Abstract
Macrolide resistance in Streptococcus pneumoniae has been associated with two main mechanisms: target modification by Erm methyltransferases and efflux by macrolide pumps. The ketolide ABT-773, which has a 3-keto group and no l-cladinose sugar, represents a new class of drugs with in vitro activity against a variety of resistant bacteria. Several approaches were undertaken to understand how ABT-773 was able to defeat resistance mechanisms. We demonstrated tighter ribosome binding of ABT-773 than erythromycin. We also showed that ABT-773 (i) accumulated in macrolide-sensitive S. pneumoniae at a higher rate than erythromycin, (ii) was able to bind with methylated ribosomes, though at lower affinities than with wild-type ribosomes, and (iii) accumulated in S. pneumoniae strains with the efflux-resistant phenotype.
Macrolide resistance in Streptococcus pneumoniae has been associated with two main mechanisms. The first resistant strains described were of the macrolide-lincosamide-streptogramin B (MLS) phenotype (18). These strains have a posttranslational modification of 23S rRNA due to the methylation of A2058 (Escherichia coli numbering system) by Erm methyltransferase, resulting in a conformational change in the ribosomes. Resistance to macrolides may be constitutively expressed or induced by sub-MICs of the macrolide (18, 20). The second mechanism of resistance recently found in S. pneumoniae, and referred to as the M phenotype, results from the active efflux of 14- and 15-membered macrolides (16). The macrolide efflux pump protein in S. pneumoniae is encoded by the gene mefE (17). The prevalence of M phenotype resistance in clinical isolates of S. pneumoniae has been reported to be a significant portion (60 to 75%) of the resistance population in the United States (8, 14, 15, 16).
The ketolide ABT-773, which has a 3-keto group and no l-cladinose sugar, represents a new class of drugs with in vitro activity against a variety of resistant phenotypes (1). The purpose of this study was to understand how ABT-773 is able to defeat these resistance mechanisms. Several approaches were undertaken, including the measurement of drug-ribosome binding kinetics, drug uptake and efflux rates, and drug effect on cell-free protein translation.
MATERIALS AND METHODS
Chemicals and radioisotopes.
Todd-Hewitt broth and yeast extract were obtained from Difco Laboratories (Detroit, Mich.) Miller's M9 medium was prepared with M9 salts purchased from Life Technologies LTD (Paisley, Scotland) but without vitamins or amino acids, as described elsewhere (10). Luciferin reagents (GenLux kit) were purchased from Wallac (Turku, Finland). All other chemicals, including carbonyl cyanide m-chlorophenylhydrazone (CCCP), were purchased from Sigma Chemical Co. (St. Louis, Mo.). [N-methyl-14C]erythromycin (55 mCi/mmol) was obtained from NEN Life Science Products (Boston, Mass.), and unlabeled erythromycin was made in-house. [N-methyl-14C]ABT-773 (27 mCi/mmol) was prepared in-house by coupling 6-0-allyl ketolide and 3-[U-14C]bromoquinoline according to a patent procedure (12). The 3-[U-14C]bromoquinoline was obtained from Chem Syn Laboratories (Lenexa, Kans.). Unlabeled ABT-773 was prepared in a similar manner but with unlabeled substrates.
Organisms, growth conditions, and MIC.
S. pneumoniae strains used in this study and listed in Table 1 are from the Abbott Culture Collection. The strains were clinical isolates obtained from various sources, except for strain 2486, which is ATCC 6303, and strain 5635, which is ATCC 49619. Phenotype determinations were previously reported by Shortridge et al. (15). Strain 5970 has an inducible ermB with a low residual level of ribosome methylation. When strain 5970 is grown in the presence of 0.1 to 1 μg of erythromycin per ml (inducer), the methylated ribosome population increases from ≤5 up to 75%. Strains 6395 and 1813 carry ermB genes which are constitutively expressed at high levels. In vivo ribosome methylation levels and erm induction were measured by the previously published method (20). Strains used in transport experiments were grown in Todd-Hewitt broth, plus 0.5% (wt/vol) yeast extract (THB+Y) at 37°C to a cell density of about 1 × 108 to 3.8 × 108 CFU/ml (A600 = 0.4).
TABLE 1.
