manag. Sci. 2017;73:2559–2568. doi: 10.1002/ps.4655. [DOI] [PubMed] [Google Scholar]
- 47.Iriti M., Faoro F. Abscisic acid is involved in chitosan-induced resistance to tobacco necrosis virus (TNV) Plant Physiol. Biochem. 2008;46:1106–1111. doi: 10.1016/j.plaphy.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Alazem M., He M.H., Moffett P., Lin N.S. Abscisic acid induces resistance against bamboo mosaic virus through Argonaute2 and 3. Plant Physiol. 2017;174:339–355. doi: 10.1104/pp.16.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tylewicz S., et al. Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science. 2018;360:212–215. doi: 10.1126/science.aan8576. [DOI] [PubMed] [Google Scholar]
- 50.Wang X., et al. Salicylic acid regulates plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell. 2013;25:2315–2329. doi: 10.1105/tpc.113.110676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scalschi L., et al. Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 2015;81:304–315. doi: 10.1111/tpj.12728. [DOI] [PubMed] [Google Scholar]
- 52.Slabaugh E., Davis J.K., Haigler C.H., Yingling Y.G., Zimmer J. Cellulose synthases: new insights from crystallography and modeling. Trends Plant Sci. 2014;19:99–106. doi: 10.1016/j.tplants.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Hong Z., Zhang Z., Olson J.M., Verma D.P. A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell. 2001;13:769–779. doi: 10.1105/tpc.13.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crowell E.F., et al. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21:1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez R., Lindeboom J.J., Paredez A.R., Emons A.M., Ehrhardt D.W. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 56.Kulich I., et al. Exocyst subunit EXO70H4 has a specific role in callose synthase secretion and silica accumulation. Plant Physiol. 2018;176:2040–2051. doi: 10.1104/pp.17.01693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benschop J.J., et al. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Nühse T.S., Stensballe A., Jensen O.N., Peck S.C. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Kline K.G., Barrett-Wilt G.A., Sussman M.R. In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl. Acad. Sci. Unit. States Am. 2010;107:15986–15991. doi: 10.1073/pnas.1007879107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakagami H., et al. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010;153:1161–1174. doi: 10.1104/pp.110.157347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nühse T.S., Bottrill A.R., Jones A.M., Peck S.C. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007;51:931–940. doi: 10.1111/j.1365-313X.2007.03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reiland S., et al. Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF) Proc. Natl. Acad. Sci. Unit. States Am. 2011;108:12955–12960. doi: 10.1073/pnas.1104734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reiland S., et al. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sugiyama N., et al. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 2008;4:193. doi: 10.1038/msb.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calonge T.M., Arellano M., Coll P.M., Perez P. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 2003;47:507–518. doi: 10.1046/j.1365-2958.2003.03312.x. [DOI] [PubMed] [Google Scholar]
- 66.Ishiguro J., Shibahara K., Ueda Y., Nakamura K. Fission yeast TOR signaling is essential for the down-regulation of a hyperactivated stress-response MAP kinase under salt stress. Mol. Genet. Genom. 2013;288:63–75. doi: 10.1007/s00438-012-0731-7. [DOI] [PubMed] [Google Scholar]
- 67.Qadota H., et al. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 68.Speicher T.L., Li P.Z., Wallace I.S. Phosphoregulation of the plant cellulose synthase complex and cellulose synthase-like proteins. Plants. 2018;7 doi: 10.3390/plants7030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen S., Ehrhardt D.W., Somerville C.R. Vol. 107. 2010. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase; pp. 17188–17193. (Proceedings of the National Academy of Sciences). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao D., et al. Changes in nitric oxide levels and their relationship with callose deposition during the interaction between soybean and Soybean mosaic virus. Plant Biol. 2018;20:318–326. doi: 10.1111/plb.12663. [DOI] [PubMed] [Google Scholar]
- 71.Kadota Y., et al. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 72.Xu B., et al. A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol. 2017;215:77–84. doi: 10.1111/nph.14599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leslie M.E., Rogers S.W., Heese A. Increased callose deposition in plants lacking DYNAMIN-RELATED PROTEIN 2B is dependent upon POWDERY MILDEW RESISTANT 4. Plant Signal. Behav. 2016;11 doi: 10.1080/15592324.2016.1244594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arora N.K. Springer; 2019. Impact of Climate Change on Agriculture Production and its Sustainable Solutions; pp. 95–96. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.