S. pneumoniae strains used and in vitro antibiotic activity
| Strain | Genotype | MICa (μg/ml) of:
|
|
|---|---|---|---|
| ABT-773 | Erythromycin | ||
| 2486 | Susceptible | ≤0.002 | 0.03 |
| 5635 | Susceptible | ≤0.002 | 0.03 |
| 5649 | mefb | 0.06 | 8.0 |
| 5970c | ermB | 0.008 | >128 |
| 6395d | ermB | 0.5 | >128 |
| 1813d | ermB | 1 | >128 |
MIC determined by broth microdilution reference method (11).
Gene designation for macrolide efflux pump.
Strain containing ermB methyltransferase gene with low residual ribosome methylation levels.
Strain containing ermB methyltransferase gene with high residual ribosome methylation levels.
MICs were determined by the broth microdilution reference method (11). Growth was measured at 530 nm at 24 h after drug treatment, with logarithmic-phase growing cells from an inoculum size of 5 × 105 CFU/ml.
Macrolide-ketolide transport studies.
Cells were harvested in mid-log-growth phase by centrifugation at 4,000 rpm for 10 min in a Jouan CR412 tabletop refrigerated centrifuge. The pellet was washed once with Miller's M9 medium (1% M9 salt, 0.2% glucose, 0.05% NaCl, 0.1 mM CaCl2, 1 mM MgSO4) and then resuspended in the same medium at 1/10 the original volume or 1 × 109 to 3.8 × 109 CFU/ml. Concentrated cells were incubated at room temperature (23°C) with 0.2 μg of [14C]ABT-773 or [14C]erythromycin per ml for various times.
The induction of MLS resistance was accomplished using S. pneumoniae strain 5970 grown at 37°C in the presence of 0.2 μg of erythromycin per ml (20). After induction, cells were collected and then resuspended in Miller's M9 medium without inducing agent.
In order to collapse the membrane potential in strains containing the macrolide efflux pump, Mef, cells were treated with 0.1 mM CCCP 10 min prior to the addition of radiolabeled macrolide or ketolide. Initial uptake rates were determined from a linear regression analysis of data points taken at 15- to 30-s intervals after label addition.
Radiolabeled cell samples (0.1 to 0.2 ml) were taken in triplicate at various times and separated from the medium by centrifugation (13,000 × g for 1 min) through 0.15 ml of silicone oil (550 + 556, 6:7 [vol/vol]; Boss Products) in 0.4-ml-capacity microcentrifuge tubes (2, 4, 7). The short time intervals were achieved by mixing label with cells which were in the microcentrifuge ready for spinning through oil. The microcentrifuge tubes were quick-frozen in a methanol-dry ice bath, and the tips containing the cell pellets were clipped off into scintillation vials containing 0.5 ml of water. The cells were resuspended in the water and 10 ml of Instagel-XF (Packard) was added. Samples were counted in a liquid scintillation counter. CFU were determined by serial dilution of culture in THB+Y and plating on tryptic soy agar plates with 5% sheep blood. Data were expressed as picomoles of antibiotic per 109 CFU after subtraction of a zero point (cells at 4°C with label, immediately centrifuged through silicone oil).
In vivo drug-ribosome on rates.
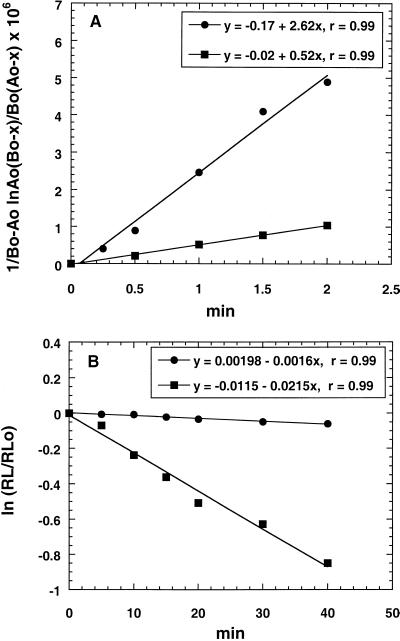
Cells were harvested and resuspended in Miller's M9 medium at 1/20 the original volume. Cells (50 μl) were placed in microcentrifuge tubes containing silicone oil prior to addition of an equal volume of radiolabeled erythromycin or ABT-773 (final drug concentration, 0.2 μg/ml). Cells plus label at 23°C were separated from medium at short time intervals by centrifugation. Cell pellets were processed as outlined above, and data were normalized to 109 CFU/ml. Drug forward rate constants were determined by plotting 1/(Bo − Ao) [ln Ao(Bo − x)/Bo(Ao − x)] versus t, where Ao is the concentration of free ribosomes estimated to be 90 nM from maximum attainable level of erythromycin accumulation in 109 CFU/ml, Bo is the concentration of free erythromycin (0.27 μM) or ABT-773 (0.26 μM) at time zero, and x is the cell-associated drug at time t. The slope is equal to the forward rate constant, k1 (5).
In vivo drug-ribosome off rates and dissociation constants.
Cells were harvested and resuspended in Miller's M9 medium at 1/10 the original volume. The concentrated cells were labeled with 0.2 μg of either [14C]ABT-773 or [14C]erythromycin per ml for 30 min (drug saturation of ribosomes). The cells were centrifuged at 4,000 rpm for 5 min (Jouan CR412 centrifuge), and the supernatant was replaced at one-fifth the original volume with fresh Miller's M9 containing 0.2 μg of unlabeled macrolide or ketolide per ml. Cell samples of 0.1 ml (prior to drug displacement) or 0.2 ml (after drug displacement) were removed and processed as outlined above. Drug off rates were determined by plotting ln(RL/RLo) versus time (in minutes) of displacement, where RL is cell-associated label at various times after drug displacement and RLo is cell associated-label at the first measured point after drug displacement. The slope of the curve is equal to the off rate per min (k−1) (19). Half-life (t1/2) values were calculated from the slope (t1/2 = 0.693/slope). The dissociation constants (Kd = k−1/k1) were calculated from the off and on rates determined above.
In vitro transcription-translation assay.
The methods for preparing S-30 extract were modified from previously published procedures (5). Cells were harvested in mid-log-growth phase; washed twice with 10 mM Tris-HCl, pH 7.5, containing 15 mM magnesium acetate [Mg(OAc)2], 50 mM KCl, 6 mM 2-mercaptoethanol, 2 mM phenylmethylsulphonyl fluoride, and 1 M NH4Cl (high-salt buffer); and then washed once with the above buffer containing 60 mM NH4Cl (low-salt buffer). After the final wash, the pellet was resuspended in a small volume of low-salt buffer. Cells were taken through one freeze-thaw cycle prior to cell disruption by two passes through a French press at 12,000 lb/in2. Cell debris was removed by successive spins at 10,000 × g for 10 min, 30,000 × g for 20 min, and 30,000 × g for 30 min and removal of pellets. The supernatant from the final spin (S-30) was dialyzed against 1 liter of low-salt buffer for 3 h with two changes. The DNA plasmid containing the luciferase reporter and the S. pneumoniae ami promoter genes were engineered in-house (details for plasmid design and construction will be published elsewhere). The ribosome fraction (pellet from 100 kg with 2-h spin of S-30) from a highly resistant strain, 1813, was mixed with S-100 fraction (supernatant from 100 kg with 2-h spin of S-30) from strain 5635 and designated reconstituted S-30. The reaction mixture contained 1 μg of DNA plasmid, 0.15 A260 units of S-30 extract or reconstituted S-30, 0.2 mM complete amino acid mix, and 5 μl of premix (Promega Corp., Madison, Wis.) in a final volume of 15 μl. The reaction took place at room temperature for 2 h. The reaction mixture was then mixed with 75 μl of luciferin reagent and immediately read in a Wallac Trilux luminescence counter. Compounds in ethanol were tested at various concentrations in this assay, and data were expressed as a percent of control (no drug group).
RESULTS
Antimicrobial activity.
ABT-773 is extremely potent against both susceptible and macrolide-resistant S. pneumoniae. As shown in Table 1, ABT-773 has excellent activity against two susceptible strains and a representative macrolide efflux strain (5649) as well as three strains with ribosome methylase. The three ermB-containing strains were chosen as representatives of the low, middle and high end of the ABT-773 MIC range for methylase-positive strains, in order to investigate the differences in this population. Isolates with an ABT-773 MIC of 0.5 μg/ml or 1 μg/ml are rare; in a recent survey (1) only 3 of 1,601 (0.2%) S. pneumoniae isolates had an ABT-773 MIC of 0.5 μg/ml (MIC at which 90% of the isolates tested are inhibited = 0.03 μg/ml).
Macrolide and ketolide uptake.
The erythromycin and ABT-773 uptake rates by susceptible and resistant strains were compared. The macrolide-susceptible S. pneumoniae strain 2486 accumulated ABT-773 and erythromycin at initial rates of 37.4 and 10.8 pmol/109 CFU/min, respectively. ABT-773 accumulation reached a peak of 84 pmol/109 CFU by 5 min, and erythromycin accumulation peaked at 82 pmol/109 CFU by 20 min in strain 2486 (Fig. 1A). Similar results were obtained with another macrolide-susceptible strain, S. pneumoniae 5635. Accumulation of ABT-773 and erythromycin in the two MLS-resistant strains, 5970 and 6395, varied. In strain 5970, both antimicrobial agents accumulated to about 40 pmol/109 CFU by 10 and 30 min, respectively. In strain 6395, ABT-773 and erythromycin accumulated to 21 pmol/109 CFU by 5 min and 14 pmol/109 CFU by 20 min, respectively (Fig. 1B). Both ABT-773 and erythromycin reached the same steady-state levels (saturation of intracellular ribosomes) in strain 2486 and 5970, while variations were seen in strain 6395 due to the high level of methylated ribosomes. In all three strains, ABT-773 accumulated in cells faster than did erythromycin.
FIG. 1.
Macrolide and ketolide accumulation in susceptible and resistant strains of S. pneumoniae. Cells (1 × 109 to 3.8 × 109 CFU/ml) in Miller's M9 medium were incubated with 0.2 μg of [14C]erythromycin or [14C]ABT-773 per ml at 23°C for the times indicated. Accumulation of erythromycin (A) and ABT-773 (B) in strains 2486 (circles), 5970 (squares), and 6395 (triangles) is shown. Accumulation of erythromycin (C) and ABT-773 (D) in strain 5649 pretreated with (circles) and without (squares) 0.1 mM CCCP is also shown. Triplicate samples were spun through silicone oil as described in Materials and Methods. Results are presented as means ± standard deviations (error bars) for the triplicate samples.
In order to look at the effect of efflux on ABT-773 uptake, a strain (5649) with mef was also studied. The inactivation of the macrolide efflux pump, Mef, with CCCP resulted in a 2.6-fold increase in initial erythromycin uptake but little or no change in the initial uptake of ABT-773 in strain 5649. The initial uptake rates for ABT-773 before and after pump inactivation were 4.4- and 1.9-fold higher than those for erythromycin, respectively (Fig. 1C and D). In addition, the steady-state level of erythromycin accumulation in mef-containing strains was increased by CCCP treatment, but the level of ABT-773 accumulation was not affected (data not shown). As a control, CCCP did not change macrolide uptake profile in erm-containing strains.
Analysis of kinetic constants based on cell associated drug.
The ribosomal association and dissociation rates for the two compounds in susceptible cells were compared. The k1 for erythromycin and ABT-773 binding to ribosomes were determined in vivo in susceptible strain 2486 from initial uptake rates (Fig. 2A). An estimate of the number of antibiotic molecules per cell was determined from the maximum level of antibiotic accumulation (Fig. 1A and B) divided by cell number times Avogadro's number (6.02 × 1023). There were roughly 50,000 molecules of drug per cell in susceptible strain 2486. The number was then used to estimate the amount of free ribosomes. Intracellular volume was determined by [3H]H2O (total volume) and [14C]dextran (extracellular volume) uptake. Ribosome concentration, Ao (refer to Materials and Methods), was then calculated. The forward rate constants for ABT-773 and erythromycin were calculated to be 2.62 × 106 and 5.28 × 105 M−1 min−1, respectively. The displacement of radiolabeled ABT-773 and erythromycin in the susceptible strain 2486 with unlabeled ABT-773 provided data for determining in vivo ribosome drug off rates (Fig. 2B). The off-rates for ABT-773 and erythromycin were 0.0016 min−1 (t1/2 = 425 min) and 0.0215 min−1 (t1/2 = 32.3 min), respectively. The data for the initial uptake rates, in vivo drug on and off rates, and Kd calculated from these data are presented together in Table 2. There was a 67-fold difference in Kd between the two drugs, with ABT-773 dissociating much more slowly.
FIG. 2.
Kinetic analysis of drug association and dissociation in S. pneumoniae 2486. (A) Drug on rate determinations. Cells (1.0 × 109 CFU/ml) in Miller's M9 medium were incubated at 23°C with either 0.2 μg of [14C]erythromycin (squares) or [14C]ABT-773 (circles) per ml for short intervals up to 2 min. Labeled cells were processed as outlined in Materials and Methods. Influx of label was plotted as 1/(Bo − Ao) [ln Ao(Bo − x)/Bo(Ao − x)] versus t, where Ao is concentration of free ribosomes, Bo is concentration of free drug at time zero, and x is cell-associated drug at time t. The slope is equal to k1. (B) Drug off rate determinations. Cells (1.2 × 109 to 1.5 × 109 CFU/ml) in Miller's M9 medium were incubated at 23°C with 0.2 μg of either [14C]erythromycin (squares) or [14C]ABT-773 (circles) per ml for 30 min (saturation of binding sites). Labeled cells were harvested and suspended in fresh medium containing 0.2 μg of unlabeled ABT-773 per ml. Efflux of label at 23°C was monitored for 40 min and plotted as ln(RL/RLo) versus t, where RL is cell-associated label at time t, RLo is cell-associated label at zero time, and the slope is k−1.
TABLE 2.
Erythromycin and ABT-773 ribosome binding kineticsa
| Drug | Initial uptake (pmol/109 CFU/min) | k1 (M−1 min−1) | k−1 (min−1) | Kd (M) |
|---|---|---|---|---|
| Erythromycin | 10.8 | 5.28 × 105 | 0.0215 | 4.1 × 10−8 |
| ABT-773 | 37.4 | 2.62 × 106 | 0.0016 | 6.1 × 10−10 |
| Ratiob | 0.3 | 0.2 | 13 | 67 |
Analysis based on cellular drug influx and efflux rates as described in Materials and Methods. Kinetic constants: k1, forward rate constant; k−1, reverse rate constant; Kd, dissociation constant = k−1/k1. The strain used was S. pneumoniae 2486.
Erythromycin/ABT-773.
Transcription-translation assay.
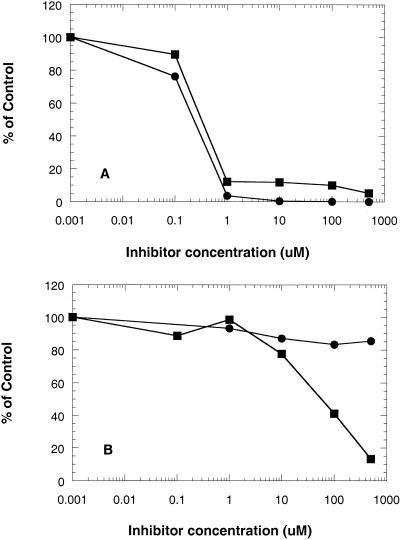
Inhibition of ribosomal function by the two compounds was measured directly in a coupled transcription-translation system. The observed inhibition activities in the coupled assays by erythromycin and ABT-773 were at the translational levels. The possibility of inhibiting the transcription reactions was excluded by Northern blot experiment in which a fragment of luciferase gene was used as a probe. No changes in mRNA levels were observed (data not shown). Using cell S-30 extract from the macrolide-susceptible strain 5635, ABT-773 and erythromycin demonstrated comparable inhibitory effects, with 50% inhibitory concentrations of 0.3 and 0.2 μM, respectively (Fig. 3A). The reconstituted S-30 containing highly methylated ribosomes was inhibited a maximum of 15% in the production of luciferase by erythromycin, while ABT-773 effectively inhibited luciferase production, with a 50% inhibitory concentration of 58.3 μM (Fig. 3B).
FIG. 3.
In vitro drug effect on transcription-translation in susceptible and resistant S. pneumoniae. Cell extracts were incubated with DNA plasmid containing the luciferase reporter gene in the presence of various concentrations of erythromycin (solid circles) or ABT-773 (solid squares) for 2 h at room temperature. The mixture was added to luciferin, and light emission read in a Wallac Trilux luminescence counter. Data were expressed as a percent of the control group (no drug). (A) Ribosomes from susceptible strain; (B) ribosomes from resistant strain with high levels of methylated ribosomes.
Erm induction.
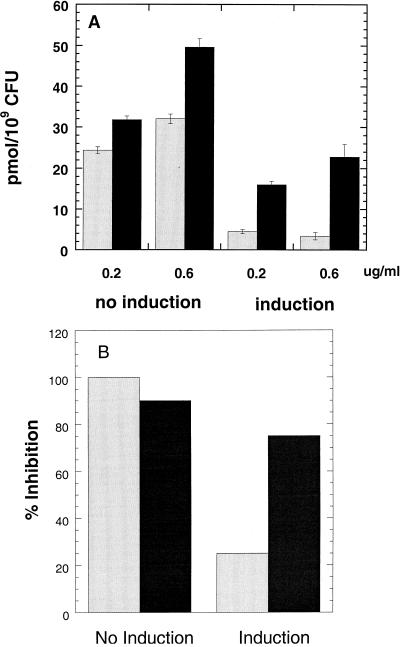
The accumulation of ABT-773 and erythromycin in S. pneumoniae 5970 before and after erm induction with erythromycin is presented in Fig. 4A. ABT-773, when tested at 0.2 and 0.6 μg/ml before erm induction, accumulated to 31.8 and 49.6 pmol/109 CFU/20 min, respectively. After erm induction, ABT-773 uptake was 16 and 22.8 pmol/109 CFU/20 min when tested at 0.2 and 0.6 μg/ml, a 50 to 54% reduction from preinduction accumulation levels. Erythromycin, tested at 0.2 and 0.6 μg/ml before induction, accumulated to 24.4 and 32 pmol/109 CFU/20 min, respectively. After induction, erythromycin uptake was 4.5 and 3.4 pmol/109 CFU/20 min, a reduction of 82 to 89% compared to the preinduction uptake.
FIG. 4.
Effect of Erm induction on whole-cell drug accumulation and cell protein synthesis. (A) Accumulation of macrolide or ketolide in S. pneumoniae 5970 with or without induction of ErmB methylase by erythromycin. Cells were grown for several generations with or without 0.2 μg of erythromycin per ml at 37°C in growth medium prior to centrifugation and resuspension in Miller's M9 medium at 3.8 × 109 CFU/ml. Cells were then incubated at 23°C with either 0.2 or 0.6 μg of [14C]erythromycin (gray bars) or [14C]ABT-773 (black bars) per ml for 20 min. Labeled cells were processed as outlined in Materials and Methods. Results are presented as means ± standard deviations (error bars) (n = 3). (B) S-30 extracts prepared from S. pneumoniae 5970 before and after Erm induction were run in the transcription-translation assay outlined in Materials and Methods. Erythromycin (gray bars) and ABT-773 (black bars) were tested at 100 μM. Results were expressed as percent inhibition with or without induction.
S-30 preparations from strain 5970 before and after erm induction were used in a transcription-translation assay. Erythromycin or ABT-773 at 100 μM completely blocked protein translation before induction. However, after induction erythromycin and ABT-773 inhibited translation 25 and 75%, respectively (Fig. 4B).
DISCUSSION
The increased potency of the novel ketolide ABT-773 against both susceptible, MLS-resistant, and M-resistant S. pneumoniae led us to investigate the parameters of drug transport, ribosome binding, and inhibition of protein synthesis in representative strains. Preliminary radioisotope studies showed that ABT-773 competed with erythromycin or itself for ribosomal binding sites on the surface of the ribosome (Z. Cao, R. Hammond, S. Pratt, A. Saiki, C. Lerner, and P. Zhong, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2135, 1999). Furthermore, RNA footprinting in domain V of 23S rRNA revealed that, similar to erythromycin, ABT-773 protected the nucleotides A2058 and A2059 (E. coli numbering system) from DMS chemical modifications (unpublished observations). These observations indicate the binding of ABT-773 to sites overlapping erythromycin binding sites on the 50S ribosomal subunit.
Macrolide transport into bacteria is a passive process, independent of energy and facilitated by the intracellular binding site, the ribosomes (2, 3, 6, 9). Thus, the more rapid accumulation of ABT-773 compared to erythromycin in susceptible strains of S. pneumoniae was dictated by the tighter binding kinetics of the ketolide compared to the macrolide. Our estimation of drug Kd based on accumulation and displacement studies assumes comparable interference of the plasma membrane with drug diffusion. The cellular drug accumulation measured in current studies is mainly due to the ribosome binding. Because the steady-state levels of drug accumulation can be simply manipulated by changing methylated ribosome populations in an inducible MLS strain (which is discussed in more detail later in the text). The possibility that methylated ribosomes could somehow change the membrane properties and interfere with macrolide uptake is minimal. Under the experimental conditions, a large difference (67-fold) in Kd between ABT-773 and erythromycin was observed. The tight binding of ABT-773 with ribosomes was also observed in the equilibrium competition binding studies using isolated ribosomes from susceptible S. pneumoniae strains (data not shown).
The transport experiment can serve as a window into available binding sites inside the cells. Resistant strain 5970, which has low basal levels of methylated ribosomes (20) appears to accumulate similar levels of both compounds even though this strain has a much higher MIC of erythromycin (>128 μg/ml) than ABT-773 (0.008 μg/ml). This contradiction can be explained by the fact that strain 5970 can be induced to higher levels of ribosome methylation by growth in the presence of erythromycin but not by ABT-773, thus the lower MIC of ABT-773 suggests that induction does not occur. Finally, strain 6395 has constitutively expressed erm and high-level-methylated ribosomes (20) and still accumulated higher levels of ABT-773 when compared to erythromycin accumulation. These data suggest some binding of ABT-773 to methylated ribosomes.
The interactions between ABT-773 and ribosomes were further investigated in ribosomal functional assays. In the transcription-translation experiments with wild-type ribosomes (Fig. 3A), both ABT-773 and erythromycin at 1 μM efficiently inhibited protein synthesis. This was inconsistent with ribosome binding affinity being directly related to the efficiency of ribosome inhibition. The transcription-translation assay apparently lacks the sensitivity at inhibitor concentrations <1 μM to reflect ribosome binding differences between the two antibiotics. The use of methylated ribosomes in the translation reactions differentiated the two drugs more clearly (Fig. 3B and 4B). Under these conditions, erythromycin lost most of its inhibitory activity, while ABT-773 was able to completely block translation at higher drug concentrations. Again, these data confirm erythromycin's complete lack of affinity with methylated ribosomes and suggest that ABT-773 had low affinity with methylated ribosomes (about 100-fold lower than wild-type ribosomes).
In order to confirm that the observed intracellular drug accumulations truly reflect the drug interactions with ribosomes, it is desirable to run the uptake experiments with cells in which the ratio of wild-type and methylated ribosomes can be regulated. Such a system was provided by the inducible MLS-resistant strain, 5970. The amount of methylated ribosomes can be estimated by the percent inhibition of in vitro protein synthesis with erythromycin, assuming the efficiencies of the translation by wild-type and methylated ribosomes are similar. Erythromycin accumulation dropped about 70 to 80% (Fig. 4A) after erm induction, which compared well with the 25% inhibition by 100 μM erythromycin in the translation assays using methylated ribosomes (Fig. 4B). These results suggested that translation with highly resistant ribosomes could be completely blocked by ABT-773 and that ABT-773 could accumulate in cells to preinduction levels if drug concentrations were high enough (uptake experiments were limited by the specific activity of the radio-labeled drug).
We have also investigated S. pneumoniae strains containing the macrolide efflux pump, Mef. The efflux resistant mechanism in this organism involves a Mef-protein (15) which can bind to macrolides and pump them out of the cell, resulting in reduced intracellular drug concentration (16, 17). In the mef-containing strains, there is competition between the ribosome and the Mef-binding sites for the intracellular drug, although efflux pumps might also intercept the drug within the membrane (13). Drugs, such as ABT-773, which possess tight ribosome binding kinetics may not bind as tightly to the efflux pump, resulting in a net influx rate which exceeds the capacity of the pump. Although we cannot conclude whether ABT-773 is recognized by the Mef-pump, we have demonstrated that this ketolide accumulated to significant levels and displayed antibacterial activity in the M-phenotype resistant strain 5649.
In summary, ABT-773 binds tightly with wild-type ribosomes and inhibits ribosomal functions in both susceptible and a broad spectrum of resistant S. pneumoniae strains, including inducible MLS, constitutive MLS, and mef-containing strains.
ACKNOWLEDGMENTS
We acknowledge Bruce Surber for providing radiolabeled ABT-773 and Claude Lerner and Anne Saiki for providing plasmids.
We acknowledge the Abbott Anti-Infective Venture for financial support.
REFERENCES
- 1.Brueggemann A B, Doern G V, Huynh H K, Wingert E M, Rhomberg P R. In vitro activity of ABT-773, a new ketolide, against recent clinical isolates of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis. Antimicrob Agents Chemother. 2000;44:447–449. doi: 10.1128/aac.44.2.447-449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capobianco J O, Goldman R C. Erythromycin and azithromycin transport into Haemophilus influenzae ATCC 19418 under conditions of depressed proton motive force (ΔμH) Antimicrob Agents Chemother. 1990;34:1787–1791. doi: 10.1128/aac.34.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capobianco J O, Goldman R C. A concurrent study of macrolide transport, binding to ribosomes and inhibition of protein synthesis in Haemophilus influenzae. In: Pandalai S G, Gayathri A, editors. Recent research developments in antimicrobial agents and chemotherapy 1. Trivandrum, India: Research Signpost; 1996. pp. 95–102. [Google Scholar]
- 4.Dette G A, Knothe H, Kaula S. Modifying effects of pH and temperature on (14C)erythromycin uptake into Staphylococcus aureus—relation to antimicrobial activity. Zentbl Bakteriol Hyg A. 1987;265:393–403. [PubMed] [Google Scholar]
- 5.Goldman R C, Kadam S K. Binding of novel macrolide structures to macrolides-lincosamides-streptogramin B-resistant ribosomes inhibits protein synthesis and bacterial growth. Animicrob Agents Chemother. 1989;33:1058–1066. doi: 10.1128/aac.33.7.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman R C, Fesik S W, Doran C C. Role of the protonated and neutral forms of macrolides in binding to ribosomes from gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 1990;34:426–431. doi: 10.1128/aac.34.3.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson J D, Hand W L, Francis J B, King-Thompson N, Corwin R W. Antibiotic uptake by alveolar macrophages. J Lab Clin Med. 1980;95:429–439. [PubMed] [Google Scholar]
- 8.Johnston N J, de Azavedo J C, Kellner J D, Low D E. Prevalence and characterization of the mechanisms of macrolide, lincosamide, and streptogramin resistance in isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2425–2426. doi: 10.1128/aac.42.9.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao J C H, Putterman M. Accumulation in gram-positive and gram-negative bacteria as a mechanism of resistance to erythromycin. J Bacteriol. 1968;95:1111–1117. doi: 10.1128/jb.95.3.1111-1117.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. p. 433. [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Supplemental tables: M100-S8. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 12.Or Y, Ma Z, Clark R. 6-0-Substituted ketolides having antibacterial activity. U.S. patent 5,866,549. 1999. [Google Scholar]
- 13.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shortridge V D, Doern G V, Brueggemann A B, Beyer J M, Flamm R K. Prevalence of macrolide resistance mechanisms in Streptococcus pneumoniae isolates from a multicenter antibiotic resistance surveillance study conducted in the United States in 1994–1995. Clin Infect Dis. 1999;29:1186–1188. doi: 10.1086/313452. [DOI] [PubMed] [Google Scholar]
- 15.Shortridge V D, Flamm R K, Ramer N, Beyer J, Tanaka S K. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 1996;26:73–78. doi: 10.1016/s0732-8893(96)00183-6. [DOI] [PubMed] [Google Scholar]
- 16.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams L T. Theory of ligand-receptor interactions. In: Williams L T, Lefkowitz R J, editors. Receptor binding studies in adrenergic pharmacology. New York, N.Y: Raven Press; 1978. pp. 27–41. [Google Scholar]
- 20.Zhong P, Cao Z, Hammond R, Chen Y, Beyer J, Shortridge V D, Phan L Y, Pratt S, Capobianco J, Reich K A, Flamm R K, Or Y-S, Katz L. Induction of ribosome methylation in MLS-resistant Streptococcus pneumoniae by macrolides and ketolides. Microb Drug Res. 1999;5:183–188. doi: 10.1089/mdr.1999.5.183. [DOI] [PubMed] [Google Scholar]